Abstract
We constructed two types of chimeric enzymes, Ch1 Amy and Ch2 Amy. Ch1 Amy consisted of a catalytic domain of Bacillus subtilis X-23 α-amylase (Ba-S) and the raw starch-binding domain (domain E) of Bacillus A2-5a cyclomaltodextrin glucanotransferase (A2-5a CGT). Ch2 Amy consisted of Ba-S and D (function unknown) plus E domains of A2-5a CGT. Ch1 Amy acquired raw starch-binding and -digesting abilities which were not present in the catalytic part (Ba-S). Furthermore, the specific activity of Ch1 Amy was almost identical when enzyme activity was evaluated on a molar basis. Although Ch2 Amy exhibited even higher raw starch-binding and -digesting abilities than Ch1 Amy, the specific activity was lower than that of Ba-S. We did not detect any differences in other enzymatic characteristics (amylolytic pattern, transglycosylation ability, effects of pH, and temperature on stability and activity) among Ba-S, Ch1 Amy, and Ch2 Amy.
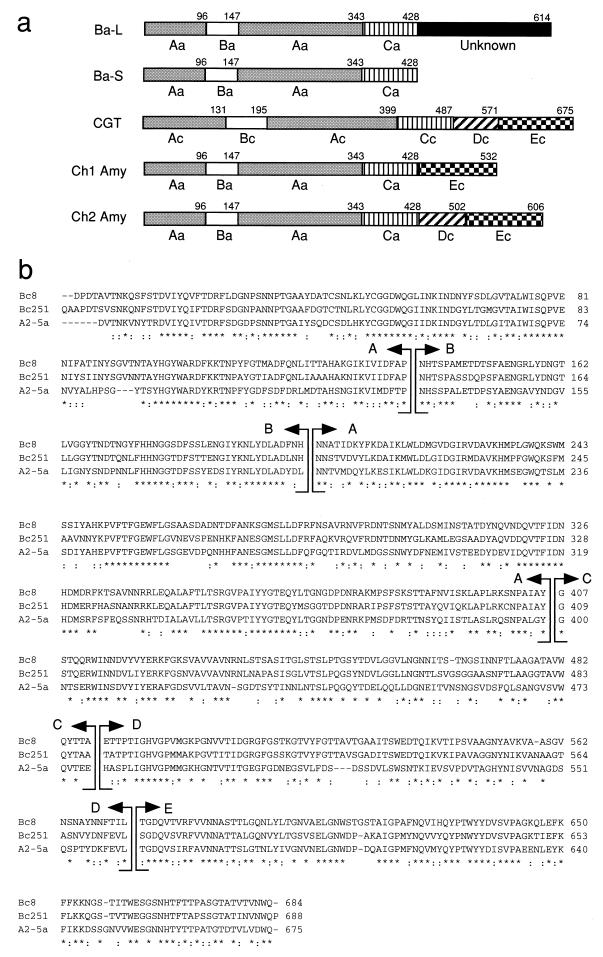
Comparison of the primary, secondary, and three-dimensional structures of enzymes in the α-amylase family has revealed that the structures of α-amylases (EC 3.2.1.1), cyclomaltodextrin glucanotransferases (CGTases) (EC 2.4.1.19), and other amylolytic enzymes are closely related (20, 21, 23, 39). The structures of these enzymes have also been studied with regard to their domain-level organization (15). Taka-amylase A (TAA; an α-amylase from Aspergillus oryzae) consists of three domains (A, B, and C) (1, 26). Domain A contains an amino-terminal (β/α)8-barrel structure, followed by a domain consisting of β-strands folded in a Greek-key motif (domain C). Domain B is inserted between the third β-strand and the third α-helix of the (β/α)8-barrel, and this domain varies greatly in both length and amino acid sequences depending on its source (13, 14). CGTases generally consist of five domains (A, B, C, D, and E). Based on the analysis of their secondary (15) and three-dimensional (17, 19, 25) structures, domains A, B, and C of CGTases correspond to the catalytic domains A, B, and C of TAA. The primary structure of domain E contains a typical motif found in other raw starch-binding proteins (38). There is little information available regarding domain D of CGTase, especially with regard to its function, except that all of the structures for domain D that have been reported so far contain similar antiparallel β-barrels (17, 19, 25).
Several chimeric enzymes have been constructed out of various amylolytic enzymes. Some of these have been studied with regard to secretion of the enzyme (16), while others have been studied with regard to changes in substrate specificities (28, 41), product specificities (29), or both (24). However, there have been few reports on hybrids of different enzyme species based on their domain-level organizations. It is generally known that hybrid proteins of different enzyme species do not express the functions expected from the original enzymes (8, 27, 36), since the original polypeptide-folding patterns are usually not maintained in the hybrid proteins. Recently, we reported that a smaller form of α-amylase from Bacillus subtilis X-23 (Ba-S [47 kDa]), which was produced proteolytically from a complete form (Ba-L [67 kDa]) by carboxyl-terminal truncation, could function as an α-amylase, with the same catalytic capacity and properties (32). By analyzing the secondary structure as well as the predicted three-dimensional structure of Ba-S, we demonstrated that Ba-S retained all of the domains (A, B, and C) which were most likely to be required for functionality as α-amylase. Furthermore, we predicted that Ba-S protein showed compact folding and that the carboxyl-terminal-region polypeptide of Ba-L existed without having direct interactions with the catalytic center (32). As to CGTase, Wind et al. (43) obtained a polypeptide containing the complete domain E and a part of domain D of CGTase from Thermoanaerobacterium thermosulfurigenes EM1. The thermal unfolding of a raw starch-binding domain of Aspergillus niger glucoamylase was found to be reversible (40). Dalmia et al. (2) reported that the raw starch-binding domains of Bacillus macerans CGTase and Aspergillus awamori glucoamylase retained their starch-binding ability when they were produced as fusion proteins with β-galactosidase in Escherichia coli. These reports strongly suggest that the raw starch-binding domain of CGTases should be an independent domain and that it retains its starch-binding ability, while maintaining its original conformation, even when the raw starch-binding domain is separated from the other four domains (A, B, C, and D).
For these reasons, we predicted that a raw starch-binding domain of CGTase could be introduced to the compactly folded Ba-S without any structural problems and that the resultant hybrid may have both α-amylase activity and raw starch-binding and -digesting abilities at the same level as with the originals. We report here the construction and characterization of chimeric enzymes made out of α-amylase from B. subtilis X-23 and CGTase from alkalophilic Bacillus sp. strain A2-5a.
MATERIALS AND METHODS
Media.
E. coli was grown in Luria-Bertani (LB) medium (34). A starch-azure plate, containing 1.5% agarose, 100 μg of ampicillin per ml, and 0.4% starch-azure on LB medium, was used to screen positive clones which had α-amylase activity.
Bacterial strain and plasmids.
E. coli TG1 [supE hsdΔ5 thi Δ(lac-proAB)/F′ (traD36 proAB+ lacIq lacZΔM15)] (34) was used as a cloning host. pUC118 (Takara, Kyoto, Japan) and pBluescript II SK(+) (Toyobo, Osaka, Japan) were used as a cloning vector and a sequencing vector, respectively. pUXA1 and pBLACG1, which bear the genes that code for B. subtilis X-23 α-amylase (Ba-L form) (DDBJ/EMBL/GenBank accession number AB015592) (32) and Bacillus A2-5a CGTase (A2-5a CGT) (DDBJ/EMBL/GenBank accession number AB015670) (33), respectively, were used as templates in the PCR.
Determination of the starting points of domains D and E of A2-5a CGT.
The amino acid sequences of A2-5a CGT and CGTases of Bacillus circulans strain no. 8 CGTase (17) and Bacillus circulans strain 251 CGTase (25), the three-dimensional structures of which have already been determined by X-ray crystallography, were aligned with “CLUSTAL W alignments in color” (Fig. 1b), and the starting points of domains D and E of A2-5a CGT were determined.
FIG. 1.
Design of chimeric enzymes made of Ba-S and A2-5a CGT. (a) Schematic diagram of Ba-L, Ba-S, A2-5a CGT, and chimeric enzymes constructed in this study. Aa, Ba, and Ca indicate domains A, B, and C of α-amylase from B. subtilis X-23, respectively. A domain of Ba-L (function unknown) is denoted by a closed bar. Ac, Bc, Cc, Dc, and Ec indicate domains A, B, C, D, and E, respectively, of Bacillus A2-5a CGTase. Numbers between domains show the last amino acid residues of each domain calculated from the amino terminus of mature enzymes. (b) Alignment of the amino acid sequences of three CGTases. Bc8, B. circulans 8 CGTase; Bc251, B. circulans 251 CGTase; A2-5a, A2-5a CGT. Identities and similarities (among I/L/F/V/M, D/Q/N, D/Q/E, N/E/K, S/A/T, R/K/H, R/K/Q, Y/H, Y/F, V/Q, V/L, and N/H) in all three sequences are denoted by asterisks and double dots, respectively. The boundaries of the domains are indicated by arrows.
Construction of chimeric genes and transformation.
Chimeric genes were constructed by PCR fusion based on the procedure of Yon and Fried (44). The expected gene products are schematically shown in Fig. 1a. PCR was carried out under the following conditions (6). Ten nanograms of DNA template, 20 pmol of primer, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM mixed deoxynucleotide triphosphates, and 2.5 U of Taq DNA polymerase were mixed in a total volume of 100 μl. For gene amplification (the first and third PCRs), PCR consisted of 30 cycles of 10 s at 94°C, 10 s at 50°C, and 1.5 min at 72°C. To make fusion genes (the second PCR), PCR consisted of 30 cycles of 1 min at 94°C, 1 min at 65°C, and 1 min at 72°C. Six different primers were used in the PCRs to construct the Ch1 Amy and Ch2 Amy genes, as follows: primer 1 was the 24-mer outer primer complementary with the antisense strand at the upstream region of the B. subtilis X-23 α-amylase gene (5′-TTTGGTACCGCAGTCAGTCTTCAA-3′); primer 2 was the 30-mer outer primer complementary with the sense strand at the downstream region of the carboxyl terminus of A2-5a CGT (5′-TTTTCTAGATCATTGCCAGTCGACGAGGAC-3′); primer 3 was the linking primer (48-mer) for constructing the Ch1 Amy gene (5′-GTCACCGGTTAATACTTCAAACTTATGAGGCGCATTTCCAATATCATC-3′); primer 4 was the linking primer (48-mer) for constructing the Ch2 Amy gene (5′-GCCGATTAGAGGAGAAGCATGCTCATGAGGCGCATTTCCAATATCATC - 3 ′ ); primer 5 was the linking primer (48-mer) for constructing the Ch1 Amy gene (5′-GATGATATTGGAAATGCGCCTCATAAGTTTGAAGTATTAACCGGTGAC-3′); and primer 6 was the linking primer (48-mer) for constructing the Ch2 Amy gene (5′-TACATGGCCGATTAGAGGAGAAGCATGAGGCGCATTTCCAATATCATC-3′). To construct the Ch1 Amy gene, the first PCR was performed with primers 1 and 3 on the template pUXA1 and primers 2 and 5 on the template pBLACG1, and the second PCR was then performed without any primers on a template of the first PCR products. To construct the Ch2 Amy gene, the first PCR was performed with primers 1 and 4 on the template pUXA1 and primers 2 and 6 on the template pBLACG1, and the second PCR was then performed without any primers on a template of the first PCR products. The second PCR products (fusion genes) were amplified in the third PCR with the outer primers 1 and 2. The resultant PCR products were purified with a Wizard PCR Preps DNA Purification System (Promega, Madison, Wis.) and digested with KpnI and XbaI, and the fusion genes were inserted into the KpnI-XbaI site of pUC118 with T4 DNA ligase. Transformation of E. coli TG-1 was performed with the ligation mixture, and the transformants were screened with a starch-azure plate on which a positive clone formed a halo. The plasmids, designated pCh1 and pCh2, which bear the Ch1 Amy and Ch2 Amy genes, respectively, were isolated by the alkaline lysis method (12).
Purification of chimeric enzymes.
E. coli TG1 carrying recombinant plasmid pCh1 or pCh2 was cultivated in LB medium containing 100 μg of ampicillin per ml at 37°C for 16 h. The cells were disrupted by sonication. The cell extract was used for purification of the chimeric enzymes as described previously (31). A raw starch adsorption step (18) was particularly effective for the purification of Ch2 Amy.
Assay.
α-Amylase activity was assayed based on the 3,5-dinitrosalicylic acid method, as described previously (11). The reaction mixture (200 μl) consisted of 0.5% soluble starch (Merck, Darmstadt, Germany) in 20 mM sodium acetate buffer (pH 5.5) and the enzyme. The reaction was stopped after 10 min of incubation at 50°C by adding 3,5-dinitrosalicylic acid reagent (200 μl). This reagent was prepared by mixing 0.4 M NaOH, 22 mM 3,5-dinitrosalicylic acid, and 1.1 M potassium sodium (+)-tartrate tetrahydrate. One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of reducing sugar as glucose per min under the assay conditions described above. A quantitative analysis of reducing sugar in the hydrolysate was performed as described above for the assay of α-amylase activity. The total carbohydrate was assayed by the phenol-sulfuric acid method (4).
SDS-PAGE and Western analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on an 8 to 16% gradient polyacrylamide gel, and immunoblotting was carried out using a polyvinylidene difluoride membrane (Millipore, Yonezawa, Japan) with a Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). Rabbit antiserum raised against Ba-L or A2-5a CGT was used as a primary antibody. The antigen-antibody complex was detected using anti-rabbit immunoglobulin G (IgG)-AP (Boehringer Mannheim GmbH) with BCIP (5-bromo-4-chloro-3-indolylphosphate) reagent (Boehringer Mannheim).
Adsorption of enzymes on raw starch.
The desired amount of enzyme was added to a suspension of 90 mg of raw maize starch in 50 mM sodium phosphate buffer (pH 6.0) to prepare 200 μl of suspension. The mixture was allowed to sit at 25°C for 10 min and then filtered through a membrane filter (0.45-μm pore size). After the raw starch was washed with the same buffer, the amount (percentage) of enzyme adsorbed (ra) was measured based on the method of Iefuji et al. (10) by the following equation: ra (%) = [(A − B)/A] × 100, where A is the enzyme activity of the original enzyme solution and B is the activity of the filtrate, including the buffer fraction used to wash the raw starch.
RESULTS
Design of chimeric enzymes made out of B. subtilis X-23 α-amylase and alkalophilic Bacillus A2-5a CGTase.
As described previously (32), Ba-S retained all of the domains (A, B, and C) which were most likely to be required for functionality as α-amylase. Furthermore, the activity of Ba-S was not affected by the addition of 186 amino acid residues (function unknown) at the carboxyl-terminal region (Fig. 1a) (32). Therefore, Ba-S was used for the catalytic part of chimeric enzymes. Domain E of CGTase is a raw starch-binding domain. Accordingly, we designed Ch1 Amy, which was composed of Ba-S and domain E of CGTase. The function of domain D, which connects domains C and E of CGTase, is unclear. Since domain E does not exist in α-amylases that do not have raw starch-binding and -digesting abilities, domain D may act as a linker region between catalytic domains (A, B, and C) and a raw starch-binding domain (E), as reported for glucoamylases (35, 42). Therefore, we also designed Ch2 Amy, which was composed of Ba-S and both D and E domains of A2-5a CGT. Sequence analyses of the Ch1 Amy gene and the Ch2 Amy gene verified that the hybrid genes expected have successfully been constructed and that no mutation in the genes has been introduced by PCRs.
Sizes of chimeric enzymes.
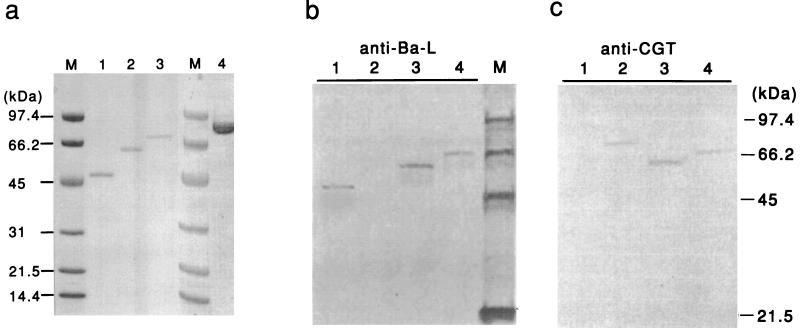
The molecular weights of Ch1 Amy and Ch2 Amy deduced from their nucleotide sequences were 59,514 and 67,701 respectively (Table 1), which were consistent with the molecular masses estimated by SDS-PAGE of the purified enzymes (Fig. 2a).
TABLE 1.
Specific activities and molar catalytic activitiesa
| Enzyme | Sp act (U/mg) | Mol wtb | Molar catalytic activity (U/nmol) |
|---|---|---|---|
| Ba-L | 362 | 67,445 | 24.4 |
| Ba-S | 514 | 47,227 | 24.2 |
| Ch1 Amy | 420 | 59,514 | 25.0 |
| Ch2 Amy | 44.7 | 67,701 | 3.03 |
Soluble starch was used as a substrate. Activity was measured based on the 3,5-dinitrosalicyclic acid method, and one unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of reducing sugar as glucose per min. Molar catalytic activities were evaluated on a molar basis.
Molecular weights were deduced from the nucleotide sequence.
FIG. 2.
SDS-PAGE (a) and Western immunoblot analysis (b and c) of purified Ba-S, chimeric enzymes, and A2-5a CGT. (a) Three micrograms of Ba-S, Ch1 Amy, and Ch2 Amy and six micrograms of A2-5a CGT were loaded onto an SDS-polyacrylamide gel, electrophoresed, and stained with Coomassie brilliant blue. Lanes: 1, Ba-S; 2, Ch1 Amy; 3, Ch2 Amy; 4, A2-5a CGT; M, molecular size marker. (b and c) After SDS-PAGE of the sample, the protein was transferred to a nitrocellulose membrane. Anti-Ba-L (b) or anti-CGT (c) antibody was used for the primary antibody. Lanes: 1, Ba-S; 2, A2-5a CGT; 3, Ch1 Amy; 4, Ch2 Amy; M, molecular size marker.
Western blot analysis.
Western blot analysis was performed to confirm immunologically that the chimeric enzymes consisted of polypeptides from Ba-S and A2-5a CGT (Fig. 2b and c). While Ba-S protein was immunoreactive to rabbit antiserum raised against Ba-L (anti-Ba-L) (Fig. 2b, lane 1) and not to that raised against A2-5a CGT (anti-CGT) (Fig. 2c, lane 1), A2-5a CGT protein was immunoreactive to anti-CGT (Fig. 2c, lane 2) and not to anti-Ba-L (Fig. 2b, lane 2). Ch1 Amy and Ch2 Amy proteins were immunoreactive to both anti-Ba-S (Fig. 2b, lanes 3 and 4, respectively) and anti-CGT (Fig. 2c, lanes 3 and 4, respectively).
Comparison of the characteristics of Ba-S and the chimeric enzymes. (i) α-Amylase activities.
Table 1 shows the specific activities and molar catalytic activities of B. subtilis X-23 α-amylases and the chimeric enzymes. While Ba-L, Ba-S, and Ch1 Amy exhibited almost identical specific activities when enzyme activity was evaluated on a molar basis, the activity of Ch2 Amy was ca. one-eighth of those of the other three enzymes.
(ii) Raw starch-binding and -digesting abilities.
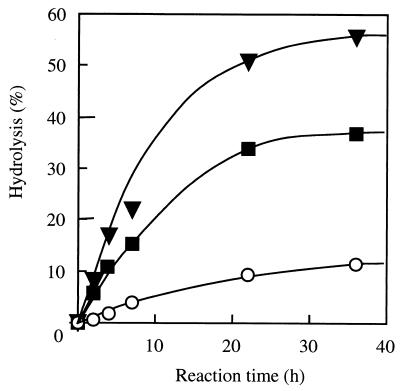
The amount (percentage) of enzyme adsorbed to raw starch was measured as described previously (32). The amounts of enzyme adsorbed to raw starch were as follows: Ba-S, 9.4%; CGT, 68.5%; Ch1 Amy, 49.8%; Ch2 Amy, 63.0%; PPA, 76.7%; and TAA, 3.2 %. For GGT, CGTase was from alkalophilic Bacillus sp. strain A2-5a, and for PPA and TAA, the α-amylases were from porcine pancreas and A. oryzae, respectively. While TAA (negative control) and Ba-S were hardly adsorbed to raw starch, Ch1 Amy and Ch2 Amy were adsorbed to raw starch. The ra of Ch2 Amy was higher than that of Ch1 Amy and was almost the same as those of A2-5a CGT and PPA (positive controls). The percentages of conversion of raw starch by Ch1 Amy (39.5% at 36 h) and Ch2 Amy (56.6% at 36 h) were also much higher than that of Ba-S (11.3% at 36 h) (Fig. 3). These results clearly indicated that the raw starch-binding and -digesting abilities of A2-5a CGT were introduced to Ba-S.
FIG. 3.
Digestion of raw maize starch by Ba-S, Ch1 Amy, and Ch2 Amy. The same amounts of enzymes, Ba-S, Ch1 Amy, and Ch2 Amy, were used on a soluble starch-hydrolyzing activity basis. A reaction mixture containing 0.3 g of raw maize starch, 37 ml of deionized water, 6 ml of 0.1 M sodium-acetate buffer (pH 5.5), 6 ml of enzyme solution (45 U/ml), and 100 μl of toluene was incubated with shaking at 30°C. At suitable intervals, the reducing sugar formed in 1 ml of the reaction mixture was measured. The degree of hydrolysis (percentage) is indicated in terms of reducing sugars as glucose per total carbohydrate. Symbols: ○, Ba-S; ■, Ch1 Amy; ▴, Ch2 Amy.
(iii) Amylolytic pattern.
Soluble starch was hydrolyzed with Ba-S and the chimeric enzymes and analyzed by paper chromatography (data not shown) as described previously (32). There were no differences between Ba-S, Ch1 Amy, and Ch2 Amy with regard to their pattern of action on soluble starch.
(iv) Transglycosylation of glucosyl residues of G3-PNP to Gn-PNP and hydroquinone.
The ratio of transglycosylation products, i.e., G4-PNP, G5-PNP, G6-PNP, G7-PNP, and G8-PNP, to reduction of the substrate (G3-PNP) and the ratio of transglycosylated hydroquinone, i.e., HQ-G1, HQ-G2, HQ-G3, and HQ-G4, to the reduction of G3-PNP were measured as described previously (32). There were no differences between Ba-S, Ch1 Amy, and Ch2 Amy (data not shown).
(v) Effects of pH and temperature on enzyme activity and stability.
The optimum pHs, optimum temperatures, and the thermal stabilities for Ba-S, Ch1 Amy, and Ch2 Amy were all the same (data not shown).
DISCUSSION
We successfully introduced raw starch-binding and -digesting abilities to B. subtilis X-23 α-amylase. This enzyme strongly induces transglycosylation in hydroquinone and kojic acid (30, 31). Therefore, introducing the ability to act on raw starch to this unique enzyme is very interesting from the standpoint of industrial application.
It is generally known that hybrid proteins are mostly unstable, and it is extremely difficult to combine the functions of each of the constituents. However, we succeeded in producing hybrid proteins which show both α-amylase activity and raw starch-binding and -digesting abilities. Three important factors might have played a role in our success. First, the folding of Ba-S protein is strong enough to retain its function as an α-amylase with or without the following extra carboxyl-terminal polypeptide (32). Second, there are several amino acid residues that may link two functional proteins, as in glucoamylases (35, 42). Indeed, we identified a flexible structure at the carboxyl terminus of the predicted three-dimensional structure of Ba-S (32). This factor may not be essential, as seen in the raw starch-digesting α-amylase from the yeast Cryptococcus sp. strain S-2 (10), which lacks a linker segment. Third, domain E of A2-5a CGT is functionally independent, as seen in the raw starch-binding domains of B. macerans CGTase and A. awamori glucoamylase (2).
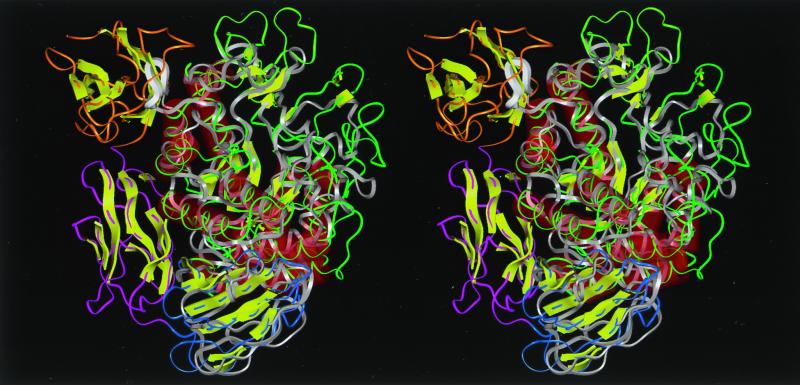
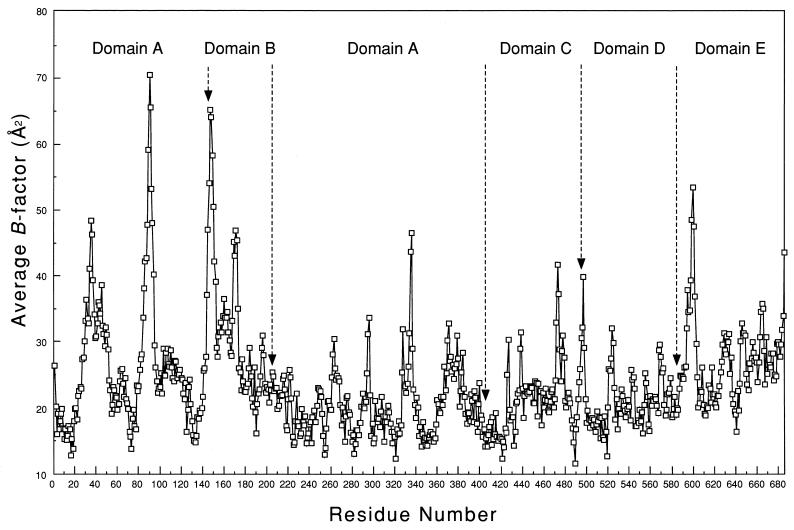
The remarkable decrease in the molar catalytic activity of Ch2 Amy (Table 1) may be due to structural distortion of the enzyme. As shown in Fig. 4, domain E of Ch1 Amy and domains D and E of Ch2 Amy could be folded in the same pattern as those of B. circulans strain 251 CGTase. We will consider the three-dimensional structure of B. circulans strain 251 CGTase instead of A2-5a CGT, since their amino acid sequences are very similar (58.4% homology) (Fig. 1). Since Ba-S and CGTases belong to the α-amylase family (20, 39), it is quite reasonable to think that the domains of chimeric enzymes maintain interactions between other domains similar to those in CGTases, and that the domain(s) following domain C (Fig. 1a) of chimeric enzymes are in a location similar to those of B. circulans 251 CGTase. When a three-dimensional structural model of Ba-S was overlaid with the three-dimensional structure of B. circulans 251 CGTase using the excellent method recently reported by Holm and Sander (9) (Fig. 4), it was obvious that the loop between β4 and α3 of Ba-S collides with domain E at the main-chain level. This structural conflict causes some conformational change in the catalytic domain and decreases the catalytic activity of Ch2 Amy. On the other hand, it is highly probable that domain E of Ch1 Amy has little effect on the catalytic domain since domain E of Ch1 Amy is most likely to occupy the location of domain D of the CGTase. Indeed, a mobile segment with a B-factor of >40 Å2 (Fig. 5) is located between domain C and domain D of the CGTase, indicating that the domain following domain C can behave in a flexible manner and settle into a suitable position with respect to the stability of the whole protein. This may explain why the molar catalytic activity of Ch1 Amy was similar to that of Ba-S (Table 1).
FIG. 4.
Stereoview of the predicted structure of Ba-S overlaid with the crystal structure of CGTase of B. circulans 251 (25) seen from the side of the (β/α)8-barrel. A structural comparison by aligning distance matrices was performed by the method of Holm and Sander (9). Molecular modeling of Ba-S was performed based on the three-dimensional structure of BSUA complexed with maltopentaose (5) using Discover-Insight II software (version 4.3; Molecular Simulation, Inc.) on an ONYX2 workstation (Silicon Graphics, Inc.). Residues 422 to 428 are not shown since the corresponding structure for BSUA could not be determined due to disorder (5). Ba-S is shown in white, and the loop between β4 and α4 of domain A (residues Pro180 to Ser187), which collides with a β-sheet structure (residues Asn635 to Pro643) in domain E of the CGTase, is shown as a thick ribbon. Green, yellow-green, blue, pink, and orange indicate domains A, B, C, D, and E of the CGTase, respectively. Red cylinders and yellow arrows represent α-helices and β-strands, respectively.
FIG. 5.
Variation of the atomic temperature factors averaged for the main-chain atoms of CGTase from B. circulans 251 (25). The data were obtained from the Protein Data Bank (entry identification: 1CDG). The average B factor of each residue is plotted against the residue number. Domain limits are indicated by arrows.
In Fig. 3, Ba-S showed a percent conversion of raw starch (11.3% at 36 h), although it does not have a raw starch-binding domain. It might be that part of the amorphous structure that exists in raw starch is digested by Ba-S. A similar phenomenon has been observed in a study using TAA (10), which also does not have a raw starch-binding domain. Ch2 Amy exhibited higher raw starch-binding and -digesting abilities than Ch1 Amy, as described previously. One interpretation for the higher raw starch digestion ability of Ch2 Amy could be that a high concentration of domain E in the Ch2 Amy reaction mixture, compared with that of Ba-S or Ch1 Amy, to adjust the amount of enzymes on a starch-hydrolyzing activity basis disrupted water aggregates surrounding raw maize starch and that this made it easier for the catalytic domain to attack the hydrated starch micelle (7, 37). Another interpretation is that domain E of Ch2 Amy was in a better location, one similar to that of the CGTase (Fig. 4), than that of Ch1 Amy for binding to the substrate and expressing its function. Thus, it is possible that domain D of CGTase may affect the location of domain E so that it is functionally optimized.
Recently, Wind et al. reported that truncation of domains D and E of the CGTase from T. thermosulfurigenes EM1 caused a remarkable decrease in its β-cyclomaltodextrin-forming activity and an increase in its saccharifying activity (43), indicating that domain D or domain E of this CGTase is related to its transglycosylation activity. In this study, we have shown that domain E of A2-5a CGT plays a role in its binding to raw starch and is functionally independent of the other four domains. We also predicted that domain D of A2-5a CGT might indirectly play a role in helping domain E act on raw starch. Maltogenic α-amylase from Bacillus stearothermophilus has been recently reported to exhibit a five-domain organization extremely similar to that of CGTases (3). Therefore, domains D and E of CGTases may not be directly related to transglycosylation activity. Indeed, we increased the transglycosylation activity of neopullulanase by increasing the hydrophobicity along the entrance path of the attacking water molecule, which is most likely used for the hydrolysis reaction (22). Further work is now in progress to clarify the factors that determine the fate of the reaction, i.e., hydrolysis or transglycosylation, in the α-amylase family (21, 39).
ACKNOWLEDGMENTS
This work was supported in part by a grant for the development of a next-generation bioreactor system from the Society for Techno-Innovation of Agriculture, Forestry, and Fisheries (STAFF).
REFERENCES
- 1.Boel E, Brady L, Brzozowski A, Derewenda Z, Dodson G, Jensen V, Petersen S, Swift H, Thim L, Woldike H. Calcium binding in α-amylases: an X-ray diffraction study at 2.1-Å resolution of two enzymes from Aspergillus. Biochemistry. 1990;29:6244–6249. doi: 10.1021/bi00478a019. [DOI] [PubMed] [Google Scholar]
- 2.Dalmia B, Schütte K, Nikolv Z. Domain E of Bacillus macerans cyclodextrin glucanotransferase: an independent starch-binding domain. Biotechnol Bioeng. 1995;47:575–584. doi: 10.1002/bit.260470510. [DOI] [PubMed] [Google Scholar]
- 3.Dauter Z, Dauter M, Brzozowski A M, Christensen S, Borchert T V, Beier L, Wilson K S, Davies G J. X-ray structure of Novamyl, the five-domain “maltogenic” α-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7 Å resolution. Biochemistry. 1999;38:8385–8392. doi: 10.1021/bi990256l. [DOI] [PubMed] [Google Scholar]
- 4.Dubois M, Gilles K A, Hamilton J K, Robers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 5.Fujimoto Z, Takase K, Doui N, Momma M, Matsumoto T, Mizuno H. Crystal structure of a catalytic-site mutant α-amylase from Bacillus subtilis complexed with maltopentaose. J Mol Biol. 1998;277:393–407. doi: 10.1006/jmbi.1997.1599. [DOI] [PubMed] [Google Scholar]
- 6.Fujiwara S, Kakihara H, Woo K B, Lejeune A, Kanemoto M, Sakaguchi K, Imanaka T. Cyclization characteristics of cyclodextrin glucanotransferase are conferred by the NH2-terminal region of the enzyme. Appl Environ Microbiol. 1992;58:4016–4025. doi: 10.1128/aem.58.12.4016-4025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashida H, Nakahara K, Kanlayakrit W, Hara T, Teramoto Y. Characteristics and function of raw-starch-affinity site on Aspergillus awamori var. kawachi glucoamylase I molecule. Agric Biol Chem. 1989;53:143–149. [Google Scholar]
- 8.Hellman J, Mäntsälä P. Construction of an Escherichia coli export-affinity vector for expression and purification of foreign proteins by fusion to cyclomaltodextrin glucanotransferase. J Biotechnol. 1992;23:19–34. doi: 10.1016/0168-1656(92)90097-s. [DOI] [PubMed] [Google Scholar]
- 9.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–602. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 10.Iefuji H, Chino M, Kato M, Iimura Y. Raw-starch-digesting and thermostable α-amylase from the yeast Cryptococcus sp. S-2: purification, characterization, cloning and sequencing. Biochem J. 1996;318:989–996. doi: 10.1042/bj3180989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imanaka T, Kuriki T. Pattern of action of Bacillus stearothermophilus neopullulanase on pullulan. J Bacteriol. 1989;171:369–374. doi: 10.1128/jb.171.1.369-374.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ish-Horowicz D, Burke J F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;14:8605–8613. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janecek S, Svensson B, Henrissat B. Domain evolution in the α-amylase family. J Mol Evol. 1997;45:322–331. doi: 10.1007/pl00006236. [DOI] [PubMed] [Google Scholar]
- 14.Jespersen H M, MacGregor E A, Henrissat B, Sierks M R, Svensson B. Starch- and glycogen-debranching and branching enzymes: prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J Protein Chem. 1993;12:791–805. doi: 10.1007/BF01024938. [DOI] [PubMed] [Google Scholar]
- 15.Jespersen H M, MacGregor E A, Sierks M R, Svensson B. Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem J. 1991;280:51–55. doi: 10.1042/bj2800051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juge N, Søgaard M, Chaix J C, Martin-Eauclaire M F, Svensson B, Marchis-Mouren G, Guo X J. Comparison of barley malt α-amylase isozymes 1 and 2: construction of cDNA hybrids by in vivo recombination and their expression in yeast. Gene. 1993;130:159–166. doi: 10.1016/0378-1119(93)90415-y. [DOI] [PubMed] [Google Scholar]
- 17.Klein C, Schulz G E. Structure of cyclodextrin glycosyltransferase refined at 2.0 Å resolution. J Mol Biol. 1991;217:737–750. doi: 10.1016/0022-2836(91)90530-j. [DOI] [PubMed] [Google Scholar]
- 18.Kometani T, Terada Y, Nishimura T, Takii H, Okada S. Purification and characterization of cyclodextrin glucanotransferase from an alkalophilic Bacillus species and transglycosylation at alkaline pHs. Biosci Biotechnol Biochem. 1994;58:517–520. [Google Scholar]
- 19.Kubota M, Matsuura Y, Sakai S, Katsube Y. Molecular structure of B. stearothermophilus cyclodextrin glucanotransferase and analysis of substrate binding site. Denpun Kagaku. 1991;38:141–146. [Google Scholar]
- 20.Kuriki T, Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus. J Gen Microbiol. 1989;135:1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- 21.Kuriki T, Imanaka T. The concept of the α-amylase family: structural similarity and common catalytic mechanism. J Biosci Bioeng. 1999;87:557–565. doi: 10.1016/s1389-1723(99)80114-5. [DOI] [PubMed] [Google Scholar]
- 22.Kuriki T, Kaneko H, Yanase M, Takata H, Shimada J, Handa S, Takada T, Umeyama H, Okada S. Controlling substrate preference and transglycosylation activity of neopullulanase by manipulating steric constraint and hydrophobicity in active center. J Biol Chem. 1996;271:17321–17329. doi: 10.1074/jbc.271.29.17321. [DOI] [PubMed] [Google Scholar]
- 23.Kuriki T, Okada S. Amylase Research Society of Japan (ed.) 1995. A new concept of the criteria for classification of various amylolytic enzymes and related enzymes; similarities in specificities and structures; pp. 87–92. Enzyme chemistry and molecular biology of amylases and related enzymes. CRC Press, Boca Raton, Fla. [Google Scholar]
- 24.Kuriki T, Stewart D C, Preiss J. Construction of chimeric enzymes out of Maize endosperm branching enzymes I and II: activity and properties. J Biol Chem. 1997;272:28999–29004. doi: 10.1074/jbc.272.46.28999. [DOI] [PubMed] [Google Scholar]
- 25.Lawson C L, Vanmontfort R, Strokopytov B, Rozeboom H J, Kalk K H, Devries G E, Penninga D, Dijkhuizen L, Dijkstra B W. Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form. J Mol Biol. 1994;236:590–600. doi: 10.1006/jmbi.1994.1168. [DOI] [PubMed] [Google Scholar]
- 26.Matsuura Y, Kusunoki M, Harada W, Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem (Tokyo) 1984;95:697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- 27.Moraes L M P, Astolfi-filho S, Oliver S G. Development of yeast strains for the efficient utilisation of starch: evaluation of constructs that express α-amylase and glucoamylase separately or as bifunctional fusion proteins. Appl Microbiol Biotechnol. 1995;43:1067–1076. doi: 10.1007/BF00166927. [DOI] [PubMed] [Google Scholar]
- 28.Mori H, Tanizawa K, Fukui T. A chimeric α-glucan phosphorylase of plant type L and H isozymes: functional role of 78-residue insertion in type L isozyme. J Biol Chem. 1993;268:5574–5581. [PubMed] [Google Scholar]
- 29.Nakano Y J, Kuramitsu H K. Mechanism of Streptococcus mutans glucosyltransferase: hybrid-enzyme analysis. J Bacteriol. 1992;174:5639–5646. doi: 10.1128/jb.174.17.5639-5646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura T, Kometani T, Takii H, Terada Y, Okada S. Acceptor specificity in the glucosylation reaction of Bacillus subtilis X-23 α-amylase towards various phenolic compounds and the structure of kojic acid glucoside. J Ferment Bioeng. 1994;78:37–41. [Google Scholar]
- 31.Nishimura T, Kometani T, Takii H, Terada Y, Okada S. Purification and some properties of α-amylase from Bacillus subtilis X-23 that glucosylates phenolic compounds such as hydroquinone. J Ferment Bioeng. 1994;78:31–36. [Google Scholar]
- 32.Ohdan K, Kuriki T, Kaneko H, Shimada J, Takada T, Fujimoto Z, Mizuno H, Okada S. Characteristics of two forms of α-amylases and structural implication. Appl Environ Microbiol. 1999;65:4652–4658. doi: 10.1128/aem.65.10.4652-4658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohdan K, Kuriki T, Takata H, Okada H. Cloning of cyclodextrin glucanotransferase gene from alkalophilic Bacillus sp. A2-5a and analysis of the raw starch-binding domain. Appl Microbiol Biotechnol. 2000;53:430–434. doi: 10.1007/s002530051637. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Semimaru T, Goto M, Furukawa K, Hayashida S. Functional analysis of the threonine-rich and serine-rich Gp-I domain of glucoamylase I from Aspergillus awamori var. kawachi. Appl Environ Microbiol. 1995;61:2885–2890. doi: 10.1128/aem.61.8.2885-2890.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibuya I, Tamura G, Shima H, Ishikawa T, Hara S. Construction of an α-amylase/glucoamylase fusion gene and its expression in Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 1992;56:884–889. doi: 10.1271/bbb.56.884. [DOI] [PubMed] [Google Scholar]
- 37.Southall S M, Simpson P J, Gilbert H J, Williamson G, Williamson M P. The starch-binding domain from glucoamylase disrupts the structure of starch. FEBS Lett. 1999;447:58–60. doi: 10.1016/s0014-5793(99)00263-x. [DOI] [PubMed] [Google Scholar]
- 38.Svensson B, Jespersen H, Sierks M R, MacGregor E A. Sequence homology between putative raw-starch binding domains from different starch-degrading enzymes. Biochem J. 1989;264:309–311. doi: 10.1042/bj2640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takata H, Kuriki T, Okada S, Takesada Y, Iizuka M, Minamiura N, Imanaka T. Action of neopullulanase: neopullulanase catalyzes both hydrolysis and transglycosylation at α-(1→4)- and α-(1→6)-glucosidic linkages. J Biol Chem. 1992;267:18447–18452. [PubMed] [Google Scholar]
- 40.Tanaka A, Karita S, Kosuge Y, Senoo K, Obata H, Kitamoto N. Thermal unfolding of the starch binding domain of Aspergillus niger glucoamylase. Biosci Biotechnol Biochem. 1998;62:2127–2132. doi: 10.1271/bbb.62.2127. [DOI] [PubMed] [Google Scholar]
- 41.Terashima M, Hosono M, Katoh S. Functional roles of protein domains on rice α-amylase activity. Appl Microbiol Biotechnol. 1997;47:364–367. doi: 10.1007/s002530050941. [DOI] [PubMed] [Google Scholar]
- 42.Williamson G, Belshaw N J, Williamson M P. O-Glycosylation in Aspergillus glucoamylase. Conformation and role in binding. Biochem J. 1992;282:423–428. doi: 10.1042/bj2820423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wind R D, Buitelaar R M, Dijkhuizen L. Engineering of factors determining α-amylase and cyclodextrin glycosyltransferase specificity in the cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1. Eur J Biochem. 1998;253:598–605. doi: 10.1046/j.1432-1327.1998.2530598.x. [DOI] [PubMed] [Google Scholar]
- 44.Yon J, Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]