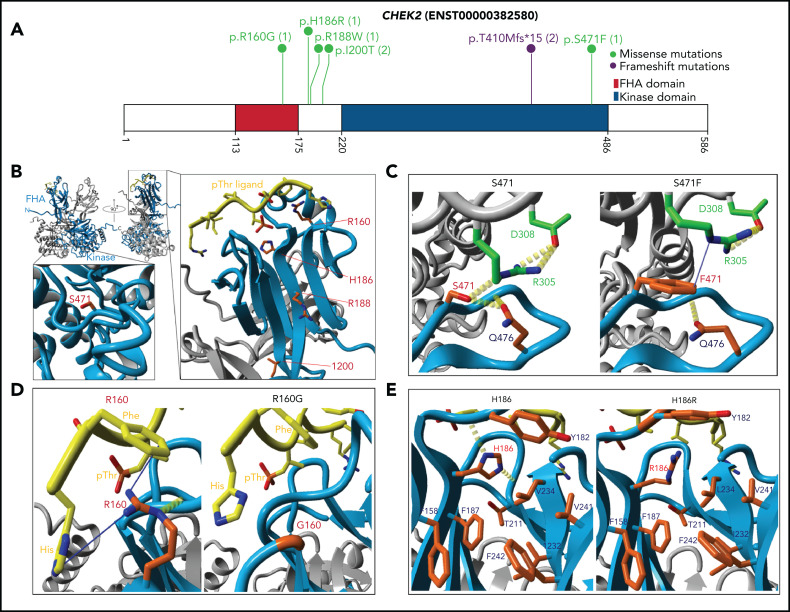
Figure 4.
CHEK2 structural model for predicting impact of variants on protein functions. (A) Location of CHEK2 P/LP germline variants, with the number of unique patients shown in parentheses. (B) 3D CHEK2 dimer model with the functional domains (FHA and kinase domain), with respective monomers in light blue and gray, and a pThr ligand in yellow. Five residues affected by P/LP missense variants identified in this study cohort (red labels) are annotated in the extended windows. (C) The S471F in the kinase domain appears to abrogate interaction with residue R305 (intermolecular), and possibly residue Q476 (intramolecular), that destabilize the dimerization of CHEK2, reducing its efficacy of transphosphorylation and activation. (D) The R160G appears critical for stabilizing 2 N-terminal residues (H1 and F2) of the ligand by 2 Cation-Pi interactions. R160 engages in an inter-β-sheet H-bond which further stabilizes the FHA-ligand interface. Alteration to a glycine likely does 2 things to it: (1) abrogates interactions between the FHA domain and ligand and (2) entropically destabilizes the structure at position 160. (E) The H186R in the FHA domain disrupts the interactions with the polar cap and the hydrophobic core, destabilizing FHA domain structure. Additional stabilizing H-bonds (in yellow) are lost, which may be critical for ligand interface integrity and association.