Abstract
Heavy metal resistance by bacteria is a topic of much importance to the bioremediation of contaminated soils and sediments. We report here the isolation of a highly cadmium-resistant Klebsiella planticola strain, Cd-1, from reducing salt marsh sediments. The strain grows in up to 15 mM CdCl2 under a wide range of NaCl concentrations and at acidic or neutral pH. In growth medium amended with thiosulfate, it precipitated significant amounts of cadmium sulfide (CdS), as confirmed by x-absorption spectroscopy. In comparison with various other strains tested, Cd-1 is superior for precipitating CdS in cultures containing thiosulfate. Thus, our results suggest that Cd-1 is a good candidate for the accelerated bioremediation of systems contaminated by high levels of cadmium.
Cadmium, a highly toxic metal and a group B1 human carcinogen, is commonly ranked among the top 10 priority pollutants by U.S. regulatory agencies. Significant amounts of this metal often infiltrate the groundwater at hazardous waste sites. A practical approach to minimizing Cd(II) levels in the subsurface systems is precipitating it with hydrogen sulfide as the highly insoluble cadmium sulfide (CdS). In reducing environments, sulfate-reducing bacteria (SRB) generate hydrogen sulfide through dissimilatory sulfate reduction. However, little is known about the heavy metal resistance and growth of these bacteria in contaminated systems. Recent studies by Poulson et al. (14) and White and Gadd (22) indicate that even low levels of free Cd(II), Zn(II), or Ni(II) ions, i.e., 20 to 200 μM, are toxic to SRB, such as Desulfovibrio and Desulfotomaculum. Our unpublished studies (P. K. Sharma and M. A. Vairavamurthy) also indicate that at least the SRB from coastal salt marshes from Shelter Island, New York, are sensitive to cadmium. However, a new Klebsiella planticola strain, Cd-1, from the same coastal environment grew to saturation in 15 mM Cd(II) ions in a minimal anaerobic medium, and after thiosulfate was added to the growth medium, it transformed significant amounts of the dissolved cadmium to CdS, up to 50 times higher than that reported for highly purified and continuously fed sulfate-reducing biofilms (22) or batch cultures of a genetically engineered Escherichia coli with overexpressed thiosulfate reductase (S. W. Bang, D. S. Clark, and J. D. Keasling, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. Q-302, 1999). In general, facultative anaerobes, such as Cd-1, are attractive candidates for accelerated bioremediation applications, as large amounts of them can be grown rapidly with various nonhazardous substrates in aerobic reactors and can then be injected into anaerobic subsurface environments. In contrast, SRB often are slow growers and also may require expensive substrates, nutrients, or cofactors for growth.
In this report, we describe the isolation and basic characteristics of strain Cd-1. We show that it can grow in high levels of Cd(II) ions under a wide range of environmental conditions, including high salinity and acidic pH. Importantly, our studies demonstrate that Cd-1 can transform high levels of cadmium to CdS in the presence of thiosulfate. We compared the growth and the ability of Cd-1 to produce CdS in media containing high levels of cadmium with those of several other metal-resistant, metal-transforming, or metal-reducing ubiquitous facultative anaerobes. These results suggest that Cd-1 is a superior candidate for the anaerobic growth and transformation of cadmium, particularly in the presence of thiosulfate.
Isolation of Cd-1.
Strain Cd-1 was isolated from near-surface sediments (up to 6 in. deep) from a coastal salt marsh on Shelter Island, New York. The isolate was enriched by adding sediment (ca. 13 g per liter) to a medium containing 0.3 M NaCl, 10 mM Na2SO4, and 0, 50, 200, or 500 μM CdCl2; 2 g of powdered Spartina alterniflora tissue per liter was added as the source of carbon and nutrients. S. alterniflora is the dominant source of organic matter sustaining the microbial populations in the salt marshes in Shelter Island. The pH of the microcosms before incubation was 5.5. They were incubated in an anaerobic chamber (Coy Laboratory Products, Inc., Grass Lake, Mich.) at 24.5°C. The headspace of the sealed microcosms contained the chamber atmosphere, i.e., H2-CO2-N2 at a ratio of 5:5:90.
After incubation for 45 days, a liquid portion from the microcosm containing 500 μM CdCl2 was transferred into a second growth medium. This new medium contained 1 mM CdCl2, 10 mM pyruvic acid (C3H3O3Na), 5 mM α-d-glucose, 100 mM NaCl, 10 mg of yeast extract per liter, and 5 mM Ches [2-(cyclohexylamino)ethanesulfonic acid], along with a 4:1 mineral solution-to-metal solution ratio (solutions described later). Following a further 10-day incubation, a portion was transferred into fresh media; the resulting second-generation enrichment was incubated for 1 day and then was serially diluted. The serial dilutions were streaked onto nutrient agar plates containing 0.5 mM CdCl2, which were then incubated both inside and outside the anaerobic chamber. Only one morphotype appeared within 24 to 72 h under both redox conditions.
Identification and characterization of Cd-1. (i) Basic characteristics.
Strain Cd-1 is a gram-negative, nonmotile, facultative anaerobe. It can grow in >6% (wt/vol) NaCl in nutrient broth both aerobically and anaerobically; thus, it is a halotolerant bacterium. It is nutritionally diverse, with the ability to utilize all but 25 of the 95 substrates on Biolog GN MicroPlate. The compounds it was unable to use are cyclodextrin, erythritol, lactulose, xylitol, d-galactonic acid lactone, d-glucosaminic acid, α-, β-, or γ-hydroxy butyric acid, itaconic acid, α-keto-butyric acid, glutaric acid, valeric acid, propionic acid, sebacic acid, l-leucine, l-ornithine, l-pyroglutamic acid, l-threonine, d- or l-carnitine, γ-amino butyric acid, urocanic acid, phenyl ethylamine, 2-amino ethanol, and 2,3-butanediol.
(ii) 16S rRNA gene analysis.
A segment of approximately 1,500 bases of Cd-1's 16S rRNA gene (nearly the entire gene) was amplified and sequenced with an Applied Biosystems 373A automated DNA sequencer (Perkin-Elmer–Applied Biosystems, Foster City, Calif.), using the Taq DyeDeoxy terminator cycle-sequencing method. The PCR amplification primers were fD1 and rP1 (21), while the DNA sequencing primers were A, C, G, H, H-complement, P, and P-complement (1). The assembled sequence, corresponding to positions 20 to 1350 of the Escherichia coli 16S rDNA sequence, was aligned with sequences for selected reference strains, i.e., those with the most similar 16S rDNA sequences, obtained from RDB and the GenBank and EMBL databases. The phylogenetic position of Cd-1 was analyzed with standard methods: distance matrix, maximum likelihood, and parsimony. Distances were calculated by the methods described by Jukes and Cantor (12). The PHYLIP version 3.5c programs (8) were used for distance matrix analyses, after which phylogenies were estimated using the algorithms of DeSoete (7) and/or Fitch and Margoliash (10). The DNAML component of the PHYLIP package was used for maximum likelihood analyses, while the PAUP program (17) was employed to construct the most parsimonious phylogenetic tree.
The 16S rDNA sequences of Cd-1 and an official strain of K. planticola (GenBank accession no. X93215) were 100% similar over the 1,327 bases that we compared, indicating that Cd-1 is a strain of K. planticola. These observations were confirmed with phylogenetic analysis; all three methods, i.e., distance matrix, maximum likelihood, and parsimony, showed a close relationship between Cd-1 and K. planticola.
(iii) Metabolic and biochemical analyses.
The Biolog System identified Cd-1 as K. planticola. The similarity index (SI) value for K. planticola and two different Cd-1 cultures was 0.514 and 0.651; it was below 0.14 for any other gram-negative strain.
The MIDI/Hewlett-Packard Microbial Identification System also placed Cd-1 under Enterobacteriaceae.
Growth of Cd-1 in Cd(II). (i) Composition of growth medium.
The anaerobic medium for strain Cd-1 contained the following per liter: 5 mM pyruvate, 1 mM α-d-glucose, 5 ml of mineral solution, 1.25 ml of metal solution, and 5 mM Trizma buffer (Sigma Chemical Co., St. Louis, Mo.). The mineral solution had the following composition per liter: 40 g of NaCl, 5 g of NH4SO4, 5 g of KCl, 5 g of β-glycerophosphate (C3H7O6PNa2), 5 g of MgCl2 · 6H2O, and 2 g of CaCl2 · 2H2O. A liter of the metal solution contained the following: 2 g of C10H14O8N2Na2 · 2H2O (pH was adjusted to 6.0 with freshly prepared KOH before adding the other compounds), 1 g of MnSO4 · H2O, 600 mg of FeCl2 · 4H2O, 200 mg of CoCl2 · 6H2O, 200 mg of (CH3COO)2Zn · 2H2O, 20 mg of CuCl2 · 2H2O, 20 mg of NiCl2 · 6H2O, and 20 mg of NaMoO4 · 2H2O. Any changes made in the medium's composition are listed elsewhere. The pH of the growth medium was around 7.1, except in experiments assessing the effect of different initial pH values on the growth in Cd(II). A 2%, i.e., 1:50, (vol/vol) inoculum was used unless indicated otherwise.
(ii) Role of NaCl.
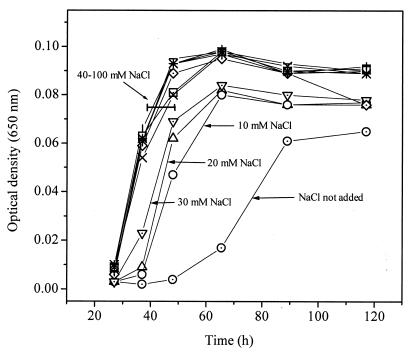
The presence of 40 to 100 mM NaCl shortened the lag phase or accelerated the growth rate in 1 mM Cd(II) (Fig. 1). However, in the absence of Cd(II), the growth rate was maximal when no NaCl was added (the growth medium contained around 3.4 mM NaCl from mineral solution; see also above). Studies with equimolar LiCl showed similar trends. These observations suggest that either an enhanced expression of Na+ extrusion systems (13) or an osmotic shock (9) protected the cells against Cd(II) toxicity. Also, less-toxic Cd—Cl coordination complexes may have been formed at a high NaCl concentration (6). Often, 100 mM NaCl was added to the growth medium, except in experiments to determine the effect of the varying NaCl concentration on Cd-1 in Cd(II).
FIG. 1.
Effect of NaCl (0 to 100 mM) on Cd-1 growth in 1 mM Cd(II). A 2-day-old anaerobic culture served as the inoculum. Repeated experiments showed similar trends.
(iii) Growth kinetics.
Under minimal fermentative conditions, Cd-1 reached the stationary phase within 1 to 2 days in 0.2 to 2.0 mM Cd(II), within 3 days in 5 mM Cd(II), and within 7 to 10 days in 7 to 15 mM Cd(II). Although the lag phase was longer with Cd(II) present, the apparent doubling time was nearly identical with or without 1 mM Cd(II), i.e., ∼3.5 h. However, in 5 mM Cd(II), the apparent doubling time was ∼10 h (doubling time values are averages from replicate cultures, and the ranges were negligible).
With or without Cd(II), Cd-1 did not grow to an optical density (OD) of >0.12 when supplied with 5 mM pyruvate and 1 mM glucose (initial OD, ∼0.008). For example, the average OD of five different cultures was 0.0801 after 28 h [0.2 to 5.0 mM Cd(II)] (Table 1). Since this maximum OD value was small, we monitored the concentrations of the viable cells to determine whether significant growth occurred. As Table 1 shows, there was an increase to about 61-fold in CFU within 30 h in Cd(II), ranging from 0.2 to 2.0 mM, and a ∼30-fold increase in 5 mM Cd(II) cultures (2× inoculum; with a 1× inoculum, a ∼10-fold increase occurred after 50 h, a time frame consistent with that observed in other experiments). Without Cd(II), a 50-fold increase in CFU was observed, a value comparable to that for cultures grown in 0.2 to 2.0 mM Cd(II) (Table 1).
TABLE 1.
The effect of Cd(II) on colony-forming ability of Cd-1a
| Cd(II) concn (mM) | OD650 (after 28 h) | Initial CFU concn (per ml)a | Final CFU concn (per ml) (after 30 h) |
|---|---|---|---|
| 0.2 | 0.082b | (1.2 ± 0.6) × 106c | (5.9 [0.7]) × 107d |
| 0.5 | 0.062 | 1.2 × 106 | (5.8 [1.2]) × 107d |
| 1.0 | 0.090 | 1.2 × 106 | 9.6 × 107 |
| 2.0 | 0.084 | 1.2 × 106 | 8.0 × 107 |
| 5.0 | 0.012 | 1.2 × 106 | 2.0 × 106e |
| 5.0 | 0.087 | 2.4 × 106f | 3.6 × 107 |
Experiment was done under fermentative conditions. The inoculum was grown overnight at 30°C under aerobic conditions in the same minimal medium (described in the text). The minimal medium lacking a carbon source for growth was used for dilution purposes, while aerobic nutrient agar was used for CFU counts.
Initial reading for all cultures was <0.008.
At this concentration, n was 4. An overnight culture was diluted 50 times.
Values are averages of duplicate data, with ranges in parentheses.
A count of 1.0 × 107 CFU per ml was observed after 50 h.
2× inoculum. Under similar conditions, cultures with 15 mM Cd(II) grew to an OD of ∼0.08 within 7 to 10 days and within 3 days when the inoculum was grown in nutrient broth.
(iv) Growth in Cd(II) at high salinity and low pH.
Since Cd-1 was isolated from coastal salt marsh sediments, we tested whether it could grow in high Cd(II) levels (0.5 to 5.0 mM) with concentrations of NaCl typical of estuarine and coastal systems (300 to 500 mM). Indeed, it grew well (final OD, ∼0.1), though at rates lower than those at the 100 mM NaCl level. We noticed that growth rates in 300 or 500 mM NaCl were lower than those in 100 mM NaCl, even in the absence of Cd(II).
Cd-1 grew well (OD, ∼0.1) in 2 mM Cd(II) at an initial external pH ranging from 4.16 to 7.18. However, its growth was faster under acidic pH, i.e., 4.16 to 5.78, in which cadmium exists as divalent cations (6). Without Cd(II), optimal growth occurred at near-neutral pH.
Transformation of cadmium to cadmium sulfide. (i) Synchrotron X-ray absorption spectroscopy.
Cd-1 cultures containing cadmium turned yellow during the stationary phase of growth, indicating that the isolate had transformed the added Cd(II) into CdS. We confirmed the identity of CdS with X-ray absorption spectroscopy using washed and freeze-dried late stationary-phase cells (harvested by centrifugation at 5,000 × g, 20 min, 4°C). Analyses included X-ray absorption near-edge structure (XANES) spectroscopy to characterize sulfur species and extended X-ray absorption fine structure (EXAFS) spectroscopy to examine the coordination environment around cadmium. We used the X-19A and X-18B beamlines at the National Synchrotron Light Source, Brookhaven National Laboratory, Upton, New York, for XANES and EXAFS studies, respectively.
(i) XANES data.
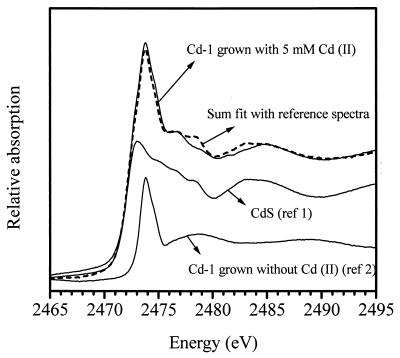
Sulfur K-edge XANES spectroscopy is a valuable technique for determining sulfur speciation because the absorption fine structure of the variety of sulfur forms are richly endowed with characteristic features, including edge energy, allowing the identification among various oxidation states and structures (18, 19). Consequently, an XANES spectrum of a sample can be deconvoluted to derive information of the different sulfur constituents in the sample. Figure 2 shows the XANES spectra of reference CdS and Cd-1 cells grown with or without 5 mM Cd(II) under fermentative conditions; the sum of the spectral fit is also shown. Deconvolution was performed according to published procedures (18). The spectra of Cd-1 show that cells grown in Cd(II) contained more reduced sulfur; the fit indicates that this difference may be attributed to CdS production. The deconvolution of the spectra indicate that CdS contributed to about 60% of reduced sulfur in cells grown with Cd(II).
FIG. 2.
Analysis of XANES spectrum of Cd-1 grown with 5 mM Cd(II) under fermentative conditions; spectra of reference cadmium sulfide (<5 μm) and Cd-1 grown without Cd(II) were used as models for deconvolution.
(ii) EXAFS data.
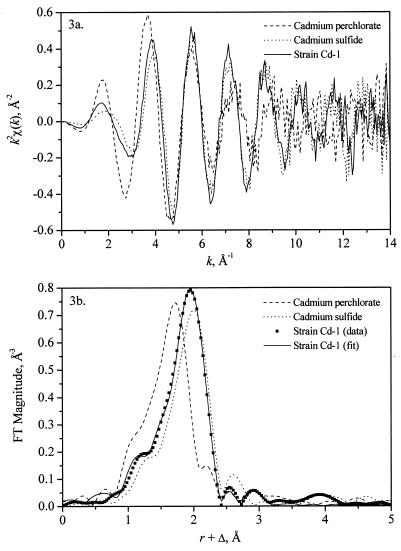
We used EXAFS to gain information on the local coordination environment of cadmium to verify its chemical identity in the Cd-1 samples. EXAFS data were analyzed by UWXAFS software (15). Figure 3a gives EXAFS data [k2-weighted χ(k), where k is the photoelectron wave vector] extracted from the averaged absorption coefficient after removing a smooth background function using the program AUTOBK. It is well established that when χ(k) is Fourier transformed over a finite k range, the result is a radial structure function exhibiting a series of peaks whose positions and amplitudes are related to the interatomic distances and the number of atoms in different coordination shells, respectively. The Fourier transform magnitudes (in the k range, from 2 to 12 Å−1) are shown in Fig. 3b. Essentially, Cd—O and Cd—S coordination numbers and distances and their disorder, together with their uncertainties, were obtained by a nonlinear least-squares fit of the theoretical EXAFS equation (16) to the data of strain Cd-1. The scattering amplitudes and phases used in the fits were extracted from the experimental standards CdS and Cd(ClO4)2 · H2O, respectively. The Cd—O and Cd—S distances in Cd(ClO4)2 and reference CdS are very different: 2.28 Å (4) and 2.52 Å (23), respectively. This difference in the first nearest-neighbor distance is manifested as the shift of the first peak in the Fourier transform magnitude of reference CdS toward higher distances. The strain Cd-1 sample is shifted to higher distances relative to Cd(ClO4)2, an independent indication that sulfur is present in the nearest environment around cadmium. Figure 3b also shows the fit to the strain Cd-1 data. The coordination numbers for the Cd—O and Cd—S bonds were 1.5 ± 1.0 (mean ± standard deviation) and 3.8 ± 1.0 Å, respectively.
FIG. 3.
Cadmium K-edge EXAFS data of strain Cd-1 grown anaerobically in 5 mM Cd(II) fitted with those of reference cadmium compounds, cadmium sulfide, and cadmium perchlorate hydrate. (a) k2-weighted χ(k); (b) fit of the Fourier-transformed data with EXAFS functions for Cd—O and Cd—S bonds.
The nature of the oxygenated cadmium ligands is still unclear. Nearly identical trends were observed for cells grown in 5 mM Cd(II) and 0.1, 0.2, or 0.5 M NaCl (the coordination numbers in this set for Cd—S bonds were 3.8 ± 1 [mean ± standard deviation], 3.8 ± 0.6, and 3.1 ± 0.6, whereas they were 1.5 ± 1, 3.8 ± 0.6, and 1.8 ± 0.7 for Cd—O bonds).
A weak and noisy EXAFS signal was observed for exponential-phase Cd-1 cells, indicating that significant cadmium was accumulated (i.e., >2 mmol per kg [dry weight] of cells, based upon lowest detection limit), primarily during the stationary phase.
(iii) Sulfide measurement.
Sulfide concentration reflecting the production of CdS by strain Cd-1 in the presence of S2O32− was determined by the methylene blue assay (Hach Company, Loveland, Ohio). The reagent produces a stable blue color within a few minutes in the presence of H2S. The estimated detection limit in deionized water matrix was ∼0.05 (± 0.02) nM, as suggested by the manufacturer. The reagent reacts positively to S2O32− as the latter dissociates to H2S at acidic pH. Therefore, we ensured the removal of S2O32− from the late-stationary phase cells before the methylene blue assay: centrifugation (5,000 × g, 20 min, 4°C) to harvest the cell pellet, followed by a rinse in deionized water. Sodium sulfide (Na2S · 9H2O) served as the source of sulfide standards; its concentration ranged from 100 to 500 μM. Absorption was measured at 665 nm with a Shimadzu UV-2101PC UV-visible light Scanning Spectrophotometer equipped with UVPC Personal Spectroscopy Software Version 3.9 (Shimadzu Scientific Instruments, Inc., Columbia, Md.). Our data indicate that strain Cd-1 produced around 1 mM CdS when grown in 5 mM Cd(II) in the presence of 5 or 10 mM S2O32− (average CdS value for 10 mM S2O32− duplicate cultures was 0.978 mM, with a standard deviation of ±0.052).
Growth of Cd-1 in other toxic metals.
Cd-1 grew within 2 to 10 days in the presence of 1 mM Cr(VI), As(V), Se(IV), Co(II), or Zn(II) and also in 0.5 mM Pb(II). Significant amounts of precipitates were formed in some cultures. For example, pink precipitates were formed in cultures with Se(IV) present indicating that the metal was reduced to elemental selenium. Precipitates also were formed in cultures with Co(II), Zn(II), or Pb(II) present. The color of the Cr(VI) media changed, but it is unclear whether significant precipitation occurred. As expected, no precipitates were formed in cultures with As(V) present.
Comparison of Cd-1 with closely related Klebsiella strains.
Comparisons of Cd-1 were made with strains of K. planticola ATCC 33531 and Klebsiella orinithinolytica ATCC 31898. Both strains grew in 5 mM Cd(II) in Cd-1's minimal medium described earlier or in Cd-1's medium amended with 5 mM NO3− or S2O32−. However, the lag phase of the ATCC strains was three to four times longer than that of Cd-1 (data not shown). The growth rate of K. planticola ATCC 33531 also was about five times slower. The lag phases and growth rates of the ATCC strains were comparable to those of Cd-1 in Cd(II)-free medium. The ATCC strains produced noticeable amounts of CdS in the presence of S2O32−; K. ornithinolytica produced around 1 mM CdS, but the amount was not determined for K. planticola. Here also, CdS production occurred primarily during the stationary phase of growth.
Comparison with other genera.
We also compared Cd-1 with many nonrelated strains, including several pseudomonads, two Shewanella strains (Shewanella oneidensis MR-1 and Shewanella putrefaciens CN32), Ralstonia eutropha ATCC 43123, Enterobacter agglomerans ATCC 27993, and Bacillus subtilis ATCC 35946. The pseudomonads included Comamonas testosteroni ATCC 1996, Pseudomonas putida ATCC 17484, and Pseudomonas aeruginosa CW-96-1 and three new isolates (Cd-2, Cd-4, and Cd-6).
Strains Cd-2 and Cd-4 were isolated from surficial sediments (top 4 in.) from the Mashomack Preserve, near a creek within 2 to 3 miles of the salt marsh from which strain Cd-1 was isolated. Biolog analysis suggested a close match of Cd-2 and Cd-4 with Pseudomonas fluorescens type C (SI, 0.575) and Pseudomonas resinovorans (SI, 0.257), respectively. Strain Cd-6 was isolated from a tetrachloroethene- to -cis-1,2-dichloroethene -dehalogenating anaerobic culture derived from tetrachloroethene-contaminated aquifer material obtained from Victoria, Tex. (V. Warikoo and P. K. Sharma, unpublished data). Cd-6 showed a close match with P. aeruginosa (Biolog SI, 0.556). All strains grew well on aerobic nutrient agar containing 1 mM CdCl2.
Past studies on metal resistance by pseudomonads mainly emphasized aerobic growth. For example, P. aeruginosa CW-96-1 was shown to grow well aerobically in up to 5 mM cadmium in a medium containing citrate and S2O32− (20). In contrast, our experiments were performed anaerobically with 5 mM CdCl2. We used Cd-1's minimal growth medium with or without 5 mM NO3− or S2O32−. Our studies showed no growth of the pseudomonads (including P. aeruginosa CW-96-1) in the presence of 5 mM cadmium under fermentative, nitrate-respiring, or thiosulfate-respiring conditions. Although S. oneidensis MR-1 grew well (OD of 0.06 within 82 h) and generated sulfide under thiosulfate-respiring conditions, it failed to grow when Cd(II) was present. Of the different strains tested, R. eutropha CH34 (3) was the only one that grew in 5 mM Cd(II) (final OD of 0.076 after 360 h). However, CH34 did not grow under thiosulfate-respiring conditions, even in the absence of cadmium. Thus, this bacterium lacks the potential to stabilize cadmium as CdS.
Cd(II) resistance in Klebsiella: a widespread phenomenon?
We found that at least three Klebsiella strains grew in high levels of Cd(II). A previous study (5) described a marine Klebsiella pneumoniae culture that grew in up to 20 mM Cd(II) under nutrient-rich aerobic conditions. However, it is not clear whether the ability to resist and grow in high Cd(II) levels is a widespread phenomenon in Klebsiella. Bhattacharyya et al. (2) screened 50 enterics, including 16 Klebsiella aerogenes (K. pneumoniae) strains, and showed that only one strain each of K. aerogenes, E. coli, and Serratia marcenes resisted 50 μM CdCl2 aerobically. Holmes et al. (11) found that the composition of the medium affects Cd(II) resistance in K. pneumoniae; the organism becomes highly sensitive to Cd(II) when some specific buffers are added to a defined growth medium [e.g., tolerance level decreases from 2 mM Cd(II) in a phosphate- or Tricine-buffered medium to 10 to 150 μM Cd(II) in a medium buffered with Tris or other buffers]. However, our strain, Cd-1, grew in mM Cd(II) levels in a Tris-buffered minimal medium.
Conclusions.
Our results clearly show the superiority of the new isolate, Cd-1, in anaerobically transforming Cd(II) to CdS, particularly when the metal is present as complexes of thiosulfate. It grew in up to 15 mM Cd(II) ions in a minimal anaerobic medium and thrived both aerobically and anaerobically under various environments, even with high salinities (>6% [wt/vol] NaCl) and pH range of 4 to 8. In fact, we are not aware of a previous study showing resistance by any eubacterium or archaebacterium to levels as high as 15 mM Cd(II) ions in a minimal anaerobic medium. Cd-1 also grew in high levels (0.5 to 1.0 mM) of various toxic metals and cocontaminants, such as Cr(VI), As(V), Se(IV), Co(II), Pb(II), and Zn(II). Thus, we believe strain Cd-1 to be a useful organism for accelerated bioremediation processes under diverse geochemical conditions.
American Type Culture Collection (ATCC) accession number.
Strain Cd-1 has been deposited in ATCC under accession no. 7008340.
Nucleotide sequence accession number.
The 16S rDNA sequence (bases 1 to 1339) of strain Cd-1 has been deposited in GenBank under accession no. AF175281.
Acknowledgments
This study was supported by the Office of Biological and Environmental Research of the U.S. Department of Energy under Prime Contract no. DE-AC02-98CH10886 with the Brookhaven National Laboratory (BNL).
We thank Mike McInerney of University of Oklahoma for helpful suggestions.
REFERENCES
- 1.Balkwill D L, Reeves R H, Drake G R, Reeves J Y, Crocker F H, King M B, Boone D R. Phylogenetic characterization of bacteria in the Subsurface Microbial Culture Collection. FEMS Microbiol Rev. 1997;20:201–216. doi: 10.1111/j.1574-6976.1997.tb00309.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya G, Chaudhuri J, Bhakta S, Mandal A. Cadmium resistance in some members of Enterobacteriaceae. Indian J Exp Biol. 1989;27:574–575. [PubMed] [Google Scholar]
- 3.Brim H, Heyndrickx M, de Vos P, Wilmotte A, Springael D, Schlegel H G, Mergeay M. Amplified rDNA restriction analysis and further genotypic characterization of metal-resistant soil bacteria and related facultative hydrogenotrophs. Syst Appl Microbiol. 1999;22:258–268. doi: 10.1016/S0723-2020(99)80073-3. [DOI] [PubMed] [Google Scholar]
- 4.Caminiti R, Johansson G. On the structures of cadmium sulfate complexes in aqueous solutions. Acta Chem Scand A. 1981;35:373–381. [Google Scholar]
- 5.Choudhury P, Kumar R. Multidrug- and metal-resistant strains of Klebsiella pneumoniae isolated from Penaeus monodon of the coastal waters of deltaic Sundarban. Can J Microbiol. 1998;44:186–189. [PubMed] [Google Scholar]
- 6.Collins Y E, Stotzky G. Factors affecting the toxicity of heavy metals to microbes. In: Beveridge T J, Doyle R J, editors. Metals and bacteria. New York, N.Y: John Wiley & Sons, Inc.; 1989. pp. 31–89. [Google Scholar]
- 7.DeSoete G. A least squares algorithm for fitting additive trees to proximity data. Psychometrika. 1983;48:621–626. [Google Scholar]
- 8.Felsentein J. PHYLIP (Phylogeny Inference Package), version 3.5c. Seattle: University of Washington; 1993. [Google Scholar]
- 9.Ferianc P, Farewell A, Nystrom T. The cadmium-stress stimulon of Escherichia coli K-12. Microbiology. 1998;144:1045–1050. doi: 10.1099/00221287-144-4-1045. [DOI] [PubMed] [Google Scholar]
- 10.Fitch W M, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 11.Holmes J D, Smith P R, Evans Gowing R, Richardson D J, Russell D A, Sodeau J R. Energy-dispersive X-ray analysis of the extracellular cadmium sulfide crystallites of Klebsiella aerogenes. Arch Microbiol. 1995;163:143–147. doi: 10.1007/BF00381789. [DOI] [PubMed] [Google Scholar]
- 12.Jukes T H, Cantor C R. Evolution of protein molecules. In: Muro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1963. pp. 21–132. [Google Scholar]
- 13.Karpel R, Alon T, Glaser G, Schuldiner S, Padan E. Expression of sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J Biol Chem. 1991;266:21753–21759. [PubMed] [Google Scholar]
- 14.Poulson S R, Colberg P J S, Drever J I. Toxicity of heavy metals (Ni, Zn) to Desulfovibrio desulfuricans. Geomicrobiol J. 1997;14:41–49. [Google Scholar]
- 15.Stern E A, Newville M, Rave B, Yacoby Y, Haskel D. The UWXAFS data analysis package: philosophy and details. Physica B. 1995;208/209:117–120. [Google Scholar]
- 16.Stern E A, Heald S M. Basic principles and applications of EXAFS. In: Koch E E, editor. Handbook on synchrotron radiation. Amsterdam, The Netherlands: North-Holland; 1983. pp. 955–1014. [Google Scholar]
- 17.Swofford D L. PAUP: phylogenetic analysis using parsimony, version 3.1.1. Champaign: Illinois Natural History Survey; 1993. [Google Scholar]
- 18.Vairavamurthy A. Using x-ray absorption to probe sulfur oxidation states in complex molecules. Spectrochim Acta Part A. 1998;54:2009–2017. [Google Scholar]
- 19.Vairavamurthy M A, Maletic D, Wang S, Manowitz B, Eglinton T, Loyons T. Characterization of sulfur-containing functional groups in sedimentary humic substances by x-ray absorption near-edge structure spectroscopy. Energy Fuels. 1997;11:546–553. [Google Scholar]
- 20.Wang C L, Michels P C, Dawson S C, Kitisakkul S, Bross J A, Keasling J D, Clark D S. Cadmium removal by a new strain of Pseudomonas aeroginosa in aerobic culture. Appl Environ Microbiol. 1997;63:4075–4078. doi: 10.1128/aem.63.10.4075-4078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisberg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White C, Gadd G M. Accumulation and effects of cadmium on sulfate-reducing bacterial biofilms. Microbiology. 1998;144:1407–1415. doi: 10.1099/00221287-144-5-1407. [DOI] [PubMed] [Google Scholar]
- 23.Wycokoff R. Crystal structures. Vol. 1. New York, N.Y: Interscience Publishers; 1964. [Google Scholar]