Abstract
We examined transfer of naphthalene-catabolic genes from donor microorganisms native to a contaminated site to site-derived, rifampin-resistant recipient bacteria unable to grow on naphthalene. Horizontal gene transfer (HGT) was demonstrated in filter matings using groundwater microorganisms as donors. Two distinct but similar plasmid types, closely related to pDTG1, were retrieved. In laboratory-incubated sediment matings, the addition of naphthalene stimulated HGT. However, recipient bacteria deployed in recoverable vessels in the field site (in situ) did not retrieve plasmids from native donors. Only when plasmid-containing donor cells and naphthalene were added to the in situ mating experiments did HGT occur.
Horizontal gene transfer (HGT) is a genetic mechanism by which microorganisms can acquire novel metabolic capabilities. Because of the potential widespread impact of HGT in a variety of environments, HGT has become the focus of increasing inquiry. HGT has been demonstrated to have diverse and far-reaching implications that include the spread of antibiotic resistance (6, 7, 20, 34), risk assessments for release of genetically engineered microorganisms (10, 45, 48), and the potential for control of environmental contaminants by microorganisms (22, 30, 44).
Because plasmids may promote their own transfer and encode advantageous metabolic traits, conjugal plasmid transfer has been studied extensively as a major mechanism of HGT in a variety of environments that range from water and sediments to biofilms and activated sludge systems (8, 9, 11, 12, 14, 32). In situ conjugal plasmid transfer in specific field sites has traditionally been difficult to document due to the variability and uncontrolled aspects of field studies. Therefore, a variety of laboratory microcosm studies and controlled field investigations have been performed in an attempt to document and elucidate factors that promote conjugal plasmid transfer. Microcosm experiments in which defined donor and/or recipient cells are added have elucidated several factors that can affect conjugal plasmid transfer. Among these, the presence of spermosphere (38) and rhizosphere (33, 37), soil sterilization and the addition of nutrients (4, 31, 43), the presence of a selective carbon source (10, 11, 29), donor and recipient cell density and cell metabolic status (29, 41), and the presence and activity of soil invertebrates (3, 5) have all been shown to influence gene transfer events in a variety of environmental samples. Two recent in situ investigations of conjugation have also been performed in field sites: manure was shown to enhance plasmid mobilization as well as survival of a Pseudomonas putida recipient strain introduced into agricultural soil (16). Also, plant growth stage influenced plasmid capture by a Pseudomonas recipient in the sugar beet phytosphere (23).
Circumstantial, or retrospective, evidence for HGT at our coal tar-contaminated field site was provided with the discovery of incongruent phylogenetic patterns between the 16S rRNA and nahAc genes in naphthalene-degrading bacterial isolates native to the site (19). Naphthalene-catabolic plasmids were subsequently isolated and characterized from these bacterial isolates. These plasmids were found to be self transmissible and homologous to pDTG1, a well-studied naphthalene-catabolic plasmid from P. putida NCIB 9816-4 (13, 40). By using the plasmid capture procedure pioneered by Bale et al. (1), the potential for horizontal gene transfer among sediment bacteria indigenous to our study site was previously demonstrated when filter matings between the native sediment bacterial community and a single cured, rifampin-resistant, site-derived recipient strain (Cg9.CR) yielded two similar but distinct types of naphthalene-catabolic plasmids (40). The present study was designed to further explore HGT of naphthalene-catabolic plasmids in our field site.
All sediment and groundwater plasmid capture experiments were performed at a coal tar-contaminated area located in Glen Falls, N.Y. Three particular locations were the focus of the present study: monitoring wells 36 (MW36) and MW4 (contaminated and uncontaminated, respectively) and contaminated surface sediments from the downgradient seep area. Characteristics and details of this site have been previously published (24–27, 42). Naphthalene-degrading strains isolated from contaminated sediment from our study site (designated Cg) have been previously described (19). Naphthalene-catabolic plasmids within these strains are designated pCg. Strains which have been cured of their naphthalene-catabolic plasmids are resistant to rifampin and therefore can serve as recipient (Nap−Rif+) strains (designated Cg_.CR) (40). All naphthalene-degrading strains were maintained on Stanier's mineral salts medium with naphthalene vapor (MSB-N) as sole carbon source (39, 40). In addition, rifampin (300 μg/ml), cycloheximide (100 μg/ml), and/or pyruvate (added to 0.1% vol/vol) (all from Sigma) were added to agar plates in the mating and soil recovery experiments described below.
Laboratory filter matings between recipient cells and groundwater microorganisms utilized recipient cells (109 cells total) grown overnight in Luria Bertani (LB) broth (35) amended with 300 μg of rifampin per ml. These were harvested by centrifugation and washed with phosphate-buffered saline (PBS) (35). Recipients were suspended in 50 μl of PBS and were kept on ice overnight during transport to our field study site where donor cells from the native groundwater community were harvested. These potential donor cells were obtained from groundwater as follows. Three well volumes (∼6 to 7 liters per well) were purged from the groundwater monitoring well at a flow rate of 300 ml/min with a peristaltic pump (Geotech Environmental, Denver, Colo.) connected by polyethylene tubing to the well-screening depth of 20.5 ft. After purging, 420 ml of groundwater was filtered through 0.2-μm-pore-size, 25-mm-diameter filters (Millipore) on site. Filters were placed onto LB plates (cell side up). The 50-μl suspension of recipients (∼109 cells) was subsequently spread onto the filter. The plates were incubated overnight, agar side down, in the dark at room temperature. Filters were removed and vortexed in 1 ml of PBS for 30 s to remove cells. Transconjugants were enumerated on MSB-N plates amended with rifampin and cycloheximide. Controls containing donor cells only were enumerated on MSB-N plates. Controls containing recipient cells only were enumerated on MSB amended with pyruvate, rifampin, and cycloheximide. Both donors and recipients were also plated onto MSB-N amended with rifampin to account for spontaneous rifampin resistance and/or experimental errors. The genotypes of putative transconjugants and cured recipient strains were compared by using enterobacterial repetitive intergenic consensus (ERIC) PCR as previously described (40, 46). Growth on naphthalene, PCR detection of nahAc, plasmid isolation, plasmid restriction fragment length polymorphism (RFLP) analysis, and Southern hybridization have all been discussed previously (40).
For the in situ sediment experiments, open-ended glass cylinders (3-cm diameter, 3-cm height, able to contain approximately 15 g of wet sediment) were inserted into contaminated sediments where contaminated water seeps to the surface at the base of a hill (26). Recipient strains, prepared as described above, were added separately to the desired density of cells per gram of wet sediment. In the treatments receiving LB broth amendments, 250 μl of 7.5× LB broth was added after adding recipients to the cylinder-enclosed sediments. For treatments receiving naphthalene amendments, approximately 0.05 g of naphthalene crystals was added to cylinder-enclosed sediments. For several treatments, donor strain Cg21 (grown overnight in LB broth, harvested by centrifugation, suspended in saline, and stored on ice until arrival at the study site) was added to a density of 106 or 107 cells/g of sediment. After adding amendments, sediments within the glass cylinders were subsequently mixed by hand with a sterile toothpick. Incubation times ranged from 2.5 h to 9 days in situ. Cylinders containing sediment plugs were then removed by hand and placed into sterile 250-ml screw-cap jars. Cells were released from sediment as follows: a 75-ml volume of 0.1% Na4P2O7 (pH 7.2) was added to the 15-g sediment plug in the jar and was vortexed for two 30-s intervals (2, 21). Sediment homogenate was then diluted in 0.1% Na4P2O7 (pH 7.2) and was plated as described above. The in situ groundwater mating attempt utilized flowthrough chambers (Margan, Inc., Atlanta, Ga.) (47) that, after being filled with approximately 38 g of autoclaved site sand and inoculated with 109 recipient cells/g of sand, were suspended in groundwater monitoring wells for 2 days prior to being retrieved, diluted, and plated on transconjugant-selective media as described above. Because our main goal was to qualitatively demonstrate HGT in the field environment using as many recipient strains as possible, experiments were logistically constrained. Our strategy was to achieve scientific quality by replicating experiments in time, not by having many replicates per treatment.
To examine the potential for horizontal gene transfer within the planktonic subsurface microbial community, cells in contaminated (MW36) and uncontaminated (MW4) groundwater were used as donors in filter matings with five cured, rifampin-resistant, site-derived recipient strains (Table 1). Based on subsequent acridine orange direct cell counts, a total of 2 × 106 cells and 2 × 107 cells were collected from MW4 and MW36, respectively. Microorganisms in aliquots of groundwater plated onto transconjugant-selective medium (no added recipient bacteria) failed to grow, indicating that the rifampin effectively inhibited native naphthalene-degrading bacteria in the groundwater. No transconjugants were obtained in the filter mating with the microorganisms native to uncontaminated groundwater and the recipients. We presume this absence of plasmid transfer from microorganisms in pristine water was due to the absence of selective pressure for naphthalene-metabolizing populations, but other causes, including fewer total cells, may also have contributed to the result.
TABLE 1.
Retrieval of two types of naphthalene-catabolic plasmids from laboratory-incubated filter matings between the microbial community native to the contaminated MW36a and recipient strains (109 cells per mating)
| Recipient | No. of transconjugants from plasmidb
|
|
|---|---|---|
| RFLPa | RFLPb | |
| Cg1.CR | 9 | 20 |
| Cg2.CR | 2 | 1 |
| Cg4.CR | 7 | 10 |
| Cg9.CR | 1 | 9 |
| Cg21.CR | 0 | 3 |
No transconjugants were obtained from filter matings with the microbial community from uncontaminated groundwater (MW4).
Total colonies resistant to rifampin and able to grow on naphthalene. RFLP patterns type a and b are described in the text and in the legend to Fig. 1.
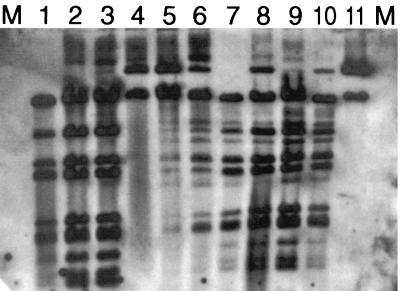
From the mating with the contaminated groundwater community, 62 potential transconjugants (Table 1) were obtained. The potential transconjugants were verified as being derived from recipient strains by ERIC repetitive extragenic palindromic (REP)-PCR (40). Each of the recipients displayed distinctive patterns consisting of 4 to 11 bands, depending on the strain. Only after ERIC REP-PCR banding patterns were found to match between each recipient and corresponding transconjugant were the latter picked and restreaked onto fresh transconjugant-selective medium in the presence and absence of naphthalene vapors. Colonies growing only in the presence of naphthalene were chosen for further analysis. All 62 of the transconjugant strains from contaminated groundwater were found to contain the predicted 700-bp nahAc gene fragment by PCR and a large plasmid, comparable in size to our original site-derived naphthalene-catabolic plasmids. Following extraction, purified plasmid DNA was digested with XhoI, separated by gel electrophoresis, and analyzed by Southern blotting and hybridizations to our pDTG1 probe (40). RFLP analysis of naphthalene-catabolic plasmids from our field site revealed two patterns (Fig. 1). RFLP pattern “a” (Fig. 1, lanes 1 to 3) consisted of seven bands (>23, 10.9, 6.0, 5.2, 3.9, 2.4, and 1.9 kb). Plasmids exhibiting RFLP pattern type “b” (Fig. 1, lanes 5 to 10) also hybridized with high affinity to our pDTG1 probe and contained additional restriction fragments of 4, 8.5, and 12 kb. RFLP types a and b confirm the patterns previously reported by Stuart-Keil et al. (40). Table 1 shows that not only were plasmids retrieved exclusively from contaminated well water, but RFLP type b seemed to be preferentially retrieved by four of the five recipient strains. Thus, using our plasmid retrieval procedure, we were able to demonstrate that the microbial community native to the contaminated groundwater contains self-transmissible naphthalene-catabolic plasmids. Stuart-Keil et al. (40) obtained similar results when the donor cells were derived from a sediment suspension from our study site.
FIG. 1.
RFLP characterization and Southern hybridization of pDTG1 to naphthalene-catabolic plasmids isolated from transconjugants retrieved from groundwater microorganisms after filter matings with recipient strains Cg1.CR, Cg2.CR, Cg9.CR, and Cg21.CR. The plasmids were digested with XhoI and were electrophoresed on a 0.7% agarose gel. M, lambda DNA cut with HindIII. Digested plasmids were loaded into the following lanes: 1, pDTG1; 2 to 7, Cg1 transconjugants; 8, Cg2 transconjugant; 9, Cg9 transconjugant; 10 and 11, Cg21 transconjugants. Lanes 1, 2, and 3 represent RFLP pattern a; lanes 5 to 10 represent RFLP pattern b. Plasmids in lanes 4 and 11 did not digest.
Three unsuccessful attempts to document plasmid transfer in the field (in situ) were completed. Experiments placed up to nine recipient bacteria at densities ranging from 106 to 109 per gram in contaminated-site sediment (twice) and in well water (once). Recipient recovery ranged from >100% for all strains after only 2.5 h to between 1.2 and >100% after 3 days, depending on the recipient strain. However, no transconjugants were recovered. Although a wide variety of factors may explain the lack of conjugation between the native microbial community and our recipient strains under field conditions, we hypothesized that three key interacting factors may prove crucial: (i) type and duration of cell-cell contact, (ii) physiological status of donors (especially as governed by carbon amendments), and (iii) substrate-specific plasmid mobilization mechanisms.
In order to more closely examine factors promoting HGT, two plasmid capture experiments were implemented by using contaminated sediments in laboratory-incubated microcosms. The first experiment examined the effects of carbon source amendment (LB broth, LB broth plus naphthalene, and sterile water) on conjugation frequency and survival of inoculated recipient strains in contaminated sediment over a period of 5 days. Recipient strains Cg1.CR, Cg2.CR, Cg4.CR, Cg5.CR, Cg8.CR, Cg9.CR, Cg12.CR, Cg16.CR, and Cg21.CR were added (106 cells/g) to 30 g of contaminated sediment in separate 250-ml widemouth ball jars. Microorganisms in sediment homogenate plated onto transconjugant-selective medium (no added recipient bacteria) failed to grow, indicating that the rifampin effectively inhibited native naphthalene-degrading bacteria. Percent recoveries of the various recipient strains ranged from 3 to 9% in the water-only treatment (5 ml per 30 g of sediment), from 1.4 to >100% in the LB broth-amended treatment (5 ml per 30 g of sediment), and from 0.2 to >100% in the LB broth plus naphthalene treatment (0.5 g in sterile test tube for vapor phase delivery). Cg1.CR and Cg21.CR were consistently among the recipients with the highest percent recoveries in all three treatments. Although recipient strains were recovered from all treatments, transconjugants were obtained only when both LB broth and naphthalene were added to the microcosms. Potential transconjugants were characterized as above. All 16 of the transconjugant strains were found to contain the nahAc gene by PCR amplification. A large plasmid, comparable in size to our original site-derived naphthalene-catabolic plasmids, was found in all of the transconjugant strains. All of the sediment-derived plasmids displayed our two basic RFLP patterns as described above: nine displayed RFLP pattern a and seven displayed RFLP pattern b of Fig. 1. Strain Cg4.CR preferentially retrieved RFLP pattern a plasmids (seven RFLP pattern a, one RFLP pattern b) while Cg2.CR (no RFLP pattern a, two RFLP pattern b) and Cg21.CR (one RFLP pattern a, three RFLP pattern b) preferentially retrieved RFLP pattern b. Cg1.CR retrieved one plasmid of each type.
The influence of incubation duration and naphthalene on conjugation frequency and recipient survival was tested in the second laboratory experiment. In an attempt to increase recipient recovery levels after an extended incubation time period, we boosted the number of Cg21.CR cells to 109/g of sediment. The presence of naphthalene (0.25 g in sterile test tube for vapor phase delivery) had little effect on recipient recovery levels throughout 11 days of incubation (Table 2), and recovery actually dropped more in the presence than in the absence of naphthalene by day 14 (P ≤ 0.05; three replicate plates). Nonetheless, 19 transconjugant colonies were obtained only after 14 days of incubation and only in the presence of naphthalene (Table 2). The transconjugants were characterized as explained above. The same two basic plasmid RFLP types were obtained: 11 RFLP pattern a and 8 RFLP pattern b. These results indicate that incubation times of 2 weeks could be required for detection of transconjugants in our microcosm studies. Time-dependent processes such as growth, motility, cell-cell contact, and cell physiological conditions all may have contributed to the conjugation events that occurred between days 11 and 14.
TABLE 2.
Influence of duration of laboratory incubation and naphthalene on survival of inoculated recipient bacteria and on frequency of conjugation between the native sediment microbial community (10 g of sediment) and recipient strain Cg21.CR inoculated at 109 cells/g
| Incubation time (days) | Recipient recoverya and transconjugant plasmid type
|
|||||
|---|---|---|---|---|---|---|
| No added naphthalene
|
Naphthalene added
|
|||||
| %R | RFLPa | RFLPb | %R | RFLPa | RFLPb | |
| 4 | 12 | 0 | 0 | 7.3 | 0 | 0 |
| 6 | 2.5 | 0 | 0 | 5.2 | 0 | 0 |
| 8 | 1.5 | 0 | 0 | 0.8 | 0 | 0 |
| 11 | 0.2 | 0 | 0 | 0.2 | 0 | 0 |
| 14 | 0.09 | 0 | 0 | 0.04 | 11 | 8 |
%R, percent recipient recovery. Percent of initial inoculum growing from a 2-g subsample diluted and plated in duplicate on Stanier's mineral salts medium supplemented with 300 μg of rifampin per ml, 100 μg of cycloheximide per ml, and 0.1% pyruvate.
We obtained transconjugants in two of our laboratory studies: after 5 days of incubation in the presence of LB broth and naphthalene and after 14 days of incubation in the presence of naphthalene only (Table 2). Thus, we returned to our field site to determine if conditions found conducive to plasmid transfer in the laboratory could trigger the process in the field. In this fourth in situ sediment experiment, the effects of added naphthalene and donor strains were tested. Recipient strains Cg21.CR and Cg1.CR, prepared as described above, were inoculated separately at 107 cells/g into glass cylinder-enclosed site sediment in the presence and absence of added naphthalene (approximately 0.05 g each). In additional treatments, to test the influence of donor populations on horizontal gene transfer in bulk sediment in situ, we added a donor strain Cg21 (from which strain Cg21.CR was derived [40]) at 106 and 107 cells/g to our Cg21.CR treatments. In another treatment focusing on the role of donor strains, we added cells native to the sediment that had been grown on naphthalene for 7 days. Cg1.CR and Cg21.CR were recovered after both 2 and 9 days. In all treatments except Cg21.CR plus Cg21 donor (106/g), recipient recovery was significantly higher (P ≤ 0.05; three replicate plates) in the presence of naphthalene. The reason for the influence of naphthalene on survival here, but not in the prior laboratory tests (Table 2), is unknown. Despite recipient recovery (ranging from 0.007 to 62%), we did not obtain transconjugants from any recipient-only treatments, nor did we obtain transconjugants from any of our treatments to which we added laboratory-incubated naphthalene-grown sediment microorganisms from a naphthalene enrichment culture. Furthermore, when donor strain Cg21 was added at 106 cells/g, no transconjugants were obtained. However, when the donor strain (Cg21) was added at 107 cells/g to sediments along with Cg21.CR recipient in the presence of added naphthalene, a single transconjugant colony was obtained. This colony was derived from Cg21.CR as verified by ERIC REP-PCR. After plasmid extraction, XhoI digestion, and Southern hybridization to pDTG1, the plasmid obtained from the Cg21 transconjugant was shown to be identical to pCg21.
We can suggest several hypotheses to explain why we obtained a transconjugant only from our treatment to which we added Cg21.CR and Cg21 donor at 107 cells/g. Because we do not know the number of potential donors added from our naphthalene enrichment, it is possible that we did not add a sufficient number of donors, thereby preventing adequate cell-cell contact with our recipients. This idea is supported by the lack of transconjugants resulting from our treatment where we added a 10-fold-lower inoculum of Cg21 donor (from 107 to 106/g). Another possibility is that potential donors from the enrichment culture were intrinsically less compatible with our added Cg21.CR than its parent Cg21 donor. In support of this idea, interstrain laboratory filter matings between Cg21 donor and phylogenetically distant recipient strains occurred 100- to 1,000-fold less frequently than matings between Cg21 donor and its cured daughter strain (40). In the field environment, the physical and physiological status of native donors will play a major role in determining the success of conjugation events with added recipient bacteria. These factors are currently not well understood, but we hypothesize that the lack of adequate cell-cell contact as well as the suboptimal physiological status of potential donors are among the most critical.
The goal of this study was to increase understanding of HGT in the field by using laboratory and field studies. The four decades of adaptation by the microbial community native to our study site is a duration that defies experimental duplication. In addition, our experiments have been limited by (i) rates of survival of inoculated recipient strains, (ii) the traits of our recipient strains, and (iii) uncertainties about the naturally occurring donor organisms. Nonetheless, we have shown that HGT can occur under favorable laboratory and field conditions and that recipient survival is necessary but not sufficient for conjugation to occur. Clearly, conjugation was a strain-specific process in sediment that was triggered by carbon, especially naphthalene, amendment. Naphthalene stimulation of conjugation may occur via a variety of possible mechanisms. The naphthalene could have stimulated growth of potential donors in the sediment environment. The growth rate of donors has been shown to be positively correlated with plasmid transfer rate under laboratory culture conditions as well as in soil microcosms (36, 41). In addition to enhancing the physiological status of the donor cells, the naphthalene could have led to an increase in donor population size. The population density of both donors and recipients obviously controls cell-cell contact, but such contact becomes important in sediment where cellular interactions can be impeded by the solid matrix. Conjugation frequencies drop several orders of magnitude when comparing filter matings to matings in soil microcosms. For example, Neilson et al. (28) demonstrated transfer frequencies of plasmid pJP4 to decrease from 10−3/parent in laboratory matings on solid agar media to 10−5/parent in sterile soil to 10−6/parent in 2,4-dichlorophenoxyacetic acid-amended nonsterile soil. Thus, an increase in donor population size would enable increased cell-cell contact with our recipients in the sediment environment. Similarly, it is also possible that naphthalene stimulated growth of the newly formed transconjugant cells, thus increasing their population size to a level detectable by our experimental system.
Alternatively, naphthalene could have acted as a molecular signal to induce plasmid transfer from the donors native to our field site to our added recipient bacteria. Plant-synthesized octopines have been shown to induce transfer of the Ti plasmid between Agrobacterium tumefaciens cells (15). However, to date, no analogous naphthalene-specific transfer mechanisms have been described. Naphthalene could also affect individual cell motility. Grimm and Harwood (17, 18) have demonstrated chemotaxis of Pseudomonas spp. to naphthalene. Perhaps travel over small distances can increase the cell-cell contact clearly crucial for HGT.
We conclude that a variety of interacting factors govern HGT of naphthalene-catabolic plasmids in our field site, most notably suitable donor cells at proper density and the presence of naphthalene. The mechanism of HGT stimulation by naphthalene awaits explanation.
Acknowledgments
This research was supported by the U.S. Air Force Office of Scientific Research (grant no. F49620-95-0346), by the National Institute of Environmental Health Sciences (grant no. ES-05950-03), and by USDA/Hatch grant no. 189434.
We are grateful to R. Lorang for assistance with laboratory microcosms, C. Bakermans for assistance with field work, R. Verity for assistance with plasmid preparations, R. Garen for the gel image, and anonymous reviewers for constructive criticism.
REFERENCES
- 1.Bale M J, Fry J C, Day M J. Transfer and occurrence of large Hg resistance plasmids in river epilithon. Appl Environ Microbiol. 1988;54:972–978. doi: 10.1128/aem.54.4.972-978.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balkwill D L, Ghiorse W C. Characterization of subsurface bacteria associated with two shallow aquifers in Oklahoma. Appl Environ Microbiol. 1985;50:580–588. doi: 10.1128/aem.50.3.580-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byzov B A, Claus H, Tretyakova E B, Ryabchenko N F, Mozgovaya I N, Zvyagintsev D G, Filip Z. Plasmid transfer between introduced and indigenous bacteria in leaf-litter, soil, and vermicompost as affected by soil invertebrates. Biol Fertil Soils. 1999;28:169–176. [Google Scholar]
- 4.Clerc S, Simonet P. Efficiency of the transfer of a pSAM2-derivative plasmid between two strains of Streptomyces lividans in conditions ranging from agar slants to non-sterile soil microcosms. FEMS Microbiol Ecol. 1996;21:157–165. [Google Scholar]
- 5.Daane L L, Molina J A E, Sadowsky M J. Plasmid transfer between spatially separated donor and recipient bacteria in earthworm-containing soil microcosms. Appl Environ Microbiol. 1997;63:679–686. doi: 10.1128/aem.63.2.679-686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 7.Davies J. Origins and evolution of antibiotic resistance. Microbiologia. 1996;12:9–16. [PubMed] [Google Scholar]
- 8.Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- 9.Digiovanni G D, Neilson J W, Pepper I L, Sinclair N A. Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl Environ Microbiol. 1996;62:2521–2526. doi: 10.1128/aem.62.7.2521-2526.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Droge M, Puehler A, Selbitschka W. Horizontal gene transfer as a biosafety issue: a natural phenomenon of public concern. J Biotechnol. 1998;64:75–90. doi: 10.1016/s0168-1656(98)00105-9. [DOI] [PubMed] [Google Scholar]
- 11.Dronen A K, Torsvik V, Goksoyr J, Top E M. Effect of mercury addition on plasmid incidence and gene mobilizing capacity in bulk soil. FEMS Microbiol Ecol. 1998;27:381–394. [Google Scholar]
- 12.Ehlers L J, Bouwer E J. RP4 plasmid transfer among species of Pseudomonas in a biofilm reactor. Water Sci Technol. 1999;39:163–171. [Google Scholar]
- 13.Evans W C, Fernley H N, Griffiths E. Oxidative metabolism of phenanthrene and anthracene by soil Pseudomonads. Biochem J. 1965;95:819–831. doi: 10.1042/bj0950819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fry J C, Day M J. Plasmid transfer in the epilithon. In: Fry J C, Day M J, editors. Bacterial genetics in natural environments. New York, N.Y: Chapman & Hall; 1990. pp. 55–80. [Google Scholar]
- 15.Fuqua W C, Winans S C. A luxR-luxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotz A, Smalla K. Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl Environ Microbiol. 1997;63:1980–1986. doi: 10.1128/aem.63.5.1980-1986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm A C, Harwood C S. Chemotaxis of Pseudomonas spp. to the polyaromatic hydrocarbon naphthalene. Appl Environ Microbiol. 1997;63:4111–4115. doi: 10.1128/aem.63.10.4111-4115.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm A C, Harwood C S. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J Bacteriol. 1999;181:3310–3316. doi: 10.1128/jb.181.10.3310-3316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho M W, Traavik T, Olsvik O, Tappeser B, Vyvyan H C, Von Weizsacker C, McGavin G C. Gene technology and gene ecology of infectious diseases. Microb Ecol Health Dis. 1998;10:33–59. [Google Scholar]
- 21.Holben W E. Isolation and purification of bacterial DNA from soil. In: Weaver R W, Angle J S, Bottomley P S, editors. Methods of soil analysis, part 2. Microbiological and biochemical properties. Madison, Wis: Soil Science Society of America; 1995. pp. 727–751. [Google Scholar]
- 22.Leahy J G, Colwell R R. Microbial degradation of hydrocarbons in the environment. Microbiol Rev. 1990;54:305–315. doi: 10.1128/mr.54.3.305-315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lilley A K, Bailey M J. The acquisition of indigenous plasmids by a genetically marked Pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl Environ Microbiol. 1997;63:1577–1583. doi: 10.1128/aem.63.4.1577-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen E L, Bilotta-Best S E, Ghiorse W C. Development of a rapid 14C based field method for assessing potential biodegradation of organic compounds in soil and sediment samples. J Microbiol Methods. 1995;21:317–327. [Google Scholar]
- 25.Madsen E L, Sinclair J L, Ghiorse W C. In situ biodegradation: microbiological patterns in a contaminated aquifer. Science. 1991;252:830–833. doi: 10.1126/science.2028258. [DOI] [PubMed] [Google Scholar]
- 26.Madsen E L, Thomas C T, Wilson M S, Sandoli R L, Bilotta S E. In situ dynamics of aromatic hydrocarbons (AHs) and bacteria capable of AH metabolism in a coal tar waste-contaminated field site. Environ Sci Technol. 1996;30:2412–2416. [Google Scholar]
- 27.Madsen E L, Mann C L, Bilotta S E. Oxygen limitations and aging as explanations for the field persistence of naphthalene in coal tar-contaminated surface sediments. Environ Toxicol Chem. 1996;15:1876–1882. [Google Scholar]
- 28.Neilson J W, Josephson K L, Pepper I L, Arnold R B, DiGiovanni G D. Frequency of horizontal gene transfer of a large catabolic plasmid (pJP4) in soil. Appl Environ Microbiol. 1994;60:4053–4058. doi: 10.1128/aem.60.11.4053-4058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Normander B, Christensen B B, Molin S, Kroer N. Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris) Appl Environ Microbiol. 1998;64:1902–1909. doi: 10.1128/aem.64.5.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters M, Heinaru E, Talpsep E, Wand H, Stottmeister U, Heinaru A, Nurk A. Acquisition of a deliberately introduced phenol degradation operon, pheBA, by different indigenous Pseudomonas species. Appl Environ Microbiol. 1997;63:4899–4906. doi: 10.1128/aem.63.12.4899-4906.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pukall R, Tschaepe H, Smalla K. Monitoring the spread of broad host and narrow host range plasmids in soil microcosms. FEMS Microbiol Ecol. 1996;20:53–66. [Google Scholar]
- 32.Ravatn R, Zehnder A J B, van der Meer J R. Low-frequency horizontal transfer of an element containing the chlorocatecol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl Environ Microbiol. 1998;64:2126–2132. doi: 10.1128/aem.64.6.2126-2132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richaume A, Smit E, Faurie G, van Elsas J D. Influence of soil type on the transfer of plasmid RP4 from Pseudomonas fluorescens to introduced recipient and to indigenous bacteria. FEMS Microbiol Ecol. 1992;101:281–292. [Google Scholar]
- 34.Salyers A A, Shoemaker N B. Resistance gene transfer in anaerobes: new insights, new problems. Clin Infect Dis. 1996;23(Suppl. 1):S36–S43. doi: 10.1093/clinids/23.supplement_1.s36. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Smets B F, Rittman B E, Stahl D A. The specific growth rate of Pseudomonas putida PAW1 influences the conjugal transfer rate of the TOL plasmid. Appl Environ Microbiol. 1993;59:3430–3437. doi: 10.1128/aem.59.10.3430-3437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smit E, Wolters A, van Elsas J D. Self-transmissible mercury resistance plasmids with gene-mobilizing capacity in soil bacterial populations: influence of wheat roots and mercury addition. Appl Environ Microbiol. 1998;64:1210–1219. doi: 10.1128/aem.64.4.1210-1219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorensen S J, Jensen L E. Transfer of plasmid RP4 in the spermosphere and rhizosphere of barley seedling. Antonie Leeuwenhoek. 1998;73:69–77. doi: 10.1023/a:1000661115753. [DOI] [PubMed] [Google Scholar]
- 39.Stanier R Y, Palleroni N J, Douderoff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 40.Stuart-Keil K G, Hohnstock A M, Drees K P, Herrick J B, Madsen E L. Plasmids responsible for horizontal transfer of naphthalene catabolism genes between bacteria at a coal tar-contaminated site are homologous to pDTG1 from Pseudomonas putida NCIB 9816-4. Appl Environ Microbiol. 1998;64:3633–3640. doi: 10.1128/aem.64.10.3633-3640.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudarshana P, Knudsen G R. Effect of parental growth on dynamics of conjugative plasmid transfer in the pea spermosphere. Appl Environ Microbiol. 1995;61:3136–3141. doi: 10.1128/aem.61.8.3136-3141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor B, Mauro D, Foxwell J, Ripp J, Taylor T. Characterization and monitoring before and after source removal at a former manufactured gas plant (MGP) disposal site. Palo Alto, Calif: Electric Power Research Institute; 1996. [Google Scholar]
- 43.Top E, Mergeay M, Springael D, Verstraete W. Gene escape model transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990;56:2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Meer J R, Werlen C, Nishino S F, Spain J C. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl Environ Microbiol. 1998;64:4185–4193. doi: 10.1128/aem.64.11.4185-4193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Elsas J D, Duarte G F, Rosado A S, Smalla K. Microbiological and molecular biological methods for monitoring microbial inoculants and their effects in the soil environment. J Microbiol Methods. 1998;32:133–154. [Google Scholar]
- 46.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6832. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weisbrod N, Ronan D, Nativ R. New method for sampling groundwater colloids under natural gradient flow conditions. Environ Sci Technol. 1996;30:3094–3101. [Google Scholar]
- 48.Wilson M, Lindow S E. Release of recombinant microorganisms. Annu Rev Microbiol. 1993;47:913–944. doi: 10.1146/annurev.mi.47.100193.004405. [DOI] [PubMed] [Google Scholar]