Abstract
In many biological systems, chemical reactions or changes in a physical state are assumed to occur instantaneously. For describing the dynamics of those systems, Markov models that require exponentially distributed inter-event times have been used widely. However, some biophysical processes such as gene transcription and translation are known to have a significant gap between the initiation and the completion of the processes, which renders the usual assumption of exponential distribution untenable. In this paper, we consider relaxing this assumption by incorporating age-dependent random time delays (distributed according to a given probability distribution) into the system dynamics. We do so by constructing a measure-valued Markov process on a more abstract state space, which allows us to keep track of the “ages” of molecules participating in a chemical reaction.
We study the large-volume limit of such age-structured systems. We show that, when appropriately scaled, the stochastic system can be approximated by a system of Partial Differential Equations (PDEs) in the large-volume limit, as opposed to Ordinary Differential Equations (ODEs) in the classical theory. We show how the limiting PDE system can be used for the purpose of further model reductions and for devising efficient simulation algorithms. In order to describe the ideas, we use a simple transcription process as a running example. We, however, note that the methods developed in this paper apply to a wide class of biophysical systems.
Keywords: stochastic transcription, translation, random time delays, multiscale analysis, survival dynamical systems, age-dependent processes, non-Markovian systems
1. Introduction
We consider biophysical systems described by a set of chemical reactions. The chemically identical molecular entities in the system are called (chemical) species. A chemical reaction refers to the event of creation, annihilation, or conversion of a number of molecules of one or more species. Here, we assume the system is well mixed spatially in that a randomly chosen molecule of a species has an equal chance to chemically interact with any other molecule of any species in the system. A Continuous Time Markov Chain (CTMC) is a natural choice to model the species copy numbers of such systems.
When modeling Chemical Reaction Networks (CRNs) stochastically using CTMCs, one assumes that every reaction occurs instantaneously after an exponentially distributed amount of time. Whenever a reaction takes place, we update the system state. A random time-change representation of the Poisson process is often used to write the trajectory equations and to analyze the system dynamics [1, 2, 3, 4]. The sample paths of the CTMC are simulated exactly using the Doob–Gillespie’s Stochastic Simulation Algorithm (SSA) [5, 6, 7] or the next reaction method by Gibson and Bruck [8].
1.1. Delays are inherent and a useful model reduction tool
It has been reported that some biological processes do not take place instantaneously. In other words, there is a time lag between the initiation and the completion of the process. Time delays are observed inherently in many biological systems, such as gene transcription [9, 10, 11] and translation [12], cell cycle in cancer treatment [13], intracellular viral dynamics [14, 15], control of infectious diseases [16], population growth [17, 18], RNA and protein folding [19, 20], and enzyme catalyzed reactions [21, 22]. Sometimes time delays are introduced purposefully as a useful means to reduce model complexity and compensate for the lack of experimental observation in both deterministic and stochastic models of biological processes.
Intermediate, ancillary processes or unobserved reactions can be replaced by time delays. For example, production of hes1 mRNA from hes1 gene has been modeled using delay differential equations where detailed mRNA synthesis and processing steps are replaced by a time delayed reaction [23]. While modeling the mammalian circadian clock, intermediate protein dynamics can be simplified as transcriptional feedback loops with time delayed variables in delay differential equations [24]. In enzyme catalyzed reactions with multiple intermediates, the production of the final product can be expressed as a distributed delay equation, which is a useful tool when measurements on multiple intermediates in the experiment are not available [25].
Introduction of time delays as a model reduction technique has also been applied in discrete stochastic models for CRNs. For instance, model complexity of unimolecular reaction networks is reduced by generating delay distributions with key model features that are derived by computing first passage times of target species [26]. In [27], the production of yellow fluorescent protein has been described using a time-delayed birth and death process where a randomly distributed time delay was generated to simplify a sequence of steps in gene activation.
1.2. Our contribution
In most previous works in this area, the focus was on investigating stochastic models for CRNs with constant or random time delays. In those models, the probability that a reaction occurs within the next short interval of time is commonly described by a propensity (also known as intensity) function of the reaction. The waiting time for non-delayed reactions is exponentially distributed [28]. In practice, the occurrence of some reactions is not only determined by the molecular counts of the reactants but also affected by the age distributions or lifetimes of the reactant molecules. For example, mRNA decay rates vary depending on the age of each mRNA. Moreover, the age of the mRNAs determines polysome size distributions and protein synthesis rates in translation ([29, 30], Chapters 3 and 5 in [31]). It was also reported that an mRNA tail length distribution depends on the average age of mRNA population and that the tail-length distribution plays a significant role in deadenylation and decay dynamics of mRNA populations [32, 33].
When time delays are used to aggregate out ancillary or unobserved processes and reduce model complexity, it makes more sense that the length of time delay depends on the age of each reactant molecule (e.g., mRNA, protein, and enzymes). Therefore, it is worthwhile to consider an individual-based age-structured stochastic model for CRNs.
In this work, we develop a way to describe CRNs with random time delays and non-delayed reaction rates incorporating the age of each reactant and making use of hazard functions in survival analysis [34, 35]. See Appendix C for some preliminaries on relevant mathematical and statistical concepts. In our approach, the hazard functions are set as constant, time-dependent, or age-dependent functions generalizing the notion of reaction rate constants in propensity functions. Our model keeps track of the age of each reactant molecule and provides a new way to express time delays in non-Markovian models. Moreover, the method also allows us to describe discrete stochastic CRNs with constant or random time delays without age dependence, as considered in previous works. We study the large-volume limit of the proposed non-Markov CRN and provide a mean-field PDE limit for the age densities by virtue of the Law of Large Numbers (LLN), as opposed to an ODE limit in the classical theory. The PDE limit is based on existing results in the literature [36, 37] and follows from the standard limit theory for measure-valued Markov processes. However, novel usage of the PDE limit can provide further approximations and pave the way for efficient simulation algorithms. For the sake of illustration, we show how the PDE limit can be used to approximate Mean First Passage Times (MFPTs) in the context of CRNs. As another by-product of the LLN, we show how an efficient (fast) hybrid simulation algorithm can be devised when a subset of the CRN is abundantly available, giving a flavor of multiscale approximation. Finally, as simple applications of our approach, we briefly discuss a prokaryotic auto-regulation and the Quasi-Steady State Approximation (QSSA) in the context of the Michaelis–Menten enzyme kinetic reactions. Numerical examples have been provided wherever deemed necessary. For the sake of ready usage of our methods, the Julia scripts used in the numerical examples have been made available via a GitHub repository [38].
The following notational conventions are adhered to throughout the paper. We use 1{A}(x) to denote the indicator (or characteristic) function of a set A, i.e., 1{A}(x) = 1 if and only if x ∈ A. Given a suitable space E, let D([0, ∞), E) (or D([0, T], E)) denote the space of E-valued càdlàg functions defined on [0, ∞) (or [0, T], for some T > 0). The set of Borel subsets of a set A will be denoted by . The set of natural numbers are denoted by . The set of real numbers is denoted by . Other notations will be introduced as and when needed.
2. The simplest model with a delay
Let us consider a simple CRN with two chemical species A and B. First, we shall describe the standard Markovian approach and then introduce an age structure to allow non-exponential holding times. The following network describes the production and the degradation of A along with a conversion from A to B
| (2.1) |
where b, τ, and d, depending on whether we are in the Markovian or non-Markovian setup, will be either reaction rate constants or hazard functions for the production of A, the conversion from A to B, and the degradation of A, respectively.
An example similar to the CRN in Equation (2.1) was investigated in some previous works with time delays [39, 40]. It is worth noting that the simplistic CRN described in Equation (2.1) can be thought of as a model reduction of a more complex CRN. For instance, a series of conversion type reactions
| (2.2) |
can be described by a single conversion reaction A →τ B with an appropriate hazard function τ. For the sake of illustration, let us assume we are in the Markovian setup so that k1, k2, … , kn are positive constants. We can interpret the CRN in Equation (2.2) as follows: One molecule of A gets transformed into a molecule of A1 after an exponentially distributed (with rate k1) amount of time. Then, the molecule of A1 gets transformed into a molecule of A2 after an exponentially distributed (with rate k2 this time) amount of time. This process goes on until the molecule finally gets transformed into a molecule of B from a molecule of An−1. Therefore, from the perspective of a single A molecule, the amount of time required for the molecule to finally get transformed into a molecule of B is the sum of those exponentially distributed amounts of times (with rates k1, k2, … , kn). Under independence, the probability distribution of the total amount of time required for a single A molecule to get transformed into a B molecule can be described by a convolution of the individual exponential distributions. Denoting the corresponding hazard function by τ, one can describe the CRN in Equation (2.2) by a single conversion reaction A →τ B. Similarly, a series of birth-death-conversion type reactions
can be approximated by a single birth type reaction ∅ →τ B with an appropriate hazard function τ. Therefore, even a simplistic model such as the CRN in Equation (2.1) covers a nontrivial class of CRNs and builds the foundation for studying more complex CRNs.
2.1. Standard Markov approach
The standard way to model the CRN in Equation (2.1) is to use a CTMC to describe the counts of molecules of the species A and B over time. In such a model, whenever each reaction fires, the consumption and the production of molecules are instantaneous. Let , denote the stochastic processes counting the copy numbers of the species A and B respectively. Here, the quantities b, τ, and d are reaction rate constants. The propensity functions corresponding to the three chemical reactions are defined as
where xi(t) denotes the number of molecules of the chemical species i at time t, for i = A, B. Define Tk to be the waiting time until the next reaction of type birth (k = b), conversion (k = τ), and death (k = d). Then, Tk is exponentially distributed with rate λk(t) for k = b, τ, d. The probability of each reaction’s occurrence is expressed in terms of the corresponding propensity function as follow:
for k = b, τ, d when Δt is small enough. Then, the trajectory equations can be written in a straightforward fashion following the random time changed representation of Poisson processes as
where R1, R2, and R3 are unit rate Poisson processes [2]. We assume we do not have any B molecules in the system initially, i.e., . Now, if we scale the stochastic processes by a scaling parameter n, e.g., volume of the system, it follows directly from the LLN for Poisson processes [41, 42] that the scaled process can be approximated by the solution to the following system of ODEs:
Notice that the birth rate b vanishes in the limit because we did not assume any scaling of b with respect to n. In general, one would assume that the overall birth rate scales linearly with n so that it is sustained in the limit.
2.2. Age-structured model
Now, let us introduce age and delay into the CRN described by Equation (2.1). We assume that the production rate of B and the degradation rate of A depend on the age of the reactant molecule of species A. We use “age” as an umbrella term to refer to the amount of elapsed time since a specific event. Thus, “age” could mean different things depending on the application area. The most straightforward way is the biological or the physical age, which we take as the time duration since the molecule was born or created. In systems where a certain reaction can fire only when a gene is activated, one could define age as the time duration since activation of the gene. In some cases, it may be desirable to define delays in terms of time duration since the initiation of a reaction. The notion of age is sufficiently general to account for those cases as well. For example, a reaction A → B in which the delay is defined purely in terms of time difference between initiation and completion of the reaction, can be replaced by the reaction system A → F → B where F is a fictitious species. The physical age of this fictitious species F is precisely the time since the initiation of the reaction A → B. Now, putting an appropriate hazard function on the reaction F → B, we can introduce a random or a deterministic delay in the reaction A → B. Therefore, for the CRN in Equation (2.1), it seems sufficient to define the age to be the physical age of the molecules of A.
When we have an age-structured model, the counts (copy numbers of the species A and B) are inherently non-Markovian unless the holding times are exponentially distributed. However, if we keep track of the ages of the molecules in addition to the counts, we can get a Markov system, albeit on a more abstract state space. A neat way to do so is to use measure-valued processes that keep track of the age distribution of the molecules over time. Moreover, the measure-valued processes are also Markovian, which allows us to make use of the already existing limit theory for Banach space-valued Markov processes. This approach to age-structured modeling in biology is not new. Our work builds on the existing literature [36, 37, 43, 44]. In the next section, we describe how the measure-valued processes can be utilized in the context of the CRN in Equation (2.1).
2.3. The measure-valued process and the limiting system
Let us denote by NA(t) and NB(t) the numbers of molecules of the chemical species A and B at time t. Then, individual molecules of A are labelled 1, 2, ⋯, NA(t). We denote the age of the i-th molecule of the species A by ai(t) for i = 1, 2, ⋯, NA(t). Similarly, we denote by bj(t) the age of the j-th molecule of the species B at time t. Now, we define a measure-valued process where and describe the age distributions of chemical species A and B at time t. To be more precise, we define
| (2.3) |
where δx is the Dirac measure, a function that takes value 1 if the argument to the function (a measurable set) contains x and zero otherwise. The components and of Xt are finite point measures with atoms placed on the individual ages of the molecules. For example, gives us the count of species A molecules with ages in the set (0.5, 11.25] at time t. In general, gives us the count of species A molecules whose ages lie in the set F at time t.
For any point measure and a measurable function f, we denote the integration of the function f with respect to the measure μ by
If μ ≔ (μ1, μ2, … , μL), for some positive integer L, is a vector of point measures and f is a measurable function, we use the notation ⟨⟨μ, f⟩⟩ to denote
Therefore, we have and where 1 stands for the identity function. The set of non-negative real numbers is denoted by . The total population size is given by
The process Xt is a Markov process on the space where T > 0 is a finite time horizon and is the space of finite, point measures on .
In order to simplify notations, we introduce maps , for i = 1, 2, 3, … , the purpose of which is to extract the i-th atom (the age of the i-th molecule) from a point measure following some partial order (e.g., “greater or equal to” relation). Therefore, gives us the age of the i-th molecule of the species A at time t. We can now write down the trajectory equations:
| (2.4) |
| (2.5) |
where Q1, Q2, Q3 are independent Poisson Point Measures (PPMs) with intensity measures ds × dθ, ds × di × dθ, and ds × di × dθ respectively, where di is a counting measure on , and ds and dθ are Lebesgue measures on . Provided the global jump rates are upper bounded by a finite quantity and the initial population size does not explode supn E[n−1NA(0)] < ∞, the trajectory equations admit a unique pathwise solution (see [37, Theorem 2.5] for a similar derivation).
Under some assumptions on the hazard functions and the initial age distribution of the A molecules, the scaled process n−1Xt converges to a deterministic, continuous function whose components and are themselves measure-valued functions satisfying
for a sufficiently large class of test functions f : (a, s) → fs(a). The convergence of the scaled stochastic process n−1Xt to the deterministic function xt can be proved using techniques similar to those in [36, 45, 37, 43, 44]. However, for the sake of completeness, a brief, intuitive argument is presented in Appendix D.
Since the measure-valued function is determined entirely by , it suffices to study . The densities yA(t, a) of the measure , when they exist, are an important quantity describing the distribution of age of the species A molecules in the large-volume mean-field limit. The density function yA should satisfy
| (2.6) |
with the initial and the boundary conditions
where fA(s) specifies the age distribution of A molecules at time t = 0. To be more precise, it is the density of the limiting measure , which we assume exists, with respect to the Lebesgue measure. Notice that the birth rate b vanishes in the limit, as in case of CTMC model, because we did not assume any scaling of the birth rate with respect to n.
Let yB denote the limiting proportion of B molecules in the system. Then, yB can be described entirely in terms of the density yA as a solution to the ODE:
| (2.7) |
with the initial condition yB(0) = 0. Luckily, the limiting system Equation (2.6) can be solved explicitly using standard analysis techniques:
where Sτ and Sd are the survival functions of the probability distributions characterized by the hazard functions τ and d respectively. Therefore, the limiting concentration of B molecules can be described by
In Figure 1, we numerically show the agreement between the theoretical limits in Equation (2.6) and (2.7) and the stochastic simulation. More specifically, we compare with stochastic simulations of and yB(t), with . As it can be verified, the approximation error vanishes in the limit. Because Xt is a Markov process, the simulation of the stochastic CRN in Equation (2.1) can be carried out by adapting the Doob–Gillespie’s SSA, which involves simulating two quantities at each step: 1) simulating the next reaction time; and 2) determining the reaction type. Note that, for the CRN in Equation (2.1), there are (2NA(t) + 1) different reactions possible at time t, even though there are only three types of reactions. The next reaction time can be simulated by drawing an exponential random variable with rate equal to the total hazard (the sum of the hazards corresponding to those (2NA(t) + 1) possible reactions). The total hazard is given by . The type of reaction is then decided by drawing a categorical random variable whose probability masses are the ratios of the individual hazards and the total hazard. This discrete event simulation algorithm is a straightforward adaptation of Doob–Gillespie’s SSA for CTMCs. However, it must be noted that the simulation of a non-Markovian CRN is computationally more expensive than the CTMCs. For the sake of completeness, a pseudocode describing the above procedure is given in Algorithm 2.1. An implementation in the Julia programming language [46] is also made available in [38].
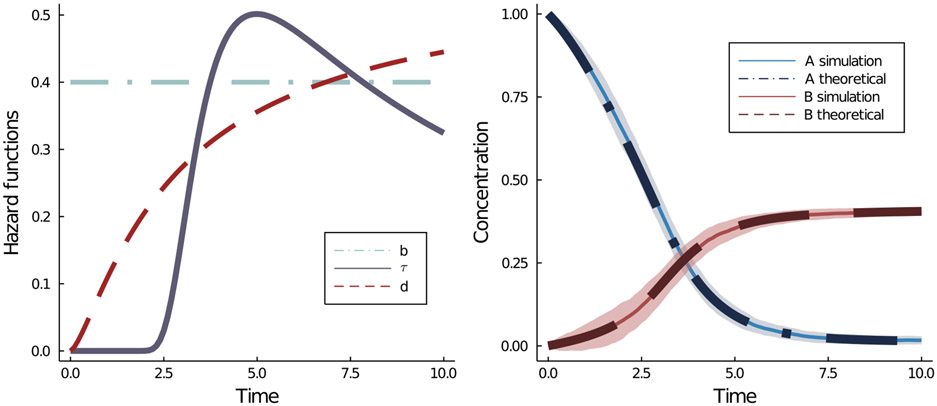
Figure 1.
(Left) The shapes of the three hazard functions in the CRN described by Equation (2.1). Here, b = 0.4. The hazard functions τ and d correspond to a Generalized Extreme Value distribution with parameters (1.25/0.30, 1.250, 0.30) and a Gamma distribution with parameters (2.5, 1.75) respectively. Here, the conversion reaction has been explicitly made a delayed one. (Right) Comparison of the theoretical limiting trajectory and the simulated trajectories of concentrations of A and B molecules. The mean of the simulated trajectories is shown in solid lines, while the theoretical mean curve (given by the PDE limit) is shown in dashed lines. The width of the ribbons indicate 1 standard derivation fluctuation around the mean. Here, n = 100, i.e., the initial number of A molecules is 100. It is evident that the theoretical limit provides a fairly accurate approximation to the scaled processes.
In Section 1, we mentioned that introduction of delay into a CRN could also serve the purpose of model reduction. Indeed, the LLN limit y ≔ (yA, yB) provides a model reduction of the original non-Markovian CRN in Equation (2.1). In the following, we discuss two other examples of usefulness of the LLN limit in the form of a PDE system. The first one approximates MFPTs, while the second one describes a faster simulation algorithm.
2.4. Mean First Passage Times
Mean First Passage Times are important quantities in the study of stochastic processes and dynamical systems. In the context of CRNs, they could arise in several ways [47, 48]. For instance, natural questions that could arise for the CRN in Equation (2.6) are how long it takes to deplete all molecules of species A or to produce the first molecule of B. One of the benefits of the LLN limit is that it can be used to approximate FPTs when the scaling parameter n is sufficiently large. The following illustrates this point.
Suppose we are interested in the time required to produce the first molecule of B. Following the exact simulation Algorithm 2.1 adapted from Doob–Gillespie’s SSA, the total hazard for the production of a B molecule is . In the large-volume limit, we can approximate this hazard by . Therefore, for a sufficiently large n, the MFPT can be approximated by
| (2.8) |
which, of course, vanishes in the limit of n → ∞. Moreover, the FPTs can be approximated by a random variable following an exponential distribution with mean m, whenever n is sufficiently large. It follows that we can use a simple likelihood function (based on the exponential distribution) for the purpose of statistical inference of the underlying parameters, provided we have observations on the FPTs. This method, called dynamic survival analysis, of estimating parameters based on timings rather than counts was recently explored in the context of an epidemiology in [34]. Dynamic survival analysis of general CRNs will be discussed elsewhere.
| Algorithm 2.1 Pseudocode for the exact simulation of the CRN in Equation (2.1). | |
|---|---|
|
|
In Figure 2, we show the accuracy of this approximation when n = 100. The approximation appears to be reasonably accurate. More importantly, this suggests we might be able to devise an efficient simulation algorithm using such approximate results. We explore this idea next.
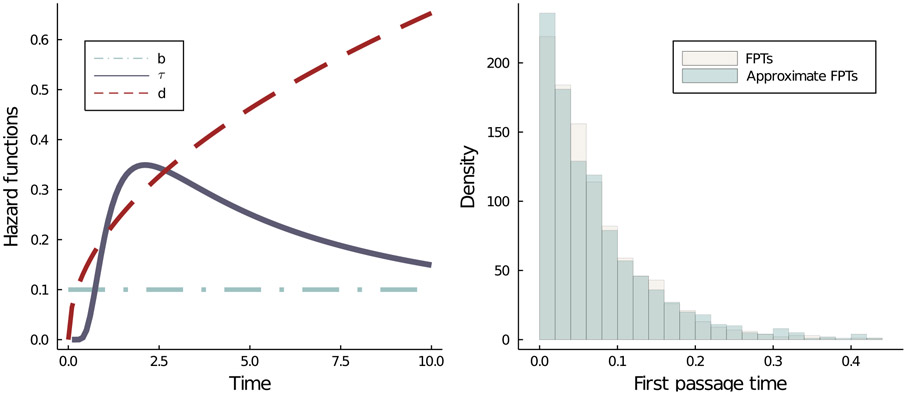
Figure 2.
(Left) The shapes of the three hazard functions in the CRN described by Equation (2.1). Here, b = 0.1. The hazard functions τ and d characterize an Inverse Gamma distribution with parameters (1.75, 4.25) and a Weibull distribution with parameters (1.5, 3.75) respectively. (Right) The density of approximate First Passage Times (FPTs) match that of the true FPTs. Here, n = 100.
2.5. Fast hybrid simulation
Consider a situation when the species A is abundantly available at the beginning of the reaction. Naturally, we expect the PDE approximation to the age density of the species A to be quite accurate, even though there will be considerable stochastic fluctuations in the copy numbers of B, at least initially. However, if we approximate the age density of A by the limiting PDE, we can also approximate the initial growth of the B molecules by a Poisson process whose time-varying intensity is driven by the PDE. We use this idea to devise a hybrid simulation algorithm, which is, again, essentially an adaptation of the Doob–Gillespie’s SSA in the sense that it only draws next reaction times from an exponential distribution whose mean depends on the solution to the PDE. For the sake of completeness, a pseudocode describing the idea is provided in Algorithm 2.2.
| Algorithm 2.2 Pseudocode for the hybrid simulation algorithm | |
|---|---|
|
|
In Figure 3, we show the accuracy of the hybrid simulation algorithm. Expectedly, the hybrid simulation is considerably faster than the full stochastic simulation of the CRN in Equation (2.1). A more elaborate comparison of performance is shown in Figure 4. However, it is worth noting that the hybrid simulation algorithm, by design, will underestimate the variance in the counting process for the species B. Therefore, one should use the hybrid simulation when it suffices to get the mean trajectory accurately. Alternatively, one can borrow ideas to estimate the variance correctly in other simulation algorithms [49, 50, 51]. Similar ideas to expedite simulations have been proposed previously. For instance, Ganguly et al. [52] propose a jump-diffusion approximation to the stochastic CRNs and provide error analysis while others [28, 53] introduce hybrid simulation methods using a piecewise deterministic Markov process.
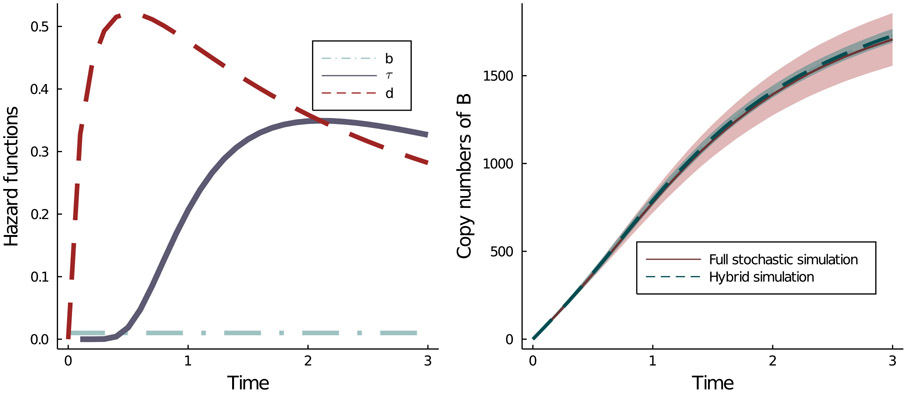
Figure 3.
An example of the hybrid simulation approach. (Left) The shapes of the three hazard functions the CRN described by Equation (2.1). (Right) Comparison of the hybrid simulation algorithm (Algorithm 2.2) with the full stochastic simulation algorithm (Algorithm 2.1). Here, the birth rate b = 0.01. The distributions characterized by τ and d are inverse gamma distribution with parameters (1.75, 4.25) and a Beta prime distribution with parameters (1.75, 1.25). The value of n in this example is 5000. The full stochastic simulation took 305.714216 seconds, while the hybrid simulation took only 62.093832 seconds on a 2.3 GHz 18-Core Intel Xeon W machine.
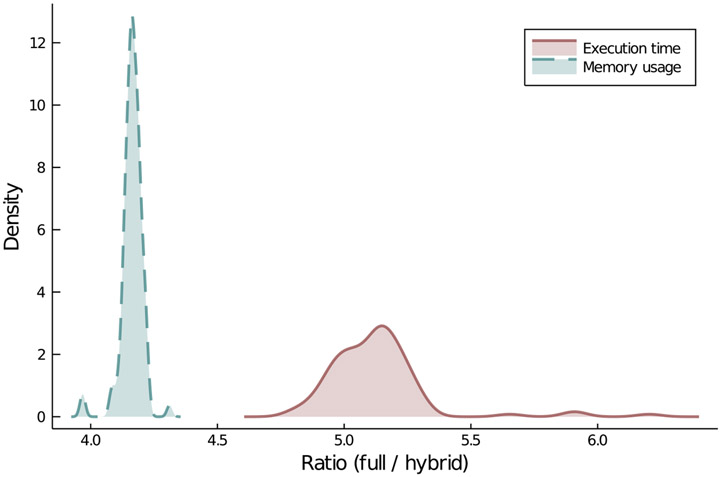
Figure 4.
Efficiency of the hybrid simulation algorithm. The figure shows the empirical density of the ratios of execution times and memory usage of the full stochastic simulation and those of the hybrid simulation algorithm described in Algorithm 2.2. It is evident that the hybrid simulation algorithm is at least five times faster in terms of execution times and at least four times more efficient in terms of memory usage. The simulation set-up is the same as Figure 3. The performance evaluation of the hybrid simulation is done using the BenchmarkTools.jl package [54] in Julia language [46]
3. Michaelis–Menten enzyme-kinetic reactions
Michaelis–Menten enzyme-catalyzed chemical reactions form an important class of CRNs particularly because of their vast applications in the industry [55, 56]. Several descriptions of this class of reactions are available in the literature. For the sake of simplicity, in what follows we shall adopt the simplest form of the Michaelis–Menten enzyme-catalyzed reactions. In this form, the CRN comprises a reversible binding of a molecule of a substrate (S) and a molecule of an enzyme (E) into a molecule of a substrate-enzyme complex, and an irreversible conversion of a molecule of the complex into a molecule of a product (P) leaving the molecule of the enzyme free. That is, the system consists of the following reactions:
| (3.9) |
In traditional models of enzyme kinetics, the quantities k1, k−1, and k2 are reaction rate constants. When modeled stochastically using a CTMC, the mean-field limit of the scaled concentrations is described by the following set of ODEs (see [57] for more details):
| (3.10) |
The [·] notation is used to denote the concentrations. The ODE system in Equation (3.10) has been studied extensively in the literature. We will adopt our measure-valued representation to incorporate potential age structure in the Michaelis–Menten CRN.
3.1. Enzyme kinetics with age structure
We assume the binding reaction depends on the age of the participating molecule of the enzyme. That is, only k1 is age-dependent; k−1 and k2 are constants. For the species E, S, C, and P, define the measure-valued stochastic processes
where NE, NS, NC, NP denote the counts of molecules of E, S, C, and P respectively. Similarly, ei, si, ci, pi denote the age of the i-th molecule of E, S, C, and P respectively. The process X ≔ (XE, XS, XC, XP) is a Markov process on the space . Please note that we need to scale the hazard function k1 corresponding to the bimolecular reaction by n−1 following the stochastic law of mass actions [1].
As before, we are interested in the large-volume limit of the scaled process n−1Xt. The scaled stochastic process n−1Xt converges to a deterministic function whose components , , , are finite measures on by virtue of the LLN.
Let yE denote the density of the measure with respect to the Lebesgue measure. Also, let yS, yC, yP denote the concentrations of the S, C, and P molecules. Then, we get the following limiting system:
| (3.11) |
with the boundary condition
and the initial condition yE(0, s) = fE(s) such that . Appropriate initial conditions for S, C, and P are also assumed. This limiting system can now be used to study the effects of delay in the binding reaction. One interesting approximation that has been widely applied in the context of Michaelis–Menten enzyme kinetic reactions is what is known as a Quasi-Steady State Approximation [58]. There are many forms of QSSAs, namely, standard QSSA (sQSSA), total QSSA (tQSSA), and reversible QSSA (rQSSA). Detailed analysis of any of the QSSAs is beyond the scope of the present work. For the purpose of illustration, we informally describe an analogue of the sQSSA here.
3.2. The standard QSSA
The QSSAs are a multiscale approximation of the Michaelis–Menten enzyme-kinetic reactions. The basic assumption behind the standard QSSA is that the substrate-enzyme complex C reaches its steady-state much quicker than the other species. In the deterministic set-up, the approximation is achieved by setting in Equation (3.11), which allows one to work with a smaller system of ODEs. Several conditions for the validity of the sQSSA have been proposed in the literature. See [57] for a detailed discussion.
Following the deterministic approach in our case, we set in Equation (3.11) to get a reduced PDE system that is analogous to the sQSSA. To be more precise, yields
which further yields an approximate system
| (3.12) |
Recall that yE solves (∂t + ∂s)yE(t, s) = −k1(s)yE(t, s)yS(t) with boundary condition yE(t, 0) = (k−1 + k2)yC(t) and initial condition yE(0, s) = fE(s). As a consequence, yE is determined by yS and yC, and can be partially solved in terms of yS and yC. Therefore, the reduced system of ODEs in Equation (3.12) is indeed autonomous and therefore, serves as an sQSSA of the CRN in Equation (3.9).
In the stochastic set-up, the QSSAs are obtained by means of the probabilistic multiscaling techniques developed in [3, 4]. The stochastic and the deterministic QSSAs mostly agree with each other with some notable differences. Please see [57] for examples of discrepancies as well as more details on the methods. Here, for paucity of space, we do not consider the stochastic QSSAs or possible discrepancies between stochastic and deterministic methods in the present age-structured models.
4. Prokaryotic auto-regulation
As another example, we consider a simple genetic network with feedback. We apply our approach using an age-dependent measure-valued process to build a model for a simple prokaryotic auto-regulation with a time delay. We modify an auto-regulation mechanism in the prokaryote gene network in [59] (Section 1.5.7). We simplify the example by approximating transcription and translation as a one-step process with a time delay and replacing repression of the gene by a protein dimer to repression by a single protein instead. For other related examples for the gene transcription and translation, see Section 2.1.1 in [60] and [61, 62, 63, 64].
Consider a genetic network with a gene (G), a protein (P), and a gene-protein complex (C). The gene activates production of protein following a hazard function bP and the protein degrades following a hazard function dP. The protein can reversibly bind with the gene to form a complex with binding hazard bC and unbinding hazard dC. Since the gene-protein complex cannot participate in the production of protein, this is auto-regulation of the gene by its complex. Schematically, the reactions are as follows:
| (4.13) |
In (4.13), we assume that the age of the gene is important. Therefore, the hazard functions bP and bC are assumed to be age-dependent whereas dC and dP are assumed to be constants. Note that after unbinding of the gene-protein complex, the age of the gene is reset to zero. On the other hand, the age of the gene is not affected by the protein production.
Denote by NG(t), NP(t), and NC(t) the total molecular counts of the gene, the protein, and the gene-protein complex at time t, respectively. For the species G, P, and C, define the measure-valued processes
where we denote the age of the i-th molecule of the species G, P, and C by gi, pi, and ci respectively. As in the case of the Michaelis–Menten enzyme kinetic reaction, we scale the hazard function bC corresponding to the bimolecular reaction by n−1 following the stochastic law of mass actions [1].
The LLN limit of the scaled process can be derived following by now familiar arguments of the previous examples. As one would expect, the scaled process n−1Xt converges to a deterministic function whose components are finite measures on . Since we assume only the age of the gene is important, we consider the limiting age density yG of the gene, which we obtain as the density, when it exists, of the measure with respect to the Lebesgue measure. Similarly, define the limiting concentrations of the product yP and the complex yC. The limiting system is then described by
| (4.14) |
with the boundary condition
and the initial condition yG(0, s) = fG(s), which specifies the initial ages of the gene. Note that the hazard function for unbinding of the gene-protein complex appears in the boundary condition since we assumed that the age of the gene is reset to zero when the complex breaks into the gene and the protein. Also, recall that bP(s) encodes a time delay in transcription and translation. For example, we may set bP(s) = r1[τ,∞)(s), which asserts that protein is produced only when the age of the gene is greater than τ with a hazard function r.
5. Discussion
Many biological processes with time delays, including CRNs, cannot be directly modeled using CTMCs due to non-exponentially distributed inter-event times of the processes. The simulation and analysis of systems with an age structure and time delays become challenging since the system dynamics are affected by the inherent randomness (stochasticity) as well as time delays. One way to simulate such stochastic systems with age structure and time delays is to modify simulation algorithms for CTMC models where the next reaction time and type are determined based on molecule counts of reactants. Bratsun et al. [39], Barrio et al. [65] and Cai [66] constructed modified SSAs, while Anderson [67] introduced a modified next reaction method to simulate discrete stochastic chemical reaction networks with delays. Notably, all of those works assume that the time lags in the delayed reactions are constant. Furthermore, in [68], Caravagna and Hillston described a non-Markovian stochastic process algebra, called Bio-PEPAd, to incorporate deterministic delays and perform formal analysis. Mura et al. [69] described how general holding time distributions can be incorporated in the programming language BlenX and studied the effect of the choice of the reaction time distributions. A stochastic simulation algorithm for non-Markovian biochemical reactions based on constraint programming is presented in [70].
CRNs with an age structure and random time delays provide a more realistic description of stochastic biophysical or chemical systems compared to the ones with fixed time delays. Unfortunately, the literature on stochastic systems with random time delays remains sparse. In a previous work by Koyama (Chapter 4 in [40]), the author investigated a stochastic kinetic network with a random time delay where a delayed reaction can be interrupted by another reaction and can fail to complete. In another work by Marquez-Lago et al. [71], the authors utilized probability distributed time delays to incorporate spatial effects such as diffusion or translocation of molecules in temporal stochastic models. In a recent work by Choi et al. [27], the authors described protein production in transcription and translation as a birth and death process with a random time delay.
In this paper, we developed a new way to incorporate an age structure and time delays in CRNs using age-dependent processes. We availed ourselves of previous theoretical works [36, 37, 43, 44] designed to study age-dependent population dynamics. We applied those stochastic models in the context of CRNs to account for the non-Markovian property due to the time delays. The use of age-dependent hazard functions not only enables us to model age-dependent time delays or reaction rates but also covers the modeling of constant and random time delays in the existing literature. We illustrated our method using simple biophysical systems in gene regulation and enzyme kinetics, but it will easily apply to general CRNs.
One potential disadvantage of the age-dependent processes is that simulation can be prohibitive since the age of each individual molecule of the chemical species of interest needs to be tracked over the entire simulation time. Therefore, we derived a large-volume limit of the age-dependent process for CRNs in the form of PDEs using the analytic methods in [36, 37, 43, 44] and used the PDE limit to construct a hybrid simulation algorithm, which, in our example, turned out to be five times faster than the full stochastic simulation. Moreover, we approximated a Mean First Passage Time efficiently utilizing the theoretical limit.
In this work, we emphasized how age-structured processes and their large-volume limits can be applied to model CRNs, in particular, biophysical or chemical systems with time delays. Many previous findings for general CRNs under Markovian assumption can be reinvestigated and extended to non-Markovian settings using age-structured processes. It would be interesting to see how the long time behavior of stochastic CRNs is affected by incorporating age structure. For example, it would be interesting to study stationary distributions of autocatalytic CRNs with switching behavior [72], to identify a class of CRNs maintaining product-form Poisson distributions for all times [73] and to find when CRNs show nonexplosive behavior [74]. Another interesting direction will be to study stability of CRNs [75] and to estimate transition times between different attractors in CRNs [76].
For the sake of simplicity, we have assumed in this paper that the molecular entities of all chemical species are abundant at the same order of magnitude so as to obtain the large-volume limit under the classical scaling. A natural extension of this work is to consider general CRNs with a wide range of molecular abundances and reaction rates where we can apply multiscale approximations to reduce model complexity [1, 3, 77]. We leave such investigation to future work. In this paper, we briefly described how an analogue of QSSA can be derived in the Michaelis-Menten enzyme-kinetic reactions. As shown in the related previous work [57, 58], both deterministic and stochastic QSSAs can be revisited with an extension of our approach to multiscale approximations in enzyme kinetics under non-Markovian setting. Another promising application of our approach seems to be in parameter inference and survival analysis of general CRNs with age structure. Given the current interests in pandemic modeling, such CRNs could lead to interesting examples in population dynamics and epidemiology. We hope to be able to pursue such work in the near future.
We conclude our discussion by briefly mentioning a class of CRNs modeled using Poisson processes with time-varying intensities. While retaining the Markov property, time-varying intensities provide a flexible way to aggregate out unobserved processes and to account for heterogeneity in the system such as cell-to-cell variability, changes in the volume or temperature of a cell affecting reaction rates [78, 67, 79]. However, the crucial difference between those models and ours is that time-varying intensities alone cannot induce a dependence structure of time delays on the initiation times of reactions whereas introduction of an age structure can. This is because time-varying intensities are a property of the system, whereas the age is a property of the individual molecule. Therefore, making the intensities depend explicitly on the individual ages of the molecules, as we do in this paper, provides a richer class of models.
Funding
WKB was supported by the National Institute of Allergy and Infectious Diseases (NIAID) grant R01 AI116770, the National Science Foundation (NSF) grant DMS-2027001 and the Ohio State University’s President’s Postdoctoral Scholars Program (PPSP). EK was supported by NIAID grant R01 AI116770 and the NSF grants DMS-2027001 and DMS-1853587. GAR was supported by the NSF grants DMS-2027001 and DMS-1853587. HWK was supported in part by the NSF under the grant DMS-1620403. The project was initiated when HWK was visiting the Mathematical Biosciences Institute (MBI) at the Ohio State University in Summer 2019. The authors acknowledge the hospitality and the support of MBI. The content of this manuscript is solely the responsibility of the authors and does not represent the official views of NSF, NIGMS, NIAID, or NIH.
A. Table of symbols
| Symbol | Meaning |
|---|---|
| The set of natural numbers | |
| The set of reals | |
| The set of non-negative reals | |
| 1{A}(x) | Indicator (characteristic) function of the set A |
| δx | Dirac delta function at x |
| The Borel σ-field of subsets of a set A | |
| The space of finite point measures on the set E | |
| D([0, T], E) | The space of E-valued càdlàg functions defined on [0, T] |
| ⟨μ, f⟩ | The integral ʃ f dμ |
B. Acronyms
- CDF
Cumulative Distribution Function
- CRN
Chemical Reaction Network
- CTMC
Continuous Time Markov Chain
- FPT
First Passage Time
- LLN
Law of Large Numbers
- MFPT
Mean First Passage Time
- ODE
Ordinary Differential Equation
- PDE
Partial Differential Equation
Probability Density Function
- PPM
Poisson Point Measure
- QSSA
Quasi-Steady State Approximation
- rQSSA
reversible QSSA
- sQSSA
standard QSSA
- SSA
Stochastic Simulation Algorithm
- tQSSA
total QSSA
C. Preliminaries
For the sake of completeness, we briefly describe some statistical and mathematical preliminaries here. Consider a continuous random variable U taking nonnegative values with Cumulative Distribution Function (CDF) GU and Probability Density Function (PDF) gU. The survival function SU of the random variable U is defined as
| (C.15) |
The hazard function hU of the random variable U is defined as
| (C.16) |
Hazard and survival functions are extensively used in survival analysis to model time to event data, e.g., time to death, time to hospitalization, time to default, time to failure etc. Intuitively, the hazard function describes the probability of failure in an infinitesimally small time period (t, t + Δt) given survival till time t. With little application of calculus, one can see that
which yields another useful relationship between the hazard function and the survival function:
where is called the cumulative hazard function. Hazard and survival functions cannot always be obtained in closed form. Probability distributions for which we can obtain them in closed form include Weibull, exponential, log-logistic distributions. The case of exponential distribution is unique in that it is the only probability distribution for which the hazard function is constant. However, a constant hazard is unrealistic in models for many biophysical systems.
D. Brief derivation of the PDE limit
In this section, we provide a brief, intuitive derivation of the PDE limit mentioned in Section 2.3. The line of argument follows the standard tightness-uniqueness route for abstract Markov processes and has been used in several prior works [36, 45, 37, 43, 44]. A rigorous proof of convergence for a general class of non-Markovian CRNs will be discussed elsewhere.
Consider the CRN in Equation (2.1) with the measure-valued process Xt as defined in Section 2.3. The components , satisfy the trajectory equations given in Equations (2.4) and (2.5). In order to study moments and martingale properties of and , it is worthwhile to check that
for a sufficiently large class of test functions f : (a, s) → fs(a).
As in the case of standard Markov model in Section 2.1, we are now interested in the large-volume limit (n → ∞) of the scaled stochastic process n−1Xt. By virtue of the LLN, if we assume i) the hazard functions are continuous, ii) the global jump rates are bounded above by a finite quantity, iii) a finite second moment condition on the initial population size supn E [n−2NA(0)2] < ∞, and iv) the initial age distribution does not explode, we have that the scaled process n−1Xt converges to a deterministic function whose components and are themselves measure-valued functions. This can be formally justified by verifying that the sequence of processes n−1Xt is tight and then, showing that the limit points (along subsequences) are unique. We can identify the limit points by studying certain martingale processes associated with the scaled processes n−1Xt. Outline of the argument is provided below.
D.1. Martingale property and tightness-uniqueness
First, under the above mentioned assumptions, we can show that the components of the scaled process n−1Xt do not explode (similar derivation in [37, Proposition 2.7]). Now, note that the trajectory equations for the processes and given in Equations (2.4) and (2.5) are driven by PPMs. Since we have
and using the compensated PPMs of the PPMs Q1, Q2, Q3, we can show the processes
are zero mean, square integrable, càdlàg martingale processes with predictable quadratic variations of the order n−1. Since we expect the predictable quadratic variations to vanish in the limit of n → ∞, the scaled process n−1Xt converges to a deterministic, continuous function xt. The tightness of the process n−1Xt can be established by verifying a criterion due to Roelly [80] in the vague topology and the Aldous–Rebolledo criteria [81]. See [36] or [37, Proposition 3.1] for similar calculations. Furthermore, thanks to the martingale representations above, we expect the limit xt to satisfy
The uniqueness of the solutions can be shown by first establishing that the solutions remain bounded on finite time intervals (recall the global jump rates are assumed bounded) and then invoking Grönwall’s lemma to show the distance between two possible solutions must vanish proving the desired uniqueness.
E. Software
The numerical results in this paper are obtained by the Julia programming language [46]. The Julia scripts (compatible with version 1.4.1) used in this paper have been made available publicly at a dedicated GitHub repository [38].
References
- [1].Ball K, Kurtz TG, Popovic L and Rempala G 2006. The Annals of Applied Probability 16 1925–1961 [Google Scholar]
- [2].Anderson DF and Kurtz TG 2011. Continuous Time Markov Chain Models for Chemical Reaction Networks Design and Analysis of Biomolecular Circuits ed Koeppl H, Setti G, di Bernardo M and Densmore D (Springer; ) pp 3–42 [Google Scholar]
- [3].Kang HW and Kurtz TG 2013. The Annals of Applied Probability 23 529–583 [Google Scholar]
- [4].Kang HW. BMC systems biology. 2012;6:143. doi: 10.1186/1752-0509-6-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gillespie DT 1976. Journal of computational physics 22 403–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gillespie DT 1977. The journal of physical chemistry 81 2340–2361 [Google Scholar]
- [7].Gillespie DT 2007. Annu. Rev. Phys. Chem 58 35–55 [DOI] [PubMed] [Google Scholar]
- [8].Gibson MA and Bruck J 2000. The journal of physical chemistry A 104 1876–1889 [Google Scholar]
- [9].Palangat M, Meier TI, Keene RG and Landick R 1998. Molecular cell 1 1033–1042 [DOI] [PubMed] [Google Scholar]
- [10].Hoyle NP and Ish-Horowicz D 2013. Proceedings of the National Academy of Sciences 110 E4316–E4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Swinburne IA and Silver PA 2008. Developmental cell 14 324–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Baron JW and Galla T 2019. Journal of the Royal Society Interface 16 20190436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhou Y, Gwadry FG, Reinhold WC, Miller LD, Smith LH, Scherf U, Liu ET, Kohn KW, Pommier Y and Weinstein JN 2002. Cancer Research 62 1688–1695 [PubMed] [Google Scholar]
- [14].Herz A, Bonhoeffer S, Anderson RM, May RM and Nowak MA 1996. Proceedings of the National Academy of Sciences 93 7247–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bai F, Huff KE and Allen LJ 2019. Journal of biological dynamics 13 47–73 [DOI] [PubMed] [Google Scholar]
- [16].Fraser C, Riley S, Anderson RM and Ferguson NM 2004. Proceedings of the National Academy of Sciences 101 6146–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kuang Y 1993. Delay differential equations: with applications in population dynamics (Academic press; ) [Google Scholar]
- [18].Giang DV, Lenbury Y and Seidman TI 2005. Journal of mathematical analysis and applications 305 631–643 [Google Scholar]
- [19].van den Berg B, Wain R, Dobson CM and Ellis RJ 2000. The EMBO journal 19 3870–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].van Meerten D, Girard G and van Duin J 2001. Rna 7 483–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Easterby JS 1981. Biochemical Journal 199 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kekenes-Huskey PM, Eun C and McCammon J 2015. The Journal of chemical physics 143 09B601_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Monk NA 2003. Current Biology 13 1409–1413 [DOI] [PubMed] [Google Scholar]
- [24].Korenčič A, Bordyugov G, Košir R, Rozman D, Goličnik M and Herzel H 2012. PloS one 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hinch R and Schnell S 2004. Journal of mathematical chemistry 35 253–264 [Google Scholar]
- [26].Barrio M, Leier A and Marquez-Lago TT 2013. The Journal of chemical physics 138 104114. [DOI] [PubMed] [Google Scholar]
- [27].Choi B, Cheng YY, Cinar S, Ott W, Bennett MR, Josić K and Kim JK 2020. Bioinformatics 36 586–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hepp B, Gupta A and Khammash M 2015. The Journal of chemical physics 142 034118. [DOI] [PubMed] [Google Scholar]
- [29].Valleriani A, Ignatova Z, Nagar A and Lipowsky R 2010. EPL (Europhysics Letters) 89 58003 [Google Scholar]
- [30].Deneke C, Lipowsky R and Valleriani A 2013. PloS one 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Deneke C. Ph. D. Thesis 2012 [Google Scholar]
- [32].Prieto S, Bernard J and Scheffler IE 2000. Journal of Biological Chemistry 275 14155–14166 [DOI] [PubMed] [Google Scholar]
- [33].Eisen TJ, Eichhorn SW, Subtelny AO, Lin KS, McGeary SE, Gupta S and Bartel DP 2020. Molecular Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].KhudaBukhsh WR, Choi B, Kenah E and Rempała GA 2020. Interface Focus 10 20190048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Calderazzo S, Brancaccio M and Finkenstädt B 2019. Bioinformatics 35 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fournier N and Méléard S 2004. The Annals of Applied Probability 14 1880–1919 ISSN 1050-5164 [Google Scholar]
- [37].Tran VC 2008. ESAIM. Probability and Statistics 12 345–386 ISSN 1292-8100 [Google Scholar]
- [38].KhudaBukhsh WR, Kang HW, Kenah E and Rempała G 2020. DelayModel: A Julia implementation of age-structured stochastic chemical reaction networks GitHub repository URL https://github.com/wasiur/DelayModel [Google Scholar]
- [39].Bratsun D, Volfson D, Tsimring LS and Hasty J 2005. Proceedings of the National Academy of Sciences 102 14593–14598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Koyama M. Ph. D. Thesis 2013 [Google Scholar]
- [41].Kurtz TG 1970. Journal of applied Probability 7 49–58 [Google Scholar]
- [42].Kurtz TG 1978. Stochastic Processes and their Applications 6 223–240 [Google Scholar]
- [43].Ferrière R and Tran VC 2009. Stochastic and deterministic models for age-structured populations with genetically variable traits CANUM 2008 (ESAIM Proc. vol 27) (EDP Sci., Les Ulis: ) pp 289–310 [Google Scholar]
- [44].Méléard S and Tran VC 2012. Stochastic Processes and their Applications 122 250–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Champagnat N, Ferrière R and Méléard S 2008. Individual-Based Probabilistic Models of Adaptive Evolution and Various Scaling Approximations Seminar on Stochastic Analysis, Random Fields and Applications V ed Dalang RC, Russo F and Dozzi M (Birkhäuser; Basel: ) pp 75–113 [Google Scholar]
- [46].Bezanson J, Edelman A, Karpinski S and Shah VB 2017. SIAM Review 59 65–98 [Google Scholar]
- [47].MacNamara S, Burrage K and Sidje RB 2008. Multiscale Modeling & Simulation 6(4) [Google Scholar]
- [48].Kim J, Dark J, Eciso G and Sindi S 2020. arXiv preprint URL https://arxiv.org/pdf/2005.05503.pdf [Google Scholar]
- [49].Franz B, Flegg MB, Chapman SJ and Erban R 2013. SIAM Journal on Applied Mathematics 73 1224–1247 [Google Scholar]
- [50].Harrison JU and Yates CA 2016. Journal of The Royal Society Interface 13 20160335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kang HW and Erban R 2019. Bulletin of mathematical biology 81 3185–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ganguly A, Altintan D and Koeppl H 2015. Multiscale Modeling & Simulation 13(4) [Google Scholar]
- [53].Gupta A and Khammash M 2019. Bulletin of Mathematical Biology 81 3121–3158 [DOI] [PubMed] [Google Scholar]
- [54].Chen J and Revels J 2016. arXiv e-prints arXiv:1608.04295 (Preprint 1608.04295) [Google Scholar]
- [55].Cornish-Bowden A 2012. Fundamentals of Enzyme Kinetics 4th ed (Wiley; ) [Google Scholar]
- [56].Segel IH 1975. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems (Wiley; ) [Google Scholar]
- [57].Kang HW, KhudaBukhsh WR, Koeppl H and Rempała G 2019. Bulletin of Mathematical Biology 81 13031336. [DOI] [PubMed] [Google Scholar]
- [58].Eilertsen J and Schnell S 2020. Mathematical Biosciences 108339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wilkinson DJ 2012. Stochastic modelling for systems biology 2nd ed (CRC press; ) [Google Scholar]
- [60].Anderson DF and Kurtz TG 2015. Stochastic analysis of biochemical systems (Springer; ) [Google Scholar]
- [61].Rempala GA, Ramos KS and Kalbfleisch T 2006. Journal of theoretical biology 242 101–116 [DOI] [PubMed] [Google Scholar]
- [62].Kim JK, Rempala GA and Kang HW 2017. Multiscale Modeling & Simulation 15 1376–1403 [Google Scholar]
- [63].Cappelletti D, Majumder AP and Wiuf C 2019. arXiv preprint arXiv:1912.00401 [Google Scholar]
- [64].Cao Z and Grima R 2019. Journal of The Royal Society Interface 16 20180967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Barrio M, Burrage K, Leier A and Tian T 2006. PLoS computational biology 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Cai X. The Journal of chemical physics. 2007;126:124108. doi: 10.1063/1.2710253. [DOI] [PubMed] [Google Scholar]
- [67].Anderson DF. The Journal of chemical physics. 2007;127:214107. doi: 10.1063/1.2799998. [DOI] [PubMed] [Google Scholar]
- [68].Caravagna G and Hillston J 2012. Theoretical Computer Science 419 26 – 49 [Google Scholar]
- [69].Mura I, Prandi D, Priami C and Romanel A 2009. Electronic Notes in Theoretical Computer Science 253 83 – 98 proceedings of Seventh Workshop on Quantitative Aspects of Programming Languages (QAPL 2009) [Google Scholar]
- [70].Chiarugi D, Falaschi M, Hermith D, Olarte C and Torella L 2015. BMC Systems Biology 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Marquez-Lago TT, Leier A and Burrage K 2010. BMC systems biology 4 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bibbona E, Kim J and Wiuf C 2020. arXiv arXiv–2001 [Google Scholar]
- [73].Anderson DF, Schnoerr D and Yuan C 2020. Journal of Mathematical Biology 1–33 [DOI] [PubMed] [Google Scholar]
- [74].Anderson DF, Cappelletti D, Koyama M and Kurtz TG 2018. Bulletin of mathematical biology 80 2561–2579 [DOI] [PubMed] [Google Scholar]
- [75].Agazzi A and Mattingly JC 2018. arXiv preprint arXiv:1810.065f7 [Google Scholar]
- [76].Agazzi A, Dembo A and Eckmann JP 2018. Annals of Applied Probability 28 1821–1855 [Google Scholar]
- [77].Kang HW, Kurtz TG and Popovic L 2014. The Annals of Applied Probability 24 721–759 [Google Scholar]
- [78].Lu T, Volfson D, Tsimring L and Hasty J 2004. Systems Biology 1 121–128 [DOI] [PubMed] [Google Scholar]
- [79].Chevalier MW and El-Samad H 2014. The Journal of Chemical Physics 141 214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].RoellyCoppoletta S 1986. Stochastics 17 43–65 [Google Scholar]
- [81].Joffe A and Metivier M 1986. Advances in Applied Probability 18 20–65 [Google Scholar]