Abstract
Background:
Previous studies showed that lifestyle behaviors (cigarette smoking, alcohol, coffee) are inversely associated with Parkinson’s disease (PD). The prodromal phase of PD raises the possibility that these associations may be explained by reverse causation.
Objective:
To examine associations of lifestyle behaviors with PD using two-sample Mendelian randomisation (MR) and the potential for survival and incidence-prevalence biases.
Methods:
We used summary statistics from publicly available studies to estimate the association of genetic polymorphisms with lifestyle behaviors, and from Courage-PD (7,369 cases, 7,018 controls; European ancestry) to estimate the association of these variants with PD. We used the inverse-variance weighted method to compute odds ratios (ORivw) of PD and 95% confidence intervals (Cl). Significance was determined using a Bonferroni-corrected significance threshold (p = 0.017).
Results:
We found a significant inverse association between smoking initiation and PD (ORivw per 1-SD increase in the prevalence of ever smoking = 0.74, 95%CI = 0.60–0.93, p = 0.009) without significant directional pleiotropy. Associations in participants ≤67 years old and cases with disease duration ≤7 years were of a similar size. No significant associations were observed for alcohol and coffee drinking. In reverse MR, genetic liability toward PD was not associated with smoking or coffee drinking but was positively associated with alcohol drinking.
Conclusion:
Our findings are in favor of an inverse association between smoking and PD that is not explained by reverse causation, confounding, and survival or incidence-prevalence biases. Genetic liability toward PD was positively associated with alcohol drinking. Conclusions on the association of alcohol and coffee drinking with PD are hampered by insufficient statistical power.
Keywords: Smoking, alcohol, coffee, Parkinson’s disease, Mendelian randomisation
INTRODUCTION
Parkinson’s disease (PD) is considered as a multifactorial disease, involving genetic susceptibility and environmental factors [1]. Epidemiologic studies have revealed a striking pattern, whereby several lifestyle behaviors, including cigarette smoking, alcohol, and coffee drinking, are inversely associated with PD [2], It remains debated whether these inverse associations are causal. The existence of a long prodromal phase in PD [3, 4] raises the possibility that they may be explained by reverse causation defined as situations where the outcome precedes and influences the exposure. For instance, the prodromal phase of PD may be characterized by a loss of appetence for cigarettes [5], and patients who later develop PD may be more successful in their attempts to quit smoking during the prodromal phase due to reduced dopamine reward [6].
Mendelian randomisation (MR) is a form of instrumental variable analysis that uses genetic variants associated with an exposure as instruments to estimate its causal association with a disease [7]. Unlike traditional epidemiologic approaches, MR analyses are not biased by confounding or reverse causation if a set of assumptions are met [8]. However, MR analyses in elderly populations may be biased due to survival bias, a form of selection bias that may arise as older persons consist of a non-random subset of the general population who have survived long enough to be included into the study [9]. This bias is a particular threat for MR studies investigating exposures that strongly affect survival [10], and is more pronounced for two-sample than one-sample MR studies [9]. MR studies that include prevalent cases may also be biased by incidence-prevalence bias, if the genetic instrument is associated with survival after disease onset [11].
As part of the Comprehensive Unbiased Risk factor Assessment for Genetics and Environment in PD (Courage-PD) consortium, we used two-sample MR to examine the association of three lifestyle behaviours (smoking, alcohol drinking, coffee drinking) with PD, and assessed whether MR findings are robust in analyses addressing the potential for survival and incidence-prevalence biases.
MATERIAL AND METHODS
Study design: two-sample Mendelian randomisation
MR uses genetic variants, mainly single nucleotide polymorphisms (SNPs), associated with an exposure to estimate its causal effect on an outcome. For SNPs to be valid instruments, three assumptions must be verified: i) SNPs should be associated with the exposure (IV1 assumption); ii) SNPs should not be directly associated with the outcome except through the exposure (IV2 assumption); and iii) SNPs should not be associated with unmeasured confounders of the exposure-outcome association (i.e., no horizontal pleiotropy, IV3 assumption) [7].
In two-sample MR, summary statistics (effect size estimates and standard errors [SE]) for the SNP-exposure and SNP-outcome associations, required to estimate the causal exposure-outcome association, come from two independent samples.
PD GWAS: Courage-PD consortium
We used summary statistics from a GWAS (NeuroChip) [12] in 23 out of 35 studies from the Courage-PD consortium (Supplementary Material). We excluded from the analyses: samples overlapping with the international Parkinson Disease Genomics Consortium (iPDGC); Asian studies (so that estimates for SNP-exposure and SNP-PD associations come from European populations); studies that included cases only; studies with less than 50 cases and 50 controls. As the role of environmental factors may be different in carriers of Mendelian PD mutations, we also excluded participants with GBA/LRRK2 mutations or with positive family history of PD. Participants’ characteristics are shown in Supplementary Table 1.
In each study, the frequency of SNPs was compared in cases and controls under an additive model using logistic regression adjusted for sex and the first four principal components. We meta-analysed summary statistics from the 23 GWAS (European ancestry: 7,369 cases, 7,018 controls) using a fixed (I2 ≤ 25%) or random (I2 > 25%) effects model (Supplementary Material).
Individual studies within each genome-wide association study had received approval from a relevant institutional review board from their country, and informed consent was obtained from participants or from a caregiver, legal guardian, or other proxy.
GWAS of exposures
We used summary statistics (betas, SEs) from published GWAS in individuals of European descent to select SNPs associated with exposures of interest at a genome-wide significant threshold (p < 5 × 10−8) and with a minor allele frequency (MAF) ≥0.01. To retain independent SNPs, we clumped them based on European ancestry reference data (1000 Genomes Project, r2 > 0.001, genomic region = 10,000 kb). For SNPs not available in Courage-PD, we selected proxies in high linkage disequilibrium (LD) with the index SNP (r2 > 0.8) according to LDlink [13] or SNIPA [14]. Summary statistics were harmonised on alleles positively associated with exposures. Ambiguous palindromic SNPs (A/T, C/G) with a MAF > 0.42 were discarded [15].
Smoking initiation and alcohol drinking
The GWAS and Sequencing Consortium of Alcohol and Nicotine use (GSCAN) provided summary statistics for smoking initiation (n = 1,232,091, 203 SNPs) and for the number of alcohol drinks per week (n = 941,280, 71 SNPs) in participants of European descent [16]. The proxy rsl 17495226 in high LD (r2 = 0.90) with the index SNP rs6050446 was used.
Information on smoking status was missing in several studies from Courage-PD; therefore, we were not able to run analyses stratified by smoking status and to examine the role of SNPs associated with smoking heaviness or age at smoking cessation or initiation in smokers [17]. To overcome this limitation, we used a lifetime smoking exposure index developed using UK Biobank data (in 462,690 European descent) that allows to perform MR using summary data from samples unstratified by smoking status. Briefly, this lifetime smoking index combines several aspects of smoking, including smoking status and, among ever smokers, duration, heaviness, and cessation, and was validated using positive outcomes (e.g., lung cancer). We selected 126 SNPs associated with this index [17].
Coffee drinking
We used a GWAS on self-reported bitter and sweet beverage consumption, including coffee (European ancestry, n = 370,000, 11 SNPs) [18].
Statistical analyses
Statistical analyses were performed using the TwoSampleMR, MRPRESSO, and simex R packages (R Foundation for Statistical Computing, Vienna, Austria). P-values are two-sided. We examined three main exposures (smoking initiation, alcohol drinking, coffee drinking). To account for multiple testing, we used a conservative approach and applied a Bonferroni corrected significance level of 0.017 (i.e., 0.05/3). P-values ≤ 0.05 but > 0.017 were considered as suggestive statistical evidence for an association [19].
Only SNPs available in 75% ( n = 17) of the studies or more were retained for our analyses. For each SNP, we computed the proportion of the variance of the exposure explained by the SNP (R2) [8].
For MR analyses of individual SNPs, we used the Wald ratio estimate (exponentiated ratio of the SNP-outcome association to the SNP-exposure association).
Our primary MR analyses based on multiple SNPs were conducted using the random-effects inverse-variance weighted (IVW) method that provides accurate estimates for SNPs that verify IV assumptions. Heterogeneity between genetic instruments was tested using the Cochran’s Q-statistic [7].
In sensitivity MR analyses, we used other approaches that relax some IV assumptions. The MR-Egger method can detect directional pleiotropy and provides corrected effect estimates but has low power and requires the InSIDE (INstrument Strength Independent of Direct Effect) assumption [20]. We used the I2 GX statistic to quantify the strength of regression dilution bias in SNP-exposure association estimates, with values < 90% indicating violation of the NOME (No Measurement Error) assumption, in which case we used the SIMEX method to correct MR-Egger estimates [21], The weighted median-method provides consistent estimates if at least 50% of the SNPs are valid instruments [22]. The weighted mode-based method uses the causal estimate from each SNP to calculate the modal estimate. The largest group of variants with the same causal estimate in the asymptotic limit are considered valid instruments [23]. MR-PRESSO allows to detect outliers (global test), to compute an estimate corrected for horizontal pleiotropy after removing them (if p-global test <0.05), and to test the difference between the original and updated estimates (distortion test) [24], In addition, as two SNPs were associated with both alcohol and coffee drinking, we repeated analyses for these phenotypes after excluding them.
PD is typically a disease of old age, and survival bias may bias genetic associations and MR estimates in any direction in studies in older populations for exposures that are associated with survival into old age (Supplementary Figure 1) [9, 10]. In some situations, increasing age is associated with increasing bias; in addition, bias is likely to be more pronounced in two-sample than one-sample MR studies. As smoking and alcohol drinking are strongly associated with survival, we performed analyses stratified by median age at study of cases and controls (67 years) and examined whether MR estimates observed overall were consistent with those seen in younger participants in whom survival bias is unlikely as mortality rates are low in this group [25–27].
The consortium included prevalent and incident patients. If genetic variants have a stronger effect on survival in PD patients than controls, genetic associations may be biased [11]. We examined whether MR estimates in cases with shorter disease duration (median ≤ 7 years) compared to controls were consistent with those obtained overall.
Finally, we tested for reverse causation by performing a reverse MR analysis where the exposure was PD genetic susceptibility and the outcome was smoking initiation, alcohol, or coffee drinking. We identified top SNPs (and corresponding association estimates) from the largest PD iPDGC GWAS as exposure instrumental variables [28], and extracted summary statistics for these SNPs from the GWAS for lifestyle behaviours [16–18]. For smoking initiation, we used summary statistics without 23andMe (N = 599,289). In this analysis, we estimated the effect of genetic liability toward PD on lifestyle behaviours in order to examine whether genetic liability toward PD is associated with smoking initiation, alcohol drinking, or coffee drinking. ORs are scaled to 1-unit increase in log odds of the prevalence of PD [29].
Statistical power
For each exposure, we computed the proportion of variance explained by the SNPs, the F-statistic as a measure of instrument strength [15], and statistical power for a type-I error rate of 5% and 1.7% (Supplementary Table 2) [30].
Data availability
Results can be reproduced using Supplementary Tables 3 and 4; no additional data available.
RESULTS
The associations of SNPs with exposures and PD are shown in Supplementary Tables 3 and 4. Supplementary Table 5 shows the number of SNPs retained for each exposure and F-statistics.
Smoking and PD
Of the SNPs positively associated with smoking traits, two (1.1%) were positively associated with PD at p< 0.05 for smoking initiation, and five (4.4%) for the lifetime smoking index; six (3.3%) were inversely associated with PD for smoking initiation, and five (4.4%) for the lifetime smoking index (Supplementary Table 3).
Smoking initiation was significantly and inversely associated with PD (ORIVW = 0–74, 95% Cl = 0.60–0.93, p = 0.009; Table 1, Supplementary Figure 2). The weighted median approach also showed a significant inverse association (p = 0.008) while the weighted mode approach yielded slightly lower point estimates with wider CIs. The MR-Egger method did not show significant directional pleiotropy.
Table 1.
Effect of genetically-predicted smoking on PD
| Exposure | Odds ratio (95% Cl) | P |
|---|---|---|
|
| ||
| Smoking initiation (per 1-SD increase in the prevalence of ever smoking) | ||
| IVW (p-heterogeneity = 0.39) | 0.74 (0.60–0.93) | 0.009 |
| Weighted median | 0.64 (0.47–0.89) | 0.008 |
| Weighted mode | 0.58 (0.24–1.42) | 0.23 |
| MR Egger (p-pleiotropy = 0.59; I2GX = 0.66) | 0.59 (0.24–1.45) | 0.25 |
| Corrected MR Egger | 0.56 (0.22–1.39) | 0.26 |
| MR-PRESSO (p-pleiotropy = 0.24) | - | - |
| Lifetime smoking index (per l-unit)a | ||
| IVW (p-heterogeneity = 0.007) | 0.54 (0.29–1.00) | 0.050 |
| Weighted median | 0.37 (0.16–0.84) | 0.017 |
| Weighted mode | 0.29 (0.06–1.34) | 0.12 |
| MR Egger (p-pleiotropy = 0.93; I2GX = 0.64) | 0.60 (0.05–6.77) | 0.68 |
| Corrected MR Egger | 0.60 (0.06–6.47) | 0.68 |
| MR-PRESSO (p-pleiotropy = 0.010, p-distortion = 0.77)b | 0.51 (0.29–0.89) | 0.021 |
Cl, confidence interval; IVW, inverse variance weighted.
In the UK Biobank, the lifetime smoking index had a mean of 0.359 and a standard deviation (SD) of 0.694; a 1-SD increase in the lifetime smoking index is equivalent to an individual smoking 20 cigarettes a day for 15 years and stopping 17 years ago or an individual smoking 60 cigarettes a day for 13 years and stopping 22 years ago. The ORIVW for a 1-SD increase is 0.670.694 =0.76.
Outliers: rs202645.
In sensitivity analyses, there was a suggestive inverse association for the lifetime smoking index (ORIVW = 0.54, 95% Cl = 0.29–1.00, P = 0.050; Table 1, Supplementary Figure 2) that became more significant after excluding two outliers identified by MR-PRESSO (ORMR-PRESSO = 0.51, 95% Cl = 0.29–0.89, p = 0.021). The weighted median method yielded an inverse association that was statistically significant (p = 0.017). MR-Egger did not show significant directional pleiotropy.
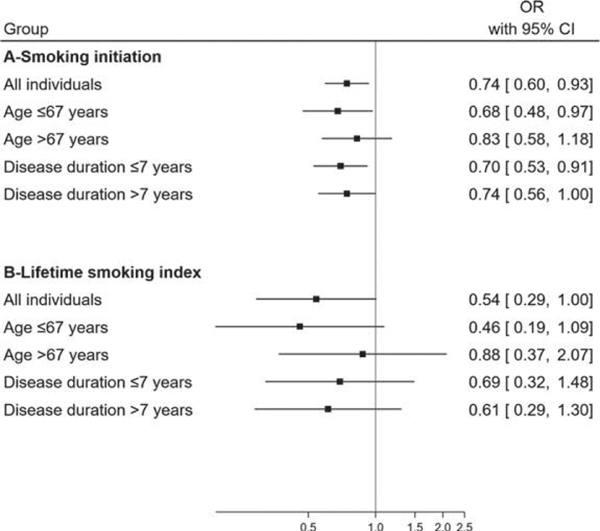
Both for smoking initiation and the lifetime smoking index, associations tended to be stronger in younger individuals (smoking initiation: ORIVW = 0.68, 95% Cl = 0.48–0.97; lifetime smoking index: ORIVW = 0.46, 95% Cl = 0.19–1.09) than in older ones (smoking initiation: ORIVW = 0.83, 95% CI = 0.58–1.18; lifetime smoking index: ORIVW = 0.88, 95% Cl = 0.37–2.07) and were consistent with those seen overall (Figure 1, Supplementary Table 6). Associations were of a similar size in patients with shorter and longer disease duration and comparable to associations seen overall (Figure 1, Supplementary Table 7).
Fig. 1.
Forest plot showing MR estimates for the association of smoking initiation and a lifetime smoking index with PD, overall and after stratification by age at study and disease duration in cases. OR, odds ratio; Cl, confidence interval.
Alcohol drinking and PD
Of the SNPs positively associated with alcohol drinking, two (3.2%) were positively and three (4.8%) were inversely associated with PD at p < 0.05 (Supplementary Table 3).
There was no significant association between alcohol drinking and PD (ORIVW = 0.68, 95% Cl = 0.39–1.18, p = 0.17; Table 2, Supplementary Figure 2). Exclusion of two SNPs associated with coffee drinking led to similar conclusions. In stratified analyses, there was no significant association between alcohol drinking and PD in younger individuals and patients with shorter disease duration (Supplementary Tables 6 and 7). There was a trend towards an inverse association only in prevalent PD cases (ORIVW = 0.53, 95% Cl = 0.27–1.02, p = 0.057).
Table 2.
Effect of genetically-predicted alcohol drinking on PD
| Exposure | Odds ratio (95% Cl) | P |
|---|---|---|
|
| ||
| Alcohol drinking (per 1-SD increase of ln(drinks per week)) | ||
| IVW (p-heterogeneity = 0.062) | 0.68 (0.39–1.18) | 0.17 |
| Weighted median | 0.86 (0.42–1.75) | 0.67 |
| Weighted mode | 0.86 (0.41–1.77) | 0.68 |
| MR Egger (p-pleiotropy = 0.94; I2GX = 0.96) | 0.70 (0.30–1.61) | 0.40 |
| MR-PRESSO (p-pleiotropy = 0.042, p-distortion = 0.73a) | 0.77 (0.46–1.29) | 0.33 |
| Alcohol drinking: after exclusion of 2 SNPs associated with coffee drinking: rs 1260326 and rs 2472297 (per 1-SD increase of ln(drinks per week) | ||
| IVW (p-heterogeneity=0.11) | 0.78 (0.45–1.35) | 0.37 |
| Weighted median | 0.88 (0.44–1.76) | 0.72 |
| Weighted mode | 0.85 (0.46–1.58) | 0.61 |
| MR Egger (p-pleiotropy = 0.98; I2GX = 0.96) | 0.78 (0.34–1.78) | 0.56 |
| MR-PRESSO (p-pleiotropy = 0.089) | - | - |
Cl, confidence interval; IVW, inverse variance weighted.
Outliers: rs9607814.
MR analyses using a single SNP in the ADH1B gene (rs1229984) showed no association with PD (ORWaldratio = 0.93, 95% Cl = 0.45–1.91, p = 0.85; Supplementary Table 3); this gene plays an important role in explaining alcohol drinking variance and analyses based on this gene are less likely to be biased by pleiotropy due to the known role of alcohol dehy-drogenase in alcohol metabolism.
Coffee drinking and PD
Of the SNPs positively associated with coffee drinking, one (9.1%) was positively, and another (9.1%) was inversely associated with PD at p < 0.05 (Supplementary Table 3).
There was a positive and non-statistically significant association of genetically-predicted coffee drinking with PD (ORIVW = 1.69, 95% Cl = 0.51–5.63, p = 0.40; Table 3, Supplementary Figure 2). However, after excluding two SNPs associated with alcohol drinking, the association decreased (ORIVW = 1–09,95% Cl = 0.26–4.45 ,p = 0.91). Stratified analyses showed no associations (Supplementary Tables 6 and 7).
Table 3.
Effect of genetically-predicted coffee drinking on PD
| Exposure | Odds ratio (95% Cl) | P |
|---|---|---|
|
| ||
| Coffee drinking (per ln(cups per day)) | ||
| IVW (p-heterogeneity = 0.017) | 1.69 (0.51–5.63) | 0.40 |
| Weighted median | 1.51 (0.50–4.51) | 0.47 |
| Weighted mode | 1.45 (0.52–4.03) | 0.50 |
| MR Egger (p-pleiotropy = 0.73; I2GX = 0.97) | 2.50 (0.21–29.59) | 0.49 |
| MR-PRESSO (p-pleiotropy = 0.035, p-distortion = 0.86)a | 1.86(0.67–5.11) | 0.26 |
| Coffee drinking: after exclusion of 2 SNPs associated with alcohol drinking: rs 1260326 and rs 2472297 (per ln(cups per day) | ||
| IVW (p-heterogeneity = 0.053) | 1.09 (0.26–4.45) | 0.91 |
| Weighted median | 1.06 (0.32–3.53) | 0.93 |
| Weighted mode | 1.11 (0.31–4.01) | 0.88 |
| MR Egger (p-pleiotropy = 0.35; I2GX = 0.96) | 3.86 (0.22–66.63) | 0.38 |
| MR-PRESSO (p-pleiotropy = 0.12) | - | - |
Cl, confidence interval; IVW, inverse variance weighted.
Outlier: rs574367.
MR analyses based on SNPs in two genes that are known to play an important role in caffeine metabolism (AHR-rs4410790, CYP7A2-rs2472297) showed no association with PD (rs4410790: ORwaldratio = 0.79, 95% Cl = 0.21–2.98, p = 0.72; rs2472297: ORWaldratio = 2.09, 95% Cl = 0.47–9.16, p = 0.33; Supplementary Table 3).
Reverse MR
Supplementary Table 8 shows the SNPs and association estimates used for reverse MR analyses. Table 4 shows the results of reverse MR analyses. There was no association between genetic liability toward PD and smoking or coffee drinking. For alcohol drinking, genetic liability toward PD was positively associated with alcohol drinking using the IVW method.
Table 4.
Reverse Mendelian randomisation using PD-associated SNPs from iPDGC [28] as genetic instruments
| Method | OR (95%CI) | P | p-het. | p-pleio. |
|---|---|---|---|---|
|
| ||||
| Smoking initiation | 64 SNPs | |||
| IVW | 1.01 (0.99–1.02) | 0.18 | <0.001 | |
| Weighted median | 1.01 (0.99–1.02) | 0.36 | ||
| Weighted mode | 1.01 (0.99–1.03) | 0.35 | ||
| MR-Egger | 1.02 (0.99–1.05) | 0.15 | 0.36 | |
| MR-PRESSO | - | - | <0.001a | |
| Lifetime smoking index | 65 SNPs | |||
| IVW | 1.00(0.99–1.01) | 0.83 | <0.001 | |
| Weighted median | 1.00(0.99–1.01) | 0.38 | ||
| Weighted mode | 0.99(0.98–1.01) | 0.092 | ||
| MR-Egger | 1.01 (0.99–1.02) | 0.34 | 0.23 | |
| MR-PRESSO | 1.00(0.99–1.01) | 0.56 | <0.001b | |
| Alcohol drinking | 64 SNPs | |||
| IVW | 1.02(1.01–1.03) | 0.002 | <0.001 | |
| Weighted median | 1.01 (0.99–1.02) | 0.14 | ||
| Weighted mode | 1.01 (0.99–1.02) | 0.21 | ||
| MR-Egger | 1.02 (0.99–1.04) | 0.097 | 0.85 | |
| MR-PRESSO | 1.01 (1.00–1.02) | 0.042 | <0.001c | |
| Coffee drinking | 65 SNPs | |||
| IVW | 1.00(0.99–1.01) | 0.15 | <0.001 | |
| Weighted median | 1.00(0.99–1.01) | 0.82 | ||
| Weighted mode | 1.00(0.99–1.01) | 0.65 | ||
| MR-Egger | 1.00(0.99–1.01) | 0.69 | 0.21 | |
| MR-PRESSO | 1.00(0.99–1.01) | 0.25 | <0.001d | |
OR, odds ratio per 1-unit increase in log odds of the prevalence of PD; IVW, inverse-variance weighted (random-effect); Cl, confidence interval; p-het., p for heterogeneity (IVW); p-pleio., p for pleio- tropy (MR-Egger and MR-PRESSO).
No outlier detected.
Outliers: rsl2497850, rsl2600861, and rs62053943; p-distortion = NS.
Outliers; rs62053943, rs6476434, rs6854006, rs7134559, and rs823118; p-distortion = 0.041.
Outliers; rs2248244 and rs61169879; p-distortion = NS.
MR-PRESSO identified 5 outliers; the corrected MR estimate after excluding these outliers was still in favour of a positive association.
DISCUSSION
Based on data from the Courage-PD consortium and after exclusion of samples overlapping with iPDGC, our findings add further evidence in favour of an inverse association between smoking initiation and PD but not for alcohol and coffee drinking.
Smoking
According to observational studies, ever smokers have a ~40% reduced risk of developing PD [31]. It has been argued, however, that this association may be due to reverse causation [5] as PD patients may stop smoking more easily during the PD prodromal phase than other persons due to reduced responsiveness to nicotine [6]. Others argued that the association seen in long-term ex-smokers [1] and the dose-effect relationship seen both in ex- and current-smokers are not in favour of this hypothesis [32].
Previous MR studies examined the association between smoking and PD using iPDGC data. One study (9,581 cases, 33,245 controls) assessed several risky behaviours in relation with PD [33], and reported a significant inverse association for smoking initiation (213 SNPs; OR per log odds of ever smoking = 0.71, 95% Cl = 0.57–0.90). In another study (37,688 cases with some overlap with the previous study, 18,618 UK Biobank proxy-cases, 1.4 million controls), there was a trend towards an inverse association between ever-smoking and PD that was not statistically significant (OR per log odds of ever-smoking = 0.94, 95% Cl = 0.88–1.01) [28]. Recently, another MR study used the same dataset to assess the role of several tobacco behaviours (smoking initiation, continuation, heaviness, and age at initiation). They showed that smoking continuation (current versus former smokers) was inversely associated with PD (OR per doubling of odds for smoking continuation = 0.64, 95%CI = 0.46–0.89, p = 0.008) but smoking initiation, heaviness, or age at initiation were not. However, they used only 87 SNPs for smoking initiation and analyses were not stratified by smoking status for analyses of smoking characteristics among smokers [34], The iPDGC PD MR Portal, a platform which offers MR analyses for a total of 5,839 GWAS versus the largest iPDGC GWAS available with some overlap with the two previous studies, also reports some evidence to support a protective effect of current smoking using a genetic instrument that included 12 SNPs [35]. These studies are not independent as they relied on overlapping datasets and did not examine associations in younger subjects or in those with shorter disease duration to assess whether survival or incidence-prevalence bias may have distorted MR estimates.
Our study replicates these findings and add further evidence in favour of an association between smoking initiation and lower PD risk; in addition, our findings show that genetic liability toward PD is not associated with smoking initiation which does not support the hypothesis of reverse causation for smoking. We further show that survival into old age and incidence-prevalence bias are unlikely explanations for the inverse association between smoking and PD. Since smoking is a major risk factor of mortality, differential survival could potentially bias the association between smoking-related SNP and PD; however, the observation that associations in younger participants were consistent with those seen overall is against this hypothesis and supports an association between smoking and PD [9,25–27]. In addition, the finding that associations tended to be weaker in older participants than in younger ones is in agreement with some previous observational studies that showed age-dependent associations between smoking and PD with no association in the oldest age groups [36,37]. This pattern could be interpreted in two different ways: i) survival bias may have diluted association estimates in older subjects; ii) alternatively, there may exist age-related etiologic heterogeneity in PD, and smoking may play a weaker role in PD at older ages where other factors may be more important. There was no marked difference in ORs according to disease duration, thus suggesting that incidence-prevalence bias is unlikely.
The consistency of MR findings with those from traditional epidemiologic studies and reports of smoking-by-gene interactions [38, 39] support a causal association between smoking and PD. However, the underlying mechanisms remain poorly understood. Although cigarette smoke includes a wide range of chemical components, a role for nicotine in PD was suggested by studies showing that it reduced neuronal damage in culture systems and protected against nigrostriatal dopaminergic damage in parkinsonian animal models [40]. In addition, among five compounds present in cigarette smoke (anabasine, cotinine, hydroquinone, nicotine, nomicotine), nicotine and hydroquinone inhibited alpha-synuclein aggregation [41].
Previous randomised clinical trials of nicotine for motor symptoms in PD failed to show an effect [42–44], but their design may have been hampered by small sizes and short follow-up. In addition, these trials were conducted in PD patients and examined disease progression rather than prevention. Additional well-designed neuroprotection trials are needed.
Alcohol drinking
A meta-analysis of the relation between alcohol and PD reported an inverse association in case- control studies but not in cohort studies, with marked heterogeneity across studies [45]. Two of the cohort studies reported positive associations, while three reported inverse associations, but none of them was statistically significant.
A previous MR study that examined several risky behaviours in relation with PD using iPDGC data (9,581 cases, 33,245 controls) did not provide evidence in favour of an association for the number of drinks per week (70 SNPs; OR=1.15, 95% CI = 0.87–1.53) or alcohol consumption (7 SNPs; OR = 1.39, 95% Cl = 0.11–17.56) [33]. In contrast, another MR study that used the most recent iPDGC dataset showed an inverse and significant association with the number of drinks per weeks (OR = 0.79, 95%CI = 0.65–0.96,p = 0.021; 33 SNPs) [28]. We did not find a significant association with alcohol; however, given the low power of the study to detect an association with this exposure, we cannot draw firm conclusions.
Alternatively, reverse MR showed that genetic liability toward PD is positively associated with alcohol drinking, thus suggesting that persons at higher PD risk are more prone to drink alcohol, independently of whether they actually develop PD. This finding warrants further investigation and replication in further studies.
Coffee drinking
Previous studies, both case-control and cohort, have shown an inverse association between coffee drinking or caffeine intake and PD, with a dose-effect relationship, that was present after adjustment for smoking or in never-smokers [46].
A previous MR study that assessed several risky behaviours in relation with PD using iPDGC data (9,581 cases, 33,245 controls) did not provide evidence in favour of an association for the number of coffee cups per day (4 SNPs; OR = 1.03, 95% Cl = 0.65–1.63). We did not find a significant association with coffee drinking, but given the low power of the study to detect an association with this exposure, we cannot draw firm conclusions. However, we did find that genetic liability toward PD was not associated with coffee drinking which does not support the hypothesis of reverse causation for coffee drinking.
Strengths and weaknesses of the study
Strengths of this study include the assessment of several lifestyle behaviours and use of data from a large consortium in which PD cases were carefully assessed by experienced movement disorders specialists. The MR design represents another strength as it avoids bias from reverse causation and confounding [8]. This approach, however, relies on a number of assumptions. We cannot exclude that our findings might have been affected by weak instrument bias. However, in MR analyses, the F-statistic was >10 for all exposures and bias from weak instruments is expected to be towards the null in a two-sample setting [47]. Pleiotropy is an important concern for MR analyses, and recent guidelines recommend using multiple methods that make different assumptions to assess the robustness of findings [15]. We used a number of approaches developed to address this issue, including the weighted median and mode, MR-Egger, and MR-PRESSO. Population stratification is unlikely to be a major concern for our analyses, as we restricted analyses within the Courage-PD consortium to individuals of European descent, cases were compared to controls from each site, and analyses were adjusted for principal components. Another strength of our study is that individual data on age at study and disease duration were available, which allowed us to run stratified analyses. Although stratification inevitably leads to a loss of statistical power, we were able to examine whether associations were consistent in younger participants and in those with shorter disease duration. As PD is a disease of old age, it is possible that genetic associations are biased by survival bias, but analyses in younger participants are unlikely to be biased as they did not reach the age where mortality rates are high [9].
The main limitation of our study pertains to its statistical power. Our power calculations showed that our sample size was sufficient to detect ORs in the range of 0.4–0.6 for a type-1 error rate of 1.7% to 5%, for smoking and alcohol but not for coffee drinking. The highest power was noted for smoking initiation; the study was underpowered to detect weaker associations for coffee and alcohol drinking, hence limiting our ability to draw firm conclusions for these exposures. Please note that our study examined a limited number of exposures (selected a priori based on existing literature) and is not exploratory, and that the Bonferroni correction is conservative. Our aim was to assess whether MR findings were consistent with those from observational studies using different sets of genetic instruments and methods, and to perform subgroup sensitivity analyses to examine the robustness of our findings. One limitation of our analyses for smoking is that we were not able to stratify analyses by smoking status and to compare results in ever and never smokers using genetic instruments for smoking intensity or cessation [48]. We addressed this issue by using a lifetime smoking exposure index that takes into account several aspects of smoking and was developed in order to allow two-sample MR using data unstratified by smoking status [17].
CONCLUSIONS AND FUTURE RESEARCH
Using an independent dataset, our study confirms previous MR findings adds further evidence in favour of a protective effect of smoking on PD and shows that this association is not explained by survival or incidence-prevalence bias. For alcohol and coffee drinking, larger studies and stronger genetic instruments are needed.
The number of PD cases is predicted to double between 2015–2040 [49], and the identification of neuroprotective strategies is a major goal. Our findings may help prioritise neuroprotective approaches for PD. Further research is necessary to improve genetic instruments for some of the exposures examined here, to understand the pathways involved in these associations, to determine whether these findings are corroborated in non-European populations, whether PD patients subgroups should be more specifically targeted, and whether there are critical periods of exposure.
ADDITIONAL COURAGE-PD INVESTIGATORS
Sophia N Pchelina (Saint Petersburg, Russia), Thomas Brücke (Wien, Austria), Marie-Anne Loriot (Paris, France), Claire Mulot (Paris, France), Yves Koudou (Villejuif, France), Jean-Christophe Corvol (Paris, France), Georgia Xiromerisiou (Larissa, Greece), Christos Koros (Athens, Greece), Matina Maniati (Athens, Greece), Maria Bozi (Athens, Greece), Micol Avenali (Pavia, Italy), Margherita Canesi (Milan, Italy), Giorgio Sacilotto (Milan, Italy), Michela Zini (Milan, Italy), Roberto Cilia (Milan, Italy), Francesca Del Sorbo (Milan, Italy), Nicoletta Meucci (Milan, Italy), Letizia Straniero (Milan, Italy), Rosanna Asselta (Milan, Italy), Radha Procopio (Catanzaro, Italy), Aldo Quattrone (Catanzaro, Italy), Manabu Funayama (Tokyo, Japan), Aya Ikeda (Tokyo, Japan), Takashi Matsushima (Tokyo, Japan), Yuanzhe Li (Tokyo, Japan), Hiroyo Yoshino (Tokyo, Japan), Zied Landoulsi (Luxembourg, Luxembourg), Rubén Femández-Santiago (Barcelona, Spain), Nicholas Wood (London, UK), Huw R Morris (London, UK).
Supplementary Material
ACKNOWLEDGMENTS
We thank the UK Biobank, GWAS and Sequencing Consortium of Alcohol and Nicotine use (GSCAN), the Bitter and sweet beverage consumption GWAS consortium and the Psychiatric Genetics Consortium for providing summary statistics for these analyses.
This study used data from the Courage-PD consortium, conducted under a partnership agreement between 35 studies. The Courage-PD consortium is supported by the EU Joint Program for Neurodegenerative Disease research (JPND; https://www.neurodegenerationresearch.eu/initiatives/annual-calls-for-proposals/closed-calls/risk-factors-2012/risk-factor-call-results/courage-pd/).
CD is the recipient of a doctoral grant from Université Paris-Saclay, France.
MS was supported by the grants from the German Research Council (DFG/SH 599/6-1), MSA Coalition, and Michael J Fox Foundation (USA Genetic Diversity in PD Program: GAP-India Grant ID: 17473).
ABS, DGH, and CE are funded by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, project ZOl AG000949.
ER is funded by the Canadian Consortium on Neurodegeneration in Aging.
SK is funded by MSWA.
PT is the recipient of an Estonian Research Council Grant PRG957.
EMV is funded by the Italian Ministry of Health (Ricerca Corrente 2021).
SB and JC are supported by grants from the National Research Foundation of South Africa (Grant Number: 106052); the South African Medical Research Council (Self-Initiated Research Grant); and Stellenbosch University, South Africa; they also acknowledge the support of the NRF-DST Centre of Excellence for Biomedical Tuberculosis Research; South African Medical Research Council Centre for Tuberculosis Research; Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town.
PP and MDF have received funding from the Spanish Ministry of Science and Innovation (SAF2013-47939-R).
KW and NLP are funded by the Swedish Research Council, grant numbers K2002-27X-14056-02B, 521-2010-2479, 521-2013-2488, 2017-02175.
NLP is funded by the National Institutes of Health, grant numbers ES10758 and AG 08724.
CR is funded by the Märta Lundkvist Foundation, Swedish Brain Foundation, Karolinska Institutet Research Fund.
ACB from the Swedish Brain Foundation, Swedish Research Council, Karolinska Institutet Research Funds.
MT is funded by the Parkinson’s UK.
PG GEN sample collection was funded by the MRC and UK Medical Research Council (CEC, KEM).
The sponsors had no role in the study design, data collection, data analysis, data interpretation, the writing of the report, or the decision to submit the paper for publication.
CONFLICT OF INTEREST
PM reports grants from Fonds National de Recherche (FNR), grants from German Research Council (DFG), during the conduct of the study; and Patrick May is co-founder of MeGeno S.A., Eschsur-Alzette, Luxembourg, a company for personal genomics.
DRB worked as a staff scientist at Megeno S.A. ABS reports grants from Department of Defense, during the conduct of the study; grants from Michael J Fox Foundation, outside the submitted work.
WP reports personal fees from Grünenthal, personal fees from Abb Vie, personal fees from AOP Orphan, personal fees from Zambon, personal fees and other from Boehringer Ingelheim, personal fees from Stada, personal fees from UCB Pharma, outside the submitted work.
AEL reports personal fees from Abb Vie, personal fees from AFFiRis, personal fees from Janssen, personal fees from Biogen, personal fees from Merck, personal fees from Sun Pharma, personal fees from Corticobasal Solutions, personal fees from Sunovion, personal fees from Paladin, personal fees from Lilly, personal fees from Medtronic, personal fees from Theravance, personal fees from Lundbeck, personal fees from Retrophin, personal fees from Roche, personal fees from PhotoPharmics, outside the submitted work.
AB reports grants from France Parkinson + FRC, grants from ANR - EPIG - Agence nationale de recherche, grants from ANR - JPND - Agence nationale de recherche, grants from RDS (Roger de Spoelberch Foundation), grants from France Alzheimer, grants from Institut de France, grants from ANR - EPIG, grants from FMR (maladies rares), outside the submitted work.
JCC reports grants from the Michael J Fox Foundation, Sanofi, and served in advisory boards for Air Liquide, Biogen, Denali, Ever Pharma, Idorsia, Prevail Therapeutic, Theranexus, UCB, outside the submitted work.
MCCH reports grants from France Parkinson, grants from ANR −Agence nationale de recherche, (MetDePaDi, Synapark), grant from ANR – JPND (TransNeuro), Agence rationale de recherche, Grant; grants from Fondation de France, grants from the Michael J Fox Foundation, outside the submitted work.
KB reports grants from MJFF, grants from BMBF, personal fees from Zambon, UCB, Abbvie, grants from University of Tuebingen, outside the submitted work.
EMV reports speaking honoraria from Zambon; served as expert panelist for the International Parkinson and Movement Disorder Society; serves as Associate Editor of Journal of Medical Genetics, Section Editor of Pediatric Research, Member of the Editorial Board of Movement Disorders Clinical Practice; grants from the Italian Ministry of Health, CARIPLO Foundation, Pierfranco and Luisa Mariani Foundation, outside the submitted work.
NH reports grants from - Japan Agency for Medical Research and Development (AMED), grants from - Japan Society for the Promotion of Science (JSPS), grants from - Ministry of Education Culture,Sports,Science and Technology Japan; Grant-in-Aid for Scientific Research on Innovative Areas, personal fees and other from Dai-Nippon Sumitomo Pharma Co., Ltd, personal fees and other from Takeda Pharmaceutical Co., Ltd., personal fees and other from Kyowa Kirin Co., Ltd., personal fees and other from GSK K.K, personal fees and other from Nippon Boehringer Ingelheim, Co., Ltd, personal fees and other from FP Pharmaceutical Corporation, personal fees and other from Eisai Co., Ltd., personal fees and other from Kissei Pharmaceutical Company, personal fees and other from Nihon Medi-physics Co., Ltd, personal fees and other from Novartis Pharma K.K, personal fees and other from Biogen Idec Japan Ltd, personal fees and other from AbbVie, from Medtronic, Inc., other from Boston Scientific Japan, personal fees and other from Astellas Pharma Inc., grants and other from Ono Pharmaceutical Co., Ltd, other from Nihon Pharmaceutical Co., Ltd, other from Asahi Kasei Medical Co., Ltd, other from Mitsubishi Tanabe Pharma Corporation, personal fees and other from Daiichi Sankyo Co., other from OHARA Pharmaceutical Co., Ltd, other from Meiji Seika Pharma, personal fees from Sanofi K.K., personal fees from Pfizer Japan Inc., personal fees from Alexion Pharmaceuticals, personal fees from Mylan N.V, personal fees from MSD K.K, personal fees from Lund Beck Japan, other from Hisamitsu Pharmaceutical Co., Inc, outside the submitted work.
KN reports grants from - Japan Society for the Promotion of Science (JSPS), outside the submitted work.
PK reports other from Centre Hospitalier de Luxembourg; University of Luxembourg, grants from Fonds National de Recherche (FNR), from null, outside the submitted work.
BPCW reports grants from ZonMW, grants from Hersenstichting, grants from uniQure, other from uniQure, grants from Gossweiler Fund, grants from Rad-boud university medical centre, outside the submitted work.
BRB reports grants from Netherlands Organization for Health Research and Development, grants from Michael J. Fox Foundation, grants from Parkinson Vereniging, grants from Parkinson Foundation, grants from Gatsby Foundation, grants from Verily Life Sciences, grants from Horizon 2020, grants from Topsector Life sciences and Health, grants from Stichting Parkinson Fonds, grants from UCB, grants from Abbvie, during the conduct of the study; personal fees from Biogen, personal fees from Abbvie, personal fees from Walk with Path, personal fees from UCB, personal fees from Abbvie, personal fees from Zambon, personal fees from Bial, personal fees from Roche, outside the submitted work; and Serves as editor-in-chief of the Journal of Parkinson’s Disease and serves on the editorial board of Practical Neurology and Digital Biomarkers.
MT (M.Toft) reports grants from Research Council of Norway, during the conduct of the study; grants from South-Eastern Norway Regional Health Authority, grants from Michael J. Fox Foundation, outside the submitted work.
LP reports grants from Norwegian Health Association, grants from South-Eastern Norway Regional Health Authority, outside the submitted work.
JJF reports grants from GlaxoSmithKline, grants from Grunenthal, grants from Fundação MSD (Portugal), grants from TEVA, grants from MSD, grants from Allergan, grants from Novartis, grants from Medtronic, grants from GlaxoSmithKline, grants from Novartis, grants from TEVA, grants from Lundbeck, grants from Solvay, grants from BIAL, grants from Merck-Serono, grants from Merz, grants from Ipsen, grants from Biogen, grants from Acadia, grants from Allergan, grants from Abbvie, grants from Sunovion Pharmaceuticals, personal fees from Faculdade de Medicina de Lisboa, personal fees from CNS - Campus Neurológico Sénior, personal fees from BIAL, personal fees from Novartis, outside the submitted work.
ET received honoraria for consultancy from TEVA, Bial, Prevail Therapeutics, Boehringer Ingelheim, Roche and BIOGEN and has received funding for research from Spanish Network for Research on Neurodegenerative Disorders (CIBERNED)- Instituto Carlos III (ISCIII), and The Michael J. Fox Foundation for Parkinson’s Research (MJFF).
KW reports grants from Swedish Research Council, during the conduct of the study.
NLP reports grants from Swedish Research Council, during the conduct of the study.
AP reports grants from Parkinsonfonden (The Swedish Parkinson Foundation), grants from ALF (Swedish Government), grants from Region Skåne, Sweden, grants from Hans-Gabriel och Trolle Wacht-meister Stiftelse för Medicinsk Forskning, Sweden, during the conduct of the study; personal fees from Elsevier, outside the submitted work.
MT (M. Tan) reports grants from Parkinson’s UK, other from Michael J Fox Foundation, other from University College London, outside the submitted work.
RK reports grants from Fonds National de Recherche (FNR), grants from German Research Council (DFG), non-financial support from Abbvie, Zambon, during the conduct of the study; personal fees from University of Luxembourg; Luxembourg Institute of Health; Centre Hospitalier de Luxembourg, grants from Fonds National de Recherche, Luxembourg (FNR), grants from Fonds National de Recherche, Luxembourg (FNR), grants from Fonds National de Recherche (FNR), Luxembourg/German Research Council (DFG), grants from Fonds National de Recherche, Luxembourg (FNR), personal fees from Desitin/Zambon, personal fees from Abbvie GmbH, personal fees from Medtronic GmbH, outside the submitted work.
TG reports personal fees from UCB Pharma, personal fees from Novartis, personal fees from Teva, personal fees from MedUpdate, grants from The Michael J Fox Foundation for Parkinson’s Research, grants from Bundesministerium für Bildung und Forschung (BMBF), grants from Deutsche Forschungsgemeinschaft (DFG), other from “Joint Programming for Neurodegenerative Diseases” (JPND) program, funded by the European Commission, outside the submitted work; in addition, Dr. Gasser has a patent Patent Number: EP1802749 (A2) KASPP (LRRK2) gene, its production and use for the detection and treatment of neurodegenerative disorders issued.
Footnotes
SUPPLEMENTARY MATERIAL
The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/JPD-212851.
REFERENCES
- [1].Ascherio A, Schwarzschild MA (2016) The epidemiology of Parkinson’s disease: Risk factors and prevention. Lancet Neurol 15, 1257–1272. [DOI] [PubMed] [Google Scholar]
- [2].Bellou V, Beibasis L, Tzoulaki I, Evangelou E, Ioannidis JP (2016) Environmental risk factors and Parkinson’s disease: An umbrella review of meta-analyses. Parkinsonism Relat Disord 23, 1–9. [DOI] [PubMed] [Google Scholar]
- [3].Elbaz A (2016) Prodromal symptoms of Parkinson’s disease: Implications for epidemiological studies of disease etiology. Rev Neurol (Paris) 172, 503–511. [DOI] [PubMed] [Google Scholar]
- [4].Savica R, Boeve BF, Mielke MM (2018) When do α-synucleinopathies start? An epidemiological timeline: A review. JAMA Neurol 75, 503–509. [DOI] [PubMed] [Google Scholar]
- [5].Ritz B, Rhodes SL (2010) After half a century of research on smoking and PD, where do we go now? Neurology 74, 870–871. [DOI] [PubMed] [Google Scholar]
- [6].Ritz B, Lee PC, Lassen CF, Arah OA (2014) Parkinson disease and smoking revisited: Ease of quitting is an early sign of the disease. Neurology 83, 1396–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burgess S, Butterworth A, Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G (2008) Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat Med 27, 1133–1163. [DOI] [PubMed] [Google Scholar]
- [9].Smit RAJ, Trompet S, Dekkers OM, Jukema JW, le Cessie S (2019) Survival bias in Mendelian randomization studies: A threat to causal inference. Epidemiology 30, 813–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Boef AG, le Cessie S, Dekkers OM (2015) Mendelian randomization studies in the elderly. Epidemiology 26, el5–16. [DOI] [PubMed] [Google Scholar]
- [11].Ellenberg JH (1994) Differential postmorbidity mortality in observational studies of risk factors for neurologic disorders. Neuroepidemiology 13, 187–194. [DOI] [PubMed] [Google Scholar]
- [12].Blauwendraat C, Faghri F, Pihlstrom L, Geiger JT, Elbaz A, Lesage S, Corvol JC, May P, Nicolas A, Abramzon Y, Murphy NA, Gibbs JR, Ryten M, Ferrari R, Bras J, Guerreiro R, Williams J, Sims R, Lubbe S, Hernandez DG, Mok KY, Robak L, Campbell RH, Rogaeva E, Traynor BJ, Chia R, Chung SJ, International Parkinson’s Disease Genomics Consortium C-PDC, Hardy JA, Brice A, Wood NW, Houlden H, Shulman JM, Morris HR, Gasser T, Kruger R, Heutink P, Sharma M, Simon-Sanchez J, Nalls MA, Singleton AB, Scholz SW (2017) NeuroChip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol Aging 57, 247 e249–247 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Machiela MJ, Chanock SJ (2015) LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmuller G (2015) SNiPA: An interactive, genetic variant-centered annotation browser. Bioinformatics 31, 1334–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, Hartwig FP, Holmes MV, Minelli C, Relton CL, Theodoratou E (2019) Guidelines for performing Mendelian randomization investigations. Wellcome Open Res 4, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X, andMe Research T, Psychiatry HA-I, Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, Hottenga JJ, Huang H, Jang SK, Jansen PR, Ling Y, Magi R, Matoba N, McMahon G, Mulas A, Orru V, Palviainen T, Pandit A, Reginsson GW, Skogholt AH, Smith JA, Taylor AE, Turman C, Willemsen G, Young H, Young KA, Zajac GJM, Zhao W, Zhou W, Bjomsdottir G, Boardman JD, Boehnke M, Boomsma DI, Chen C, Cucca F, Davies GE, Eaton CB, Ehringer MA, Esko T, Fiorillo E, Gillespie NA, Gudbjartsson DF, Haller T, Harris KM, Heath AC, Hewitt JK, Hickie IB, Hokanson JE, Hopfer CJ, Hunter DJ, Iacono WG, Johnson EO, Kamatani Y, Kardia SLR, Keller MC, Kellis M, Kooperberg C, Kraft P, Krauter KS, Laakso M, Lind PA, Loukola A, Lutz SM, Madden PAF, Martin NG, McGue M, McQueen MB, Medland SE, Metspalu A, Mohlke KL, Nielsen JB, Okada Y, Peters U, Polderman TJC, Posthuma D, Reiner AP, Rice JP, Rimm E, Rose RJ, Runarsdot- tir V, Stallings MC, Stancakova A, Stefansson H, Thai KK, Tindle HA, Tyrfingsson T, Wall TL, Weir DR, Weis- ner C, Whitfield JB, Winsvold BS, Yin J, Zuccolo L, Bierut LJ, Hveem K, Lee JJ, Munafo MR, Saccone NL, Wilier CJ, Comelis MC, David SP, Hinds DA, Jorgenson E, Kaprio J, Stitzel JA, Stefansson K, Thorgeirsson TE, Abecasis G, Liu DJ, Vrieze S (2019) Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 51, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wootton RE, Richmond RC, Stuijfzand BG, Lawn RB, Sallis HM, Taylor GMJ, Hemani G, Jones HJ, Zammit S, Davey Smith G, Munafo MR (2019) Evidence for causal effects of lifetime smoking on risk for depression and schizophrenia: A Mendelian randomisation study. Psychol Med 50, 2435–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhong VW, Kuang A, Danning RD, Kraft P, van Dam RM, Chasman DI, Comelis MC (2019) A genome-wide association study of bitter and sweet beverage consumption. Hum Mol Genet 28, 2449–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Larsson SC, Traylor M, Malik R, Dichgans M, Burgess S, Markus HS, CoStream Consortium obotlGoAsP (2017) Modifiable pathways in Alzheimer’s disease: Mendelian randomisation analysis. BMJ 359, j5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burgess S, Thompson SG (2017) Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 32, 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR (2016) Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the 12 statistic. Int J Epidemiol 45, 1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bowden J, Davey SG, Haycock PC, Burgess S (2016) Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol 40, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hartwig FP, Davey Smith G, Bowden J (2017) Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 46, 1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Verbanck M, Chen CY, Neale B, Do R (2018) Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50, 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Elbaz A, Alperovitch A (2002) Bias in association studies resulting from gene-environment interactions and competing risks. Am J Epidemiol 155, 265–272. [DOI] [PubMed] [Google Scholar]
- [26].Schooling CM (2018) Selection bias in population-representative studies? A commentary on Deaton and Cartwright. Soc Sci Med 210, 70. [DOI] [PubMed] [Google Scholar]
- [27].Schooling CM (2019) Biases in GWAS - the dog that did not bark. bioRxiv, 709063. [Google Scholar]
- [28].Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, Bras J, Young E, von Coelln R, Simón-Sánchez J, Schulte C, Sharma M, Krohn L, Pihlstrøm L, Siitonen A, Iwaki H, Leonard H, Faghri F, Gibbs JR, Hernandez DG, Scholz SW, Botia JA, Martinez M, Corvol JC, Lesage S, Jankovic J, Shulman LM, Sutherland M, Tienari P, Majamaa K, Toft M, Andreassen OA, Bangale T, Brice A, Yang J, Gan-Or Z, Gasser T, Heutink P, Shulman JM, Wood NW, Hinds DA, Hardy JA, Morris HR, Gratten J, Visscher PM, Graham RR, Singleton AB, 23 and Me Research Team; System Genomics of Parkinson’s Disease Consortium; International Parkinson’s Disease Genomics Consortium (2019) Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol 18,1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Burgess S, Labrecque JA (2018) Mendelian randomization with a binary exposure variable: Interpretation and presentation of causal estimates. Eur J Epidemiol 33, 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brion MJ, Shakhbazov K, Visscher PM (2013) Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 42,1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Noyce AJ, Bestwick JP, Silveira-Moriyama L, Hawkes CH, Giovannoni G, Lees AJ, Schrag A (2012) Meta-analysis of early nonmotor features and risk factors for Parkinson disease. Ann Neurol 72, 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gallo V, Vineis P, Cancellieri M, Chiodini P, Barker RA, Brayne C, Pearce N, Vermeulen R, Panico S, Bueno-de-Mesquita B, Vanacore N, Forsgren L, Ramat S, Ardanaz E, Arriola L, Peterson J, Hansson O, Gavrila D, Sacerdote C, Sieri S, Kühn T, Katzke VA, van der Schouw YT, Kyrozis A, Masala G, Mattiello A, Pemeczky R, Middleton L, Saracci R, Riboli E (2019) Exploring causality of the association between smoking and Parkinson’s disease. Int J Epidemiol 48, 912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Grover S, Lill CM, Kasten M, Klein C, Del Greco MF, Konig IR (2019) Risky behaviors and Parkinson disease: A mendelian randomization study. Neurology 93, el412-el424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dominguez-Baleon C, Ong JS, Scherzer CR, Renteria ME, Dong X (2021) Understanding the effect of smoking and drinking behavior on Parkinson’s disease risk: A Mendelian randomization study. Sci Rep 11,13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Noyce AJ, Bandres-Ciga S, Kim J, Heilbron K, Kia D, Hemani G, Xue A, Lawlor DA, Smith GD, Duran R, Gan-Or Z, Blauwendraat C, Gibbs JR, 23 and Me Research Team5 IPsDGC, Hinds DA, Yang J, Visscher P, Cuzick J, Morris H, Hardy J, Wood NW, Nalls MA, Singleton AB(2019) The Parkinson’s disease Mendelian randomization research portal. Mov Disord 34, 1864–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tzourio C, Rocca WA, Breteler MM, Baldereschi M, Dartigues JF, Lopez-Pousa S, Manubens-Bertran JM, Alpérovitch A (1997) Smoking and Parkinson’s disease. An age-dependent risk effect? The EUROPARKINSON Study Group. Neurology 49,1267–1272. [DOI] [PubMed] [Google Scholar]
- [37].Ritz B, Ascherio A, Checkoway H, Marder KS, Nelson LM, Rocca WA, Ross GW, Strickland D, Van Den Eeden SK, Gorell J (2007) Pooled analysis of tobacco use and risk of Parkinson disease. Arch Neurol 64, 990–997. [DOI] [PubMed] [Google Scholar]
- [38].Lee PC, Ahmed I, Loriot MA, Mulot C, Paul KC, Bronstein JM, Ritz B, Elbaz A (2018) Smoking and Parkinson disease: Evidence for gene-by-smoking interactions. Neurology 90, e583–e592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chuang YH, Lee PC, Vlaar T, Mulot C, Loriot MA, Hansen J, Lill CM, Ritz B, Elbaz A (2017) Pooled analysis of the HLA-DRB1 by smoking interaction in Parkinson disease. Ann Neurol 82, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Quik M, Perez XA, Bordia T (2012) Nicotine as a potential neuroprotective agent for Parkinson’s disease. Mov Disord 27, 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hong DP, Fink AL, Uversky VN (2009) Smoking and Parkinson’s disease: Does nicotine affect alpha-synuclein fibrillation? Biochim Biophys Acta 1794, 282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ebersbach G, Stöck M, Müller J, Wenning G, Wissel J, Poewe W (1999) Worsening of motor performance in patients with Parkinson’s disease following transdermal nicotine administration. Mov Disord 14, 1011–1013. [DOI] [PubMed] [Google Scholar]
- [43].Vieregge A, Sieberer M, Jacobs H, Hagenah JM, Vieregge P (2001) Transdermal nicotine in PD: A randomized, double-blind, placebo-controlled study. Neurology 57, 1032–1035. [DOI] [PubMed] [Google Scholar]
- [44].Villafane G, Thiriez C, Audureau E, Straczek C, Kerschen P, Cormier-Dequaire F, Van Der Gucht A, Gurruchaga JM, Quéré-Came M, Evangelista E, Paul M, Defer G, Damier P, Remy P, Itti E, Fenelon G (2018) High-dose transdermal nicotine in Parkinson’s disease patients: A randomized, open-label, blinded-endpoint evaluation phase 2 study. Eur J Neurol 25,120–127. [DOI] [PubMed] [Google Scholar]
- [45].Jimenez-Jimenez FJ, Alonso-Navarro H, Garcia-Martin E, Agundez JAG (2019) Alcohol consumption and risk for Parkinson’s disease: A systematic review and meta-analysis. J Neurol 266, 1821–1834. [DOI] [PubMed] [Google Scholar]
- [46].Qi H, Li S (2014) Dose-response meta-analysis on coffee, tea and caffeine consumption with risk of Parkinson’s disease. Geriatr Gerontol Int 14,430–439. [DOI] [PubMed] [Google Scholar]
- [47].Pierce BL, Burgess S (2013) Efficient design for Mendelian randomization studies: Subsample and 2-sample instrumental variable estimators. Am J Epidemiol 178,1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gage SH, Davey Smith G, Ware JJ, Flint J, Munafo MR (2016) G=E: What GWAS can tell us about the environment. PLoS Genet 12, el005765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dorsey ER, Bloem BR (2018) The Parkinson pandemic-a call to action. JAMA Neurol 75, 9–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Results can be reproduced using Supplementary Tables 3 and 4; no additional data available.