Abstract
Identifying individuals with early mild cognitive impairment (EMCI) can be an effective strategy for early diagnosis and delay the progression of Alzheimer’s disease (AD). Many approaches have been devised to discriminate those with EMCI from healthy control (HC) individuals. Selection of the most effective parameters has been one of the challenging aspects of these approaches. In this study we suggest an optimization method based on five evolutionary algorithms that can be used in optimization of neuroimaging data with a large number of parameters. Resting-state functional magnetic resonance imaging (rs-fMRI) measures, which measure functional connectivity, have been shown to be useful in prediction of cognitive decline. Analysis of functional connectivity data using graph measures is a common practice that results in a great number of parameters. Using graph measures we calculated 1155 parameters from the functional connectivity data of HC (n = 72) and EMCI (n = 68) extracted from the publicly available database of the Alzheimer’s disease neuroimaging initiative database (ADNI). These parameters were fed into the evolutionary algorithms to select a subset of parameters for classification of the data into two categories of EMCI and HC using a two-layer artificial neural network. All algorithms achieved classification accuracy of 94.55%, which is extremely high considering single-modality input and low number of data participants. These results highlight potential application of rs-fMRI and efficiency of such optimization methods in classification of images into HC and EMCI. This is of particular importance considering that MRI images of EMCI individuals cannot be easily identified by experts.
Introduction
Alzheimer’s disease (AD) is the most common type of dementia, with around 50 million patients worldwide [1,2]. AD is usually preceded by a period of mild cognitive impairment (MCI) [3,4]. Identifying the subjects with MCI could be an effective strategy for early diagnosis and delay the progression of AD towards irreversible brain damage [5–7]. While researchers were fairly successful in diagnosis of AD, researchers were significantly less successful in diagnosis of MCI [8–11]. In particular, detection of early stages of MCI (EMCI) has been proven to be very challenging [12–14]. Therefore, in this study we propose a novel method based on evolutionary algorithms to select a subset of graph features calculated from functional connectivity data to discriminate between healthy participants (HC) and EMCI.
It has been shown that the brain goes through many functionally, structurally and physiologically changes prior to any obvious behavioral symptoms in AD [15–17]. Therefore, many approaches have been devised based on biomarkers to distinguish between HC, and different stages of MCI, and AD [18–20]. For example, parcellation of structural magnetic resonance imaging (MRI) data has been used in many studies as brain structure changes greatly in AD [21–24]. Further, in two studies, we showed that T1-weighted MRI (structural MRI; sMRI) can be used in classification of AD and MCI. Indeed, the majority of early studies looking at classification of AD and HC was done on sMRI [22]. This is mostly due to costs and accessibility of sMRI data [23].
While structural neuroimaging has shown some success in early detection of AD, functional neuroimaging has proven to be a stronger candidate [25–27]. Functional MRI (fMRI) allows for the examination of brain functioning while a patient is performing a cognitive task. This technique is especially well suited to identifying changes in brain functioning before significant impairments can be detected on standard neuropsychological tests, and as such is sensitive to early identification of the disease processes [28,29]. While fMRI requires participants to perform a task, resting-state fMRI (rs-fMRI) is capable of measuring the spontaneous fluctuations of brain activity without any task, hence it is less sensitive to individual cognitive abilities [30–32].
One important feature of rs-fMRI is the ability to measure functional connectivity changes [33,34], which has been shown to be a prevalent change in AD [35–38]. Furthermore, it is shown that the increased severity of cognitive impairment is associated with increasing alteration in connectivity patterns, suggesting that disruptions in functional connectivity may contribute to cognitive dysfunction and may represent a potential biomarker of impaired cognitive ability in MCI. In particular, research has highlighted that longitudinal alterations of functional connectivity are more profound in earlier stages as opposed to later stages of the disease [39]. Therefore, analysis of functional connectivity can provide an excellent opportunity in identification of early states of AD.
As functional connectively analysis inherently relies of networks of activity, researchers have used graph theory measures to investigate the global, as well as local, characteristics of different brain areas [40–43] Click or tap here to enter text. This method has been used successfully in a wide range of application in both healthy participants and patients [44] Click or tap here to enter text. such as depression [45,46] Click or tap here to enter text., Parkinson’s disease [47] Click or tap here to enter text., as well as AD [48] Click or tap here to enter text. Graph theories provides us with a way to study AD [48–53] and comprehensively compare functional connectivity organization of the brain between patients and controls [43–45] Click or tap here to enter text. and importantly between different stages of AD [54,55] Click or tap here to enter text. This method can also unveil compensatory mechanisms, thus revealing brain functional differences in participants with comparable level of cognitive ability [56–59] Click or tap here to enter text.
Graph theory analysis of rs-fMRI data, however, leads to a large number of parameters. Therefore, to reduce computational complexity, it is essential to select an optimal subset of features that can lead to high discrimination accuracy [60,61]. Feature selection is particularly complicated due to the non-linear nature of classification methods: more parameters do not necessarily lead to better performance, and there is also a dependency of parameters [62,63]. Therefore, it is extremely important to utilize a suitable optimization method that can deal with nonlinear high-dimensional search spaces.
Evolutionary algorithms (EA) are biologically-inspired algorithms that are extremely effective in optimization algorithms with large search spaces [64–66]. These methods, in contrast with many other search methods such as complete search, greedy search, heuristic search and random search [67,68], do not suffer from stagnation in local optima and/or high computational cost [69,70]. Feature selection has been used to improve the quality of the feature set in many machine learning tasks, such as classification, clustering and time-series prediction [71]. Classification and time-series prediction are particularly relevant to many neurodegenerative diseases: classification can be used in identification of those with brain damage [72,73] and time-series prediction can be used in estimation of disease progression [74,75].
EA has been used in characterization and diagnosis of AD [76–78]. Such methods have achieved reasonably high accuracy in classification of AD and HC (70–95%). They, however, have been unsuccessful in classification of the MCI patients [79]. Therefore, in this study, we devised a method that achieves higher accuracy in the classification of HC and EMCI participants compared to the past-published research. We used MRI and rs-fMRI data of a group of healthy participants and those with EMCI. We applied graph theory to extract a collection of 1155 parameters. This data is then given to five different EA methods to select an optimum subset of parameters. These selected parameters are subsequently given to an artificial neural network to classify the data into two groups of HC and EMCI. We aimed at identifying the most suitable method of optimization based on accuracy and training time, as well as identifying the most informative parameters.
Methods
Participants
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD).
Data for 140 participants were extracted from the ADNI [80–82]. See Table 1 for the details of the data. EMCI participants had no other neurodegenerative diseases except MCI. The EMCI participants were recruited with memory function approximately 1.0 SD below expected education adjusted norms [83]. HC subjects had no history of cognitive impairment, head injury, major psychiatric disease, or stroke.
Table 1. Demographics of the data for participants included in this study.
| EMCI | HC | P | |
|---|---|---|---|
| n | 68 | 72 | |
| Female (n [%]) | 38 [55.88] | 38 [52.77] | |
| Age (mean [SD]) | 71.73 [7.80] | 69.97 [5.60] | 0.297 |
| MMSE (mean [SD]) | 28.61 [1.60] | 28.40 [4.60] | 0.302 |
| CDR | 0.5 or 1 | 0 | < 0.001 |
notes: CDR: Clinical dementia rating, MMSE: Mini-mental state exam, HC: Healthy control, EMCI: Early mild cognitive impairment.
Proposed method
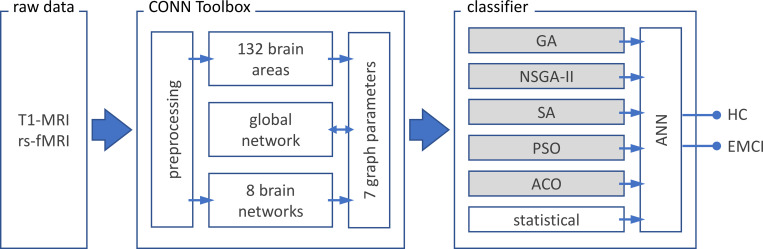
sMRI and rs-fMRI data was extracted from the ADNI database [82]. The data is given to CONN toolbox [84] in MATLAB v2018 (MathWorks, California, US). CONN is a tool for preprocessing, processing, and analysis of functional connectivity data. Preprocessing consisted of reducing subject motion, image distortions, and magnetic field inhomogeneity effects and application of denoising methods for reduction of physiological effects and other sources of noise. The processing stage consisted of extraction of functional connectivity and graph theory measures. In this stage, through two pipelines, a collection of 1155 parameters are extracted (see below) [84,85]. These parameters are then given to one of the dimension reduction methods (five EA and one statistical method) to select a subset of features. The selected features are finally given to an artificial neural network to classify the data into two categories of HC and EMCI. The classification method was performed via a 90/10 split; 90% of the data was used for the training and 10% of the data was used for validation. See Fig 1 for the summary of the procedure of the method.
Fig 1. Procedure of the proposed method.
T1-MRI (sMRI) and resting-state fMRI (rs-fMRI) data of healthy participants (HC; n = 72) and patients with early mild cognitive impairment (EMCI; n = 68) are extracted from ADNI database 82. Preprocessing, parcellation of brain area (132 regions based on AAL and Harvard-Oxford atlas) and extraction of the functional connectivity (8 network parameters with a total of 32 nodes), as well as the 7 graph parameters are done using CONN toolbox 84. Subsequently the global network is calculated based on the network parameters. The 1155 ([132 brain regions + 32 nodes of brain networks + 1 global network] × 7 graph parameters) extracted parameters are given to one of the optimization methods to select the best subset of parameters that lead to best classification method. Optimization methods consisted of five evolutionary algorithms (boxes with grey shading) and one statistical algorithm. The outputs of these methods are given to an artificial neural network (ANN) with two hidden layers to classify the data into HC and EMCI. AAL: Automated anatomical atlas; GA: Genetic algorithm; NSGA-II: Nondominated sorting genetic algorithm II; ACO: Ant colony optimization; SA: Simulated annealing; PSO: Particle swarm optimization; seven graph features: Degree centrality, betweenness centrality, path length, clustering coefficient, local efficiency, cost and global efficiency.
Data acquisition and processing
Brain structural sMRI data with 256×256×170 voxels and 1×1×1 mm3 voxel size were extracted for all subjects. MRI data preprocessing steps consisted of non-uniformity correction, segmentation into grey matter, white matter and cerebrospinal fluid (CSF) and spatial normalization to MNI space.
Rs-fMRI data were obtained using an echo-planar imaging sequence on a 3T Philips MRI scanner. Acquisition parameters were: 140 time points, repetition time (TR) = 3000 ms, echo time (TE) = 30 ms, flip angle = 80°, number of slices = 48, slice thickness = 3.3 mm, spatial resolution = 3×3×3 mm3 and in plane matrix = 64×64. FMRI images preprocessing steps consisted of motion correction, slice timing correction, spatial normalization to MNI space, low frequency filtering to keep only (0.01–0.1 Hz) fluctuations.
CONN toolbox [84,85] is used to process the sMRI and rs-fMRI data. The output of this toolbox is 1155 values consisting of: (a) 132 distinct brain areas according to Automated Anatomical Labeling (AAL) and Harvard-Oxford atlases, (b) eight brain networks containing 32 nodes and (c) a global network parameter that is the average of seven graph parameters [86–88]. All these values are multiplied by seven graph parameters, see below. See supplementary data for details of these parameters. The sMRI images were used to register the functional images and improve the analysis of the rs-fMRI data.
Functional connectivity
Functional connectivity, also called “resting state” connectivity, is a measure for the temporal correlations among the blood-oxygen-level-dependent (BOLD) signal fluctuations in different brain areas [89–91]. The functional connectivity matrix is the correlation, covariance, or the mutual information between the fMRI time series of every two brain regions, which is stored in an n×n matrix for each participant, where n is the number of brain regions obtained by atlas parcellation [91]. To extract functional connectivity between different brain areas we used Pearson correlation coefficients formula as following [84,92]:
where S is the BOLD time series at each voxel (for simplicity all-time series are considered central to zero means), R is the average BOLD time series within an ROI, r is the spatial map of Pearson correlation coefficients, and Z is the seed-based correlations (SBC) map of Fisher-transformed correlation coefficients for this ROI [93].
Graph parameters
We used the graph theory technique to study topological features of functional connectivity graphs across multiple regions of the brain [86–94]. Graph nodes represented brain regions and edges represented interregional resting-state functional connectivity. The graph measurements in all of the ROIs are defended using nodes = ROIs, and edges = suprathreshold connections. For each subject, graph adjacency matrix A is computed by thresholding the associated ROI to-ROI Correlation (RRC) matrix r by an absolute (e.g., z>0.5) or relative (e.g., highest 10%) threshold. Then, from the resulting graphs, some measurements can be computed addressing topological properties of each ROI within the graph as well as of the entire network of ROIs. The adjacency matrix is employed for estimating common features of graphs including (1) degree centrality (the number of edges that connect a node to the rest of the network) (2) betweenness centrality (the proportion of shortest paths between all node pairs in the network that pass through a given index node), (3) average path length (the average distance from each node to any other node), (4) clustering coefficient (the proportion of ROIs that have connectivity with a particular ROI that also have connectivity with each other), (5) cost (the ratio of the existing number of edges to the number of all possible edges in the network), (6) local efficiency (the network ability in transmitting information at the local level), (7) global efficiency (the average inverse shortest path length in the network; this parameter is inversely related to the path length) [95].
Dimension reduction methods
We used five EA to select the most efficient set number of features. These algorithms are as follows:
Genetic algorithm (GA): GA is one of the most advanced algorithms for feature selection [96]. This algorithm is based on the mechanics of natural genetics and biological evolution for finding the optimum solution. It consists of five steps: selection of initial population, evaluation of fitness function, pseudo-random selection, crossover, and mutation [97]. For further information refer to supplementary Methods section. Single point, double point, and uniform crossover methods are used to generate new individuals. In this study we used 0.3 and 0.1 as mutation percentage and mutation rate, respectively; 20 members per population, crossover percentage was 14 with 8 as selection pressure [74,98].
Nondominated sorting genetic algorithm II (NSGA-II): NSGA is a method to solve multi-objective optimization problems to capture a number of solutions simultaneously [99]. All the operators in GA are also used here. NSGA-II uses binary features to fill a mating poll. Nondomination and crowding distance are used to sort the new members. For further information refer to supplementary Methods section. In this study the mutation percentage and mutation rate were set to 0.4 and 0.1, respectively; population size was 25, and crossover percentage was 14.
Ant colony optimization algorithm (ACO): ACO is a metaheuristic optimization method based on the behavior of ants [100]. This algorithm consists of four steps: initialization, creation of ant solutions (a set of ants build a solution to the problem being solved using pheromones values and other information), local search (improvement of the created solution by ants), and global pheromone update (update in pheromone variables based on search action followed by ants) [101]. ACO requires a problem to be described as a graph: nodes represent features and edges indicate which features should be selected for the next generation. In features selection, the ACO tries to find the best solutions using prior information from previous iterations. The search for the optimal feature subset consists of an ant traveling through the graph with a minimum number of nodes required for satisfaction of stopping criterion [102]. For further information refer to supplementary Methods section. We used 10, 0.05, 1, 1 and 1 for the number of ants, evaporation rate, initial weight, exponential weight, and heuristic weight, respectively.
Simulated annealing (SA): SA is a stochastic search algorithm, which is particularly useful in large-scale linear regression models [103]. In this algorithm, the new feature subset is selected entirely at random based on the current state. After an adequate number of iterations, a dataset can be created to quantify the difference in performance with and without each predictor [104,105]. For further information refer to supplementary Methods section. We set initial temperature and temperature reduction rate with 10 and 0.99, respectively.
Particle swarm optimization (PSO): PSO is a stochastic optimization method based on the behavior of swarming animals such as birds and fish. Each member finds optimal regions of the search space by coordinating with other members in the population. In this method, each possible solution is represented as a particle with a certain position and velocity moving through the search space [106–108]. Particles move based on cognitive parameter (defining the degree of acceleration towards the particle’s individual local best position, and global parameter (defining the acceleration towards the global best position). The overall rate of change is defined by an inertia parameter. For further information refer to supplementary Methods section. In this paper simulation we use 20 as the warm size, cognitive and social parameters were set to 1.5 and inertia as 0.72.
Statistical approach: To create a baseline to compare dimension reduction methods based on evolutionary algorithms, we also used the statistical approach to select the features based on the statistical difference between the two groups. We compared the 1155 parameters using two independent-sample t-test analyses. Subsequently we selected the parameters based on their sorted p values.
Classification method
For classification of EMCI and HC we used a multi-layer perceptron artificial neural network (ANN) with two fully-connected hidden layers with 10 nodes each. Classification method was performed via a 10-fold cross-validation. We used Levenberg-Marquardt Back propagation (LMBP) algorithm for training [109–111] and mean square error as a measure of performance. The LMBP has three steps: (1) propagate the input forward through the network; (2) propagate the sensitivities backward through the network from the last layer to the first layer; and finally (3) update the weights and biases using Newton’s computational method [109]. In the LMBP algorithm the performance index F(x) is formulated as:
where e is vector of network error, and x is the vector matrix of network weights and biases. The network weights are updated using the Hessian matrix and its gradient:
Where J represent Jacobian matrix. The Hessian matrix H and its gradient G are calculated using:
where the Jacobian matrix is calculated by:
where am−1 is the output of the (m−1)th layer of the network, and Sm is the sensitivity of F(x) to changes in the network input element in the mth layer and is calculated by:
where wm+1 represents the neuron weight at (m+1)th layer, and n is the network input [109].
Results
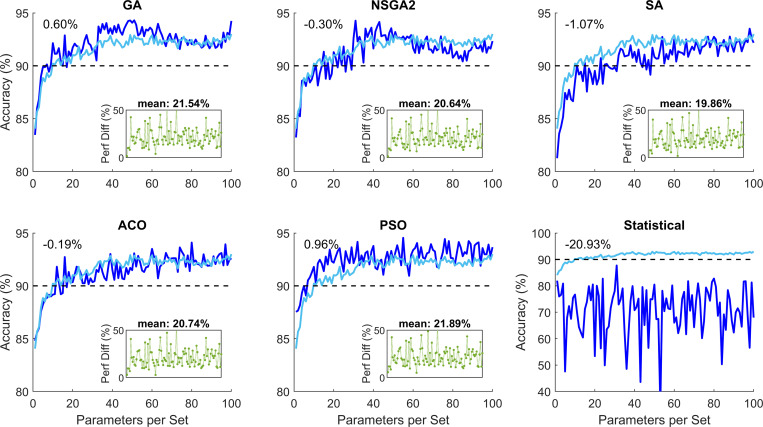
The preprocessing and processing of the data was successful. We extracted 1155 graph parameters per participant (see S1–S11 Figs). This data was used for the data optimization step. Using the five EA optimization methods and the statistical method, we investigated the performance of the classification for different numbers of subset of parameters. Fig 2 shows the performance of these methods for 100 subsets of parameters with 1 to 100 parameters. These plots are created based on 200 repetitions of the EA algorithms. To investigate the performance of the algorithms with more repetitions, we ran the same algorithms with 500 repetitions. These simulations showed no major improvement of increased repetition (maximum 0.84% improvement; see S11 Fig).
Fig 2. Classification performance of the five evolutionary algorithm (EA) methods and the statistical method for parameter subsets with 1 to 100 elements.
The light blue color shows the average of the five EV algorithms. The number on the top left-hand corner represents the difference between the relevant plot and the mean performance of the EA methods. The green plot subplot in each panel represents superiority of the relevant EA as compared to the statistical method for different 100 subsets. The percentage value above the subplot shows the mean superior performance for the 100 subsets compared to the statistical method. These plots show that the EA performed significantly better than the statistical method. GA: Genetic algorithm; NSGA2: Nondominated sorting genetic algorithm II; ACO: Ant colony optimization; SA: Simulated annealing; PSO: Particle swarm optimization.
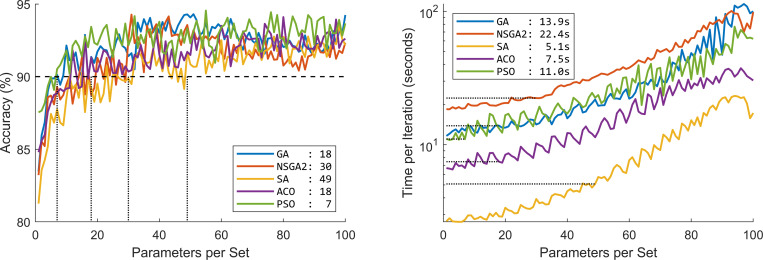
A threshold of 90% was chosen as the desired performance accuracy. Statistical modeling performance constantly less than this threshold. The five EA methods achieved this performance with varying number of parameters. Fig 3 shows the accuracy percentage and the optimization speed of the five EA methods.
Fig 3.
Performance of the five evolutionary algorithms (EA) in terms of (a) percentage accuracy and (b) optimization speed. The values in the legend of panel (a) show the minimum number of parameters required to achieve minimum 90% accuracy. The values in the legend of panel (b) show the minimum optimization speed to achieve minimum 90% accuracy based on panel (a). GA: Genetic algorithm; NSGA2: Nondominated sorting genetic algorithm II; ACO: Ant colony optimization; SA: Simulated annealing; PSO: Particle swarm optimization.
There is growing body of literature showing gender differences. It has been shown that women are more likely to suffer from AD. Therefore, to investigate whether our analysis method performs better on a particular gender or not, we split the data into two groups of female and male participants. Our analysis showed that there is no meaningful difference between the two groups (see S2 Table).
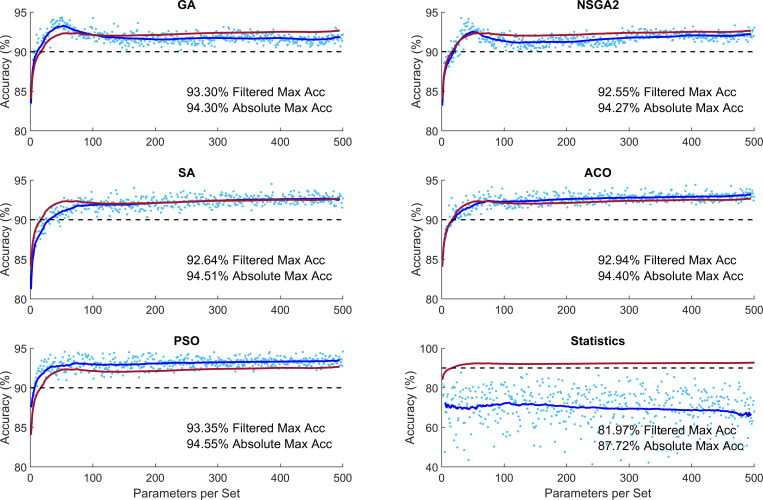
To investigate whether increasing number of parameters would increase performance, we performed similar simulations with maximum 500 parameters in each subset. This analysis showed that the performance of the optimization methods plateaus without significant increase from 100 parameters (Fig 4). This figure shows that performance of the optimization methods was between 92.55–93.35% and 94.27–94.55% for filtered and absolute accuracy, respectively. These accuracy percentages are significantly higher than 81.97% and 87.72% for filtered and absolute accuracy in the statistical classification condition.
Fig 4. Performance of different optimization methods for increased number of parameters per subset.
The light blue dots indicate the performance of algorithms for each subset of parameters. The dark blue curve shows the moving average of the samples with window of ±20 points (Filtered Data). The red curve shows the mean performance of the five evolutionary algorithms. GA: Genetic algorithm; NSGA2: Nondominated sorting genetic algorithm II; ACO: Ant colony optimization; SA: Simulated annealing; PSO: Particle swarm optimization.
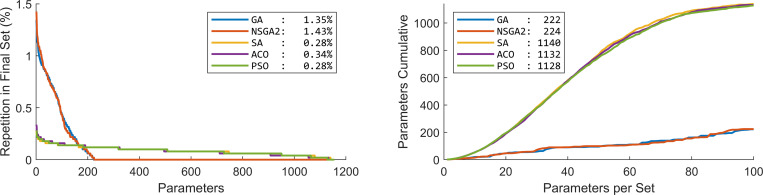
To investigate the contribution of different parameters in the optimization of classification we looked at the distribution of parameters in the 100 subsets calculated above (Fig 5). GA and NSGA showed that the majority of the subsets consisted of repeated parameters: out of the 1155 parameters only about 200 of the parameters were selected in the 100 subsets. SA, ACO and PSO, on the other hand, showed a more diverse selection of parameters: almost all the parameters appeared in at least one of the 100 subsets.
Fig 5. Distribution of different parameters over the 100 subsets of parameters.
(a) Percentage of presence of the 1155 parameters. In the Statistical method, which is not present in the plot, the first parameter was repeated in all the 100 subsets. Numbers in the legend show the percentage repetition of the most repeated parameter. (b) Cumulative number of unique parameters over the 100 subsets of parameters. This plot shows that GA and NSGA2 concentrated on a small number of parameters, while the SA, ACO and PSO selected a more diverse range of parameters in the optimization. Numbers in the legend show the number of utilized parameters in the final solution of the 100 subsets of parameters. GA: Genetic algorithm; NSGA2: Nondominated sorting genetic algorithm II; ACO: Ant colony optimization; SA: Simulated annealing; PSO: Particle swarm optimization.
To identify the parameters that are most predominantly involved in classification of HC and EMCI, we extracted the list of the five most indicative brain regions and networks, Table 2. These are selected based on the total number of times that they appeared in the 100 simulations using the five EA’s.
Table 2. Summary of the five most indicative brain areas and networks in the classification of healthy (HC) and early mild cognitive impairment (EMCI).
| Brain Area | Brain Networks | |||
|---|---|---|---|---|
| Method | Area | Graph Param. | Network | Graph Param. |
| GA | SFG | Local Efficiency | Global Network | Global Efficiency |
| Insular Cortex | Local Efficiency | Global Network | Local Efficiency | |
| Frontal Pole | Degree Centrality | Global Network | Clustering Coefficient | |
| Middle Frontal Gyrus | Betweenness Centrality | Global Network | Average Path Length | |
| Inferior Frontal Gyrus; pars triangularis | Clustering Coefficient | Global Network | Betweenness Centrality | |
| NSGA-II | Insular Cortex | Local Efficiency | Global Network | Betweenness Centrality |
| SFG | Local Efficiency | Global Network | Local Efficiency | |
| Frontal Pole | Degree Centrality | Global Network | Cost | |
| Middle Frontal Gyrus | Betweenness Centrality | Global Network | Clustering Coefficient | |
| Precentral Gyrus | Local Efficiency | Global Network | Global Efficiency | |
| SA | SFG | Local Efficiency | Language–IFG | Degree Centrality |
| IFG; pars triangularis | Degree Centrality | Visual–Occipital | Global Efficiency | |
| Lingual Gyrus | Betweenness Centrality | Visual–Lateral | Average Path Length | |
| Thalamus | Cost | Dorsal Attention–FEF | Betweenness Centrality | |
| MTG; temporooccipital part | Average Path Length | Language–pSTG | Degree Centrality | |
| ACO | Occipital Pole | Local Efficiency | Fronto-parietal–PPC | Degree Centrality |
| SFG | Global Efficiency | Dorsal Attention–FEF | Local Efficiency | |
| Middle Frontal Gyrus | Global Efficiency | Visual–Occipital | Cost | |
| Inferior Temporal Gyrus; posterior division | Degree Centrality | Default Mode–LP | Cost | |
| Intracalcarine Cortex | Betweenness Centrality | Dorsal Attention–FEF | Clustering Coefficient | |
| PSO | Occipital Pole | Local Efficiency | Dorsal Attention–FEF | Local Efficiency |
| SFG | Local Efficiency | Visual–Medial | Global Efficiency | |
| Frontal Medial Cortex | Betweenness Centrality | Dorsal Attention–FEF | Local Efficiency | |
| Supplementary Motor Cortex | Local Efficiency | Language–pSTG | Clustering Coefficient | |
| Lingual Gyrus | Betweenness Centrality | Salience–Anterior Insula | Degree Centrality |
Notes: FEF: Frontal-eye-field, IFG: Inferior frontal gyrus, LP: Lateral parietal, PPC: Posterior parietal cortex, MTG: Middle temporal gyrus, pSTG: Posterior superior temporal gyrus, SFG: Superior frontal gyrus.
Discussions
Using CONN toolbox, we extracted 1155 graph parameters from rs-fMRI data. The optimization methods showed superior performance over statistical analysis (average 20.93% superiority). The performance of the EA algorithms did not differ greatly (range 92.55–93.35% and 94.27–94.55% for filtered and absolute accuracy, respectively) with PSO performing the best (mean 0.96% superior performance) and SA performing the worst (mean 1.07% inferior performance), (Fig 2). The minimum number of required parameters to guarantee at least 90% accuracy differed quite greatly across the methods (PSO and SA requiring 7 and 49 parameters, respectively). The processing time to achieve at least 90% accuracy also differed across the EA methods (SA and NSGA2 taking 5.1s and 22.4s per optimization) (Fig 3). Increased number of parameters per subset did not increase the performance accuracy of the methods greatly (Fig 4).
Classification of data into AD and HC has been investigated extensively. Many methods have been developed using different modalities of biomarkers. Some of these studies achieved accuracies greater than 90% [112]. Classification of earlier stages of AD, however, has been more challenging; only a handful of studies have achieved accuracy higher than 90%, Table 3. The majority of these studies implemented convolutional and deep neural networks that require extended training and testing durations with many input data. For example, Payan et al. (2015) applied convolutional neural networks (CNN) on a collection of 755 HC and 755 MCI and achieved accuracy of 92.1% [113]. Similarly, Wang et al. (2019) applied deep neural networks to 209 HC and 384 MCI data and achieved accuracy of 98.4% [114] (see also [115–118]). Our method achieved an accuracy of 94.55%. To the best of our knowledge, between all the studies published to date, this accuracy level is the second highest accuracy after Wang et al (2019) [114].
Table 3. Summary of the studies aiming at categorization of healthy (HC) and mild cognitive impairment (MCI) using different biomarkers and classification methods.
Only best performance of each study is reported for each group of participants and classification method. Further details of the following studies are in S1 Table.
| HC | MCI | ||||||
|---|---|---|---|---|---|---|---|
| Study ↑ | Cit. | Method | Modalities | n | Cat. | n | Acc% |
| Wolz et al (2011) | [119] | LDA | MRI | 231 | sMCI | 238 | 68 |
|
|
cMCI | 167 | 84 | ||||
| Zhang et al (2011) | [120] | SVM | MRI+FDG-PET+ CSF | 231 | SMCI | 238 | 82 |
| LDA | PMCI | 167 | 84 | ||||
| Liu et al (2012) | [121] | SRC | MRI | 229 | MCI | 225 | 87.8 |
| Gray et al (2013) | [122] | RF | MRI+PET+CSF+genetic | 35 | MCI | 75 | 75 |
| Liu et al (2013) | [123] | SVM + LLE | MRI | 137 | sMCI | 92 | 69 |
| cMCI | 97 | 81 | |||||
| Wee et al (2013) | [124] | SVM | MRI | 200 | MCI | 200 | 83.7 |
| Guerrero et al (2014) | [125] | SVM | MRI | 134 | EMCI | 229 | 65 |
| MRI | 175 | cMCI | 116 | 82 | |||
| Payan & Montana (2015) | [113] | CNN | MRI | 755 | MCI | 755 | 92.1 |
| Prasad et al (2015) | [127] | SVM | DWI | 50 | EMCI | 74 | 59.2 |
| LMCI | 38 | 62.8 | |||||
| Suk et al (2015) | [127] | DNN | MRI+PET | 52 | MCI | 99 | 90.7 |
| Shakeri et al (2016) | [128] | DNN | MRI | 150 | EMCI | 160 | 56 |
| MRI | LMCI | 160 | 59 | ||||
| Aderghal, Benois-Pineau et al (2017) | [129] | CNN | MRI | 228 | MCI | 399 | 66.2 |
| Aderghal, Boissenin et al (2017) | [130] | CNN | MRI | 228 | MCI | 399 | 66 |
| Billones et al (2017) | [115] | CNN | MRI | 300 | MCI | 300 | 91.7 |
| Guo et al (2017) | [131] | SVM | rs-fMRI | 28 | EMCI | 32 | 72.8 |
| LMCI | 32 | 78.6 | |||||
| Korolev et al (2017) | [132] | CNN | MRI | 61 | LMCI | 43 | 63 |
| EMCI | 77 | 56 | |||||
| Wang et al (2017) | [116] | CNN | MRI | 229 | MCI | 400 | 90.6 |
| Li & Liu (2018) | [133] | CNN | MRI | 229 | MCI | 403 | 73.8 |
| Qiu et al (2018) | [117] | CNN | MRI+MMSE+ LM |
303 | MCI | 83 | 90.9 |
| Senanayake et al (2018) | [134] | CNN | MRI+NM | 161 | MCI | 193 | 75 |
| Altaf et al (2018) | [135] | SVM | MRI | 90 | MCI | 105 | 79.8 |
| Ensemble | MRI | MCI | 75 | ||||
| KNN | MRI | MCI | 75 | ||||
| Tree | MRI | MCI | 78 | ||||
| SVM | clinical+MRI | MCI | 83 | ||||
| Ensemble | clinical+MRI | MCI | 82 | ||||
| KNN | clinical+MRI | MCI | 86 | ||||
| Tree | clinical+MRI | MCI | 80 | ||||
| Forouzannezhad et al (2018) | [118] | SVM | MRI | 248 | EMCI | 296 | 73.1 |
| MRI | LMCI | 193 | 63 | ||||
| PET | LMCI | 73.6 | |||||
| PET+MRI | LMCI | 76.9 | |||||
| PET+MRI | EMCI | 75.6 | |||||
| PET+MRI+NTS | LMCI | 91.9 | |||||
| PET+MRI+NTS | EMCI | 81.1 | |||||
| Hosseini Asl et al (2018) | [136] | CNN | MRI | 70 | MCI | 70 | 94 |
| Jie, Liu, Shen et al (2018) | [137] | SVM | rs-fMRI | 50 | EMCI | 56 | 78.3 |
| Jie, Liu, Zhang et al (2018) | [138] | SVM | rs-fMRI | 50 | MCI | 99 | 82.6 |
| Raeper et al (2018) | [118] | SVM + LDA | MRI | 42 | EMCI | 42 | 80.9 |
| Basaia et al (2019) | [140] | CNN | MRI | 407 | cMCI | 280 | 87.1 |
| sMCI | 533 | 76.1 | |||||
| Forouzannezhad et al (2019) | [141] | DNN | MRI | 248 | EMCI | 296 | 61.1 |
| MRI | LMCI | 193 | 64.1 | ||||
| PET | EMCI | 58.2 | |||||
| PET | LMCI | 66 | |||||
| MRI+PET | EMCI | 68 | |||||
| MRI+PET | LMCI | 71.7 | |||||
| MRI+PET+NTS | EMCI | 84 | |||||
| MRI+PET+NTS | LMCI | 84.1 | |||||
| Wang et al (2019) | [114] | DNN | MRI | 209 | MCI | 384 | 98.4 |
| Wee et al (2019) | [142] | CNN | MRI | 300 | LMCI | 208 | 69.3 |
| EMCI | 314 | 51.8 | |||||
| 242 | MCI | 415 | 67.6 | ||||
| Kam et al (2020) | [143] | CNN | rs-fMRI | 48 | EMCI | 49 | 76.1 |
| Fang et al (2020) | [144] | GDCA | MRI+PET | 251 | EMCI | 79.2 | |
| Forouzannezhad et al (2020) | [145] | GP | MRI | 248 | EMCI | 296 | 75.9 |
| MRI | LMCI | 193 | 62.1 | ||||
| MRI+PET | EMCI | 75.9 | |||||
| MRI+PET | LMCI | 78.1 | |||||
| MRI+PET+DI | EMCI | 78.8 | |||||
| MRI+PET+DI | LMCI | 79.8 | |||||
| PET | LMCI | 76.1 | |||||
| Jiang et al (2020) | [146] | CNN | MRI | 50 | EMCI | 70 | 89.4 |
| Kang et al (2020) | [147] | CNN | DTI | 50 | EMCI | 70 | 71.7 |
| CNN | MRI | EMCI | 73.3 | ||||
| DTI+MRI | EMCI | 94.2 | |||||
| Yang et al (2021) | [148] | SVM | rs-fMRI | 29 | EMCI | 29 | 82.8 |
| LMCI | 18 | 87.2 | |||||
| our method | EA + ANN | rs-fMRI | 68 | EMCI | 72 | 94.5 |
Notes: ↑ table sorted based on the year of publication. Acc: Classification accuracy percentage between MCI and HC groups; ANN: Artificial neural networks; Cat.: Category of MCI; Cit.: Citation; cMCI: MCI converted to AD; CNN: Convolutional neural networks; DI: Demographic information; DNN: Deep neural network; DTI: Diffusion tensor imaging; DWI: Diffusion-weighted imaging; EA: Evolutionary algorithms; EMCI: Early-MCI; GDCA: Gaussian discriminative component analysis; GP: Gaussian process; KNN: K nearest neighbors; LDA: Linear discriminative analysis; LLE: Locally linear embedding; LM: Logical memory; LMCI: Late-MCI; NTS: Neuropsychological test scores; MMSE: Mini-mental state examination (MMSE); NM: Neuropsychological measures; PET: Positron emission therapy; rs-fMRI: Resting-state fMRI; sMCI: Stable MCI; NM: Neuropsychological measures; SRC: Sparse representation-based classifier; SVM: Support vector machine.
Research has shown that having a combination of information from different modalities supports higher classification accuracies. For example, Forouzannezhad et al. (2018) showed that a combination of PET, MRI and neuropsychological test scores (NTS) can improve performance by more than 20% as compared to only PET or MRI [118]. In another study, Kang et al. (2020) showed that a combination of diffusion tensor imaging (DTI) and MRI can improve accuracy by more than 20% as compared to DTI and MRI alone [147]. Our analysis, while achieving superior accuracy compared to a majority of the prior methods, was based on one biomarker of MRI, which has a lower computational complexity than multi-modality data.
Interpretability of the selected features is one advantage of the application of evolutionary algorithms as the basis of the optimization algorithm. This is in contrast with algorithms based on CNN or deep neural networks (DNN) that are mostly considered as black boxes [149]. Although research has shown some progress in better understanding the link between the features used by the system and the prediction itself in CNN and DNN, such methods remain difficult to verify [150,151]. This has reduced trust in the internal functionality and reliability of such systems in clinical settings [152]. Our suggested method clearly selects features based on activity of distinct brain areas, which are easy to interpret and understand [78–153]. This can inform future research by bringing the focus to brain areas and the link between brain areas that are more affected by mild cognitive impairment.
Our analysis showed that dorsal attention network is altered in EMCI, confirming past literature [154–156]. Dorsal attention network in addition to the ventral attention network form the human attention system [157]. The dorsal attention network employs dorsal fronto-parietal areas, including intraparietal sulcus (IPS) and frontal eye fields (FEF). It is involved in mediation of goal-directed process and selection for stimuli and response. Specifically, our data highlighted the role of the FEF in the dorsal attention network. This is in line with past literature showing the role of FEF in cognitive decline [158]. Our data also revealed the importance of superior frontal gyrus (SFG) in cognitive decline [159,160]. SFG is thought to contribute to higher cognitive functions and particularly to working memory (WM) [161]. Additionally, SFG interconnects multiple brain areas that are involved in a diverse range of cognitive tasks such as cognitive control and motor behavior [162].
In terms of graph parameters, our results showed importance of local efficiency, betweenness centrality and degree centrality in classification of EMCI and HC. Local efficiency is a parameter for the transformation of information in a part of the network. This parameter indicates the efficiency between two nodes and represents the efficiency in exchange of information through a network edge [87,163]. Reduction of this parameter has been linked with cognitive decline in past literature [164]. Betweenness centrality for any given node (vertex) measures the number of shortest paths between pairs of other nodes that pass through this node, reflecting how efficiently the network exchanges the information at the global level. Betweenness centrality is high for nodes that are located on many short paths in the network and low for nodes that do not participate in many short paths [164]. Finally, degree centrality reflects the number of instantaneous functional connections between a region and the rest of the brain within the entire connectivity matrix of the brain. It can assess how much a node influences the entire brain and integrates information across functionally segregated brain regions [165] (see also [166]). Our data showed that changes in these parameters can effectively contribute in classification of early-MCI patients from healthy controls.
We implemented five of the most common evolutionary algorithms. They showed similar overall optimization performance ranging between 92.55–93.35% and 94.27–94.55% for filtered and absolute accuracy, respectively. They, however, differed in optimization curve, optimization time and diversity of the selected features. PSO could guarantee a 90% accuracy with only 7 features. SA on the other hand required 49 features to guarantee a 90% accuracy. Although SA required more features to guarantee a 90% accuracy, it was the fastest optimization algorithm with only 5.1s for 49 features. NSGA-II on the other hand, required 22.4s to guarantee a 90% accuracy. These show the diversity of the algorithms and their suitability in different applications requiring highest accuracy, least number of features or fastest optimization time [71,76,167].
One distinct characteristic of GA and NSGA-II was the more focused search amongst features as compared to the other methods. GA and NSGA-II selected 222 and 224 distinct features in the first 100 parameter sets, respectively, while the other methods covered almost the whole collection of features, covering more than 97.6%. Notably GA and NSGA-II showed “curse of dimensionality” (also known as “peaking phenomenon”) with optimal number of features around 50 parameters [168–171]. Therefore, perhaps the features selected by GA and NSGA-II are more indicative of distinct characteristics of the differences between HC and EMCI.
Our analysis was conducted on a sample of 140 patients. This number of datapoint in the context of classification using ANN and CNN is relatively small. For instance, Wang et al (2019) 114 used 593 samples. Having additional samples can provide more reliable results. Therefore, future research should aim to explore a larger dataset.
In this study, we proposed a method for classification of the EMCI and HC groups using graph theory. These results highlight the potential application of graph analysis of functional connectivity and efficiency of evolutionary algorithm in combination with a simple perceptron ANN in the classification of images into HC and EMCI. We proposed a fully automatic procedure for predication of early stages of AD using rs-fMRI data features. This is of particular importance considering that MRI images of EMCI individuals cannot be easily identified by experts. Further development of such methods can prove to be a powerful tool in the early diagnosis of AD.
Supporting information
The colors indicate t-value for one-sample t-test statistics.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Comparison of classification performance for 200 repetitions (light blue) and 500 repetitions (dark blue) for different optimization algorithms per parameter set. The subplots show the difference between 200 and 500 repetitions, showing small superior performance for 500 repetitions. This is an indication that the algorithms converted within the first 200 repetitions.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Acknowledgments
Authors would like to thank Oliver Herdson for proofreading and his helpful comments.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/study-design/ongoing-investigations/.
Data Availability
Data used in the results presented in the study are available from the Alzheimer’s Disease Neuroimaging Institute (ADNI) http://adni.loni.usc.edu/. For the complete list of the images and participants’ code used in this study see supplementary data table. Using these codes, other researchers can access the complete dataset. Other researchers will have access to this data in the exact same manner as the authors. To access the data researchers can log into the ADNI website and follow these steps: Download > Image Collections > Advanced Search > Search > Select the scans > Add to collection > CSV download > Advanced download. Furthermore, the processed data can be accessed as a MATLAB file via Open Science Framework data repository https://doi.org/10.17605/OSF.IO/BE94G. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/study-design/ongoing-investigations/.
Funding Statement
NO The authors received no specific funding for this work.
References
- 1.Scheltens P. et al. Alzheimer’s disease. The Lancet 388, 505–517 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Nichols E. et al. Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. The Lancet Neurology 18, 88–106 (2019). doi: 10.1016/S1474-4422(18)30403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braak H. & Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathologica 82, 239–259 (1991). doi: 10.1007/BF00308809 [DOI] [PubMed] [Google Scholar]
- 4.Edmonds E. C. et al. Early versus late MCI: Improved MCI staging using a neuropsychological approach. Alzheimer’s and Dementia 15, 699–708 (2019). doi: 10.1016/j.jalz.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platero C. & Tobar M. C. Predicting Alzheimer’s conversion in mild cognitive impairment patients using longitudinal neuroimaging and clinical markers. Brain Imaging and Behavior (2020) doi: 10.1007/s11682-020-00366-8 [DOI] [PubMed] [Google Scholar]
- 6.Petersen R. C. et al. Practice guideline update summary: Mild cognitive impairment report of theguideline development, dissemination, and implementation. Neurology 90, 126–135 (2018). doi: 10.1212/WNL.0000000000004826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nestor P. J., Scheltens P. & Hodges J. R. Advances in the early detection of Alzheimer’s disease. Nature Medicine 10, S34–S41 (2004). doi: 10.1038/nrn1433 [DOI] [PubMed] [Google Scholar]
- 8.Khazaee A., Ebrahimzadeh A. & Babajani-Feremi A. Application of advanced machine learning methods on resting-state fMRI network for identification of mild cognitive impairment and Alzheimer’s disease. Brain Imaging and Behavior 10, 799–817 (2016). doi: 10.1007/s11682-015-9448-7 [DOI] [PubMed] [Google Scholar]
- 9.Zhou Q. et al. An optimal decisional space for the classification of alzheimer’s disease and mild cognitive impairment. IEEE Transactions on Biomedical Engineering 61, 2245–2253 (2014). doi: 10.1109/TBME.2014.2310709 [DOI] [PubMed] [Google Scholar]
- 10.Petersen R. C. et al. Mild cognitive impairment: A concept in evolution. Journal of Internal Medicine 275, 214–228 (2014). doi: 10.1111/joim.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Marco M., Beltrachini L., Biancardi A., Frangi A. F. & Venneri A. Machine-learning Support to Individual Diagnosis of Mild Cognitive Impairment Using Multimodal MRI and Cognitive Assessments. Alzheimer Disease & Associated Disorders 31, 278–286 (2017). doi: 10.1097/WAD.0000000000000208 [DOI] [PubMed] [Google Scholar]
- 12.Wang B. et al. Early Stage Identification of Alzheimer’s Disease Using a Two-stage Ensemble Classifier. Current Bioinformatics 13, 529–535 (2018). [Google Scholar]
- 13.Zhang T. et al. Classification of early and late mild cognitive impairment using functional brain network of resting-state fMRI. Frontiers in Psychiatry 10, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hojjati S. H., Ebrahimzadeh A. & Babajani-Feremi A. Identification of the early stage of alzheimer’s disease using structural mri and resting-state fmri. Frontiers in Neurology 10, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bature F., Guinn B. A., Pang D. & Pappas Y. Signs and symptoms preceding the diagnosis of Alzheimer’s disease: A systematic scoping review of literature from 1937 to 2016. BMJ Open 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reitz C. & Mayeux R. Alzheimer disease: Epidemiology, diagnostic criteria, risk factors and biomarkers. Biochemical Pharmacology 88, 640–651 (2014). doi: 10.1016/j.bcp.2013.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubois B. et al. Preclinical Alzheimer’s disease: Definition, natural history, and diagnostic criteria. Alzheimer’s and Dementia vol. 12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan C. C., Yu J. T. & Tan L. Biomarkers for preclinical alzheimer’s disease. Journal of Alzheimer’s Disease 42, 1051–1069 (2014). doi: 10.3233/JAD-140843 [DOI] [PubMed] [Google Scholar]
- 19.Schmand B., Huizenga H. M. & van Gool W. A. Meta-analysis of CSF and MRI biomarkers for detecting preclinical Alzheimer’s disease. Psychological Medicine 40, 135–145 (2010). doi: 10.1017/S0033291709991516 [DOI] [PubMed] [Google Scholar]
- 20.Frisoni G. B. et al. Strategic roadmap for an early diagnosis of Alzheimer’s disease based on biomarkers. The Lancet Neurology 16, 661–676 (2017). doi: 10.1016/S1474-4422(17)30159-X [DOI] [PubMed] [Google Scholar]
- 21.Mueller S. G. et al. Hippocampal atrophy patterns in mild cognitive impairment and alzheimer’s disease. Human Brain Mapping 31, 1339–1347 (2010). doi: 10.1002/hbm.20934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamani J., Sadr A. & Javadi A. A Large-scale Comparison of Cortical and Subcortical Structural Segmentation Methods in Alzheimer’ s Disease: a Statistical Approach. bioRxiv (2020) doi: 10.1101/2020.08.18.256321 [DOI] [PubMed] [Google Scholar]
- 23.Pini L. et al. Brain atrophy in Alzheimer’s Disease and aging. Ageing Research Reviews 30, 25–48 (2016). doi: 10.1016/j.arr.2016.01.002 [DOI] [PubMed] [Google Scholar]
- 24.Frisoni G. B., Fox N. C., Jack C. R., Scheltens P. & Thompson P. M. The clinical use of structural MRI in Alzheimer disease. Nature Reviews Neurology 6, 67–77 (2010). doi: 10.1038/nrneurol.2009.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yassa M. A. et al. High-resolution structural and functional MRI of hippocampal CA3 and dentate gyrus in patients with amnestic Mild Cognitive Impairment. NeuroImage 51, 1242–1252 (2010). doi: 10.1016/j.neuroimage.2010.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sperling R. The potential of functional MRI as a biomarker in early Alzheimer’s disease. Neurobiology of Aging 32, S37–S43 (2011). doi: 10.1016/j.neurobiolaging.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wierenga C. E. & Bondi M. W. Use of functional magnetic resonance imaging in the early identification of Alzheimer’s disease. Neuropsychology Review 17, 127–143 (2007). doi: 10.1007/s11065-007-9025-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pievani M., de Haan W., Wu T., Seeley W. W. & Frisoni G. B. Functional network disruption in the degenerative dementias. The Lancet Neurology 10, 829–843 (2011). doi: 10.1016/S1474-4422(11)70158-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teipel S. et al. Multimodal imaging in Alzheimer’s disease: Validity and usefulness for early detection. The Lancet Neurology 14, 1037–1053 (2015). doi: 10.1016/S1474-4422(15)00093-9 [DOI] [PubMed] [Google Scholar]
- 30.Lee M. H., Smyser C. D. & Shimony J. S. Resting-state fMRI: A review of methods and clinical applications. American Journal of Neuroradiology 34, 1866–1872 (2013). doi: 10.3174/ajnr.A3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vemuri P., Jones D. T. & Jack C. R. Resting state functional MRI in Alzheimer’s disease. Alzheimer’s Research and Therapy 4, 1–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fox M. D. & Greicius M. Clinical applications of resting state functional connectivity. Frontiers in Systems Neuroscience 4, (2010). doi: 10.3389/fnsys.2010.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greicius M. D., Krasnow B., Reiss A. L. & Menon V. Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proceedings of the National Academy of Sciences of the United States of America 100, 253–258 (2003). doi: 10.1073/pnas.0135058100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheline Y. I. & Raichle M. E. Resting state functional connectivity in preclinical Alzheimer’s disease. Biological Psychiatry 74, 340–347 (2013). doi: 10.1016/j.biopsych.2012.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H. Y. et al. Resting brain connectivity: Changes during the progress of Alzheimer disease. Radiology 256, 598–606 (2010). doi: 10.1148/radiol.10091701 [DOI] [PubMed] [Google Scholar]
- 36.Zhou J. et al. Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer’s disease. Brain 133, 1352–1367 (2010). doi: 10.1093/brain/awq075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dennis E. L. & Thompson P. M. Functional brain connectivity using fMRI in aging and Alzheimer’s disease. Neuropsychology Review 24, 49–62 (2014). doi: 10.1007/s11065-014-9249-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jalilianhasanpour R., Beheshtian E., Sherbaf F. G., Sahraian S. & Sair H. I. Functional Connectivity in Neurodegenerative Disorders: Alzheimer’s Disease and Frontotemporal Dementia. Topics in Magnetic Resonance Imaging 28, 317–324 (2019). doi: 10.1097/RMR.0000000000000223 [DOI] [PubMed] [Google Scholar]
- 39.Zhan Y. et al. Longitudinal Study of Impaired Intra- and Inter-Network Brain Connectivity in Subjects at High Risk for Alzheimer’s Disease. Journal of Alzheimer’s Disease 52, 913–927 (2016). doi: 10.3233/JAD-160008 [DOI] [PubMed] [Google Scholar]
- 40.Bullmore E. & Sporns O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nature Reviews Neuroscience 10, 186–198 (2009). doi: 10.1038/nrn2575 [DOI] [PubMed] [Google Scholar]
- 41.Rubinov M. & Sporns O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage 52, 1059–1069 (2010). doi: 10.1016/j.neuroimage.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 42.van den Heuvel M. P. & Sporns O. Network hubs in the human brain. Trends in Cognitive Sciences 17, 683–696 (2013). doi: 10.1016/j.tics.2013.09.012 [DOI] [PubMed] [Google Scholar]
- 43.Farahani F. v., Karwowski W. & Lighthall N. R. Application of graph theory for identifying connectivity patterns in human brain networks: A systematic review. Frontiers in Neuroscience 13, 1–27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanken T. F. et al. Connecting brain and behavior in clinical neuroscience: A network approach. Neuroscience and Biobehavioral Reviews 130, 81–90 (2021). doi: 10.1016/j.neubiorev.2021.07.027 [DOI] [PubMed] [Google Scholar]
- 45.Yun J. Y. & Kim Y. K. Graph theory approach for the structural-functional brain connectome of depression. Progress in Neuro-Psychopharmacology and Biological Psychiatry 111, 110401 (2021). doi: 10.1016/j.pnpbp.2021.110401 [DOI] [PubMed] [Google Scholar]
- 46.Amiri S., Arbabi M., Kazemi K., Parvaresh-Rizi M. & Mirbagheri M. M. Characterization of brain functional connectivity in treatment-resistant depression. Progress in Neuro-Psychopharmacology and Biological Psychiatry 111, 110346 (2021). doi: 10.1016/j.pnpbp.2021.110346 [DOI] [PubMed] [Google Scholar]
- 47.Beheshti I. & Ko J. H. Modulating brain networks associated with cognitive deficits in Parkinson’s disease. Molecular Medicine 27, (2021). doi: 10.1186/s10020-021-00284-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai Z. et al. Identifying and mapping connectivity patterns of brain network hubs in Alzheimer’s disease. Cerebral Cortex 25, 3723–3742 (2015). doi: 10.1093/cercor/bhu246 [DOI] [PubMed] [Google Scholar]
- 49.Tijms B. M. et al. Alzheimer’s disease: connecting findings from graph theoretical studies of brain networks. Neurobiology of Aging 34, 2023–2036 (2013). doi: 10.1016/j.neurobiolaging.2013.02.020 [DOI] [PubMed] [Google Scholar]
- 50.Brier M. R. et al. Functional connectivity and graph theory in preclinical Alzheimer’s disease. Neurobiology of Aging 35, 757–768 (2014). doi: 10.1016/j.neurobiolaging.2013.10.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He Y. & Evans A. Graph theoretical modeling of brain connectivity. Current Opinion in Neurology 23, 341–350 (2010). doi: 10.1097/WCO.0b013e32833aa567 [DOI] [PubMed] [Google Scholar]
- 52.Bassett D. S. & Bullmore E. T. Human brain networks in health and disease. Current Opinion in Neurology 22, 340–347 (2009). doi: 10.1097/WCO.0b013e32832d93dd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stam C. J. Modern network science of neurological disorders. Nature Reviews Neuroscience 15, 683–695 (2014). doi: 10.1038/nrn3801 [DOI] [PubMed] [Google Scholar]
- 54.Hojjati S. H., Ebrahimzadeh A., Khazaee A. & Babajani-Feremi A. Predicting conversion from MCI to AD using resting-state fMRI, graph theoretical approach and SVM. Journal of Neuroscience Methods 282, 69–80 (2017). doi: 10.1016/j.jneumeth.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 55.Khazaee A., Ebrahimzadeh A. & Babajani-Feremi A. Classification of patients with MCI and AD from healthy controls using directed graph measures of resting-state fMRI. Behavioural Brain Research 322, 339–350 (2017). doi: 10.1016/j.bbr.2016.06.043 [DOI] [PubMed] [Google Scholar]
- 56.Behfar Q. et al. Graph theory analysis reveals resting-state compensatory mechanisms in healthy aging and prodromal Alzheimer’s disease. Frontiers in Aging Neuroscience 12, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregory S. et al. Operationalizing compensation over time in neurodegenerative disease. Brain 140, 1158–1165 (2017). doi: 10.1093/brain/awx022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao Z. et al. Abnormal cortical networks in mild cognitive impairment and alzheimer’s disease. PLoS Computational Biology 6, (2010). doi: 10.1371/journal.pcbi.1001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cabeza R. et al. Maintenance, reserve and compensation: the cognitive neuroscience of healthy ageing. Nature Reviews Neuroscience 19, 701–710 (2018). doi: 10.1038/s41583-018-0068-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blum A. L. & Langley P. Selection of relevant features and examples in machine learning. Artificial Intelligence 97, 245–271 (1997). [Google Scholar]
- 61.Reunanen J. Overfitting in making comparisons between variable selection methods. Journal of Machine Learning Research 3, 1371–1382 (2003). [Google Scholar]
- 62.John G. H., Kohavi R. & Pfleger K. Irrelevant Features and the Subset Selection Problem. in Machine Learning Proceedings 1994. 121–129 (Elsevier, 1994). doi: [DOI] [Google Scholar]
- 63.Chu C., Hsu A. L., Chou K. H., Bandettini P. & Lin C. P. Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. NeuroImage 60, 59–70 (2012). doi: 10.1016/j.neuroimage.2011.11.066 [DOI] [PubMed] [Google Scholar]
- 64.Jaderberg M. et al. Population based training of neural networks. arXiv (2017). [Google Scholar]
- 65.del Ser J. et al. Bio-inspired computation: Where we stand and what’s next. Swarm and Evolutionary Computation 48, 220–250 (2019). [Google Scholar]
- 66.Gu S., Cheng R. & Jin Y. Feature selection for high-dimensional classification using a competitive swarm optimizer. Soft Computing 22, 811–822 (2018). [Google Scholar]
- 67.Dash M. & Liu H. Feature selection for classification. Intelligent Data Analysis 1, 131–156 (1997). [Google Scholar]
- 68.Liu Huan & Yu Lei. Toward integrating feature selection algorithms for classification and clustering. IEEE Transactions on Knowledge and Data Engineering 17, 491–502 (2005). [Google Scholar]
- 69.Telikani A., Tahmassebi A., Banzhaf W. & Gandomi A. H. Evolutionary Machine Learning: A Survey. ACM Computing Surveys 54, 1–35 (2022). [Google Scholar]
- 70.Zawbaa H. M., Emary E., Grosan C. & Snasel V. Large-dimensionality small-instance set feature selection: A hybrid bio-inspired heuristic approach. Swarm and Evolutionary Computation 42, 29–42 (2018). [Google Scholar]
- 71.Xue B., Zhang M., Browne W. N. & Yao X. A Survey on Evolutionary Computation Approaches to Feature Selection. IEEE Transactions on Evolutionary Computation 20, 606–626 (2016). [Google Scholar]
- 72.Soumaya Z., Drissi Taoufiq B., Benayad N., Yunus K. & Abdelkrim A. The detection of Parkinson disease using the genetic algorithm and SVM classifier. Applied Acoustics 171, 107528 (2021). [Google Scholar]
- 73.Bi X. A., Hu X., Wu H. & Wang Y. Multimodal Data Analysis of Alzheimer’s Disease Based on Clustering Evolutionary Random Forest. IEEE Journal of Biomedical and Health Informatics 24, 2973–2983 (2020). doi: 10.1109/JBHI.2020.2973324 [DOI] [PubMed] [Google Scholar]
- 74.Johnson P. et al. Genetic algorithm with logistic regression for prediction of progression to Alzheimer’s disease. BMC Bioinformatics 15, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bi X. A., Xu Q., Luo X., Sun Q. & Wang Z. Analysis of progression toward Alzheimer’s disease based on evolutionary weighted random support vector machine cluster. Frontiers in Neuroscience 12, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma M., Pradhyumna S. P., Goyal S. & Singh K. Machine Learning and Evolutionary Algorithms for the Diagnosis and Detection of Alzheimer’s Disease. in Data Analytics and Management. Lecture Notes on Data Engineering and Communications Technologies (eds. Khanna A., Gupta D., Z, P., S, B. & O, C.) 229–250 (Springer, 2021). doi: 10.1007/978-981-15-8335-3_20 [DOI] [Google Scholar]
- 77.Dessouky M. & Elrashidy M. Feature Extraction of the Alzheimer’s Disease Images Using Different Optimization Algorithms. Journal of Alzheimer’s Disease & Parkinsonism 6, (2016). [Google Scholar]
- 78.Kroll, J. P., Eickhoff, S. B., Hoffstaedter, F. & Patil, K. R. Evolving complex yet interpretable representations: Application to Alzheimer’s diagnosis and prognosis. 2020 IEEE Congress on Evolutionary Computation, CEC 2020—Conference Proceedings (2020) doi: 10.1109/CEC48606.2020.9185843 [DOI]
- 79.Beheshti I., Demirel H. & Matsuda H. Classification of Alzheimer’s disease and prediction of mild cognitive impairment-to-Alzheimer’s conversion from structural magnetic resource imaging using feature ranking and a genetic algorithm. Computers in Biology and Medicine 83, 109–119 (2017). doi: 10.1016/j.compbiomed.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 80.Jack C. R. et al. Introduction to the recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s and Dementia 7, 257–262 (2011). doi: 10.1016/j.jalz.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jack C. R. et al. Update on the Magnetic Resonance Imaging core of the Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s and Dementia 6, 212–220 (2010). doi: 10.1016/j.jalz.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jack C. R. et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging 27, 685–691 (2008). doi: 10.1002/jmri.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aisen P. S., Petersen R. C., Donohue M. & Weiner M. W. Alzheimer’s Disease Neuroimaging Initiative 2 Clinical Core: Progress and plans. Alzheimer’s and Dementia 11, 734–739 (2015). doi: 10.1016/j.jalz.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nieto-Castanon A. Handbook of fcMRI methods in CONN. (2020). [Google Scholar]
- 85.Whitfield-Gabrieli S. & Nieto-Castanon A. Conn: A Functional Connectivity Toolbox for Correlated and Anticorrelated Brain Networks. Brain Connectivity 2, 125–141 (2012). doi: 10.1089/brain.2012.0073 [DOI] [PubMed] [Google Scholar]
- 86.Khazaee A., Ebrahimzadeh A. & Babajani-Feremi A. Identifying patients with Alzheimer’s disease using resting-state fMRI and graph theory. Clinical Neurophysiology 126, 2132–2141 (2015). doi: 10.1016/j.clinph.2015.02.060 [DOI] [PubMed] [Google Scholar]
- 87.Latora V. & Marchiori M. Efficient behavior of small-world networks. Physical Review Letters 87, 198701-1–198701–4 (2001). doi: 10.1103/PhysRevLett.87.198701 [DOI] [PubMed] [Google Scholar]
- 88.Achard S. & Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Computational Biology 3, 0174–0183 (2007). doi: 10.1371/journal.pcbi.0030017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Biswal B., Zerrin Yetkin F., Haughton V. M. & Hyde J. S. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magnetic Resonance in Medicine 34, 537–541 (1995). doi: 10.1002/mrm.1910340409 [DOI] [PubMed] [Google Scholar]
- 90.Damoiseaux J. S. et al. Reduced resting-state brain activity in the “default network” in normal aging. Cerebral Cortex 18, 1856–1864 (2008). [DOI] [PubMed] [Google Scholar]
- 91.Fox M. D. et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proceedings of the National Academy of Sciences of the United States of America 102, 9673–9678 (2005). doi: 10.1073/pnas.0504136102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jafri M. J., Pearlson G. D., Stevens M. & Calhoun V. D. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. NeuroImage 39, 1666–1681 (2008). doi: 10.1016/j.neuroimage.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joel S. E., Caffo B. S., van Zijl P. C. M. & Pekar J. J. On the relationship between seed-based and ICA-based measures of functional connectivity. Magnetic Resonance in Medicine 66, 644–657 (2011). doi: 10.1002/mrm.22818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.delEtoile J. & Adeli H. Graph Theory and Brain Connectivity in Alzheimer’s Disease. Neuroscientist 23, 616–626 (2017). doi: 10.1177/1073858417702621 [DOI] [PubMed] [Google Scholar]
- 95.Fornito A., Andrew Z. & Edward B. Fundamentals of brain network analysis. (Academic Press, 2016). [Google Scholar]
- 96.Tsai C. F., Eberle W. & Chu C. Y. Genetic algorithms in feature and instance selection. Knowledge-Based Systems 39, 240–247 (2013). [Google Scholar]
- 97.Goldenberg D. E. Genetic algorithms in search, optimization and machine learning. (Addison Wesley, 1989). [Google Scholar]
- 98.Vandewater L., Brusic V., Wilson W., Macaulay L. & Zhang P. An adaptive genetic algorithm for selection of blood-based biomarkers for prediction of Alzheimer’s disease progression. BMC Bioinformatics 16, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deb K., Pratap A., Agarwal S. & Meyarivan T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Transactions on Evolutionary Computation 6, 182–197 (2002). [Google Scholar]
- 100.Dorigo M., Caro G. di & Gambardella L. M. Ant Algorithms for Discrete Optimization. Artificial Life 5, 137–172 (1999). doi: 10.1162/106454699568728 [DOI] [PubMed] [Google Scholar]
- 101.Akhtar A. Evolution of Ant Colony Optimization Algorithm—A Brief Literature Review. arXiv (2019). [Google Scholar]
- 102.Kalami Heris S. M. & Khaloozadeh H. Ant Colony Estimator: An intelligent particle filter based on ACO ℝ. Engineering Applications of Artificial Intelligence 28, 78–85 (2014). [Google Scholar]
- 103.Kirkpatrick S., Gelatt C. D. & Vecchi M. P. Optimization by Simulated Annealing. Science 220, 671–680 (1983). doi: 10.1126/science.220.4598.671 [DOI] [PubMed] [Google Scholar]
- 104.Anily S. & Federgruen A. Simulated Annealing Methods With General Acceptance Probabilities. Journal of Applied Probability 24, 657–667 (1987). [Google Scholar]
- 105.Bertsimas D. & Tsitsiklis J. Simulated annealing. Statistical Science 8, 10–15 (1993). [Google Scholar]
- 106.Kennedy J. & Eberhart R. Particle swarm optimization. in Proceedings of ICNN’95—International Conference on Neural Networks vol. 4 1942–1948 (IEEE, 1995). [Google Scholar]
- 107.Wang X., Yang J., Teng X., Xia W. & Jensen R. Feature selection based on rough sets and particle swarm optimization. Pattern Recognition Letters 28, 459–471 (2007). [Google Scholar]
- 108.Team Y. Particle swarm optimization in MATLAB. (2015). [Google Scholar]
- 109.Lv C. et al. Levenberg-marquardt backpropagation training of multilayer neural networks for state estimation of a safety-critical cyber-physical system. IEEE Transactions on Industrial Informatics 14, 3436–3446 (2018). [Google Scholar]
- 110.de Rubio J. J. Stability Analysis of the Modified Levenberg-Marquardt Algorithm for the Artificial Neural Network Training. IEEE Transactions on Neural Networks and Learning Systems 1–15 (2020) doi: 10.1109/TNNLS.2020.3015200 [DOI] [PubMed] [Google Scholar]
- 111.Hagan M. T. & Menhaj M. B. Training feedforward networks with the Marquardt algorithm. IEEE Transactions on Neural Networks 5, 989–993 (1994). doi: 10.1109/72.329697 [DOI] [PubMed] [Google Scholar]
- 112.Wen J. et al. Convolutional neural networks for classification of Alzheimer’s disease: Overview and reproducible evaluation. Medical Image Analysis 63, 101694 (2020). doi: 10.1016/j.media.2020.101694 [DOI] [PubMed] [Google Scholar]
- 113.Payan, A. & Montana, G. Predicting Alzheimer’s disease a neuroimaging study with 3D convolutional neural networks. ICPRAM 2015 - 4th International Conference on Pattern Recognition Applications and Methods, Proceedings 2, 355–362 (2015).
- 114.Wang H. et al. Ensemble of 3D densely connected convolutional network for diagnosis of mild cognitive impairment and Alzheimer’s disease. Neurocomputing 333, 145–156 (2019). [Google Scholar]
- 115.Billones, C. D., Demetria, O. J. L. D., Hostallero, D. E. D. & Naval, P. C. DemNet: A Convolutional Neural Network for the detection of Alzheimer’s Disease and Mild Cognitive Impairment. IEEE Region 10 Annual International Conference, Proceedings/TENCON 3724–3727 (2017) doi: 10.1109/TENCON.2016.7848755 [DOI]
- 116.Wang S., Shen Y., Chen W., Xiao T. & Hu J. Automatic Recognition of Mild Cognitive Impairment from MRI Images Using Expedited Convolutional Neural Networks. in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) vol. 10613 LNCS 373–380 (2017). [Google Scholar]
- 117.Qiu S. et al. Fusion of deep learning models of MRI scans, Mini–Mental State Examination, and logical memory test enhances diagnosis of mild cognitive impairment. Alzheimer’s and Dementia: Diagnosis, Assessment and Disease Monitoring 10, 737–749 (2018). doi: 10.1016/j.dadm.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Forouzannezhad, P., Abbaspour, A., Cabrerizo, M. & Adjouadi, M. Early Diagnosis of Mild Cognitive Impairment Using Random Forest Feature Selection. in 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS) vol. 53 1–4 (IEEE, 2018).
- 119.Wolz R. et al. Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PLoS ONE 6, 1–9 (2011). doi: 10.1371/journal.pone.0025446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang D., Wang Y., Zhou L., Yuan H. & Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage 55, 856–867 (2011). doi: 10.1016/j.neuroimage.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu M., Zhang D. & Shen D. Ensemble sparse classification of Alzheimer’s disease. NeuroImage 60, 1106–1116 (2012). doi: 10.1016/j.neuroimage.2012.01.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gray K. R., Aljabar P., Heckemann R. A., Hammers A. & Rueckert D. Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. NeuroImage 65, 167–175 (2013). doi: 10.1016/j.neuroimage.2012.09.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu X., Tosun D., Weiner M. W. & Schuff N. Locally linear embedding (LLE) for MRI based Alzheimer’s disease classification. NeuroImage 83, 148–157 (2013). doi: 10.1016/j.neuroimage.2013.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wee C. Y., Yap P. T. & Shen D. Prediction of Alzheimer’s disease and mild cognitive impairment using cortical morphological patterns. Human Brain Mapping 34, 3411–3425 (2013). doi: 10.1002/hbm.22156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guerrero R., Wolz R., Rao A. W. & Rueckert D. Manifold population modeling as a neuro-imaging biomarker: Application to ADNI and ADNI-GO. NeuroImage 94, 275–286 (2014). doi: 10.1016/j.neuroimage.2014.03.036 [DOI] [PubMed] [Google Scholar]
- 126.Prasad G., Joshi S. H., Nir T. M., Toga A. W. & Thompson P. M. Brain connectivity and novel network measures for Alzheimer’s disease classification. Neurobiology of Aging 36, S121–S131 (2015). doi: 10.1016/j.neurobiolaging.2014.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Suk H. il , Lee S. W. & Shen D. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Structure and Function 220, 841–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shakeri M., Lombaert H., Tripathi S. & Kadoury S. Deep spectral-based shape features for Alzheimer’s disease classification. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 10126 LNCS, 15–24 (2016). [Google Scholar]
- 129.Aderghal, K., Benois-Pineau, J., Afdel, K. & Gwenaëlle, C. FuseMe: Classification of sMRI images by fusion of deep CNNs in 2D+e projections. ACM International Conference Proceeding Series Part F1301, (2017).
- 130.Aderghal K., Boissenin M., Benois-Pineau J., Catheline G. & Afdel K. Classification of sMRI for AD Diagnosis with Convolutional Neuronal Networks: A Pilot 2-D+ ϵ Study on ADNI. in MultiMedia Modeling, Lecture Notes in Computer Science (eds. Amsaleg L., Gu ðmundsson G., Gurrin C., Jónsson B. & Satoh S.) 690–701 (Springer International Publishing, 2017). doi: 10.1007/978-3-319-51811-4 [DOI] [Google Scholar]
- 131.Guo H., Zhang F., Chen J., Xu Y. & Xiang J. Machine learning classification combining multiple features of a hyper-network of fMRI data in Alzheimer’s disease. Frontiers in Neuroscience 11, 1–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Korolev S., Safiullin A., Belyaev M. & Dodonova Y. Residual and plain convolutional neural networks for 3D brain MRI classification. arXiv 835–838 (2017). [Google Scholar]
- 133.Li F. & Liu M. Alzheimer’s disease diagnosis based on multiple cluster dense convolutional networks. Computerized Medical Imaging and Graphics 70, 101–110 (2018). doi: 10.1016/j.compmedimag.2018.09.009 [DOI] [PubMed] [Google Scholar]
- 134.Senanayake, U., Sowmya, A. & Dawes, L. Deep fusion pipeline for mild cognitive impairment diagnosis. in 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) 1394–1997 (IEEE, 2018). doi: 10.1109/ISBI.2018.8363832 [DOI]
- 135.Altaf T., Anwar S. M., Gul N., Majeed M. N. & Majid M. Multi-class Alzheimer’s disease classification using image and clinical features. Biomedical Signal Processing and Control 43, 64–74 (2018). [Google Scholar]
- 136.Hosseini Asl E. et al. Alzheimer rsquo s disease diagnostics by a 3D deeply supervised adaptable convolutional network. Frontiers in Bioscience 23, 584–596 (2018). [DOI] [PubMed] [Google Scholar]
- 137.Jie B., Liu M. & Shen D. Integration of temporal and spatial properties of dynamic connectivity networks for automatic diagnosis of brain disease. Medical Image Analysis 47, 81–94 (2018). doi: 10.1016/j.media.2018.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jie B., Liu M., Zhang D. & Shen D. Sub-Network Kernels for Measuring Similarity of Brain Connectivity Networks in Disease Diagnosis. IEEE Transactions on Image Processing 27, 2340–2353 (2018). doi: 10.1109/TIP.2018.2799706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Raeper R., Lisowska A. & Rekik I. Cooperative Correlational and Discriminative Ensemble Classifier Learning for Early Dementia Diagnosis Using Morphological Brain Multiplexes. IEEE Access 6, 43830–43839 (2018). [Google Scholar]
- 140.Basaia S. et al. Automated classification of Alzheimer’s disease and mild cognitive impairment using a single MRI and deep neural networks. NeuroImage: Clinical 21, 101645 (2019). doi: 10.1016/j.nicl.2018.101645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Forouzannezhad, P., Abbaspour, A., Li, C., Cabrerizo, M. & Adjouadi, M. A Deep Neural Network Approach for Early Diagnosis of Mild Cognitive Impairment Using Multiple Features. Proceedings - 17th IEEE International Conference on Machine Learning and Applications, ICMLA 2018 1341–1346 (2019) doi: 10.1109/ICMLA.2018.00218 [DOI]
- 142.Wee C. Y. et al. Cortical graph neural network for AD and MCI diagnosis and transfer learning across populations. NeuroImage: Clinical 23, 101929 (2019). doi: 10.1016/j.nicl.2019.101929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kam T. E., Zhang H., Jiao Z. & Shen, Di. Deep Learning of Static and Dynamic Brain Functional Networks for Early MCI Detection. IEEE Transactions on Medical Imaging 39, 478–487 (2020). doi: 10.1109/TMI.2019.2928790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fang C. et al. Gaussian discriminative component analysis for early detection of Alzheimer’s disease: A supervised dimensionality reduction algorithm. Journal of Neuroscience Methods 344, 108856 (2020). doi: 10.1016/j.jneumeth.2020.108856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Forouzannezhad P. et al. A Gaussian-based model for early detection of mild cognitive impairment using multimodal neuroimaging. Journal of Neuroscience Methods 333, 108544 (2020). doi: 10.1016/j.jneumeth.2019.108544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jiang J., Kang L., Huang J. & Zhang T. Deep learning based mild cognitive impairment diagnosis using structure MR images. Neuroscience Letters 730, 134971 (2020). doi: 10.1016/j.neulet.2020.134971 [DOI] [PubMed] [Google Scholar]
- 147.Kang L., Jiang J., Huang J. & Zhang T. Identifying Early Mild Cognitive Impairment by Multi-Modality MRI-Based Deep Learning. Frontiers in Aging Neuroscience 12, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang P. et al. Fused Sparse Network Learning for Longitudinal Analysis of Mild Cognitive Impairment. IEEE Transactions on Cybernetics 51, 233–246 (2021). doi: 10.1109/TCYB.2019.2940526 [DOI] [PubMed] [Google Scholar]
- 149.Reyes M. et al. On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities. Radiology: Artificial Intelligence 2, e190043 (2020). doi: 10.1148/ryai.2020190043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Amorim, J. P., Abreu, P. H., Reyes, M. & Santos, J. Interpretability vs. Complexity: The Friction in Deep Neural Networks. Proceedings of the International Joint Conference on Neural Networks 1–7 (2020) doi: 10.1109/IJCNN48605.2020.9206800 [DOI]
- 151.Pereira S. et al. Enhancing interpretability of automatically extracted machine learning features: application to a RBM-Random Forest system on brain lesion segmentation. Medical Image Analysis 44, 228–244 (2018). doi: 10.1016/j.media.2017.12.009 [DOI] [PubMed] [Google Scholar]
- 152.Fan F., Xiong J. & Wang G. On Interpretability of Artificial Neural Networks. arXiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Beheshti I., Demirel H. & Matsuda H. Classification of Alzheimer’s disease and prediction of mild cognitive impairment-to-Alzheimer’s conversion from structural magnetic resource imaging using feature ranking and a genetic algorithm. Computers in Biology and Medicine 83, 109–119 (2017). doi: 10.1016/j.compbiomed.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 154.Chhatwal J. P. et al. Preferential degradation of cognitive networks differentiates Alzheimer’s disease from ageing. Brain 141, 1486–1500 (2018). doi: 10.1093/brain/awy053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li R. et al. Attention-related networks in Alzheimer’s disease: A resting functional MRI study. Human Brain Mapping 33, 1076–1088 (2012). doi: 10.1002/hbm.21269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang Z. et al. Functional degeneration in dorsal and ventral attention systems in amnestic mild cognitive impairment and Alzheimer’s disease: An fMRI study. Neuroscience Letters 585, 160–165 (2015). doi: 10.1016/j.neulet.2014.11.050 [DOI] [PubMed] [Google Scholar]
- 157.Corbetta M. & Shulman G. L. Control of goal-directed and stimulus-driven attention in the brain. Nature Reviews Neuroscience 3, 201–215 (2002). doi: 10.1038/nrn755 [DOI] [PubMed] [Google Scholar]
- 158.Qian S., Zhang Z., Li B. & Sun G. Functional-structural degeneration in dorsal and ventral attention systems for Alzheimer’s disease, amnestic mild cognitive impairment. Brain Imaging and Behavior 9, 790–800 (2015). doi: 10.1007/s11682-014-9336-6 [DOI] [PubMed] [Google Scholar]
- 159.Zhao Q., Lu H., Metmer H., Li W. X. Y. & Lu J. Evaluating functional connectivity of executive control network and frontoparietal network in Alzheimer’s disease. Brain Research 1678, 262–272 (2018). doi: 10.1016/j.brainres.2017.10.025 [DOI] [PubMed] [Google Scholar]
- 160.Li M. et al. Alterations in resting-state functional connectivity of the default mode network in amnestic mild cognitive impairment: An fMRI study. BMC Medical Imaging 17, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Boisgueheneuc F. du et al. Functions of the left superior frontal gyrus in humans: A lesion study. Brain 129, 3315–3328 (2006). doi: 10.1093/brain/awl244 [DOI] [PubMed] [Google Scholar]
- 162.Li W. et al. Subregions of the human superior frontal gyrus and their connections. NeuroImage 78, 46–58 (2013). doi: 10.1016/j.neuroimage.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 163.Rubinov M. & Sporns O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage 52, 1059–1069 (2010). doi: 10.1016/j.neuroimage.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 164.Zhou Y. & Lui Y. W. Small-World Properties in Mild Cognitive Impairment and Early Alzheimer’s Disease: A Cortical Thickness MRI Study. ISRN Geriatrics 2013, 1–11 (2013). doi: 10.1155/2013/542080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guo Z. et al. Abnormal degree centrality in Alzheimer’s disease patients with depression: A resting-state functional magnetic resonance imaging study. Experimental Gerontology 79, 61–66 (2016). doi: 10.1016/j.exger.2016.03.017 [DOI] [PubMed] [Google Scholar]
- 166.Javadi A.-H. et al. Hippocampal and prefrontal processing of network topology to simulate the future. Nature Communications 8, 14652 (2017). doi: 10.1038/ncomms14652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Šprogar M., Šprogar M. & Colnarič M. Autonomous evolutionary algorithm in medical data analysis. Computer Methods and Programs in Biomedicine 80, (2005). doi: 10.1016/s0169-2607(05)80004-5 [DOI] [PubMed] [Google Scholar]
- 168.Trunk G. A Problem of Dimensionality: A Simple Example. IEEE Transactions on Pattern Analysis and Machine Intelligence PAMI-1, 306–307 (1979). doi: 10.1109/tpami.1979.4766926 [DOI] [PubMed] [Google Scholar]
- 169.Hughes G. F. On the Mean Accuracy of Statistical Pattern Recognizers. IEEE Transactions on Information Theory 14, 55–63 (1968). [Google Scholar]
- 170.Zollanvari A., James A. P. & Sameni R. A Theoretical Analysis of the Peaking Phenomenon in Classification. Journal of Classification 37, 421–434 (2020). [Google Scholar]
- 171.Jain A. K., Duin R. P. W. & Mao J. Statistical pattern recognition: A review. IEEE Transactions on Pattern Analysis and Machine Intelligence 22, 4–37 (2000). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The colors indicate t-value for one-sample t-test statistics.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Comparison of classification performance for 200 repetitions (light blue) and 500 repetitions (dark blue) for different optimization algorithms per parameter set. The subplots show the difference between 200 and 500 repetitions, showing small superior performance for 500 repetitions. This is an indication that the algorithms converted within the first 200 repetitions.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Data Availability Statement
Data used in the results presented in the study are available from the Alzheimer’s Disease Neuroimaging Institute (ADNI) http://adni.loni.usc.edu/. For the complete list of the images and participants’ code used in this study see supplementary data table. Using these codes, other researchers can access the complete dataset. Other researchers will have access to this data in the exact same manner as the authors. To access the data researchers can log into the ADNI website and follow these steps: Download > Image Collections > Advanced Search > Search > Select the scans > Add to collection > CSV download > Advanced download. Furthermore, the processed data can be accessed as a MATLAB file via Open Science Framework data repository https://doi.org/10.17605/OSF.IO/BE94G. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/study-design/ongoing-investigations/.