Abstract
A modified nested reverse transcriptase PCR (RT-PCR) method was used to detect the expression of nitrogenase genes in meso-oligotrophic Lake George, New York. Net (>20-μm pore size) plankton samples collected from two sites (Dome Island and Hague Marina) were extracted for total RNA and genomic DNA to determine the identity of diazotrophic organisms that were present and those that were actively expressing nitrogenase genes. Phylogenetic analysis of individual sequences cloned from PCR amplifications showed that there were phylogenetically diverse groups of bacteria that possessed a nifH gene, including representatives of unicellular and filamentous cyanobacteria, the α- and γ-subdivisions of the division Proteobacteria (α- and γ-proteobacteria), and a previously undefined group of bacteria. The phylotypes cloned from RT-PCR amplifications, which were actively expressing nifH transcripts, clustered with the unicellular and filamentous cyanobacteria, α-proteobacteria, and the novel bacterial cluster. No bacterial sequences were found which clustered with sequences from cluster II (alternative nitrogenases), III (nitrogenases in strict anaerobes), or IV (nifH-like sequences). These results indicate that there were several distinct groups of nitrogen-fixing microorganisms in the net plankton from both sampling sites and that most of the groups had representative phylotypes that were actively expressing nitrogenase genes.
Many aquatic communities are deficient in fixed inorganic nitrogen (4, 11). Nitrogen-fixing microorganisms can obtain nitrogen from atmospheric dinitrogen (N2) and are important since they can alleviate nitrogen limitation of productivity of aquatic and terrestrial environments (4, 21). Nitrogen-fixing cyanobacteria often form blooms in nitrogen-limited lakes and estuaries.
Nitrogen fixation is catalyzed by the enzyme nitrogenase. Nitrogenase is highly conserved among diverse N2-fixing organisms (13). The phylogenetic analysis of molecular sequences of nifH, which encodes the Fe protein component of nitrogenase, yields tree topologies that are largely similar to 16S rRNA phylogeny (23) and are useful for identifying unknown diazotrophs (24).
Recently, nitrogenase gene sequences (nifH) have been amplified and sequenced from a number of environments, including rice roots, soils, and oceans, and invertebrates, such as zooplankton and termites (1, 10, 12, 17, 19, 22, 25). However, the mere presence of nitrogenase genes does not indicate that bacteria are actively fixing nitrogen. Particularly in nutrient-limited aquatic environments, it is important to know whether nitrogen-fixing microorganisms that are present are actually expressing the nitrogenase enzyme. Although 15N or acetylene-reduction techniques are available for detecting nitrogen fixation activity, they involve incubation of samples, can have limited sensitivity, and do not provide information on which microorganisms are actively fixing nitrogen. Culturing techniques have been used to determine the type of individual species present, but these techniques yield biased results and a misrepresentation of the types of bacterial species that are active in the environment (10, 16).
The reverse transcriptase PCR (RT-PCR) makes it possible to assay for cells that are actively expressing specific genes at the time of sampling, and it has been used recently to detect expression of genes in the environment, including nifH (5, 9). In parallel, PCR of DNA obtained from the same samples can confirm the presence of nitrogen fixers as well as detect microorganisms that have the nitrogen fixation genes but that are not expressing nitrogenase at the time of sampling. Comparison of sequences obtained by RT-PCR and PCR can therefore be used to investigate the diversity of organisms expressing genes under different environmental conditions and in different habitats (5).
Several nutrients are often present in low concentrations in aquatic environments, and it is usually difficult to determine the specific nutrient(s) limiting productivity (4). Lake George is a large meso-oligotrophic lake in northern New York State. During the summer season, the lake is stratified with levels of nitrate, ammonium, and soluble reactive phosphorus that are typically below the detection limit in the epilimnion (7). While Lake George, like many freshwater systems, has been assumed to be phosphorus limited, both nitrogen and phosphorus are in short supply, making Lake George a good candidate for the study of factors regulating the expression of nitrogenase. Furthermore, Lake George is a long narrow lake divided almost equally into two subbasins, with the major outflow from the northernmost extent of the north basin (14). Previous studies had suggested that the south basin had higher concentrations of chlorophyll and productivity than the north basin (2), although more recent analysis indicated only moderate differences that were not statistically different (8). There are apparently differences in composition of plankton communities between the two basins (15). The primary objective of this study was to determine if there were nitrogen-fixing microorganisms in the net plankton of Lake George and if these microorganisms were actively expressing nitrogenase, indicating that nitrogen may have been limiting their growth.
Net plankton were collected from two sampling sites located in Lake George (Hague Marina and Dome Island) on 1 June 1998. One-liter net plankton samples were collected with a zooplankton net (20-μm mesh size) from a vertical tow at a depth of 20 meters. A 500-ml sample of the net concentrate was diluted in Lake George water and filtered through a 0.45-μm-pore-size mixed-cellulose-ester membrane (Millipore Corporation, Bedford, Mass.). Samples were then resuspended in 500 μl of buffer QRL1 (Qiagen, Valencia, Calif.) and homogenized with an electric pestle for 30 s. Samples were stored at −80°C.
Genomic DNA from the filter samples was extracted using a slight modification of the method of Giovannoni et al. (3), as described by Braun et al. (1). Net plankton samples were initially stored in buffer QRL1 (Qiagen) and then extracted with phenol-chloroform. The DNA was precipitated with ammonium acetate (3 M, pH 5.2) and ethanol. The precipitated DNA was resuspended in a solution containing 10 mM Tris (pH 8.0) and 1 mM EDTA.
DNA was also extracted from a number of cultivated, but not axenic, cyanobacterial isolates from Lake George in order to determine if they contained nif genes that were related to the cyanobacterial nifH genes detected in the net plankton. DNA was extracted from colonies grown on agar plates, using the method of Zehr et al. (25).
Total RNA was extracted from the filters using the RNeasy minikit (Qiagen), purified with an RNeasy mini-spin column (Qiagen) according to the manufacturer's protocol, and resuspended in 50 μl of H2O. DNA in the samples was digested using RQ1 DNase (Promega, Madison, Wis.) for 30 min at 37°C. The DNase enzyme was removed from the sample using the RNeasy minikit protocol.
Two degenerate oligonucleotide PCR primers were designed to amplify an approximately 460-bp segment of the nifH gene. This fragment brackets the nifH1 (corresponding to Azotobacter vinelandii nucleotide positions 639 to 655; 5′-TGY GAY CCN AAR GCN GA-3′) and nifH2 (A. vinelandii positions 1000 to 984; 5′-AND GCC ATC ATY TCN CC-3′) primer sites designed by Zehr and McReynolds (26), and it is similar to the amplified region obtained using primers designed by Ohkuma et al. (10). The additional pair of primers nifH4 (A. vinelandii positions 546 to 562; 5′-TTY TAY GGN AAR GGN GG-3′) and nifH3 (A. vinelandii positions 1018 to 1002; 5′-ATR TTR TTN GCN GCR TA-3′) were designed for nested PCR based on conserved sequences outside of nifH1 and nifH2. All four of these primers were degenerate (Y = T or C; R = A or G; D = A, G, or T; and N = A, C, G, or T). The nested PCR proved to be less affected by sample inhibition than in single-stage PCR, with substantially increased sensitivity. The primer sites were conserved throughout nifH genes in clusters I, II, III, and IV.
Reverse transcription reactions were performed in mixtures containing 28 μl of diethyl pyrocarbonate-treated H2O, 10 μl of 5× avian myeloblastosis virus buffer, 1 μl of a deoxynucleoside triphosphate (dNTP) mixture (a 10 mM concentration of each dNTP), and 1 pmol of primer nifH3. The reaction mixtures were exposed to UV light (254 nm) for 20 min to prevent contamination. One microliter of avian myeloblastosis virus RT (Promega) was then added along with 1 μl of DNase-treated RNA. Reaction mixtures were incubated at 42°C for 30 min.
After reverse transcription, 1 μl of the cDNA was added to 49 μl of the first-round PCR mixture (4 mM MgCl2, 10× reaction buffer, 10 mM dNTPs, 100 pmol each of nifH3 and nifH4 primers, and 2.5 U of Taq polymerase). The PCR was carried out with 30 cycles of denaturation at 95°C (1 min), annealing at 55°C (1 min), and extension at 72°C (1 min). The second round of the nested PCR was performed with 1 μl of the first-round product in a mixture of 4 mM MgCl2, 10× reaction buffer, 10 mM dNTPs, 100 pmol each of nifH1 and nifH2 primers, and 2.5 U of Taq polymerase, with 30 cycles of the same temperature and time conditions as in the first step of the nested PCR. DNA samples were amplified by nested PCR under the same conditions as the RT-PCR but without the reverse transcription step.
Two types of negative controls confirmed that the RT-PCR results were from RNA and not from contaminating DNA. The first control used direct nested PCR of the RNA samples, and the second consisted of treating the RNA samples with RNase and subjecting them to nested RT-PCR (see Fig. 3 and 4). The results of these two experiments showed that the amplification products were derived from nifH transcripts in the total RNA sample and not amplification from contaminant genomic DNA. Thus, the RT-PCR method appears to be a useful assay for nifH mRNA.
FIG. 3.
Test for DNA contamination using RNA samples subjected to RNase treatment. Samples indicated with an asterisk were treated with RNase. RT and PCR positive control reaction mixtures consisted of Trichodesmium sp. strain IMS101 RNA and DNA, respectively.
FIG. 4.
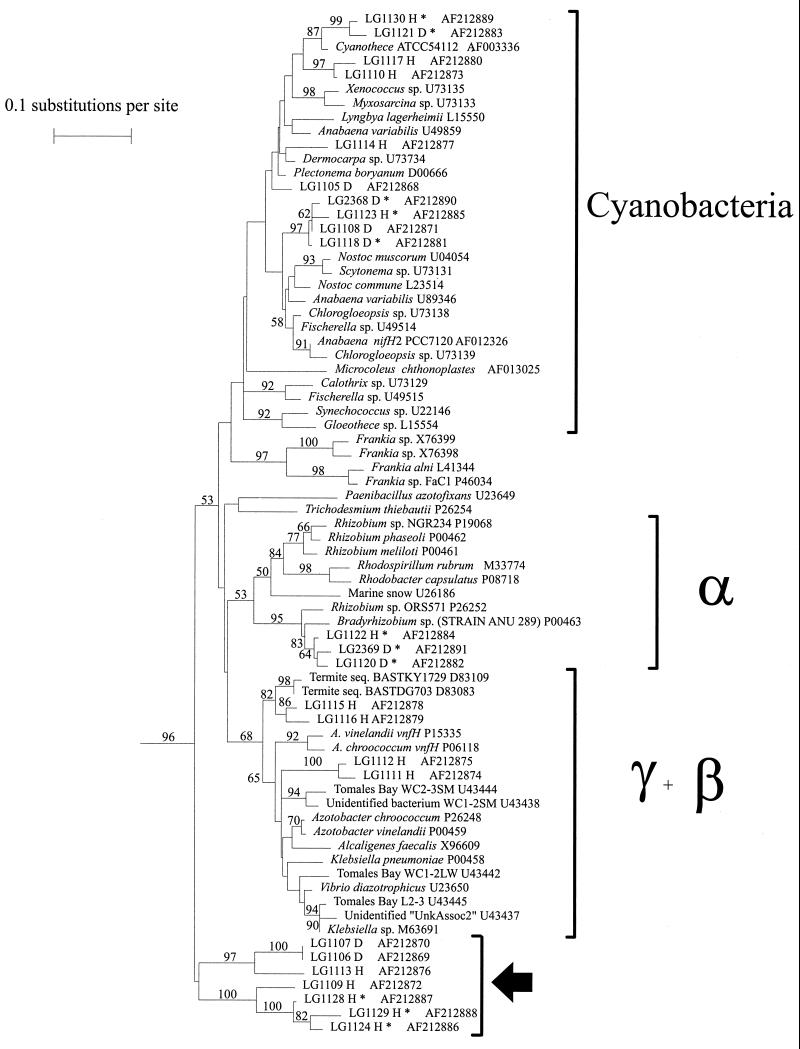
Cluster I nifH gene sequences recovered from stations in the north (Hague Marina [H]) and south (Dome Island [D]) basins of Lake George. ∗, sequences recovered from RNA by RT-PCR. The large arrow indicates a deeply branching clade outside of the proteobacteria and cyanobacteria.
After the second round of PCR amplification, the amplified fragments were gel purified and cloned into a pGEM-T vector (Promega). Clones were screened by restriction digestion to detect those with the correct size insert (approximately 359 bp). Recombinants were randomly picked from each ligation to obtain equal numbers of clones from each sample type (Hague Marina RNA, Hague Marina DNA, Dome Island DNA, and Dome Island RNA). DNA isolated from the selected clones was sequenced on both strands, by the Sanger dideoxynucleotide chain termination method.
The amplified partial Fe protein gene sequences were translated and aligned using Genetic Data Environment software (Ribosomal Database Project) (6). The amino acid sequences were aligned with representative nitrogenase sequences obtained from GenBank. Distances between pairs of sequences were calculated using the distance correction of Tajima and Nei (18), followed by the construction of phylogenetic trees by neighbor joining using TREECON for Windows software (20).
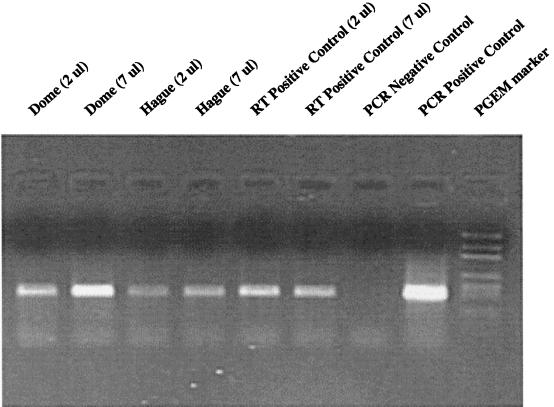
The expected 359-bp fragment was amplified from all samples following reverse transcription and PCR (Fig. 1). Increasing the amount of added RNA resulted in increased amplification product (Fig. 1, lanes 2 and 4).
FIG. 1.
RT-PCR of net plankton samples obtained from two sampling sites in Lake George, N.Y. The amount of RNA used in the reverse transcription samples is indicated following the sample description. RT and PCR positive control reaction mixtures consisted of Trichodesmium sp. strain IMS101 RNA and DNA, respectively.
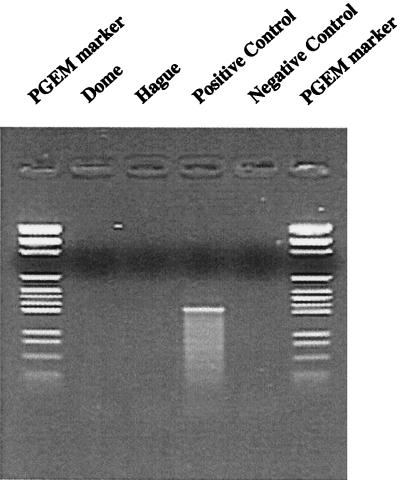
The RNA samples were tested for the presence of contaminating DNA using nested PCR without the reverse transcription step. The expected-size fragment was amplified only in the positive control lane containing target DNA (Fig. 2). No amplification product was obtained from the Dome Island or Hague Marina RNA samples without the reverse transcription step.
FIG. 2.
Direct nested PCR of RNA samples used for RT-PCR to test for DNA contamination. The PCR positive control reaction mixture consisted of Trichodesmium sp. strain IMS101 DNA.
The second test used to confirm the lack of DNA contamination was based on treating the RNA samples with RNase followed by RT-PCR (Fig. 3). No amplification product was detected in the RNA samples subjected to RNase treatment (Fig. 3).
The results of the phylogenetic analysis of the nifH genes are shown in Fig. 4 and summarized in Table 1. The Lake George set of sequences consisted of 14 unique nifH sequences obtained from RNA and 14 unique sequences obtained from DNA from each site, for a total of 28 sequences from Dome Island and 28 sequences from Hague Marina (Table 2).
TABLE 1.
Composition of sequence types in clone libraries of amplified nifH genes from Lake George net plankton
| Location | No. of sequences from:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| α-Proteobacteria
|
γ-Proteobacteria
|
Unicellular cyanobacteria
|
Filamentous cyanobacteria
|
Novel bacterial cluster
|
||||||
| DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | |
| Dome Island | 0 | 2 | 0 | 0 | 0 | 1 | 6 | 11 | 8 | 0 |
| Hague Marina | 0 | 6 | 4 | 0 | 8 | 3 | 0 | 2 | 2 | 3 |
TABLE 2.
Identification of nifH sequences obtained from Lake George samples
| Sample location and type | No. of clones | Representative sequence in Fig. 4 | GenBank accession no. |
|---|---|---|---|
| Hague Marina | |||
| DNA | 6 | LG1110 | AF212873 |
| 1 | LG1109 | AF212872 | |
| 1 | LG1111 | AF212874 | |
| 1 | LG1112 | AF212875 | |
| 1 | LG1113 | AF212876 | |
| 1 | LG1114 | AF212877 | |
| 1 | LG1115 | AF212878 | |
| 1 | LG1116 | AF212879 | |
| 1 | LG1117 | AF212880 | |
| RNA | 6 | LG1122 | AF212884 |
| 2 | LG1123 | AF212885 | |
| 3 | LG1130 | AF212889 | |
| 1 | LG1124 | AF212886 | |
| 1 | LG1128 | AF212887 | |
| 1 | LG1129 | AF212888 | |
| Dome Island | |||
| DNA | 5 | LG1105 | AF212868 |
| 7 | LG1106 | AF212869 | |
| 1 | LG1107 | AF212870 | |
| 1 | LG1108 | AF212871 | |
| RNA | 10 | LG1118 | AF212881 |
| 1 | LG2368 | AF212890 | |
| 1 | LG2369 | AF212891 | |
| 1 | LG1120 | AF212882 | |
| 1 | LG1121 | AF212883 |
The nifH sequences obtained from Dome Island clustered in nine different phylogenetic groups (Fig. 4). The four sequences derived from the PCR assay clustered in three different groups that included the cyanobacterial clade and a novel, previously undefined cluster, which is a deeply branching cluster outside of cluster I. The five sequences derived from the RT-PCR clustered with cyanobacterial sequences and sequences from the α-subdivision of the division Proteobacteria (α-proteobacteria) (Fig. 4). Corresponding sequences from PCR and RT-PCR were found to cluster together among the cyanobacterial sequences.
The 28 nifH sequences obtained from Hague Marina contained 15 different sequence types. Nine sequence types were obtained from PCR and six sequence types were obtained from RT-PCR. The DNA-derived sequences tended to cluster with sequences from cyanobacteria, γ-proteobacteria, and the novel cluster. The RT-PCR sequences clustered with sequences from cyanobacteria, α-proteobacteria, and the novel cluster (Fig. 4). The novel nifH sequences obtained from Hague Marina PCR and RT-PCR clustered together (Fig. 4).
As shown in Table 1, the nifH genes obtained from both sample sites clustered among the α- and γ-proteobacteria as well as the cyanobacteria (Fig. 4). Additional sequences derived from both sampling sites formed a divergent group of sequences that clustered together with a high bootstrap value. These sequences were detected in both PCR and RT-PCR amplifications. This specific set of sequences clustered independently of any other nifH clade (clusters II, III, and IV) (Fig. 4). No sequences derived from the Lake George net plankton samples were detected in cluster II, III, or IV.
Phylogenetic analysis of the nifH sequences obtained from both RT-PCR and PCR showed that nitrogen-fixing bacteria were present and were expressing nifH. The sequences obtained in this study were not identical to any previously published nifH sequences. The greatest similarity found was 99% similarity to a previously sequenced nifH gene from the Pacific Ocean (25). All of the sequences obtained in this study were from cluster I, and thus, no alternative (second alternative, non-molybdenum- or non-vanadium-containing) nitrogenase or Archaea nifH sequences were detected. Sequences from cluster III, which includes nif sequences from anaerobic bacteria, were not detected either, despite the fact that invertebrate plankton were collected and that cluster III sequences previously have been found to be associated with invertebrates (1, 25). It is possible that the anaerobe nif sequences were present but with only low relative abundance and, therefore, were not detected in this study.
The finding of unicellular-cyanobacterium nifH RNA and DNA sequences in the net plankton samples was unexpected. Unicellular cyanobacteria would be expected to pass through the plankton net during sample collection. Gleothece-like cells have been observed in Lake George, and a Dermocarpa-like cyanobacterium has been recovered in culture (J. L. Collier, unpublished data). The presence of these cyanobacterial nifH sequences suggests that cyanobacteria were present in aggregates and that they were expressing nitrogenase.
We tested unicellular cyanobacteria cultivated from Lake George for nifH. Interestingly, although the cyanobacterial isolates did not contain the same cyanobacterial nifH genes as detected in Lake George by PCR, bacteria associated with the isolates contained α-proteobacterial nifH genes that clustered with nifH sequences from Lake George. These types of bacteria may have been associated with the aggregated cyanobacteria collected in the net plankton.
More filamentous-cyanobacterium nifH sequences than unicellular-cyanobacterium sequences were recovered from Dome Island, by RT-PCR as well as PCR. These sequences were a fairly divergent group within the cyanobacteria but were probably most closely related to filamentous heterocystous cyanobacteria. In contrast, sequences obtained from Hague Marina DNA included a higher percentage of unicellular-cyanobacterium nifH sequences than filamentous-cyanobacterium sequences, but fewer sequences were obtained by RT-PCR (Table 1). Filamentous-cyanobacterium nifH sequences were not detected in the Hague Marina DNA samples. This could be due to a lower relative abundance of the filamentous cyanobacteria at this site or the relatively small number of sequences examined in this study.
Other types of nitrogen-fixing bacteria that were detected in the net plankton samples expressed genes that were related to α- and γ-proteobacterial nif genes. The bacteria containing these genes were most likely associated with small invertebrates (i.e., zooplankton), small particles, or phytoplankton aggregates that were collected in the net. Some of the sequences found in this study are related to sequences recently reported for termite-associated bacteria (10). For example, clones LG1115 and LG1116 cluster most closely with sequences obtained from termites and are 86% identical to the termite-associated nifH sequences (Fig. 4). Braun et al. (1) reported nifH sequences amplified from microbial enrichments initiated with marine planktonic invertebrates that grouped with cluster I sequences, branching closely to the same termite-associated nifH sequences as do sequences LG1115 and LG1116. These sequences obtained from the Lake George net plankton may have been obtained from bacteria associated with invertebrate zooplankton.
Sequences that group with LG1107 and LG1109 form a deeply branching cluster of bacteria. The high bootstrap value, in addition to the deep branching, supports the conclusion that this set of sequences represents a new phylogenetic group of N2 fixers. This clade is closest to the proteobacterial clade shown in Fig. 4 and does not represent cluster II, III, or IV sequences (data not shown). Though it is difficult to determine the type of bacteria from which these sequences were derived, it is clear that these sequences were not artifacts.
Conclusions.
The results presented in this paper demonstrate the effective use of a nested RT-PCR approach to detect bacteria expressing nifH from environmental samples. Many nitrogen-fixing bacteria were detected among the Lake George samples, and cyanobacteria, α-proteobacteria, and a novel diazotrophic proteobacterial clade expressed nifH transcripts. Furthermore, all of the bacteria detected had type I nitrogenase, and no sequences in group II, III, or IV were found. While nifH expression does not necessarily indicate that the bacteria were actively fixing N2, it does provide information on the bacteria that could have been fixing nitrogen and also suggests that nitrogen-fixing conditions existed for these phylotypes at the time of sampling. It is also interesting that these microorganisms expressed nitrogenase in a typical phosphorus-limited environment, suggesting that the microorganisms may have been limited by multiple nutrients or that microorganisms were limited by different nutrients in the same environment. Future use of this nested RT-PCR approach can be used to identify organisms actively expressing nitrogenase genes and also to learn more about the environmental factors controlling nitrogenase expression and nitrogen fixation in aquatic environments.
Nucleotide sequence accession numbers.
All sequences obtained in this study were submitted to GenBank with accession numbers AF212868 to AF212891.
Acknowledgments
This work was supported by NSF grants OCE-9503593 and IBN-9629314 and the Department of Energy.
We thank L. Richardson for assistance in the field.
REFERENCES
- 1.Braun S, Proctor L, Zani S, Mellon M T, Zehr J P. Molecular evidence for zooplankton-associated nitrogen-fixing anaerobes based on amplification of the nifH gene. FEMS Microbiol Ecol. 1999;28:273–279. [Google Scholar]
- 2.Ferris J J, Clesceri N L. A description of the trophic status and nutrient loading for Lake George, New York. North American project—a study of U.S. water bodies. Corvallis, Oreg: U.S. Environmental Protection Agency; 1977. [Google Scholar]
- 3.Giovannoni S J, DeLong E F, Schmidt T M, Pace N R. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hecky R E, Kilham P. Nutrient limitation of phytoplankton in freshwater and marine environments: a review of recent evidence on the effects of enrichment. Limnol Oceanogr. 1988;33:796–822. [Google Scholar]
- 5.Kowalchuk G A, Naoumenko Z S, Derikx P J L, Felske A, Stephen J R, Arkhipchenko I A. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl Environ Microbiol. 1999;65:396–403. doi: 10.1128/aem.65.2.396-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Momen B, Eichler L W, Boylen C W, Zehr J P. Application of multivariate statistics in detecting temporal and spatial patterns of water chemistry in Lake George, New York. Ecol Model. 1996;91:183–192. [Google Scholar]
- 8.Momen B M, Eichler L W, Boylen C W, Zehr J P. Are recent watershed disturbances associated with temporal and spatial changes in water quality of Lake George, New York, USA? Environ Manag. 1997;21:725–732. doi: 10.1007/s002679900062. [DOI] [PubMed] [Google Scholar]
- 9.Noda S, Ohkuma M, Usami R, Horikoshi K, Kudo T. Culture-independent characterization of a gene responsible for nitrogen fixation in the symbiotic microbial community in the gut of the termite Neotermes koshunensis. Appl Environ Microbiol. 1999;65:4935–4942. doi: 10.1128/aem.65.11.4935-4942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohkuma M, Noda S, Usami R, Horikoshi K, Kudo T. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:2747–2752. doi: 10.1128/aem.62.8.2747-2752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paerl H W. Physiological ecology and regulation of N2 fixation in natural waters. Adv Microb Ecol. 1990;8:305–344. [Google Scholar]
- 12.Picard C, Ponsonnet C, Paget E, Nesme X, Simonet P. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1992;58:2717–2722. doi: 10.1128/aem.58.9.2717-2722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postgate J R, Eady R R. The evolution of biological nitrogen fixation. In: Bothe H, de Bruijn F J, Newton W E, editors. Nitrogen fixation: hundred years after. Stuttgart, Germany: Gustav Fischer; 1988. pp. 31–40. [Google Scholar]
- 14.Shuster E L, LaFleur R G, Boylen C W. The hydrologic budget of Lake George, southeastern Adirondack mountains of New York. Northeast Geol. 1994;16:94–108. [Google Scholar]
- 15.Siegfried C A. Phytoplankton of Lake George: seasonal and geographic patterns. In: Boylen C W, editor. The Lake George ecosystem. Proceedings of the Lake George Research Symposium. III. Lake George, N.Y: The Lake George Association; 1981. [Google Scholar]
- 16.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steppe T F, Olson J B, Paerl H W, Litaker R W, Belnap J. Consortial N-2 fixation—a strategy for meeting nitrogen requirements of marine and terrestrial cyanobacterial mats. FEMS Microbiol Ecol. 1996;21:149–156. [Google Scholar]
- 18.Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- 19.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Genetic diversity of N2-fixing bacteria associated with rice roots by molecular evolutionary analysis of a nifD library. Can J Microbiol. 1995;41:235–240. doi: 10.1139/m95-032. [DOI] [PubMed] [Google Scholar]
- 20.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 21.Vitousek P M, Howarth R W. Nitrogen limitation on land and in the sea: how can it occur? Biogeochemistry. 1991;13:87–115. [Google Scholar]
- 22.Widmer F, Shaffer B T, Porteous L A, Seidler R J. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade Mountain Range. Appl Environ Microbiol. 1999;65:374–380. doi: 10.1128/aem.65.2.374-380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young J P W. Phylogenetic classification of nitrogen-fixing organisms. In: Stacey G, Evans H J, Burris R H, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 43–86. [Google Scholar]
- 24.Zehr J P, Capone D G. Problems and promises of assaying the genetic potential for nitrogen fixation in the marine environment. Microb Ecol. 1996;32:263–281. doi: 10.1007/BF00183062. [DOI] [PubMed] [Google Scholar]
- 25.Zehr J P, Mellon M T, Zani S. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl Environ Microbiol. 1998;64:3444–3450. doi: 10.1128/aem.64.9.3444-3450.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zehr J P, McReynolds L A. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1989;55:2522–2526. doi: 10.1128/aem.55.10.2522-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]