Abstract
Non-Structural Protein 6 (NSP6) has a protecting role for SARS-CoV-2 replication by inhibiting the expansion of autophagosomes inside the cell. NSP6 is involved in the endoplasmic reticulum stress response by binding to Sigma receptor 1 (SR1). Nevertheless, NSP6 crystal structure is not solved yet. Therefore, NSP6 is considered a challenging target in Structure-Based Drug Discovery. Herein, we utilized the high quality NSP6 model built by AlphaFold in our study. Targeting a putative NSP6 binding site is believed to inhibit the SR1-NSP6 protein-protein interactions. Three databases were virtually screened, namely FDA-approved drugs (DrugBank), Northern African Natural Products Database (NANPDB) and South African Natural Compounds Database (SANCDB) with a total of 8158 compounds. Further validation for 9 candidates via molecular dynamics simulations for 100 ns recommended potential binders to the NSP6 binding site. The proposed candidates are recommended for biological testing to cease the rapidly growing pandemic.
Keywords: COVID-19, Docking, Molecular dynamics, NSP6, DrugBank, Natural products
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19) has become a global pandemic after firstly reported in Wuhan, China in late December 2019 (Chen et al., 2020a). COVID-19 is due to a novel betacoronavirus SARS-CoV-2 causing severe pneumonia (Olwenyi et al., 2020) and leading to >145 million infected cases and >3 million deaths worldwide since the start of the pandemic (https://covid19.who.int/, accessed on April 22, 2021). Other betacoronaviruses cause severe respiratory illnesses such as SARS-CoV-1 which shares approximately 79.5% of the genome with SARS‐CoV-2 but some different mechanisms between them in the infected host cells may illustrate the aggressiveness and the virus transmission speed of the novel SARS‐CoV‐2 over SARS-CoV-1 (Rossi et al., 2020; Neuman et al., 2006; Neuman and Buchmeier, 2016; Chen et al., 2020b; MA and Veesler, 2019; Caldaria et al., 2020). According to SARS-CoV-2 structure, it has a spherical shape determined by electron microscopy with 50–200 nm diameter in addition to 9-12-nm long glycoprotein spikes protruded from the outer envelope (Zhu et al., 2020; Mittal et al., 2020). Inside the viral envelope, RNA genome comprises ∼29.900 nucleotides containing several open reading frames (ORFs) that encode different types of proteins having critical functions in the viral pathogenesis such as non-structural proteins (NSPs). NSPs include 16 proteins encoded by ORF1a and ORF1b and have in general critical roles in forming the viral replication complex (Rastogi et al., 2020; Wu et al., 2020; Kim et al., 2020). One of the most challenged NSPs of coronaviruses in target-based drug discovery is non-structural protein 6 (NSP6), as until now, no crystal structure has been resolved, but only theoretical models can be studied (Lubin et al., 2020).
NSP6, which has a size of approximately 34 kDa, is an integral membrane protein of eight transmembrane helices and a highly conserved C terminus (Pandey et al., 2020; van der Hoeven et al., 2016). Interestingly, NSP6 is associated with NSP3 and NSP4 in the assembly of replication-transcription complexes (RTCs) by stimulating the formation of double-membrane vesicles (DMVs) from endoplasmic reticulum (ER) (Pandey et al., 2020; van der Hoeven et al., 2016; Zhang et al., 2020). In addition, NSP6 is involved in protecting the viral production inside host cells by limiting the expansion of autophagosomes. Although the number of autophagosomes produced by NSP6 is higher than that induced by starvation, they have a limited ability to expand due to their smaller size (Pandey et al., 2020; Morais et al., 2020; Cottam et al., 2014). Interestingly, some evidence demonstrates that NSP6 has an important role in ER stress response by interacting with the ER sigma receptor (Santerre et al., 2020; DE Gordon et al., 2020a).
It is obvious that NSP6 is a critical protein to target with therapeutics for limiting COVID-19 breakout thus some studies have tried to find out ligands which can interact with the predicted NSP6 and inhibit its function. According to the critical interaction between NSP6 and SR-1, Gordon et al. (DE Gordon et al., 2020a) have identified 6 compounds with a significant antiviral activity that interact with SR-1 blocking its binding to NSP6 after a subset screening of 69 compounds through multiple viral assays (Terracciano et al., 2021). From these compounds: Clemastine, Haloperidol and Hydroxychloroquine are FDA-approved drugs, PB28 is a preclinical compound in addition Siramesine and Cloperastine are drugs in clinical trial phase. In addition, it has been reported that 13 FDA-approved drugs could inhibit SARS-CoV-2 activity by interacting with SR-1 in A549-ACE2 cells (Terracciano et al., 2021; DE Gordon et al., 2020b). Furthermore, antihistamines, such as Diphenhydramine, Hydroxyzine and Azelastine, have been found to exert direct antiviral activity against SARS-CoV-2 through their high affinity to SR-1 in-vitro. Some studies tried to find compounds that interact directly with NSP6 to eliminate its functions. Pandey et al. (2020) have suggested that two SR-1 binding drugs, Dextromethorphan and Haloperidol, influence alteration of the tertiary structure of SARS-CoV-2 NSP6. Lundin et al. (2014) reported K22 as a small compound inhibitor of NSP6 and has an antiviral activity for many coronaviruses. Besides, virtual screening of 75 FDA-approved antiviral drugs against NSP6 and several SARS-CoV-2 targets recommended catechin and other 6 therapeutic agents (Mishra et al., 2021).
NSP6 shows increasing interest as it could play a vital role in SARS-COV2 mutations. Nevertheless, its significant role was proved in in-vitro studies. These findings encouraged us to focus on NSP6 as a potential target for drug repurposing to tackle this disease (Sun et al., 2022; Benvenuto et al., 2020).
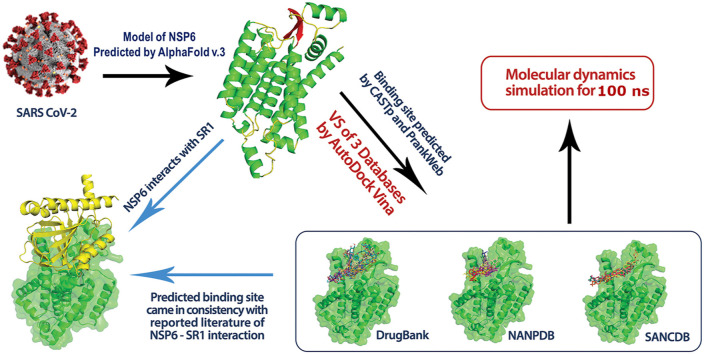
In this study, we used the model of NSP6 built by AlphaFold to predict a putative binding site for a virtual screening effort. Our prediction of the binding site via CASTp and PrankWeb came in-coherence with the literature data since it is responsible for SR1-NSP6 assembly. Therefore, it is believed when targeting the SR1-NSP6 interface, the SR1-NSP6 assembly and protein-protein interaction would be inhibited. Based on that, we carried out a prospective virtual screening to repurpose candidates from FDA-approved drugs, Northern and South African Natural Products databases. To confirm the binding of some hits, molecular dynamics simulations of five FDA-approved drugs and three natural product candidates displayed stability with NSP6 compared to the apo form. Such repurposing approach would recommend hits with known toxicity and pharmacokinetics profiles for further in-vitro anti-SARS-CoV-2 assays.
2. Methodology
2.1. Multiple sequence alignment of different NSP6 proteins
Some relevant beta coronavirus species express NSP6 protein, such as, SARS-CoV-1, MERS1, Bat coronavirus HKU9, Bat coronavirus HKU5, Bat coronavirus HKU4, Human coronavirus HKU1, Human coronavirus OC43, Murine coronavirus and Bovine coronavirus (Neuman, 2016). To explore their sequence similarity with SARS-CoV-2 NSP6, we conducted a multiple sequence alignment (MSA) and phylogenic tree analysis. The sequences were downloaded from Uniprot (UniProt: the universal protein knowledgebase, 2021) and underwent MSA through Muscle website (Madeira et al., 2019) employing ClustalW for alignment (Larkin et al., 2007). Then, Jalview (version 2.11.1.4) was used to view the results of alignment and phylogenetic tree (Waterhouse et al., 2009). Due to the lack of any structural information about these NSP6 proteins serving as a template for homology modeling for SARS-CoV-2 NSP6, we used a “template-free modeling” approach to model a SARS-CoV-2 NSP6 protein.
2.2. Structure modeling of NSP6
There are many attempts to predict SARS-CoV-2 NSP6 via FeigLab, Zhang and Alphafold as up till now there is no 3D structure of the protein (Lubin et al., 2020). Alphafold model showed superior quality compared to C–I-TASSER platform and FeigLab based on MolProbity scores due to some poor geometries, massive atomic clashes and bad backbone dihedral angles (Heo and Feig, 2020). Moreover, AlphaFold can predict protein structures by DeepMind algorithm based on deep neural network learning which is successfully able to predict the number of difficult proteins in SARS-CoV-2 including NSP6 (Senior et al., 2020; Li et al., 2015). Therefore, we used the NSP6 model predicted by DeepMind algorithm AlphaFold version 3, the updated version by Alphafold in 4 Augustus 2020, for SARS-CoV-2 NSP6 (https://deepmind.com/research/open-source/computational-predictions-of-protein-structures-associated-with-COVID-19). In addition, the quality of the model was assessed using SAVES v6.0 (https://saves.mbi.ucla.edu/) evaluating the stereochemical quality of a protein structure by analyzing residue-by-residue geometry and overall structure geometry.
2.3. Binding site prediction and virtual screening
To predict the binding site of NSP6, we used CASTp (Tian et al., 2018) (http://sts.bioe.uic.edu/castp/index.html?4jii) and PrankWeb (https://prankweb.cz/). The best predicted binding site came in-coherence with the reported literature signifying the site of SR1-NSP6 interaction and assembly (DE Gordon et al., 2020a; Alsulami et al., 2021). Accordingly, we carried out virtual screening for three different databases, namely: DrugBank, African Natural Compounds Database (SANCDB) and Northern African Natural Products Database (NANPDB). NANPDB was downloaded from (http://african-compounds.org/nanpdb/) with 4922 natural products from Northern African sources derived from plants, animals, fungi, and bacteria (Ntie-Kang et al., 2017). The SANCDB comprises 1012 natural compounds from Southern African sources, which were also downloaded (https://sancdb.rubi.ru.ac.za/) (Hatherley et al., 2015). For DrugBank, it consists of FDA approved-drugs with a total of 2224 molecules which were also downloaded from (http://redpoll.pharmacy.ualberta.ca/drugbank/) (Wishart et al., 2006). All molecules were downloaded as “sdf” format, prepared as reported earlier (Elghoneimy et al., 2021), then converted into pdbqt file format using openbabel (O'Boyle et al., 2011) and Python script (prepare_receptor4.py) provided by the MGLTools 1.5.6 (Sanner, 1999).
Virtual screening of the databases was performed using Autodock Vina. The NSP6 model was prepared for docking by adding H-atoms to the whole structure and then converted to pdbqt format by MGL tools 1.5.6 (Sanner, 1999). Consequently, the grid box was determined using MGL tools 1.5.6 by selecting the atoms in the chosen predicted binding site (mentioned in the results section). The grid box size is 30 Å × 30 Å × 34 Å with a grid spacing of 1 Å. Finally, the best compounds were selected based on docking scores (binding affinity scores ΔG) and then visualized using Pymol (Seeliger and de Groot, 2010) and Discovery Studio visualizer– Biovia (BIOVIA and Syst è mes, 2020) to study the interaction between the selected compounds and NSP6 protein.
2.4. Molecular dynamics simulations and MMGBSA analysis
The apo and the complex structures acquired from the molecular docking phase were subjected to MD simulations using GROMACS v2020.4 (Abraham et al., 2015). The protein and ions were defined by Amber force field 99SB-ILDN (Lindorff-Larsen et al., 2010) and ligands were parameterized by generalized amber force field (GAFF) (Wang et al., 2004). The partial atomic charges were calculated by AM1-BCC charge method (Jakalian et al., 2002). The structures were inserted in a cubic box and solvated by TIP3P water molecules (Jorgensen et al., 1983) and neutralized by adding sufficient number of counterions. The long-term electrostatic interactions were calculated by Particle Mesh Ewald scheme (Darden et al., 1993) with distance cutoff set to 10 Å. LINCS algorithm (Hess, 2008) was applied to restrain bonds involving hydrogen atoms. The integration timestep was set to 2 fs. Random velocities by Boltzmann distribution were assigned to each system, and eventually they were equilibrated for 1 ns in NVT ensemble at 300 K with Berendsen thermostat (Berendsen et al., 1984). To ensure stability during the equilibration, complex systems were subjected to an extra equilibration stage for 2 ns in NPT ensemble at P = 1 bar supplied by the Berendsen barostat (Berendsen et al., 1984). A 100 ns dynamic simulation was performed in the NPT ensemble using the Berendsen barostat pressure coupling algorithm.
Generalized Molecular Mechanics (MM/GBSA) has become a widely accepted thermodynamic method for calculating bind free energy by balancing accuracy and computational efficiency. These are more accurate (and less computationally expensive) than most scoring functions. To better understand the binding affinities and binding energies of protein/ligand complexes, the gmx_mmpbsa tool (Vald é s-Tresanco et al., 2021) was used to perform MM/GBSA analysis FDA approved drugs based on their stability throughout the simulation process. A total of 2000 frames were extracted throughout the simulations, then MM/GBSA was calculated for the aforementioned complexes. An average calculation of all the frames was considered.
Free binding energy between ligand and protein is calculated based on the equation:
| ΔGbinding = Gcomplex – Greceptor – Gligand |
Gcomplex: Energy of protein-ligand complex,
Greceptor: Energy of protein only,
Gligand: Energy of unbound ligand.
2.5. Pharmacokinetics and drug-likeness profile predictions
The pharmacokinetics and drug-likeness properties of the selected natural product candidates were predicted using SwissADME (Daina et al., 2017).
3. Results and discussion
3.1. Multiple sequence alignment of different NSP6 proteins
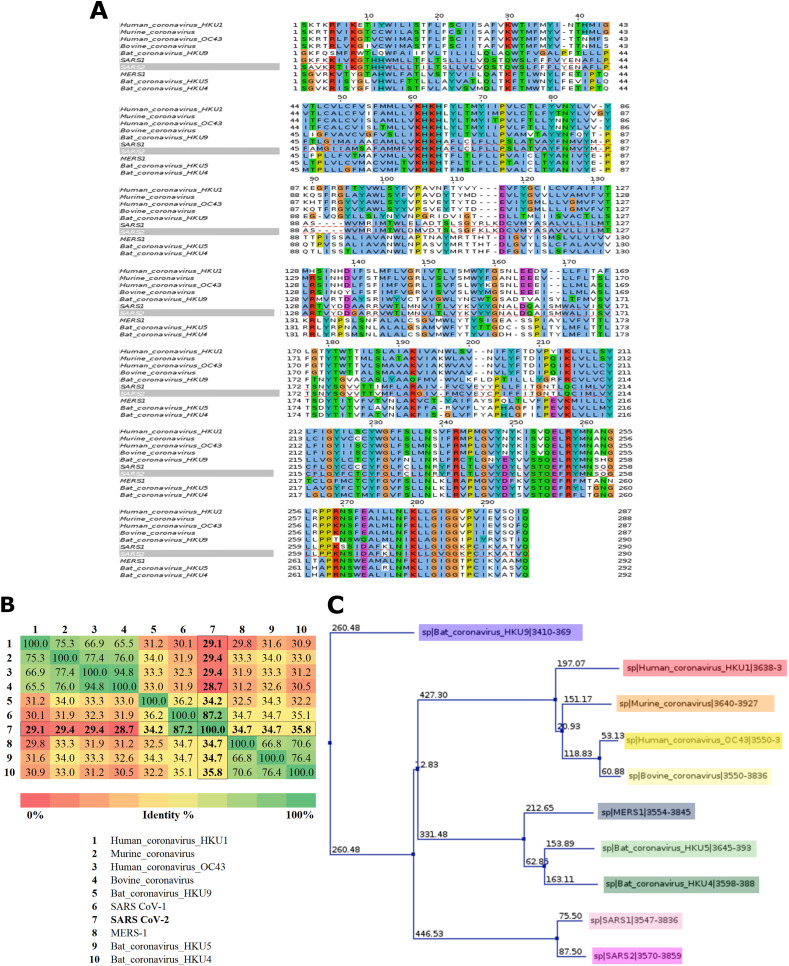
Since NSP6 of SARS-CoV-2 is a novel protein without a 3D structure, so we explored the similarity of NSP6 among relevant 10 beta coronavirus species to explore its sequence compared to closely related species. The multiple sequence alignment (MSA) and phylogenic tree indicate that NSP6 of both SARS-CoV-1 and SARS-CoV-2 is the most similar with 87.2% identity, as shown in Fig. 1 A–C, while the rest species display a low percentage identity with SARS-CoV-2 NSP6 ranging from 28.7%–35.8% Fig. 1B. Nevertheless, none of these NSP6 protein structures were solved yet and, hence, there is a lack of template information for comparative modeling of SARS-CoV-2 NSP6. Therefore, we utilized a “template-free modeling” approach to construct a model for the SARS-CoV-2 NSP6 protein.
Fig. 1.
(A) Multiple Sequence Alignment (MSA) for NSP6 Protein from 10 different beta-Coronaviruses utilized by ClustalW (Larkin et al., 2007) using muscle (Madeira et al., 2019). The color-coding of ClustalW indicates the physicochemical properties of amino acid residues. Hydrophobic, polar, positive- and negative-charged residues are highlighted blue, green, red, and magenta, respectively. Glycine, proline, and aromatic residues are shaded orange, yellow and cyan, respectively. Non-conserved amino acid residues are highlighted in white. (B) Percentage identity matrix for NSP6 among the 10 beta-Coronaviruses. (C) The phylogenetic tree of MSA.
3.2. Structure modeling of NSP6
NSP6 model is predicted by Alphafold employing deep-learning approach and conventional neural network. This approach can predict the distance and torsion distribution of protein-based on training schemes of PDB structures after getting the amino acid sequence and MSA features of a protein. Besides, Alphafold uses an algorithm based on what is called “template-free modeling” which can predict challenging proteins that lack structural similarity with other proteins (Senior et al., 2020).
AlphaFold model of NSP6 reveals 290 amino acids mainly formulates one C-terminal, two anti-parallel beta-sheets, sixteen turns and fourteen α helices from which eight transmembrane helices can be determined by TMHMM Fig. 2 A and supplementary data 1). To assess the quality of the Alphafold model, we used SAVES v6.0 server (https://saves.mbi.ucla.edu/). The Ramachandran Plot shows 94.7% of residues (252 from 290 residues) are in the most favored regions and the rest 5.3% of residues (14 from 290 residues) are found in additional allowed regions indicating no outliers Fig. 2B. According to the ERRAT server, the overall quality factor is 99.27 Fig. 2C. Globally, these matrices indicate the high quality model of NSP6.
Fig. 2.
3D structure of the generated model and structural validation using Ramachandran plot and statistics. (A) NSP6 model predicted by AlphaFold. (B) Ramachandran plot shows 94.7% of residues in most favored regions, and 5.3% in allowed regions. (C) ERRAT output with overall quality factor 99.270.
3.3. Binding site prediction and virtual screening
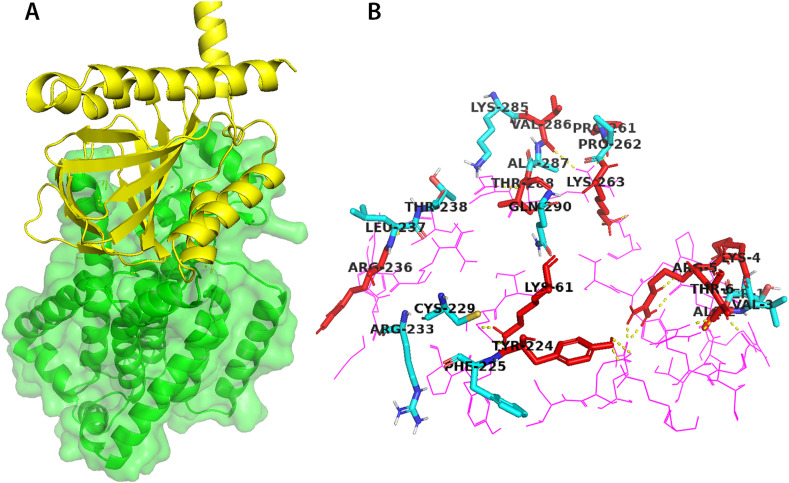
Our prediction of the binding site of the NSP6 model is based on the consensus predictions of both CASTp server and PrankWeb (supplementary data 2). The binding site is formed by the following amino: SER-1, LYS-4, ARG-5, THR-6, LYS-8, GLY-9, THR-10, HIS-12, TRP-13, MET-58, LYS-61, LYS-63, TYR-224, LEU-259, LEU-260, PRO-261, PRO-262, LYS-263, ASN-264, SER-265, ASP-267, LYS-270, LEU-271, LYS-274, THR-288, and GLN-290. Interestingly, based on literature, such binding site prediction came in-coherence with that involved for SR1-NSP6 interaction (Alsulami et al., 2021) (https://sars3d.com/model/nsp6). Based on the inspection, the amino acids involved in the interface of SR1-NSP6 and responsible for the interactions are LYS-4, ARG-5, THR-6, LYS-61, TYR-224, PRO-261, PRO-262, LYS-263, and THR-288 as seen in Fig. 3 .
Fig. 3.
Protein-protein interaction of SR1 and NSP6. (A) SR1 and NSP6 protein-protein interaction as an output of docking experiment (Alsulami et al., 2021), as shown in yellow and green cartoons, respectively. (B) a closer view of NSP6-SR1 interaction. Represented as violet sticks of Sigma Receptor 1 and amino acids of NSP 6 involved in the binding site.
We virtually screened the DrugBank, NANPDB and SANCDB using AutoDock Vina against the predicted binding site of NSP6. Top candidates of the score-ordered rank of the screened databases can be found in (supplementary data 3). The postulated binding poses of the best-scored molecules from the three databases show optimum binding at the predicted binding site of NSP6, as seen in Fig. 4 which would hinder SR1-NSP6 interactions.
Fig. 4.
Overall binding modes of molecules generated by molecular docking of the top 14 FDA-approved drugs and top 5 natural products with the selected region in the NSP6 protein. (A) Docking poses of 14 top-scored FDA-approved drugs (Venetoclax, Glecaprevir, Digitoxin, Dactinomycin, Oritavancin, Ledipasvir, Ergotamine, Irinotecan, Apixaban, Siponimod, Dihydroergocristine, Acetyldigitoxin). (B) Docking poses of 5 top-scored natural products from NANPDB (Euphoroscopin, Arjunin, Chebulagic acid, 3′-O-methyl-4-O-(3″,4″-di-O-galloyl-alpha-L-rhamnopyranosyl), and Owerreine). (C) Docking poses of 5 top-scored natural products from SANCDB (Cephalostatin 2, Cephalostatin 11, Cephalostatin 3, Cephalostatin 16, and Cephalostatin 19). All poses appeared to occupy the predicted binding site and were speculated to inhibit SR1-NSP6 protein-protein interactions.
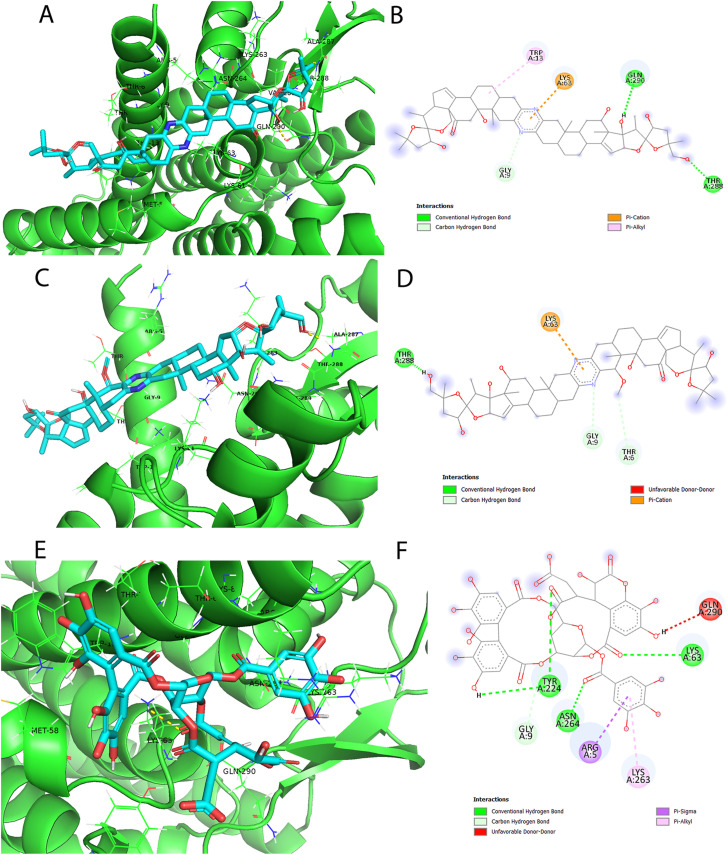
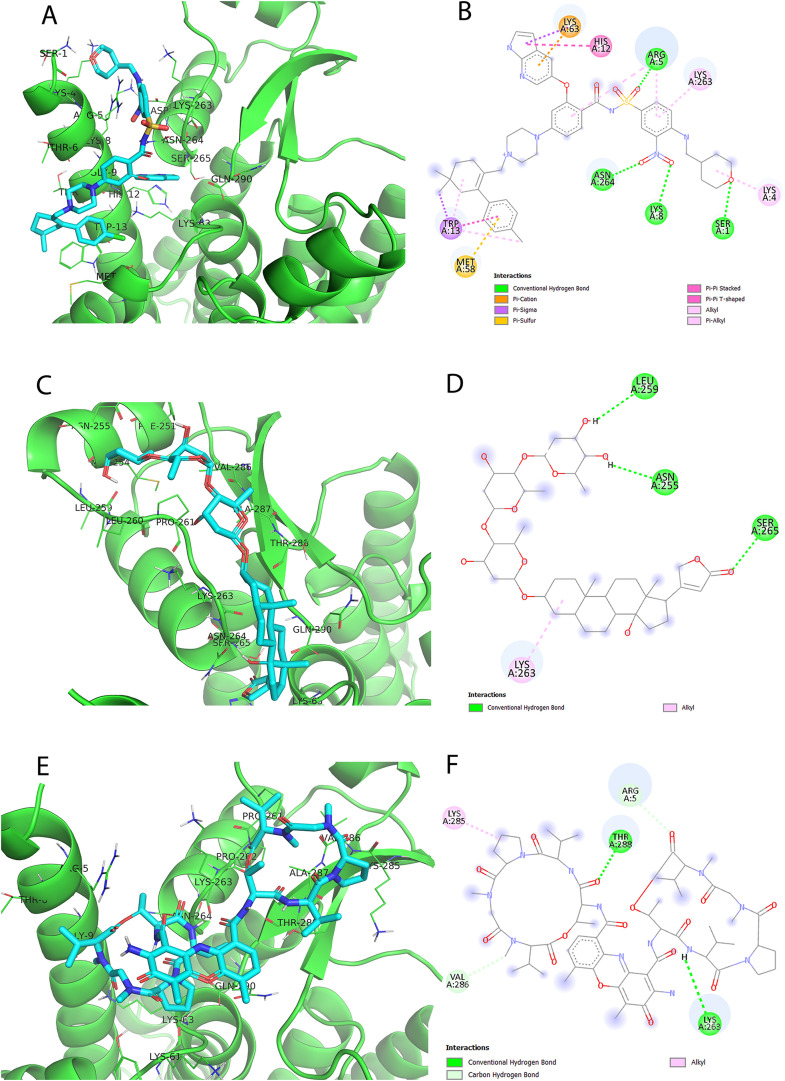
Elucidating the docking pose of Cephalostatin 2 (SA1 from SANCDB) illustrates H-bond interactions with both THR-288 and GLN-290 of NSP6 binding site. Besides, other favorable interactions such as pi-cation with LYS-63, pi-alkyl with TRP-13, and hydrophobic interactions with GLY-9, TRP-13, LYS-63, and GLN-290 can be observed in Fig. 5 A and B. Likewise, Cephalostatin 11 (SA2 from SANCDB) docking pose displays H-bond interactions with THR-288, pi-cation with LYS-63 as well as hydrophobic interactions with THR-6, GLY-9 and LYS-63 displayed in Fig. 5C and D. Both Cephalostatin 11 and Cephalostatin 2 are products of Cephalodiscus gilchristi organism and have roles in cell growth inhibitory activities (Moser, 2008; Pettit et al., 1994; GR Pettit et al., 1988).
Fig. 5.
Generated binding modes of the selected natural products with the selected region in the NSP6 protein. Cephalostatin 2 (SA1) (A, B), Cephalostatin 11 (SA2) (C, D) and Chebulagic acid (NA) (E, F), were presented as cyan sticks in the predicted binding site of NSP-6 in three- and two-dimensions. Their docking scores are −10.3, −10.1 and −9.2 (kcal/mol), respectively.
An example of NANPDB, Chebulagic acid (NA1) docking pose exhibits H-bond interactions with LYS-63, TYR-224, and ASN-264. Also, other favorable interactions such as hydrophobic interactions with ARG-5, GLY-9, LYS-63, TYR-224, LYS-263, ASN-264, and GLN-290 can be detected as shown in Fig. 5E and F. Chebulagic acid is extracted from the fruit of Terminalia chebula Retz with Topoisomerase 1 and Lipoxygenase inhibitory activities (Reddy et al., 2009). Also, it was reported to possess in-vitro anti-SARS CoV-2 activity (Du et al., 2021).
Revealing five examples of FDA-approved drugs, the docking pose of Venetoclax (DB11581 from DrugBank) shows H-bond interactions with SER-1, ARG-5, LYS-8, and ASN-264, as well as other favorable interaction types. For instance, its docking pose displays pi-cation interactions with LYS-63, pi-sigma with TRP-13 and LYS-63, pi-sulfur with MET-58, pi-pi stacking with HIS-12 and TRP-13, pi-alkyl with LYS-4, ARG-5, TRP-13 and LYS-263 as shown in Fig. 6 A and B. Venetoclax, is a potent B-cell lymphoma-2 (Bcl2) inhibitor which is primarily approved for patients with chronic lymphocytic leukemia (CLL) with or without 17p deletion and small lymphocytic lymphoma (SLL). Recently in April 2020, it was approved to be indicated in combination therapy for Acute Myeloid Leukemia (AML).
Fig. 6.
The binding modes of the selected FDA-approved drugs. Docking poses of FDA-approved drugs, namely: Venetoclax (DB11581) (A, B), Digitoxin (DB01396) (C, D), Dactinomycin (DB00970) (E, F). Their docking scores are −9.7, −9.1, −9 (kcal/mol), respectively.
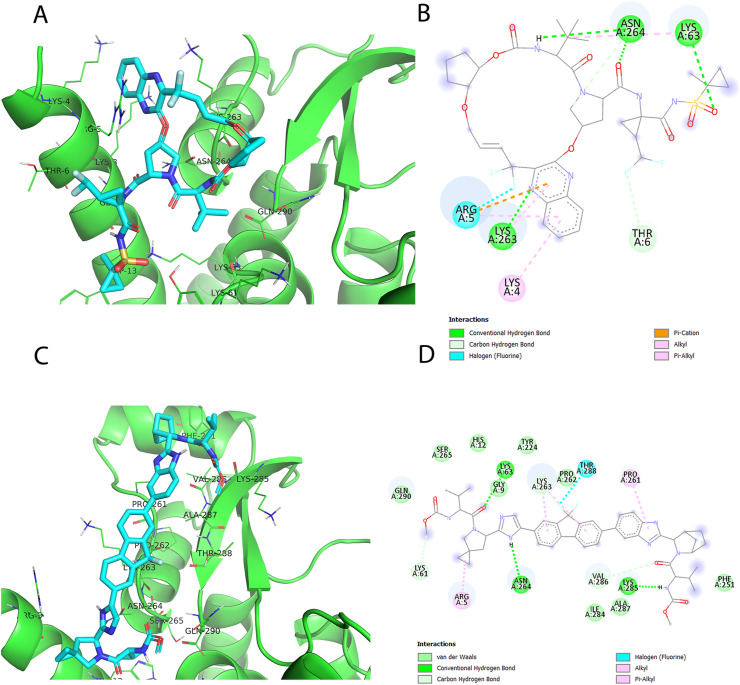
Digitoxin (DB01396 from DrugBank) docking pose demonstrates H-bond interactions with ASN-255, LEU-259, and SER-265 displayed in Fig. 6C and D. Digitoxin is a cardiac glycoside that can sometimes interchange with Digoxin (Belz et al., 2001). Originally, it is extracted from the leaves of Digitalis lanata and indicated for dysrhythmia patients as it affects Sodium–Potassium ATPase enzyme that regulates the quantity of Sodium and Potassium inside the cell (Belz et al., 2001).
Also, Dactinomycin (DB00970 from DrugBank) docking pose exhibits H-bond interactions with LYS-263 and THR-288, as well as hydrophobic interactions with ARG-5, LYS-263, VAL-286, and THR-288 as shown in Fig. 6 E and F. Dactinomycin is an antineoplastic antibiotic originally derived from Streptomyces parvulus. It is indicated for the treatment of solid tumors in children and choriocarcinoma in adult women. Its mechanism of action is through intercalation between two adjacent guanine-cytosine base pairs blocking the process of DNA transcription and RNA synthesis (Kwok et al., 2017).
Likewise, Glecaprivir (DB13879 from DrugBank) docking pose exhibits H-bond interactions with LYS-63, LYS-263, and ASN-264, and hydrophobic interactions with ARG-5, THR-6, LYS-63, LYS-263, and ASN-264. Also, other favorable types of interactions can be detected for Glecaprivir, such as pi-cation with ARG-5 as shown in Fig. 7 A and B. Similarly, ledipasvir (DB09027 from DrugBank) docking pose shows H-bond interactions with LYS-63, ASN-264, and LYS-285. Also, hydrophobic interactions with ARG-5, LYS-61, LYS-263, VAL-286, ASN-264, and GLN-290 can be observed as seen in Fig. 7C and D. Interestingly both Glecaprevir and Ledipasvir are antiviral drugs via inhibiting RNA viral replication. Glecaprevir is an inhibitor of HCV NS3-4A protease and was approved for chronic hepatitis c virus (HCV) genotype 1–6 (Salam and Akimitsu, 2013) in combination therapy with pibrentasvir. Unlike Glecaprevir, Ledipasvir targets Non-Structural Protein 5A which plays a vital role in viral replication as well (Kamal and Kamal, 2018).
Fig. 7.
Docking poses of FDA-approved drugs, namely: Glecaprevir (DB13879) (A, B) and Ledipasvir (DB09027) (C, D). Their docking scores are −9.2 and −8.8 (kcal/mol), respectively.
Generally, such postulated binding poses of DrugBank, SANCDB and NANPDB display favorable binding events at the predicted binding site of NSP6 which is believed to inhibit SR1-NSP6 protein-protein interactions.
3.4. Molecular dynamics simulations
For further assessment, we evaluated the ligand-protein stability via performing molecular dynamics (MD) simulation of the representative five DrugBank docking poses, two SANPDB docking poses and one NANPDB pose for 100 ns. Besides, a MD simulation for the NSP6 apo form was conducted as a reference for comparison purposes. This ended up with a total of 9 MD simulations for 100 ns each.
The stability of each system throughout the simulation was evaluated by estimating the root mean-squared deviation (RMSD), root mean square fluctuation (RMSF), and radius of gyration (Rg). Also, the number of H-bond interactions of ligand-protein complexes was also determined.
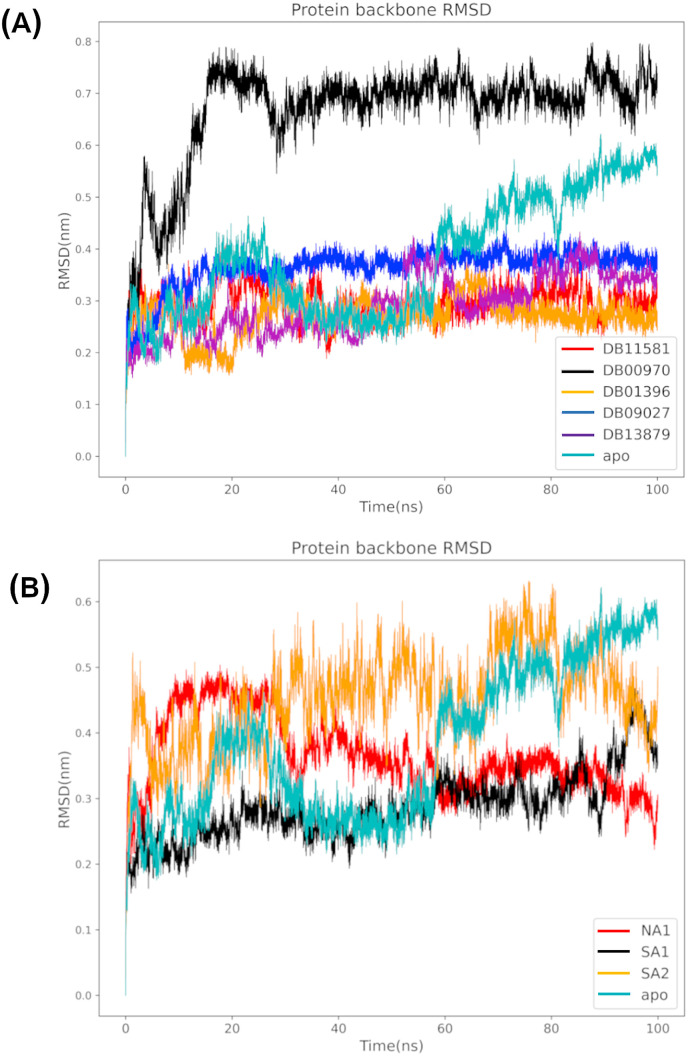
The calculated RMSD profiles of the natural and FDA approved drugs are shown in Fig. 8 A and B. The apo system displayed a significant fluctuation throughout simulation course with an average RMSD value of 0.44 nm. FDA approved drugs with exception of DB00970 (Dactinomycin) showed a stable protein RMSD profile with an average value of 0.27, 0.36, 0.30 and 0.29 nm for DB01396 (Digitoxin), DB09027 (Ledepasvir), DB11581 (Venetoclax) and DB13879 (Glecaprevir) respectively. Notably, DB00970 (Dactinomycin) demonstrated significant conformational changes in relation to the initial structure, with an average value of 0.67 nm reaching equilibrium at 20 ns. The analysis of a representative structure revealed two notable features that influenced the overall protein RMSD profile: I) the N-terminus helix was partially unfolded, resulting in the loss of key hydrophobic interactions and hydrogen bonding between the inhibitor and ARG5; and II) the short helix (residues 84–109) displayed significant spatial displacement with respect to its initial position. Natural products, on the other hand, exhibited less stable profile compared to FDA approved inhibitors as evidenced by substantial fluctuations through the simulation process.
Fig. 8.
The root mean squared deviation of the protein backbone atoms during the MD simulation of (A) FDA approved and (B) natural compounds.
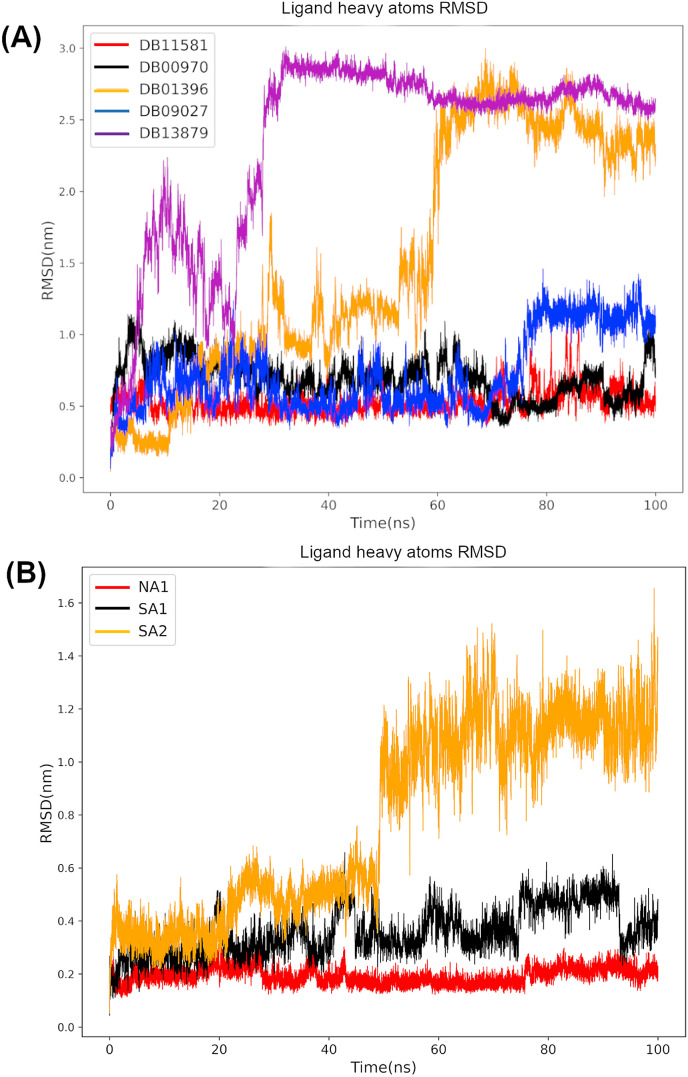
Fig. 9 demonstrates that DB00970 (Dactinomycin) and DB11581 (Venetoclax) inhibitors exhibited remarkable binding mode stability among all FDA-approved inhibitors. However, the binding modes of DB01396 (Digitoxin) and DB13879 (Glecaprevir) have changed dramatically, with average values of 1.54 and 2.34 nm, respectively. We also noticed that the binding mode of DB09027 (Ledepasvir) has partially changed at the end of the simulation course, reaching a maximum RMSD value of 1.46 nm as displayed in Fig. 9. NA1 (Chebulagic acid) and SA1 (Cephalostatin 2) natural products have a very stable binding mode throughout the simulation, with average RMSD values of 0.36 and 0.19 nm, respectively, demonstrating the reliability of the generated pose from molecular docking. On the other hand, SA2 (Cephalostatin 11) compound displayed major binding mode changes at the end of the simulation course with a maximum RMSD value of 1.65 nm.
Fig. 9.
Assessment of protein-ligand complexes stability throughout the simulation process the computed root mean squared deviation (RMSD) of candidates from FDA approved drugs (A) and natural compounds (B).
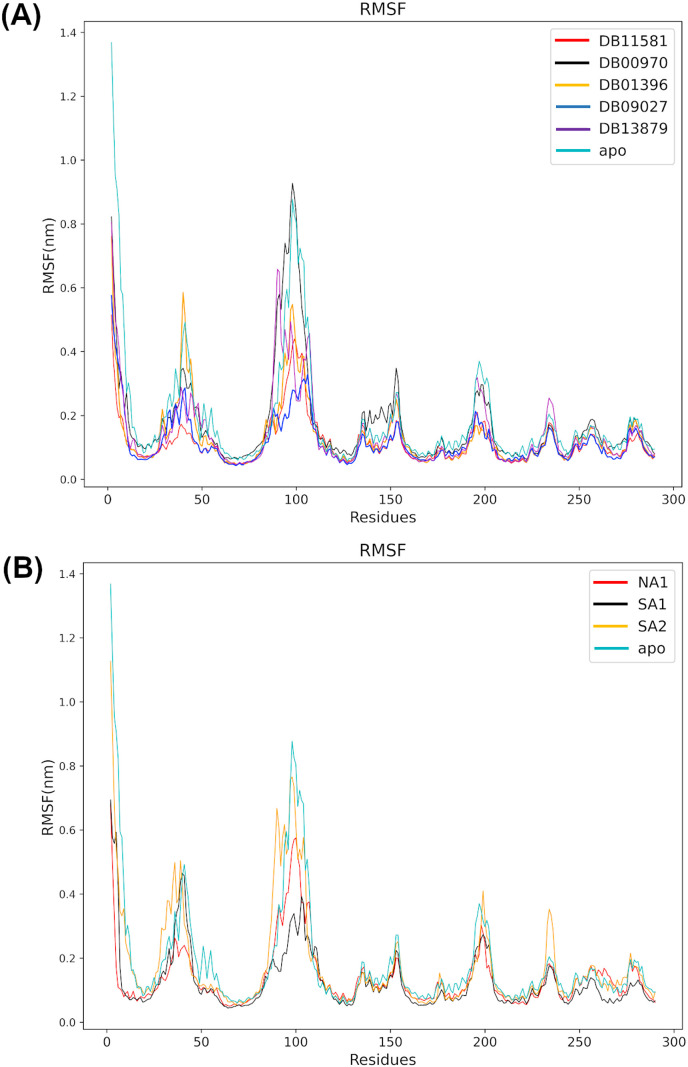
The root mean square fluctuations (RMSF) of all systems were used to assess the flexibility of the protein backbone as displayed in Fig. 10 . We observed common flexibility patterns in all systems in regions with residue numbers 30–50, 87–108, and 193–202. The flexibility of these regions was significantly reduced in systems with FDA approved drugs, except for the NSP6-DB00970 system that demonstrated high flexibility in the region 87–108 like the apo system. Natural products complex systems, on the other hand, demonstrated greater flexibility in previously indicated regions than FDA approved inhibitors, except for the NSP6-NA1 system and NSP6-SA1, which demonstrated less flexibility in regions 30–50 and 87–108, respectively.
Fig. 10.
Protein flexibility assessment using root mean square fluctuations (RMSF) throughout the simulation process. The protein backbone atoms root mean-squared fluctuations (RMSF) acquired from candidates from FDA approved drugs (A) and natural compounds (B).
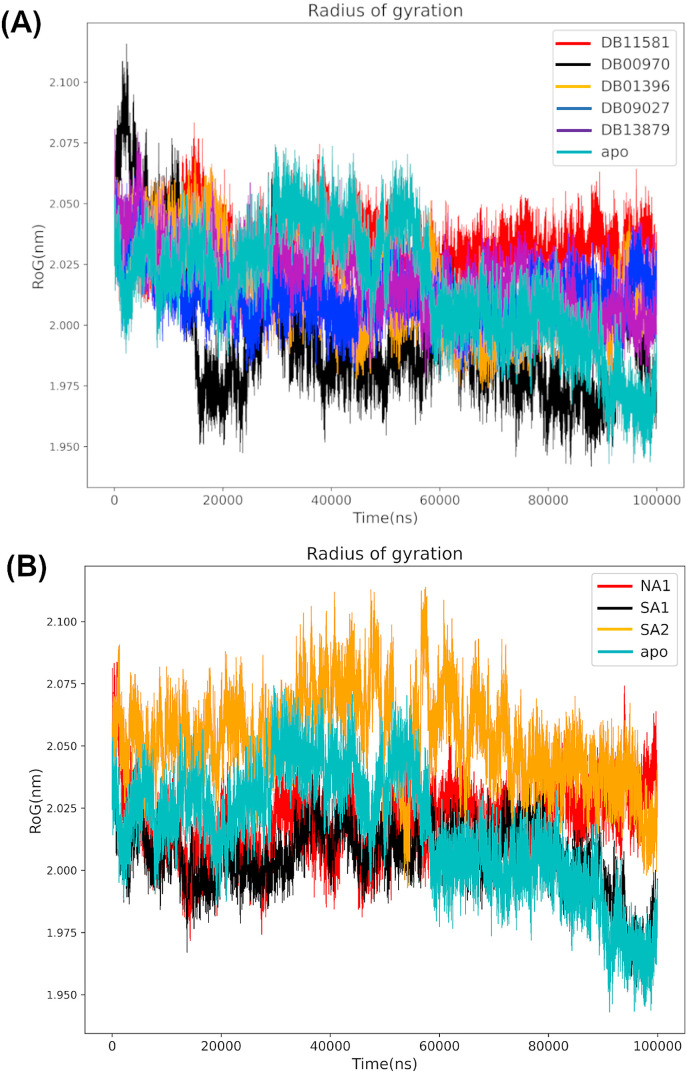
The radius of gyration was used to explore the effect of the selected compounds on the protein compactness (Fig. 11 ) throughout the simulation course. The NSP6 apo system displayed several fluctuations, indicating less stability and compactness. Chebulagic acid (NA1) and Cephalostatin 2 (SA1) showed better protein compactness than Cephalostatin 11 (SA2), which is consistent with protein backbone RMSD analysis. Furthermore, the systems with FDA approved inhibitors displayed relatively stable protein compactness profile, with an exceptional profile for the NSP6-DB00970, which was the most compacted system among all systems with an average value of 1.99 nm compared to NSP6-DB11581 with an average value of 2.03 nm.
Fig. 11.
Protein compactness evaluation by radius of gyration (Rg). Protein compactness using the radius of gyration (Rg) including the apo form and candidates from FDA approved drugs (A) and natural products (B).
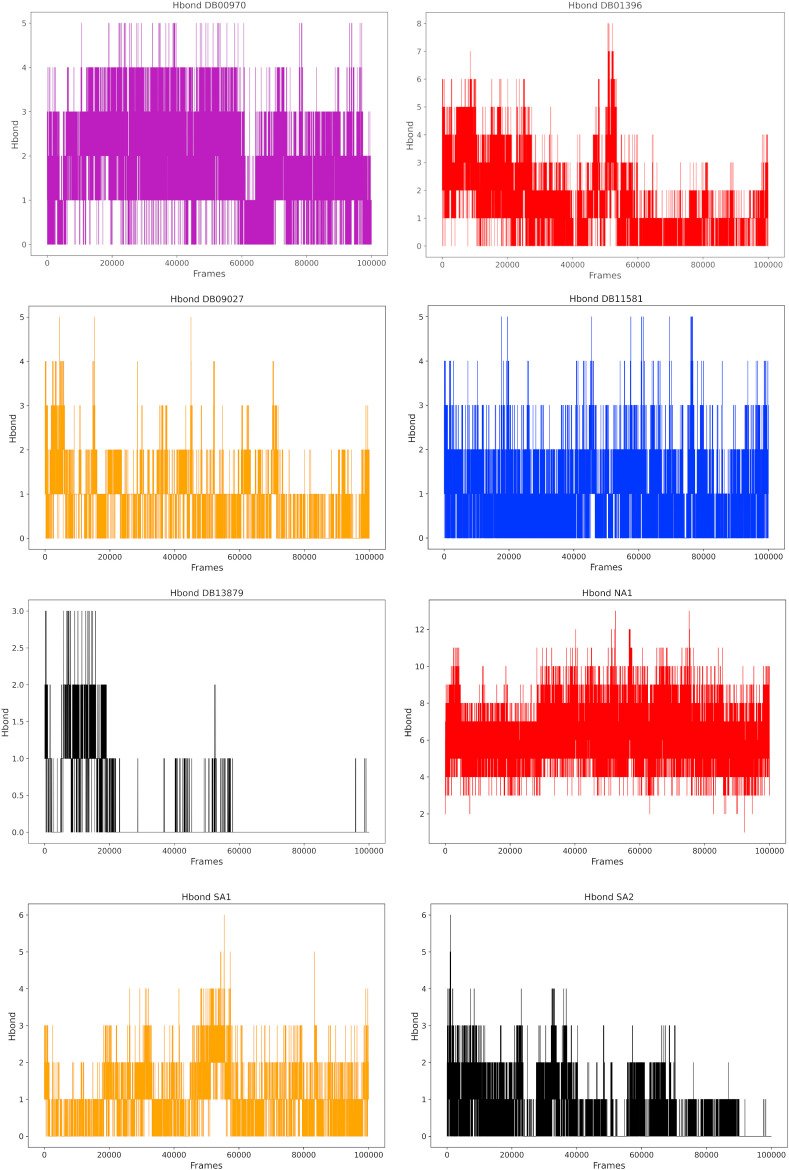
The H-bond interactions between the protein and the ligand are essential for ligand affinity and stability. The higher the number of H-bonds, the greater the stability and affinity of the ligand-protein complex. The number of H-bonds formed between the protein and its ligands were estimated by the Gromacs gmx hbond module, and the results are shown in Fig. 12 . Chebulagic acid (NA) displayed the highest number of H-bonds (10) among the tested natural products. The number of H-bonds for Cephalostatin 2 (SA1) and Cephalostatin 11 (SA2) are 4 and 6, respectively, throughout the simulation course. For the FDA-approved drugs, Venetoclax (DB11581), Digitoxin (DB01396), and Dactinomycin (DB00970), reached H-bond count with the surrounding residues of 5, 7, and 5, respectively. The relative lower number of H-bond count of Glecaprivir (DB13879) and Ledipasvir (DB09027) can be explained by involvement of other types of interactions between the ligand and protein, such as the hydrophobic interactions, as discussed earlier.
Fig. 12.
Evaluation of protein-ligand hydrogen bonds. H-bond count calculated between the protein and candidates from natural products and FDA approved drugs.
Due to the already known pharmacokinetics and toxicity, repurposing approved drugs as NSP6 inhibitors may significantly reduce the time to approval as a treatment for SARSCoV2. So, we performed the MMGBSA analysis on the FDA approved drugs that showed better stability throughout the MD simulation run.
Based on the promising results from the previous assessment and metrics, we focused on NSP6-DB00970, NSP6-SA1, NSP6-NA1, and NSP6-DB11581 systems compared to the unliganded NSP6 for further conformational sampling analysis. We investigated the conformational sampling of each system using principal component analysis (PCA) by evaluating their dominant modes of motion. The eigenvalues were computed by diagonalizing the covariance matrix of protein alpha carbon atoms. We concentrated our analysis on the first two principal components (PCs), which represent the most dominant protein motions in each system that accounts for 73%, 64%, 53%, 51%, and 44% of the motion variance for NSP6, NSP6-DB00970, NSP6-SA1, NSP6-NA1, and NSP6-DB11581 systems, respectively.
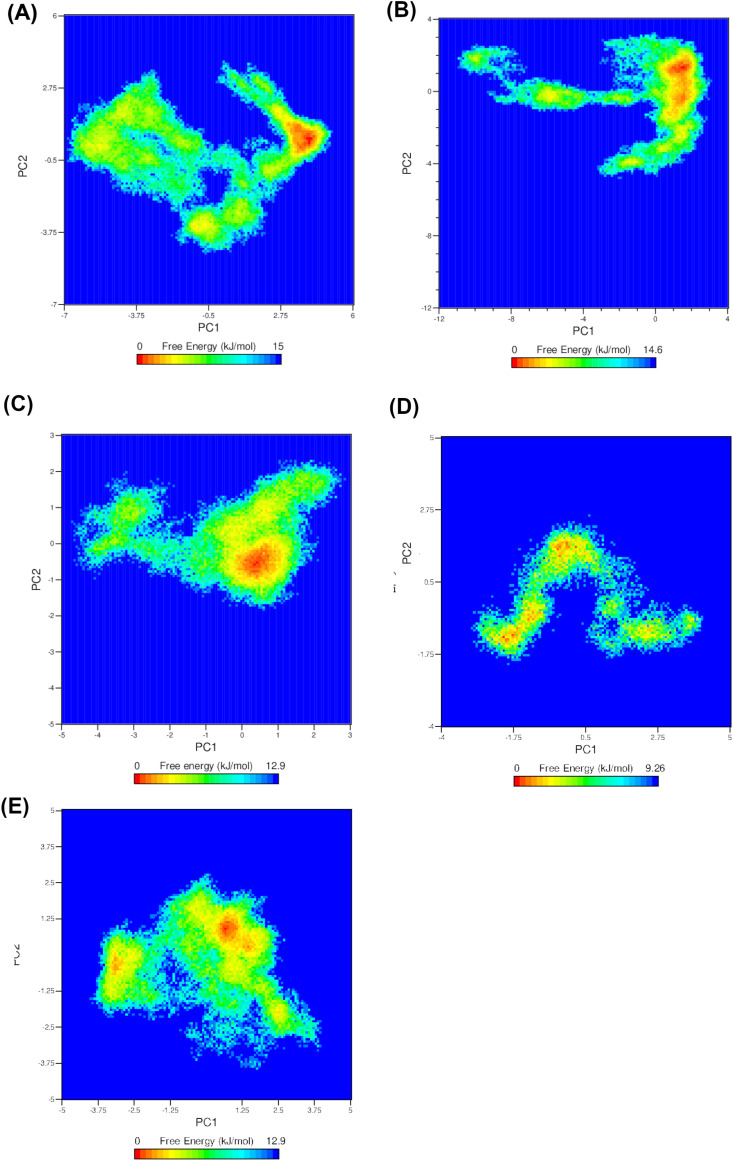
To mechanistically explore the effect of ligand binding on conformational landscape, we calculated the free energy landscape (FEL) as a function of the first two PCs acquired from PCA analysis as shown in Fig. 13 . FEL can be used to effectively describe conformational redistributions triggered by binding events (Pandey et al., 2020, 2021). The deeper color (towards the red color) in the plot reveals lower-energy conformational states. Interestingly, NSP6 unliganded form showed one major global minimum in the conformational subspace (red basin) with the occasion of less stable multiple local minima (yellow valleys), as shown in Fig. 13A. On the other hand, the FDA-approved drugs complex systems displayed wide and single basin implying one major ensemble of favorable conformations, as seen in Fig. 13B-C. Like the unliganded FEL behavior, the natural products, SA1 and NA1 complex systems with NSP6 exhibited distinct multiple low energy basins highlighting the presence of diverse ensembles of flexible and low energy conformations during 100 ns simulation, as shown in Fig. 13 D-E. These results thus clearly highlight that such compounds binding to NSP6 can alter its conformational subspace towards low energy conformations, and therefore, modulate its function.
Fig. 13.
Free Energy Landscape (FEL) of NSP6 and top selected complex systems. (A) NSP6, (B) NSP6-DB00970, (C) NSP6-DB11581, (D) NSP6-SA1 and (E) NSP6-NA1 systems.
The MM-GBSA method was used to estimate the binding free energies of selected complexes obtained from MD simulations. The calculated binding energies are shown in Table 1 . DB11581 (Venetoclax) had the best binding energy among other inhibitors, with a value of −47.24 kcal/mol, confirming its high affinity for NSP6, which is also consistent with its high binding energy obtained from molecular docking (−9.7 kcal/mol). This is due to its high van der Waal and electrostatic interaction energies of −58.04 ± 5.34 and −28.8 ± 9.2, respectively, which resulted in a strong G gas of −86.9 ± 10.9. Following that, DB00970 had relatively high binding free values of −29.62 ± 3.16 kcal/mol. DB09027 demonstrated week affinity towards NSP6 with binding energy of -17.04 ± 3.48 kcal/mol.
Table 1.
Binding Energies of target complexes. All values are given in kcal/mol.
| Inhibitor | ΔE (VdW) | ΔE (Ele) | ΔE (GB) | ΔE (SURF) | ΔG (Gas) | ΔG (Solv) | Total |
|---|---|---|---|---|---|---|---|
| DB11581 (Venetoclax) | −58.04 ± 5.34 | −28.8 ± 9.2 | 46.14 ± 8.1 | −6.6 ± 0.8 | −86.9 ± 10.9 | 39.6 ± 7.8 | −47.24 ± 6.4 |
| DB00970 (Dactinomycin) | −40.15 ± 3.71 | −60.10 ± 8.27 | 76.01 ± 7.28 | −5.38 ± 0.36 | −100.25 ± 8.42 | 70.63 ± 7.24 | −29.62 ± 3.16 |
| DB09027 (Ledipasvir) | −24.87 ± 4.79 | −15.98 ± 6.97 | 26.57 ± 7.02 | −2.75 ± 0.53 | −40.85 ± 8.42 | 23.81 ± 6.83 | −17.04 ± 3.48 |
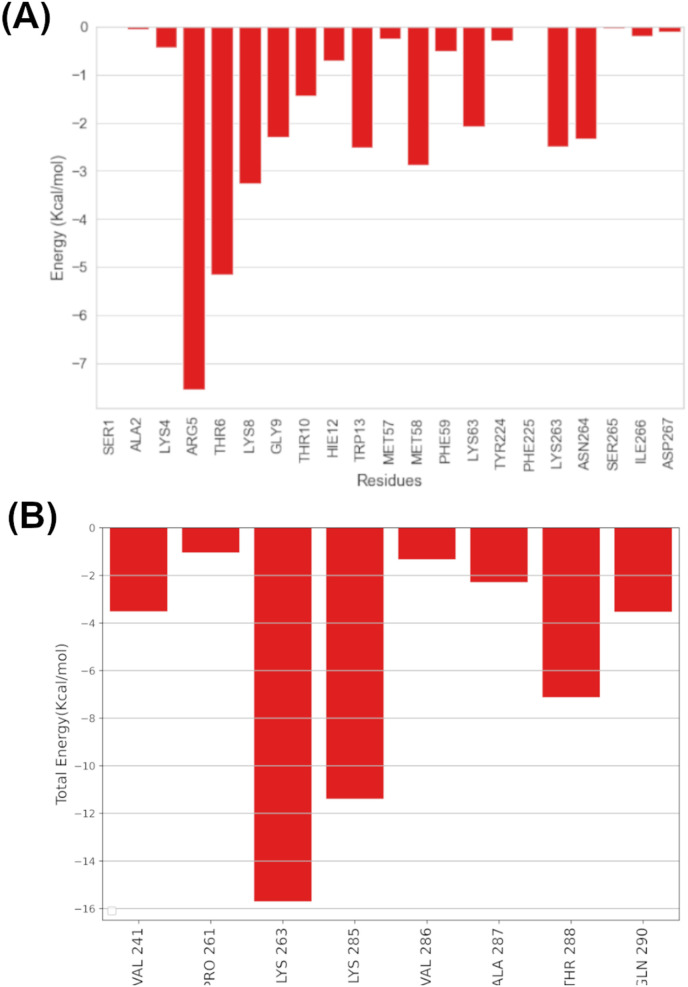
The per-residue energy decomposition of the NSP6-DB11518 (Venetoclax) complex system revealed that ARG5, THR6, LYS8, GLY9, TRP13, MET58, LYS63, LYS263 and ASN264, among other residues, had significant binding energies and contributed significantly to the binding of DB11581 as seen in Fig. 14 . Notably, ARG5 interacted strongly with the binder, with an energy value of −11.321.80 and strong Vdw and electrostatic contributions of −5.330.74 and −4.15 3.1 kcal/mol, respectively. These strong interactions are consistent with the docking pose in which the ARG5 side chain formed a hydrogen bond with the oxygen atom of Methyl 3-nitrophenyl sulfone, as well as two pi-Alkyl interactions with salicylamide and Methyl 3-nitrophenyl aromatic moieties. In addition, THR6 displayed a strong VdW interaction with DB11581. It is worth to be noted that THR6 was not observed in the docking pose but contributed to the ligand binding with an energy value of −7.75 kcal/mol implying its role in increasing stability of DB11581 inside the binding site. LYS8, MET85, TRP13 and LYS263 showed fewer binding energies with values of −4.9, −4.3, −3.7 and −3.7 kcal/mol, respectively. LYS63, GLY9 and ASN264 displayed binding energies of −3.1, −3.4 and −3.5 kcal/mol.
Fig. 14.
Per-residue energy decomposition analysis of (A) Venetoclax (DB11581) and (B) Dactinomycin (DB00970).
Free energy decomposition analysis of NSP6-DB00970 (Fig. 14 B) revealed key interactions with surrounding residues involved in inhibitor stability, including I) strong electrostatic interactions with LYS263 and LYS285 with energy values of −25.7 and −13.0 kcal/mol, respectively: II) van der Waals interactions with VAL241, LY263, LYS285, ALA287, THR288, GLN290 with energy values of −1.9, −2.4, −2.3, −1.2, −3.1 and −2.0 kcal/mol respectively. The only common residue found to interact with both inhibitors DB00970 and DB11581 is LYS263, with no observable interactions with the N-terminus helix with DB0070 as seen in the docking pose, particularly ARG5. This is due to the helix partial unfolding, which results in the loss of residual interactions with DB00970, particularly ARG5. Despite the strong interactions of DB00970 with surrounding residues, which resulted in the highest ΔGgas with a value of −100.25 ± 8.42 Kcal, DB00970 displayed an unfavorable polar contribution (ΔEGB) and solvation energy (ΔGsolv) of 76.01 ± 7.28 kcal/mol and 70.63 ± 7.24 when compared to DB11581, resulting in a higher binding affinity for DB11581. All these interactions with favorable binding energies explain the docking score obtained with DB11581 (Venetoclax) and DB00970 (Dactinomycin) provides more insights into its high in-silico affinity throughout MD simulation.
3.5. Pharmacokinetics and drug-likeness profile predictions
Since the pharmacokinetics properties and toxicity profiles of the FDA-approved drugs are well documented, this section is dedicated for the selected natural product candidates only. To demonstrate drug-likeness with respect to bioavailability, automated SwissADME server was used to predict the pharmacokinetic profile of the selected natural products candidates, namely, Cephalostatin 2, Cephalostatin 11 and Chebulagic acid. Such predictions give a general overview about the effectiveness of drug-candidates for treating a certain disease. However, further biological studies and validation experiments should be performed.
According to pharmacokinetics properties (Table 2 ), SwissADME server provides predictive models to estimate individual ADME behaviors of the putative compounds through the predictions of passive human gastrointestinal (GI) absorption and blood–brain barrier (BBB) permeation by specific methods such as BOILED-Egg (Daina et al., 2017; Daina and Zoete, 2016). The three putative natural products showed low BBB permeability which are critical for the protection of central nervous system (CNS) from any possible compounds’ adverse effects. In addition, all these compounds exhibited low gastrointestinal absorption and this finding was consistent with drug-likeness properties (Table 3 ) as Cephalostatin 2, Cephalostatin 11 and Chebulagic acid showed three violations for Lipinski rule and one violation for Veber rule.
Table 2.
In-silico predictions of the pharmacokinetics properties for the putative natural products.
| Compound | GI absorption | BBB | Pgp substrate |
|---|---|---|---|
| Cephalostatin2 | Low | No | Yes |
| Cephalostatin11 | Low | No | Yes |
| Chebulagic acid | Low | No | Yes |
| Compound | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor |
|---|---|---|---|---|---|
| Cephalostatin2 | No | No | Yes | No | No |
| Cephalostatin11 | No | No | Yes | No | No |
| Chebulagic acid | No | No | No | No | No |
GI absorption: human gastrointestinal absorption, BBB: blood–brain barrier permeation, Pgp: permeability glycoprotein, CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4 are the isoforms of CYP450.
Table 3.
In-silico predictions of the drug-likeness properties for the putative natural products.
| Compound | Lipinski #violations | Veber #violations |
PAINS #alerts |
Bioavailability Score |
|---|---|---|---|---|
| Cephalostatin2 | 3 | 1 | 0 | 0.17 |
| Cephalostatin 11 | 3 | 1 | 0 | 0.17 |
| Chebulagic acid | 3 | 1 | 0 | 0.11 |
Lipinski #violations show the number of violations of Lipinski rule as: lipophilicity (logP) ≤ 5, molecular weight ≤500, number of hydrogen bond donors ≤5 and number of hydrogen bond acceptors ≤10. Veber #violations show the number of violations of Veber rule summarized as: NRB ≤10 and TPSA ≤140 Å2. PAINS #alerts: counts the number of pan-assay interference compound. All calculations were done using SwissADME (Daina et al., 2017).
An Insight into compounds behavior with CYP family is doubtlessly of great importance as inhibition of these isoenzyme impact pharmacokinetic related drug-drug interactions (Hollenberg, 2002). As shown in Table 2, the three natural compounds did not show inhibitory actions on CYP450 isoforms except for the two southern African natural products Cephalostatin 2 and Cephalostatin 11 exhibited inhibitory effects with CYP2C9.
Permeability glycoprotein (Pgp) is an efflux pump powered by ATP and plays an important role in control the drug transportation through cell membranes protecting critical systems such as CNS (Sharom, 2011). Thus, the assessment of Pgp substrate parameter gives us a crucial insight whether the drug is a substrate to the Pgp or not (Daina et al., 2017). The three compounds of interest tend to be a substrate to Pgp as shown in (Table 2).
Pleasantly, all compounds did not exhibit any alert to be PAINS (pan-assay interference compounds) (Baell and Holloway, 2010) as shown in Table 3.
4. Conclusion
In the current emerging situation of the pandemic, many efforts were made to investigate possible therapeutics that could be indicated for SARS CoV-2 infection. Our focus is to target NSP6 protein via structure-based virtual screening via repurposing existing libraries, such as FDA approved drugs (DrugBank), Northern African Natural Products Database (NANPDB) and South African Natural Compounds Database (SANCDB). However, this task is challenging due to the lack of crystal structures of NSP6. Therefore, we considered the NSP6 model predicted by Alphafold for our investigation. Next, we predicted a putative binding site using a consensus output of both CASTp and PrankWeb. Interestingly, the predicted binding site came in consistency with the reported literature indicating the site of Sigma receptor1 (SR1)-NSP6 protein-protein interaction.
Then, virtual screening was performed using AutoDock Vina for the three databases including 2224 compounds from DrugBank, 4922 NANPDB and 1012 SANCDB. Based on the docking scores and visual inspections, we selected five candidates from DrugBank, (Venetoclax, Digitoxin, Dactinomycin, Glecaprevir, and Ledipasvir), and three candidates from both SANCDB and NANPDB as Cephalostatin 2, Cephalostatin 11 and Chebulagic acid. For additional in-silico validation, we performed 8 molecular dynamics (MD) simulations for 100 ns for these candidates complexed with NSP6, and compared to the apo form. The results showed high stability of most of these candidates in the NSP6 binding site. The calculated MM-GBSA binding free energy of Venetoclax presented the highest in-silico affinity among the top five FDA-approved drugs towards NSP6. Furthermore, the per-residue energy decomposition emphasized the significance of the interaction with ARG5, and subsequently, possible inhibition of the NSP6-SR1 protein-protein interactions. Furthermore, in-silico predictions of the pharmacokinetics and drug-likeness properties of the selected natural product candidates displayed encouraging results.
In conclusion, this study provides a pipeline on how to target a challenging target of SARS-CoV2 via virtual repurposing of FDA-approved drugs and natural products. Also, the recommended hits are endorsed for further biological assessment against NSP6 to cease the rapidly growing pandemic.
CRediT authorship contribution statement
Ahmed Abdelkader: Methodology, Visualization, Writing – original draft. Amal A. Elzemrany: Methodology, Visualization, Writing – original draft. Mennatullah El-Nadi: Methodology, Visualization, Writing – original draft. Sherif A. Elsabbagh: Methodology, Visualization, Writing – original draft. Moustafa A. Shehata: Methodology, Visualization, Writing – original draft. Wagdy M. Eldehna: Writing – review & editing. Mohamed El-Hadidi: Writing – review & editing. Tamer M. Ibrahim: Supervision, Conceptualization, Methodology, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors gratefully acknowledge the High-Performance Cluster of the University of Tuebingen and the cluster of Bibliotheca Alexandrina High-Performance Computing for granting access to perform the molecular dynamics simulations.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2022.06.008.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software. 2015 1-2 19-25. [Google Scholar]
- Alsulami A.F., Thomas S.E., Jamasb A.R., Beaudoin C.A., Moghul I., et al. SARS-CoV-2 3D database: understanding the coronavirus proteome and evaluating possible drug targets. Briefings Bioinf. 2021;22:769–780. doi: 10.1093/bib/bbaa404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell J.B., Holloway G.A. New substructure filters for removal of Pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Belz G.G., Breithaupt-Grögler K., Osowski U. Treatment of congestive heart failure--current status of use of digitoxin. Eur. J. Clin. Invest. 2001;31(Suppl. 2):10–17. doi: 10.1046/j.1365-2362.2001.0310s2010.x. [DOI] [PubMed] [Google Scholar]
- Benvenuto D., Angeletti S., Giovanetti M., Bianchi M., Pascarella S., et al. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020;81:e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J., Postma Jv, van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- BIOVIA. Systèmes Dassault. Dassault Systèmes; San Diego: 2020. Discovery Studio Visualizer, v21.1.0.20298. [Google Scholar]
- Caldaria A., Conforti C., Di Meo N., Dianzani C., Jafferany M., et al. COVID-19 and SARS: differences and similarities. Dermatol. Ther. 2020;33 doi: 10.1111/dth.13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Zoete V. A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11:1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- De Gordon, Jang G.M., Bouhaddou M., Xu J., Obernier K., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gordon, Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370 doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R., Cooper L., Chen Z., Lee H., Rong L., et al. Discovery of chebulagic acid and punicalagin as novel allosteric inhibitors of SARS-CoV-2 3CL(pro) Antivir. Res. 2021;190 doi: 10.1016/j.antiviral.2021.105075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elghoneimy L.K., Ismail M.I., Boeckler F.M., Hme Azzazy, Ibrahim T.M. Facilitating SARS CoV-2 RNA-Dependent RNA polymerase (RdRp) drug discovery by the aid of HCV NS5B palm subdomain binders: in silico approaches and benchmarking. Comput. Biol. Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GR Pettit M Inoue, Kamano Y., Dufresne C., Christie N., et al. Isolation and structure of the hemichordate cell growth inhibitors cephalostatins 2, 3, and 4. J. Chem. Soc., Chem. Commun. 1988:865–867. [Google Scholar]
- Hatherley R., Brown D.K., Musyoka T.M., Penkler D.L., Faya N., et al. SANCDB: a South African natural compound database. J. Cheminf. 2015;7:29. doi: 10.1186/s13321-015-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo L., Feig M. bioRxiv; 2020. Modeling of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Proteins by Machine Learning and Physics-Based Refinement. [Google Scholar]
- Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theor. Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- Hollenberg P.F. Characteristics and common properties of inhibitors, inducers, and activators of CYP enzymes. Drug Metab. Rev. 2002;34:17–35. doi: 10.1081/dmr-120001387. [DOI] [PubMed] [Google Scholar]
- Jakalian A., Jack D.B., Bayly C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Kamal S.M. In: Hepatitis C in Developing Countries. Kamal S.M., editor. Academic Press; 2018. Chapter 6 - hepatitis C treatment in the era of direct-acting antiviral agents: challenges in developing countries; pp. 209–246. [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.011. 914-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok K.K., Vincent E.C., Gibson J.N. In: Pharmacology and Therapeutics for Dentistry. seventh ed. Dowd F.J., Johnson B.S., Mariotti A.J., editors. Mosby; 2017. 36 - antineoplastic drugs; pp. 530–562. [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li J., Adhikari B., Cheng J. An improved integration of template-based and template-free protein structure modeling methods and its assessment in CASP11. Protein Pept. Lett. 2015;22:586–593. doi: 10.2174/0929866522666150520145717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin J.H., Zardecki C., Dolan E.M., Lu C., Shen Z., et al. bioRxiv; 2020. Evolution of the SARS-CoV-2 Proteome in Three Dimensions (3D) during the First Six Months of the COVID-19 Pandemic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Dijkman R., Bergström T., Kann N., Adamiak B., et al. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Tortorici, Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Ym, Lee J., Buso N., Gur T., et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra C.B., Pandey P., Sharma R.D., Malik M.Z., Mongre R.K., et al. Identifying the natural polyphenol catechin as a multi-targeted agent against SARS-CoV-2 for the plausible therapy of COVID-19: an integrated computational approach. Briefings Bioinf. 2021;22:1346–1360. doi: 10.1093/bib/bbaa378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A., Manjunath K., Ranjan R.K., Kaushik S., Kumar S., et al. COVID-19 pandemic: insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais I.J., Polveiro R.C., Souza G.M., Di Bortolin, Sassaki F.T., et al. The global population of SARS-CoV-2 is composed of six major subtypes. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-74050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser B.R. Review of cytotoxic cephalostatins and ritterazines: isolation and synthesis. J. Nat. Prod. 2008;71:487–491. doi: 10.1021/np070536z. [DOI] [PubMed] [Google Scholar]
- Neuman B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antivir. Res. 2016;135:97–107. doi: 10.1016/j.antiviral.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Buchmeier M.J. Supramolecular architecture of the coronavirus Particle. Adv. Virus Res. 2016;96:1–27. doi: 10.1016/bs.aivir.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., et al. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntie-Kang F., Telukunta K.K., Döring K., Simoben C.V., Am A.F., et al. NANPDB: a resource for natural products from Northern African sources. J. Nat. Prod. 2017;80:2067–2076. doi: 10.1021/acs.jnatprod.7b00283. [DOI] [PubMed] [Google Scholar]
- O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., et al. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olwenyi O.A., Dyavar S.R., Acharya A., Podany A.T., Fletcher C.V., et al. Immuno-epidemiology and pathophysiology of coronavirus disease 2019 (COVID-19) J. Mol. Med. (Berl.) 2020;98:1369–1383. doi: 10.1007/s00109-020-01961-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Prasad K., Prakash A., Kumar V. Insights into the biased activity of dextromethorphan and haloperidol towards SARS-CoV-2 NSP6: in silico binding mechanistic analysis. J. Mol. Med. (Berl.) 2020;98:1659–1673. doi: 10.1007/s00109-020-01980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Rane J.S., Chatterjee A., Kumar A., Khan R., et al. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: an in silico study for drug development. J. Biomol. Struct. Dyn. 2021;39:6306–6316. doi: 10.1080/07391102.2020.1796811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit G.R., Xu J.P., Williams M.D., Christie N.D., Doubek D.L., et al. Isolation and structure of cephalostatins 10 and 11. J. Nat. Prod. 1994;57:52–63. doi: 10.1021/np50103a007. [DOI] [PubMed] [Google Scholar]
- Rastogi M., Pandey N., Shukla A., Singh S.K. SARS coronavirus 2: from genome to infectome. Respir. Res. 2020;21:318. doi: 10.1186/s12931-020-01581-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy D.B., Reddy T.C., Jyotsna G., Sharan S., Priya N., et al. Chebulagic acid, a COX-LOX dual inhibitor isolated from the fruits of Terminalia chebula Retz., induces apoptosis in COLO-205 cell line. J. Ethnopharmacol. 2009;124:506–512. doi: 10.1016/j.jep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Rossi G.A., Sacco O., Mancino E., Cristiani L., Midulla F. Differences and similarities between SARS-CoV and SARS-CoV-2: spike receptor-binding domain recognition and host cell infection with support of cellular serine proteases. Infection. 2020;48:665–669. doi: 10.1007/s15010-020-01486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salam K.A., Akimitsu N. Hepatitis C virus NS3 inhibitors: current and future perspectives. BioMed Res. Int. 2013 doi: 10.1155/2013/467869. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. [PubMed] [Google Scholar]
- Santerre M., Arjona S.P., Allen C.N., Shcherbik N., Sawaya B.E. Why do SARS-CoV-2 NSPs rush to the ER? J. Neurol. 2020;1–10 doi: 10.1007/s00415-020-10197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- Sharom F.J. The P-glycoprotein multidrug transporter. Essays Biochem. 2011;50:161–178. doi: 10.1042/bse0500161. [DOI] [PubMed] [Google Scholar]
- Sun X., Liu Y., Huang Z., Xu W., Hu W., et al. SARS-CoV-2 non-structural protein 6 triggers NLRP3-dependent pyroptosis by targeting ATP6AP1. Cell Death Differ. 2022;29:1240–1254. doi: 10.1038/s41418-021-00916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano R., Preianò M., Fregola A., Pelaia C., Montalcini T., et al. Mapping the SARS-CoV-2-host protein-protein interactome by affinity purification mass spectrometry and proximity-dependent biotin labeling: a rational and straightforward route to discover host-directed anti-SARS-CoV-2 therapeutics. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22020532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–w367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–d489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Tresanco M.S., Valdés-Tresanco M.E., Valiente P.A., Moreno E. gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theor. Comput. 2021;17:6281–6291. doi: 10.1021/acs.jctc.1c00645. [DOI] [PubMed] [Google Scholar]
- van der Hoeven B., Oudshoorn D., Koster A.J., Snijder E.J., Kikkert M., et al. Biogenesis and architecture of arterivirus replication organelles. Virus Res. 2016;220:70–90. doi: 10.1016/j.virusres.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lan Y., Sanyal S. Membrane heist: coronavirus host membrane remodeling during replication. Biochimie. 2020;179:229–236. doi: 10.1016/j.biochi.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.