Abstract
Species diversity, phylogenetic affiliations, and environmental occurrence patterns of thiosulfate-oxidizing marine bacteria were investigated by using new isolates from serially diluted continental slope and deep-sea abyssal plain sediments collected off the coast of New England and strains cultured previously from Galapagos hydrothermal vent samples. The most frequently obtained new isolates, mostly from 103- and 104-fold dilutions of the continental slope sediment, oxidized thiosulfate to sulfate and fell into a distinct phylogenetic cluster of marine alpha-Proteobacteria. Phylogenetically and physiologically, these sediment strains resembled the sulfate-producing thiosulfate oxidizers from the Galapagos hydrothermal vents while showing habitat-related differences in growth temperature, rate and extent of thiosulfate utilization, and carbon substrate patterns. The abyssal deep-sea sediments yielded predominantly base-producing thiosulfate-oxidizing isolates related to Antarctic marine Psychroflexus species and other cold-water marine strains of the Cytophaga-Flavobacterium-Bacteroides phylum, in addition to gamma-proteobacterial isolates of the genera Pseudoalteromonas and Halomonas-Deleya. Bacterial thiosulfate oxidation is found in a wide phylogenetic spectrum of Flavobacteria and Proteobacteria.
Recent microbial surveys of marine offshore and deep-sea sediments have revealed considerable bacterial biodiversity in this extreme marine environment; cultivated isolates from deep-sea trench sediments include members of the actinomycetes, of the low guanine-plus-cytosine gram-positive bacteria, and of alpha- and gamma-Proteobacteria (66, 67). Heterotrophic, aerobic, or denitrifying deep-sea isolates, mostly barophiles, from the Atlantic and Pacific Oceans belong to the gamma-proteobacterial genera Colwellia, Moritella, and Shewanella (14). Anoxic deep sediment layers have yielded nitrate reducers, fermentative bacteria, and sulfate reducers (46). Isolations from deep-sea hydrothermal vents frequently focused on the microbial populations of the sulfur cycle. Reduced sulfur species of hydrothermal origin favor abundant microbial populations of mesophilic sulfur oxidizers, including autotrophic Thiomicrospira and Thiobacillus species (17, 30, 54, 78) as well as heterotrophic sulfur-oxidizing strains (18, 54).
The bacterial communities of hydrothermal vent sites and of nonhydrothermal water columns and sediments have at least one significant bacterial type in common, the loosely defined group of the heterotrophic sulfur-oxidizing bacteria (18, 69). These heterotrophic bacteria can oxidize reduced sulfur compounds, but do not depend on this reaction for growth. Two groups with different pH responses can be distinguished, the acid-producing and the base-producing thiosulfate oxidizers (54, 69). In both cases, the pH change is generally in the range of 0.5 to 1.5 pH units (54). Some base-producing strains studied in detail oxidized thiosulfate as an auxiliary electron donor to tetrathionate; this allowed them to use a greater portion of available organic carbon for biosynthesis rather than for respiration (68, 71). No CO2 fixation could be demonstrated for base-producing strains. The acid-producing hydrothermal vent strain TB66 showed increased cell yields in thiosulfate-amended cultures (29). The acid-producing strain NF18 showed thiosulfate-stimulated CO2 assimilation during growth on mineral medium (54); in this regard, it resembled the chemolithotrophic genera Thiobacillus or Thiomicrospira which oxidize thiosulfate completely to sulfate (54).
Due to their ecophysiological flexibility, thiosulfate-oxidizing heterotrophs are very widespread in the marine environment and have been isolated from a wide range of marine habitats, i.e., coastal waters of Vineyard Sound, the open North Atlantic, and the chemoclines of the Black Sea and the Cariaco Basin (31, 60, 69, 70). These isolations have rarely been linked to quantification of these bacteria and comparisons of their population density in different habitats. With the exception of incomplete sequences for three alpha- and gamma-proteobacterial thiosulfate oxidizers (strains NF18, AG33, and NF13) from the Galapagos hydrothermal vents (35), phylogenetic identifications of cultured marine isolates are, for the most part, missing.
Culture characteristics and phylogenetic affiliation of newly isolated thiosulfate-oxidizing bacteria from two North Atlantic sediment locations, and of previously isolated Galapagos hydrothermal vent strains (29, 54), were investigated in order to identify possible habitat-related common denominators as well as recurring differences in physiology and phylogenetic identity of these bacteria.
MATERIALS AND METHODS
Sampling and sediment characterization.
A 50- by 50-cm box corer was used to retrieve sediment samples of red clay from the North Atlantic abyssal plain at a depth of 4,500 m (37°29′N, 68°50′W) and to retrieve olive-colored silty sediment from the continental slope off New England at a depth of 1,500 m (39°50′N, 70°14′W). A box core with undisturbed sediment surface was used to obtain subcores (15-cm diameter, 40-cm depth) for further analysis. The cores were kept at 4°C, and sediment surfaces remained covered with ca. 0.5 cm of overlying water. The extent of the oxic zone, which defined the sampling scheme, was measured with oxygen electrodes on the ship and in the shore laboratory after 36 h, with identical results (52). Pore water was extracted within 48 h under N2 by using a small-scale pore water squeezer (53). Sulfide was measured with the methylene blue method immediately after pore water extraction (10). Organic carbon content and organic carbon (Corg)/total nitrogen (Ntotal) ratios were determined from acid-dried sediment with an EA 1108 Fisons Elemental Analyzer (Fisons Instruments, Inc., Beverly, Mass.).
Microbiological media.
Thiosulfate medium for sulfur-oxidizing bacteria contained (per liter): 25 g of NaCl, 1.0 g of (NH4)2SO4, 1.5 g of MgSO4 × 7H2O, 0.42 g of KH2PO4, 0.3 g of KCl, 0.2 g of NaHCO3, 0.29 g of CaCl2 × 2H2O, 1.58 g of Na2S2O3, 1 ml of SL-8 trace element solution (5), 1 ml of vitamin solution (2 mg liter−1 each of biotin and folic acid; 5 mg liter−1 each of niacine, panthothenate, lipoic acid, p-aminobenzoic acid, thiamine, riboflavin, pyridoxine, and cobalamin) (3). The NaHCO3 addition to the salt base imitates the natural carbonate-buffering system of seawater and was instrumental in isolating new sulfate-reducing bacteria (77) and sulfur-oxidizing bacteria (74). Two milliliters of 0.5% phenol red solution was added per liter of medium as a pH indicator. The pH was adjusted to 7.3, and the medium was filter sterilized with 0.2-μm-pore-size filters. Liquid medium was used for determining the most probable numbers (MPNs). Oxygen limitation in the MPNs, which could prevent complete oxidation of thiosulfate to sulfate (73), was ruled out by capping the MPN vials with very loosely fitting plastic caps that allowed gas exchange. Thiosulfate medium was used as liquid MPN medium or for 1.5% agar plates. Sulfate-free thiosulfate medium was prepared with 0.4 g of NH4Cl and 1.24 g of MgCl2 × 6H2O instead of the corresponding sulfates; this medium was used for quantification of sulfate produced by thiosulfate oxidation. Negative control plates did not contain thiosulfate, but otherwise contained the same salts, vitamins, trace elements, and pH indicator.
Media for MPN counts of sulfate-reducing bacteria were prepared with artificial seawater base (per liter, 25 g of NaCl, 5.67 g of MgCl2 × 6H2O, 6.8 g of MgSO4 × 7H2O, 1.47 g of CaCl2 × 2H2O, 0.19 g of NaHCO3, 0.66 g of KCl, and 0.09 g of KBr) and the following additions as described previously (77): 1 ml of nonchelated trace element mixture no. 1, 1 ml of selenite-tungstate solution, 30 ml of 1 M NaHCO3 solution, 1 ml of vitamin mixture, 1 ml of thiamine solution, 1 ml of vitamin B12 solution, and 7.5 ml of 0.2 M Na2S solution. As carbon sources, 20 mM lactate or 10 mM acetate were added. Medium was prepared anaerobically under an 80% N2-20% CO2 (vol/vol) gas phase. Glass culture tubes containing 9 ml of medium were gassed with 80% N2-20% CO2 (vol/vol) by using a gassing syringe according to the Hungate technique (77) before sealing the tubes with butyl rubber stoppers.
Quantifications and isolations.
MPN counts of sulfate-reducing and thiosulfate-oxidizing bacteria were done in parallel; sulfate-reducing bacteria are a source of reduced sulfur compounds in the sediment. MPN dilutions were performed in triplicate as previously described (1) with defined media for sulfate-reducing and for thiosulfate-oxidizing bacteria; the series included six 10-fold dilution steps. MPN dilution series for both media were inoculated in parallel with 0.1 ml of oxic surface sediments from the 0- to 1-cm layer at both sampling sites and with 0.1 ml of sediment from the microoxic 1- to 2-cm layer of the slope sediment. This definition of the oxic sediment layers was based on oxygen profiles of the freshly harvested sediment cores, prior to core sectioning and MPN inoculation. The MPNs for sulfate-reducing bacteria were judged positive if sulfide production, checked by the CuSO4 test (77), coincided with visible cell growth.
The thiosulfate MPN had to be evaluated differently. Changes of pH were used as a preliminary indicator of thiosulfate-oxidizing activity. Bacterial growth never reached the level of visible turbidity; the indicators of microbial activity were pH changes and microscopically visible cells obtained by centrifuging 100- to 500-μl portions of the medium. Over the course of 2 months, different bacterial populations developed in the liquid samples that increased or lowered the pH. An interpretation of all pH changes as positive would have been simplistic and of no value to account for the obviously diverse bacterial populations in these samples. Therefore, pure cultures were isolated individually from the highest dilutions which showed distinct pH changes. This approach allows the identification and cultivation of numerically dominant, but slow-growing or fastidious, bacteria that would be overgrown in enrichment culture (32). In order to maximize the diversity of the isolates, fully aerobic and microoxic-nitrate-reducing incubation regimens were chosen. Two 100-μl samples of each MPN sample were streaked on two thiosulfate agar plates. The first thiosulfate plate of each pair was incubated under fully aerobic conditions at 15°C. The second thiosulfate plate contained, in addition, 0.2 g of KNO3 per liter. All nitrate plates were incubated at 15°C in a gas-tight incubation jar which had been flushed for 20 min with nitrogen gas, but without removing small amounts of residual oxygen (for example, the small amount dissolved in the agar plates) by reducing agents. Strains obtained in this way are marked with an asterisk (e.g., alpha-proteobacterial strains DI4*, EI1*, and DIII4*).
Individual colonies of all strains were obtained by restreaking on agar plates and continued incubation. The plates were checked for pH changes and thiosulfate oxidation to sulfate. Instead of generalized MPN values, the dilution step from which each thiosulfate-oxidizing isolate was obtained is given subsequently. The characteristic pH changes of these isolates during growth on thiosulfate plates were not observed on parallel thiosulfate-free control plates.
Origin of vent strains.
The hydrothermal vent strains were isolated from different locations and sample materials at the Galapagos hydrothermal vents in 1979 (E. Ruby, personal communication) and were enriched on thiosulfate mineral media made with aged seawater and adjusted to pH 7.2 or 8 (54). Strain AG33 was obtained from the filtrate of a water sample at the Garden of Eden site characterized by abundant vent fauna. Strain TB66 was obtained from an unspecified Galapagos vent sample. Strain NF18 was isolated from Beggiatoa mat material and was enriched in thiosulfate medium where NH4Cl was omitted to select for nitrogen-fixing isolates (54). The aged seawater used for these thiosulfate mineral media contained small amounts of organic substrates, similar to the agar-solidified thiosulfate medium used for the North Atlantic sediment isolates.
Phenotypic testing.
Heterotrophic growth on different carbon sources was tested on thiosulfate-free agar plates with 10 mM carbon source. Growth was compared to negative controls without additional carbon source. The plates were incubated for a month at 15°C and were checked at intervals of 1 to 4 days. Growth at different temperatures (4, 15, 24, 37, and 42°C) was tested on marine 2216 heterotrophic agar plates (Difco, Detroit, Mich.); the plates were checked for growth for 10 days, but the results did not change after 3 or 4 days. The ability to use nitrate as electron acceptor was checked by stab inoculation of marine 2216 soft agar tubes (0.3% wt/vol) containing 0.1% (wt/vol) KNO3. Nitrite production was checked with alpha-naphthylamine; denitrification was considered to have occurred when gas bubbles in the agar coincided with the disappearance of nitrate according to the zinc powder test (22).
Sulfate production from thiosulfate was measured by streaking strains on sulfate-free agar slants containing 10 mM thiosulfate and phenol red as pH indicator. This strategy became necessary since attempts to grow the strains in liquid mineral medium turned out to be extremely time-consuming. pH changes indicative of thiosulfate-oxidizing activity took several months, while they took 1 to 2 weeks on agar slants. Traces of organic carbon compounds in the agar facilitated growth and accelerated the thiosulfate oxidation process. Since organic supplements in high concentration sometimes inhibited thiosulfate oxidation (see Results), oligotrophic, unsupplemented agar slants were a reasonable working compromise. Agar slants also allowed observation of the developing bacterial lawn on the agar surface in order to verify growth, whereas liquid media did not show increased turbidity or accumulation of a visible bacterial pellet. Each strain was streaked out in triplicate. The pH decrease was monitored by following the color change of the pH indicator until no further pH change occurred. Agar portions of 1 ml were excised from directly underneath the culture surface and were mixed overnight with 2 ml of sterile deionized water; the resulting 1:3 dilution of the medium was centrifuged to spin down and remove the agar before ion chromatography. After the samples for sulfate determination had been excised, the final pH of the agar slants was checked with a pH electrode. The isolate DIII1c, which did not change the pH and did not produce sulfate from thiosulfate, was included as a negative control.
Sulfate was detected by high-pressure liquid chromatography with indirect UV detection as described previously (51). A 50-μl volume of the culture medium was injected by using a Rheodyne valve (sample loop volume, 25 μl). Separation of anions was performed on a Polyspher IC AN-1 anion-exchange column (Merck). Indirect UV was measured at 254 nm in a Merck/Hitachi-L-4250-UV/VIS detector. An L-6210 intelligent pump connected to the UV/VIS detector was used. Flow rate for the isocratic eluent (1.5 mM phthalic acid, 1.38 mM Tris-hydroxymethyl-aminomethane, 300 mM boric acid, pH 4.0) was 1.3 ml min−1. Calibrations were carried out several times (n = 3) with dilutions of a MgSO4 standard (312 μM SO42−) in the range of 0 to 312 μM sulfate. The retention time for sulfate under these conditions was 11.55 min. All samples were diluted 1:25 with double-distilled water before injection into the high-pressure liquid chromatography. All sulfate determinations were done in triplicate for each culture tested.
DGGE.
The isolates were divided into similarity groups by phenotypic comparisons and by denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA gene fragments. DGGE separates double-stranded DNA fragments of identical length but different primary sequence, based on different melting domain structure and denaturation stability in a denaturing gradient. DGGE fragments of identical gel position most likely share identical sequences, although different DGGE fragments can match in their positions by chance. Pure cultures were analyzed by DGGE of PCR-amplified 16S rRNA gene fragments, using PCR primers GM5 (with guanosine-cytosine clamp) and 907R (41). DGGE was performed with a Bio-Rad Protean II system. A 20 to 70% denaturing gradient was used for all experiments. A value of 100% corresponds to 7 M urea and 40% (vol/vol) formamide (41). Electrophoresis was continued for 20 h at 100 V and 60°C. After electrophoresis, the gels were stained in aqueous ethidium bromide solution (0.5 μg liter−1) and photographed on a UV (302 nm) transillumination table with a Cybertech CS1 digital camera (Cybertech, Berlin, Germany).
Sequencing and phylogenetic analysis.
Representative strains were phylogenetically identified by 16S rRNA sequencing. With pure culture DNA as template, 16S rRNA genes were PCR-amplified as described (42) and were sequenced directly by using the Taq Dyedeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, Calif.). The sequence reactions were electrophoresed on an Applied Biosystems 373S DNA sequencer. Sequences were aligned by using the alignment editor SeqPup (23). Jukes-Cantor distance trees were calculated and bootstrapped with 200 resamplings by using the PHYLIP software package 3.57c (19). For parsimony bootstrap, 200 resamplings were performed with Paup 3.1.1 (65). The partially sequenced thiosulfate-oxidizing hydrothermal vent isolates AG33 and NF18 (35) were resequenced and included in the phylogenetic analysis. The thiosulfate-oxidizing hydrothermal vent strain TB66 (29) was newly sequenced and included in the phylogeny. Table 1 lists all isolates obtained in this study and GenBank numbers for sequenced strains. Near-complete 16S rDNA sequences used for the tree were retrieved from GenBank for the following organisms: Roseobacter denitrificans, M59063; Roseobacter litoralis, X78312; Sulfitobacter pontiacus, Y13155; Sargasso Sea isolate S34, U87407; Prionitis lanceolata gall symbiont, U37762; Octadecabacter arcticus, U73725; Octadecabacter antarcticus, U14583; Ruegeria gelatinovora, D88523; Marinosulfonomonas methylotropha, U62894; Roseovarius tolerans Y11551; Roseobacter algicola, X78315; Roseobacter gallaeciensis, Y13244; Sargasso Sea strain NFR, L15345; Antarctobacter heliothermus, Y11552; Sagittula stellata, U58356; Ruegeria atlantica, D88526; Silicibacter lacuscaerulensis, U77644; Paracoccus aminophilus, D32239; Paracoccus denitrificans, Y17511; Sargasso Sea clone SAR83, M63810; Pseudoalteromonas tetraodonis, X82139; Pseudoalteromonas haloplanktis, X67024; Alteromonas macleodii, X82145; Mariana trench sequence no. 1, D87345; Halomonas meridiana, M93356; Deleya aquamarina, M93352; Halovibrio variabilis, M93357; Deleya halophila, M93353; Halomonas elongata, M93355; Psychroflexus torquis, AF001365; Psychroflexus gondwanense, M92278; Flavobacterium salegens, M92279; Cytophaga latercula, D12665; Cytophaga lytica, M62796; Cytophaga marinoflava, D12668; Psychroserpens burtonensis, U62913; Gelidibacter algens, U62914; and Flexibacter maritimus, D14023.
TABLE 1.
List of isolates used in this studya
| Isolate | 16S rDNA sequence (GenBank. no.) |
|---|---|
| Alpha-Proteobacteria | |
| Abyssal strain AIII3 | AF254101 |
| Slope strain DI4 | AF254102 |
| Slope strain DII3 | |
| Slope strain DIII3 | |
| Slope strain DI4* | AF254104 |
| Slope strain EI2 | AF254103 |
| Slope strain DIII4* | AF254106 |
| Slope strain EI1* | AF254105 |
| Hydrothermal vent strain NF18 | AF254107 |
| Hydrothermal vent strain AG33 | AF254108 |
| Hydrothermal vent strain TB66 | AF254109 |
| Cytophaga-Flavobacterium | |
| Abyssal strain AIII4 | AF254114 |
| Abyssal strain AIII4* | |
| Abyssal strain AII4* | |
| Abyssal strain AII3 | AF254113 |
| Abyssal strain EI1* | |
| Gamma-Proteobacteria | |
| Abyssal strain AI4* | |
| Abyssal strain AIII3* | AF254110 |
| Abyssal strain AI3* | |
| Abyssal strain AI3 | |
| Abyssal strain AII2 | |
| Slope strain DII1a | AF254112 |
| Slope strain DII1b | |
| Slope strain DIII1 | |
| Slope strain DII2* | |
| Slope strain DIII1c | AF254111 |
Isolates are listed in the same order as they appear on the DGGE gel in Fig. 1.
RESULTS
Acid-producing thiosulfate-oxidizing bacteria.
The cultivation survey yielded different microbial populations from the continental slope and abyssal sediments; in general, heterotrophic, pH-increasing, and pH-decreasing thiosulfate-oxidizing bacterial isolates showed different occurrence patterns with respect to the sediment samples of their origin and the dilution from which they were isolated.
The most frequently and consistently found group of thiosulfate-oxidizing bacteria were heterotrophic isolates from 103- and 104-fold dilutions of the slope sediment which acidified the medium moderately by ca. 0.5 to 1.0 pH units (Table 2). The isolate AIII3 constitutes an exception; it was obtained from a 103-fold dilution of the abyssal plain sediment, and it acidified the medium by 1.7 to 1.8 U (Table 2). These acid-producing thiosulfate oxidizers showed phylogenetic and physiological similarities to the previously isolated hydrothermal vent thiosulfate oxidizers, and were therefore studied in more detail than the base-producing strains. Sulfate production from thiosulfate was a consistent feature of these isolates, although the extent of sulfate production varied considerably from strain to strain. Over 20 days of aerobic incubation at 15°C, they produced ca. 2 to 4.6 mM sulfate, corresponding to oxidation of approximately 10 to 23% of the original 10 mM thiosulfate in the medium to sulfate (Table 2). Parallel incubations on unsupplemented and 10 mM pyruvate-supplemented agar slants show different thiosulfate oxidation patterns for these sediment strains; in some cases, pyruvate inhibits thiosulfate oxidation and sulfate production, indicating that these strains prefer organic substrates over inorganic electron donors such as thiosulfate. On the other hand, the isolates DIII4* and EI1* show greatly increased thiosulfate oxidation after pyruvate supplementation, indicating that these organisms use thiosulfate and organic substrates in parallel (Table 2). The sediment isolates grew aerobically or by nitrate reduction. Strains DIII4* and EI1* reduced nitrate completely to nitrogen gas and grew much better in nitrate-reducing agar shake cultures than on plates incubated aerobically. The strains DII3 and DIII3 grew as aerobes, but could also reduce nitrate to nitrite (Table 2). All sediment strains grew at 15 or 24°C, but not at 37 or 42°C (Table 2). The substrate spectra of the sediment strains show common denominators (Table 3): organic acids and amino acids such as acetate, propionate, butyrate, glutamate, and proline and citric acid cycle intermediates can serve as sole carbon sources, but sugars generally cannot. The only exception observed so far is the C5-monomer of the common plant and bacterial cell wall component xylan, xylose, which supports growth of strains AIII3, DI4, DII3, and DIII3.
TABLE 2.
Comparison of sediment and hydrothermal vent isolates of the marine alpha-proteobacterial cluster
| Strain | Origin of sample | Dilution (ml−1) at point of isolation | Respiration substratea | NO3− reduction | Growth temperature (°C) | SO42− produced (mM)b | pH decrease on S2O32−c | SO42− produced (mM) without and with 10 mM pyruvated | DGGE group |
|---|---|---|---|---|---|---|---|---|---|
| AIII3 | 0-1 cm abyssal sed.e | 10−3 | O2 | 15, 24 | 4.54 | 1.7–1.8 | 4.2/5.05 | I | |
| DI4 | 0-1 cm slope sed. | 10−4 | O2 | 15, 24 | 4.46 | 0.7–1.0 | 2.41/2.49 | II | |
| DII3 | 0-1 cm slope sed. | 10−3 | O2, NO3− | NO2− | 15, 24 | 2.26 | 0.8–0.9 | 2.24/1.04 | II |
| DIII3 | 0-1 cm slope sed. | 10−3 | O2, NO3− | NO2− | 15, 24 | 1.97 | 0.6–0.7 | 2.55/1.12 | II |
| EI2 | 1-2 cm slope sed. | 10−2 | O2 | 15, 24 | 2.98 | 0.6–0.7 | 1.71/0.2 | III | |
| DI4* | 0-1 cm slope sed. | 10−4 | O2 | 15, 24 | 4.46 | 0.5–0.6 | 1.97/1.22 | III | |
| EI1* | 1-2 cm slope sed. | 10−1 | (O2), NO3− | N2 | 15, 24 | 2.28 | 0.7–0.8 | 1.14/7.31 | IV |
| DIII4* | 0-1 cm slope sed. | 10−4 | (O2), NO3− | N2 | 15, 24 | 2.14 | 0.6–0.7 | 0.44/4.76 | IV |
| NF18 | Beggiatoa mat, Galapagos vents | O2, NO3− | NO2− | 15, 24, 37, 42 | 4.38 | 1.5–1.6 | 3.51/9.29 | V | |
| AG33 | Water sample, Galapagos vents | O2, NO3− | NO2−f | 15, 24, 37, 42 | 3.45 | 1.2–1.3 | 3.80/7.22 | VI | |
| TB66 | Galapagos vent | O2 | 15, 24, 37, 42 | 4.98 | 1.5–1.6 | 4.92/9.93 | VII |
Parentheses indicate poor growth under air.
Average of three parallel determinations after 20 days incubation.
The pH of sterile controls was 7.7 and increased by 0.1 U. The control with the nonthiosulfate oxidizer DIII1c increased by 0.2 pH units.
Separate experiment; sulfate was determined after 14 days for one sample without pyruvate (left) and one with pyruvate (right).
Sed., sediment.
Traces of NO2−; no N2.
TABLE 3.
Substrate spectra for alpha-Proteobacteria
| Isolate | Growth ona:
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fo | Ac | Pp | Bu | bB | Py | La | Ci | aK | Sc | Mt | Ta | Gl | Ga | Fr | Ri | Xy | Su | Ml | Lc | Gl | Pr | Mi | Gy | Et | DGGE group | |
| AIII3 | + | + | + | + | − | + | + | − | + | + | + | − | + | − | + | + | + | − | b | b | + | + | + | + | − | I |
| DI4 | + | + | + | + | − | + | + | + | + | + | + | − | + | b | + | − | + | − | b | − | + | + | b | b | − | II |
| DII3 | + | + | + | + | − | + | + | b | + | + | + | − | b | − | − | − | + | − | b | − | + | + | − | + | − | II |
| DIII3 | + | + | + | + | − | + | + | + | + | + | + | − | b | − | − | − | + | − | b | − | + | + | − | b | − | II |
| EI2 | − | + | + | + | − | + | + | − | + | + | + | − | − | − | − | − | − | − | − | − | + | + | − | b | − | III |
| DI4* | − | + | + | + | − | + | + | − | + | + | + | − | − | − | − | − | − | − | − | − | + | + | − | b | − | III |
| EI1* | − | b | b | + | − | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | + | + | − | − | − | IV |
| NF18 | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | − | V |
| AG33 | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | − | + | + | + | + | + | + | + | − | VI |
| TB66 | + | + | + | + | + | + | + | − | + | + | + | − | b | − | + | b | − | + | + | − | + | + | + | + | − | VII |
+, good growth; +, weaker growth; b, near background growth; −, negative growth. Very poor growth occurred on all substrates for DIII4* under air during repeated tries; therefore, no data are given. Abbreviations for substrates (all 10 mM in mineral agar) are as follows: Fo, formate; Ac, acetate; Pp, propionate; Bu, butyrate; bB, beta-hydroxybutyrate; Py, pyruvate; La, lactate; Ci, citrate; aK, alpha-ketoglutarate; Sc, succinate; Mt, malate; Ta, tartrate; Gl, glucose; Ga, d-galactose; Fr, fructose; Ri, d-ribose; Xy, xylose; Su, sucrose; Ml, maltose; Lc, lactose; Gl, glutamate; Pr, l-proline; Mi, d-mannitol; Gy, glycerol; Et, ethanol.
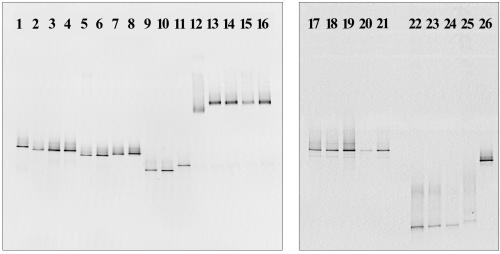
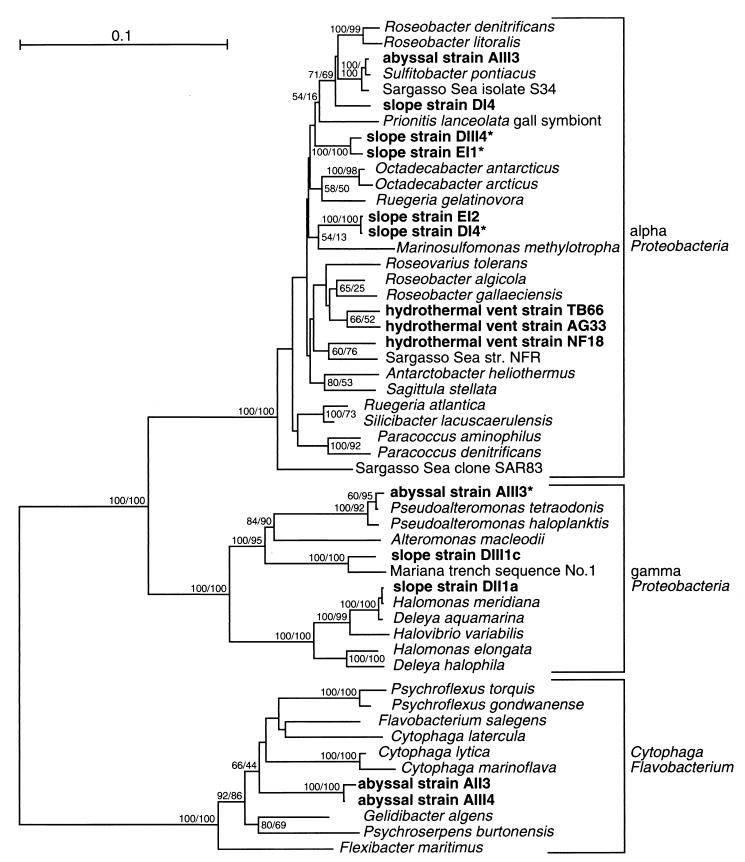
According to their DGGE patterns, the acid-producing strains from the sediments fell into four different 16S rRNA sequence groups: the abyssal isolate AIII3 (DGGE group I); the three slope isolates DI4, DII3, and DIII3 (DGGE group II); the two denitrifying slope isolates DIII4* and EI1* (DGGE group III); and the two slope isolates EI2 and DI4* (DGGE group IV) (Fig. 1, lanes 1 to 8). The DGGE band positions of the hydrothermal vent strains differed from the sediment isolates (Fig. 1, lanes 9 to 11). The acid-producing isolates from the sediments as well as the hydrothermal vents belonged to the marine alpha cluster (Fig. 2), a large phylogenetic group within the alpha-Proteobacteria consisting of mostly heterotrophic, aerobic marine bacteria. Within this large group, some smaller branches could be differentiated. Strain AIII3 was by 16S rDNA sequence almost identical to the heterotrophic thiosulfate and sulfite oxidizer S. pontiacus (62); together with the heterotrophic Sargasso Sea isolate S34 (64), they formed a clade with 100% bootstrap support. Strain DI4, representative of DGGE group II, formed a clade (71 and 69% distance and parsimony bootstrap, respectively) together with the Sulfitobacter branch and the facultative phototrophs R. litoralis and R. denitrificans (57). The two well-supported clades formed by the denitrifying slope isolates DIII4* and EI1* (DGGE group III), and the strictly aerobic strains EI2 and DI4* (DGGE group IV), were not specifically related to other members of the marine alpha-Proteobacteria (Fig. 2).
FIG. 1.
DGGE comparison of sediment isolates, including alpha-Proteobacteria (lanes 1 to 11), Flavobacteria (lanes 12 to 16), Pseudoaltermonas (lanes 17 to 21), and Halomonas isolates (lanes 22 to 25) on two 30 to 70% DGGE gels. Sediment alpha-Proteobacteria are separated as follows: AIII3, lane 1; DI4, lane 2; DII3, lane 3; DIII3, lane 4; DI4*, lane 5; EI2, lane 6; DIII4*, lane 7; and EI1*, lane 8. Hydrothermal vent alpha-Proteobacteria are separated as follows: NF18, lane 9; AG33, lane 10; and TB66, lane 11. Flavobacteria are separated as follows: AIII4, lane 12; AIII4*, lane 13; AII4*, lane 14; AII3, lane 15; and EI1*, lane 16. Pseudoalteromonas are separated as follows: AI4*, lane 17; AIII3*, lane 18; AI3*, lane 19; AI3, lane 20; and AII2, lane 21. Halomonas and others are separated as follows: DII1a, lane 22; DII1b, lane 23; DIII1, lane 24; DII2*, lane 25; and DIII1c, lane 26.
FIG. 2.
Phylogeny of deep-sea isolates and related strains, based on 16S rDNA positions 38 to 1482. The root was placed between the Flavobacteria and the Proteobacteria. Bootstrap values are given for nodes which have at least 50% bootstrap support by distance (first value) or parsimony bootstrap (second value). The scale bar corresponds to 0.1 Jukes-Cantor substitutions per nucleotide.
The three hydrothermal vent isolates, AG33, TB66, and NF18, were members of the marine alpha cluster like the sediment isolates (Fig. 2). Their higher temperature range and thiosulfate oxidation capability reflect their hydrothermal vent origin and distinguish them from the sediment isolates. The vent strains grew at 37 and 42°C, temperatures beyond the range of the sediment strains (Table 2). They produced 3.5 to 5 mM sulfate from thiosulfate, corresponding to oxidation of approximately 17 to 25% of the available thiosulfate in the medium (Table 2). They acidified the agar slants by approximately 1.5 pH units within 2 or 3 days, indicating faster thiosulfate oxidation than the sediment strains which slowly lowered the pH over approximately 2 weeks. The addition of 10 mM pyruvate at least doubled the oxidation of thiosulfate to sulfate for all three hydrothermal vent isolates (Table 2). The hydrothermal vent strains utilized a wider substrate spectrum (including sugars) than the sediment strains (Table 3) and showed visible growth after 1 to 4 days, in contrast to at least 1 week for the sediment strains.
Base-producing thiosulfate-oxidizing bacteria.
Most base-producing strains were isolated from 103- and 104-fold dilutions in the abyssal sediment. In contrast to the alpha-proteobacterial strains from the slope sediment, they did not belong to a single phylogenetic branch, but were affiliated with three different groups: the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum, the gamma-proteobacterial genus Pseudoalteromonas, and the closely intertwined gamma-proteobacterial genera Deleya and Halomonas. After growth on sulfate-free thiosulfate agar slants for 20 days under the same conditions as the acid-producing thiosulfate oxidizers, sulfate production was found for representatives of the CFB group and of Deleya-Halomonas, but not for Pseudoalteromonas.
CFB phylum.
Five isolates of the CFB phylum were obtained. Three strains came from 10−4 dilutions and one came from a 10−3 dilution of the surface 1 cm of the abyssal sediment (AIII4, AIII4*, AII4*, and AII3, respectively); a single isolate was obtained from a 10−1 dilution of the 1- to 2-cm layer of the slope sediment (EI1*). These isolates were conspicuous by slightly increasing the pH of thiosulfate plates by approximately 0.5 pH units. Strain AII3 was tested for thiosulfate oxidation and produced 2.5 mM sulfate. All isolates were strictly aerobic and did not reduce nitrate. They showed intense yellow coloration, in contrast to all other strains in this study, which were unpigmented. With the exception of abyssal strain AIII4, all isolates showed the same DGGE band (Fig. 1, lanes 12 to 16). The 16S rDNA of a representative of this common DGGE type, strain AII3, and the deviating strain AIII4 were sequenced. The isolates were closely related to each other (100% bootstrap) and belonged to a well-supported clade (92% distance and 86% parsimony bootstrap) of marine CFB species mostly from cold-water habitats, including the halophilic, antarctic species F. salegens, P. gondwanense, and P. torquis (4, 16) (Fig. 2).
Pseudoalteromonas.
Five Pseudoalteromonas isolates (AI4*, AIII3*, AI3*, AI3, and AII2) were obtained, in this order, from one 104-, three 103-, and one 102-fold dilution of the surface 1-cm layer of abyssal sediment. All Pseudoalteromonas isolates showed identical DGGE patterns (Fig. 1, lanes 17 to 21). The 16S rDNA sequence of isolate AIII3* was determined and was found to be nearly identical to the 16S rDNA sequence of P. tetraodonis (21) (Fig. 2). The Pseudoalteromonas isolates raised the pH of thiosulfate plates by 0.7 to 0.8 pH units. Strain AIII3* was tested for thiosulfate oxidation; sulfate was not found after growth on thiosulfate agar slants. The Pseudoalteromonas strains were strictly aerobic, did not reduce nitrate, and formed unpigmented colonies.
Deleya-Halomonas and others.
In contrast to the CFB and Pseudoalteromonas isolates from high dilutions of the abyssal sediment, the base-producing isolates (DII1a, DII1b, DIII1, and DII2*) were obtained from 10−1 and 10−2 dilutions of the slope sediment surface layer. These isolates differed from all others by rapidly increasing the pH of the agar plates from ca. 7.5 to more than 8.5, leading to a deep-purple coloration of the phenol red indicator, and by rapid growth and colony formation within 1 or 2 days on thiosulfate and heterotrophic plates. The isolates DII1a and DII2* were tested for thiosulfate oxidation and formed ca. 1.8 to 2 mM sulfate. The 16S rRNA sequence of strain DII1a was nearly identical to the sequences of D. aquamarina and of H. meridiana (Fig. 2), two closely related species of the phenotypically and phylogenetically intertwined genera Deleya and Halomonas which could be combined into a single genus (15). Variability was found with respect to nitrate reduction and DGGE position (Fig. 1, lanes 22 to 25), the strains DII1a and DII1b grew anaerobically and reduced nitrate to nitrite, and strain DII2* did not reduce nitrate and showed a different DGGE position.
From MPN sample DIII1, an additional strain (DIII1c) was isolated which did not change the pH of thiosulfate plates and did not produce any detectable sulfate from thiosulfate. This bacterium was the closest relative of a molecular isolate (no. 1) from sediments of the Mariana Trench (33) and showed a DGGE band distinct from the Halomonas isolates (Fig. 1, lane 26).
Sediment characteristics and sulfate reduction.
The two sample sediments differed substantially in organic carbon and C/N ratios. The slope sediment showed an organic carbon content of 1.7 to 1.8% organic C per dry weight of sediment and a Corg/Ntotal ratio of 9 to 9.5 for the upper-5-centimeter layers; the abyssal sediment had an organic carbon content of 0.58 to 0.62% and a Corg/Ntotal ratio of 6.5 to 7.5. After microbial consumption of easily degradable organic matter, clay-bound residues with low Corg/Ntotal ratios are left behind and not further degraded by bacteria (39). In comparison to the slope sediment, the abyssal sediment was more depleted of microbially utilizable substrates. The organic carbon and C/N ratios of the samples were characteristic of continental slope and abyssal sediments in the North Atlantic (12, 58) and indicate that the samples were representative for these sediment environments. The activity of the reductive sulfur cycle in these sediments was evaluated by MPN counts of sulfate-reducing bacteria and by determinations of pore water sulfate and sulfide concentrations. Sulfate-reducing bacteria (SRB) were found in the surface layers of the continental slope sediment samples that also yielded the majority of the alpha-proteobacterial isolates. The surface and the 1- to 2-cm sediment layers yielded approximately 100 cultivable SRBs ml−1 with lactate, and the 1- to 2-cm layer yielded, in addition, approximately 20 cultivable SRB ml−1 with acetate. The MPN counts were performed in the surface layers, since marine sediments often show the highest sulfate reducer densities around the oxycline near the sediment surface (at ca. 1-cm depth in the slope sediment). Here, high numbers of SRB coincide with rapid oxidation, reduction, and disproportion of thiosulfate, sometimes performed by the SRBs themselves (32). MPN counts in the abyssal sediment were unsuccessful; the population density of cultivable SRB was too low for MPN quantification. Free sulfide in the pore water and dark-gray sediment layers indicative of sulfide precipitates or pyrite were absent from both sediments. Pore water sulfate remained at seawater concentration (ca. 28 mM) and showed no sign of depletion in both cores, with bioturbation being absent. These data indicate low sulfate reduction activity and a correspondingly low supply of reduced sulfur species for thiosulfate-oxidizing bacteria.
DISCUSSION
Hydrothermal vent and sediment strains.
The acid-producing alpha-proteobacterial thiosulfate oxidizers from hydrothermal vents and sediments are phylogenetically related and physiologically similar; their differences appear to be in degree rather than in principle and show the imprint of the two environments. Under low-nutrient conditions on unsupplemented agar slants as well as under high carbohydrate load, the hydrothermal vent strains oxidized more thiosulfate to sulfate than most of the sediment strains. Visible acidification by thiosulfate oxidation as well as heterotrophic growth and colony formation could be seen after 2 days for the hydrothermal vent strains, but took 1 to 2 weeks for the sediment strains under equal conditions. The vent strains showed a larger substrate spectrum than the sediment strains, which included easily degradable sugars. Thus, the heterotrophic thiosulfate oxidizers can make versatile and quick use of the reduced sulfur and carbon sources in the hydrothermal vent environment: thiosulfate is supplied by reduced vent fluids or by chemical oxidation of metal sulfides (56). Dissolved organic carbon and particulate organic carbon levels are several hundred times more concentrated in water of vent areas, compared to normal deep-sea bottom water (8, 11). The abundant bacterial biomass growing on chimneys and sediments is also a likely source of organic nutrients (3, 28). This metabolic menu differs from the diagenetically altered and nutrient-depleted organic matter that reaches deep-sea sediments. For example, sinking organic matter retained only ca. 1% of its original content of carbohydrates, amino acids, and lipids and consisted mostly of uncharacterized refractory compounds after passage through a 1,000-m water column (76). Therefore, the limited substrate range of the thiosulfate oxidizers in the slope sediment, compared to the hydrothermal vent strains, most likely reflects the lack of undegraded, fresh substrates within the sedimentary organic carbon pool.
The reduced thiosulfate oxidation capacity of the sediment alpha-proteobacterial strains is consistent with low sulfate reduction activity and a low supply of reduced sulfur species. The microbial degradation of organic carbon from primary production and terrestrial input during sedimentation through the deep-water column severely limits organic substrate availability and, subsequently, the activity and density of anaerobic sulfate-reducing bacteria in the deep sea (2, 9).
Possible occurrence patterns.
The alpha-proteobacterial thiosulfate oxidizer populations in slope and abyssal sediments could follow nonrandom occurrence patterns. Generally, cloning and sequencing of environmental 16S rRNA genes showed the ubiquitous presence of marine alpha-Proteobacteria in the water column of all oceans (20, 40, 64) and in marine snow (49). They are especially abundant in coastal seawater. In a cloning survey, they accounted for a fifth of all bacterial clones obtained from the continental shelf near Cape Hatteras (48); in estuaries with near-marine salinity, they contributed 13 to 28% of the total amount of bacterial DNA (25). An important ecological role of several members of the marine alpha group is the decomposition of organic sulfur compounds produced mostly by eukaryotic phytoplankton (38) and salt marsh plants (45). Marine alpha group bacteria oxidize dimethylsulfoniopropionate and dimethylsulfide (26, 36) or are capable of degradation of complex plant-produced polymers such as lignin or humic substances (24). The thiosulfate oxidizers of the slope sediment could be derived from these coastal populations, although their location in outer continental slope sediments rules out active participation in coastal or pelagic cycling of organosulfur compounds (36, 74).
Members of the CFB phylum occur ubiquitously in the marine environment and are generally associated with the degradation of complex organic substrates (50). They represent one of the most frequently isolated bacterial groups from marine sediments, due to the relative ease of their isolation and their occurrence in high numbers (37, 43). The phylogenetic affiliation of the deep-sea isolates with a cluster of isolates from cold saline and marine environments (7) suggests that several related CFB species have specifically adapted to the conditions of the cold oceans, including the polar seas and the deep sea. As a caveat, an autochthonous deep-sea origin of the abyssal CFB strains is not proven; 16S rDNA sequences of CFB members have been obtained from marine snow particles (13, 49) which could carry these natural inoculates to the bottom sediments (44). A similar pattern could apply to the Pseudoalteromonas isolates. Previous marine surveys have identified Pseudoalteromonas as a frequently isolated bacterial genus in Antarctic sea ice and seawater (6) and in coastal North Atlantic waters (43). Molecular surveys of marine water columns have recovered Pseudoalteromonas 16S rDNA sequences, although not as frequently as marine alpha-proteobacterial or CFB sequences (40). The Deleya-Halomonas isolates were obtained from low dilutions of the slope sediment only (10−1 and 10−2 cells ml−1), indicating that their fast growth and easy isolation from diverse seawater, saline lake water, salted food, and marine biomass samples (34, 75) do not necessarily correspond to high cell densities in situ.
Phylogenetic diversity.
The 16S rRNA-based identification of cultured heterotrophic marine thiosulfate oxidizers puts a sharper focus on this group, with evolutionary and ecological implications. For example, the sediment isolates of the marine alpha-proteobacterial cluster, the most frequently isolated strains in this study, show an unexpected phylogenetic and physiological link with the microbial populations of hydrothermal vents. The hydrothermal vent thiosulfate oxidizers of the marine alpha-proteobacterial cluster, NF18, AG33, and TB66 (29, 54) are the evolutionary cousins of the widespread marine sediment or water column bacteria of the same group. This marine alpha-proteobacterial group is ecologically and physiologically highly diversified. In addition to the sediment and hydrothermal vent bacteria described here, this phylogenetic group also includes polar sea ice bacteria of the genus Octadecabacter (27); marine sediment-dwelling bacteria of the genus Ruegeria (55, 72); facultative phototrophs, i.e., R. litoralis and R. denitrificans (57); and organosulfur-oxidizing marine heterotrophs (24, 36). Without exception, all acid- and sulfate-producing thiosulfate oxidizers isolated in this study fall into this cluster. This strongly suggests a phylogenetic basis for the thiosulfate-oxidizing phenotype within this group.
The marine thiosulfate-oxidizing, base-producing species include members of the CFB phylum, and species of the gamma-proteobacterial genera Pseudoalteromonas and Halomonas- Deleya. A phenotypic study identified alkaliphilic, denitrifying, thiosulfate-oxidizing, tetrathionate-producing bacteria as members of the genus Deleya (61). In a phenotypic and 16S rDNA study, anaerobic tetrathionate-producing thiosulfate oxidizers were identified as members of Pseudomonas stutzeri genomovars (63). The default designation Pseudomonas (59, 68, 71) for these organisms does not reflect their actual diversity.
In short, the ability to oxidize thiosulfate and probably also other reduced sulfur compounds is widespread among different genera of heterotrophic alpha- and gamma-Proteobacteria. The frequent isolation of thiosulfate oxidizers from chemoclines of stratified marine water columns such as the Black Sea (31, 60) and the Cariaco Basin (70) and the high thiosulfate-oxidizing activity recently reported from coastal organic-matter-rich Baltic Sea sediments (47) demonstrate that these bacteria are ubiquitous catalysts in the marine sulfur cycle at oxic-anoxic interfaces.
ACKNOWLEDGMENTS
We thank the crew of the RV Oceanus and George Hampson for their expert handling of the boxcorer and their determination to obtain good samples. We also thank all cruise participants, in particular Christian Knoblauch for moral and practical support, last but not least against encroaching seasickness.
Andreas Teske was supported by DFG postdoctoral fellowship 262-1/1 and a subsequent WHOI postdoctoral fellowship.
Footnotes
Contribution no. 10187 of the Woods Hole Oceanographic Institution.
REFERENCES
- 1.American Public Health Association. Standard methods for the examination of water and wastewater, including bottom sediments and sludge. Washington, D.C.: American Public Health Association; 1969. pp. 604–609. [Google Scholar]
- 2.Battersby N S, Malcolm S J, Brown C M, Stanley S O. Sulphate reduction in oxic and suboxic North-East Atlantic sediments. FEMS Microbiol Ecol. 1985;31:225–228. [Google Scholar]
- 3.Bazylinski D A, Wirsen C O, Jannasch H W. Microbial utilization of naturally occurring hydrocarbons at the Guaymas Basin hydrothermal vent site. Appl Environ Microbiol. 1989;55:2832–2836. doi: 10.1128/aem.55.11.2832-2836.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernardet J-F, Segers P, Vancanneyt M, Berthe F, Kersters K, Vandamme P. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga aquatilis Strohl and Tait 1978.) Int. J Syst Bacteriol. 1996;46:128–148. [Google Scholar]
- 5.Biebl H, Pfennig N. Growth yields of green sulfur bacteria in mixed cultures with sulfur and sulfate reducing bacteria. Arch Microbiol. 1978;117:9–16. [Google Scholar]
- 6.Bowman J P, McCammon S A, Brown M V, Nichols D S, McMeekin T A. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman J P, McCammon S A, Lewis T, Skerratt J H, Brown J, Nichols D S, McMeekin T A. Psychroflexus torquis gen. nov., sp. nov., a psychrophilic species from Antarctic sea ice, and reclassification of Flavobacterium gondwanense (Dobson et al. 1993) as Psychroflexus gondwanense gen. nov., comb. nov. Microbiology. 1998;144:1601–1609. doi: 10.1099/00221287-144-6-1601. [DOI] [PubMed] [Google Scholar]
- 8.Brault M, Simoneit B R T, Marty J C, Saliot A. Hydrocarbons in waters and particulate material from hydrothermal environments at the East Pacific Rise, 13°N. Org Geochem. 1988;12:209–219. [Google Scholar]
- 9.Canfield D E. Sulfate reduction in deep-sea sediments. Am J Sci. 1991;291:177–188. doi: 10.2475/ajs.291.2.177. [DOI] [PubMed] [Google Scholar]
- 10.Cline J D. Spectrophotometric determinations of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 11.Comita P B, Gagosian R B, Williams P M. Suspended particulate organic material from hydrothermal vent waters at 21°N. Nature. 1984;307:450–453. [Google Scholar]
- 12.Degens E T, Mopper K. Factors controlling the distribution and early diagenesis of organic material in marine sediments. In: Riley J P, Chester R, editors. Chemical oceanography. 2nd ed. Vol. 6. London, England: Academic Press; 1976. pp. 59–113. [Google Scholar]
- 13.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 14.DeLong E F, Franks D G, Yayanos A A. Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl Environ Microbiol. 1997;63:2105–2108. doi: 10.1128/aem.63.5.2105-2108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobson S J, McMeekin T A, Franzmann P D. Phylogenetic relationships between some members of the genera Deleya, Halomonas, and Halovibrio. Int J Syst Bacteriol. 1993;43:665–673. doi: 10.1099/00207713-43-4-665. [DOI] [PubMed] [Google Scholar]
- 16.Dobson S J, Colwell R R, McMeekin T A, Franzmann P D. Direct sequencing of the polymerase chain reaction-amplified 16S rRNA gene of Flavobacterium gondwanense sp. nov., and Flavobacterium salegens sp. nov., two new species from a hypersaline antarctic lake. Int J Syst Bacteriol. 1993;43:77–83. doi: 10.1099/00207713-43-1-77. [DOI] [PubMed] [Google Scholar]
- 17.Durand P, Reysenbach A, Prieur D, Pace N. Isolation and characterization of Thiobacillus hydrothermalis sp. nov., a mesophilic obligately chemolithoautotrophic bacterium isolated from a deep-sea hydrothermal vent in Fiji Basin. Arch Microbiol. 1993;159:764–766. [Google Scholar]
- 18.Durand P, Benyagoub A, Prieur D. Numerical taxonomy of heterotrophic sulfur-oxidizing bacteria isolated from southwestern Pacific hydrothermal vents. Can J Microbiol. 1994;40:690–697. [Google Scholar]
- 19.Felsenstein J. PHYLIP (phylogeny inference package), version 3.57c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 20.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauthier G, Gauthier M, Christen R. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int J Syst Bacteriol. 1995;45:755–761. doi: 10.1099/00207713-45-4-755. [DOI] [PubMed] [Google Scholar]
- 22.Gerhard P, Murray R G E, Wood W A, Krieg N R. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. [Google Scholar]
- 23.Gilbert D. SeqPup sequence editor version 0.5. Bloomington: Biology Department, Indiana University; 1995. [Google Scholar]
- 24.Gonzalez J M, Mayer F, Moran M A, Hodson R E, Whitman W B. Sagittula stellata gen. nov. sp. nov., a lignin-transforming bacterium from a coastal environment. Int J Syst Bacteriol. 1997;47:773–780. doi: 10.1099/00207713-47-3-773. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez J M, Moran M A. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez J M, Kiene R P, Moran M A. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl Environ Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gosinck J J, Herwig R P, Staley J T. Octadecabacter arcticus gen. nov. sp. nov., and O. antarcticus, sp. nov., nonpigmented, psychrophilic gas vacuolate bacteria from polar sea ice and water. Syst Appl Microbiol. 1997;20:556–365. [Google Scholar]
- 28.Hedrick D B, Pledger R D, White D C, Baross J A. In situ microbial ecology of hydrothermal vent sediments. FEMS Microbiol Ecol. 1992;101:1–10. [Google Scholar]
- 29.Jannasch H W, Wirsen C O. The biochemical versatility of chemosynthetic bacteria at deep-sea hydrothermal vents. Bull Biol Soc Wash. 1983;6:325–334. [Google Scholar]
- 30.Jannasch H W, Wirsen C O, Nelson D C, Robertson L A. Thiomicrospira crunogena, a colorless sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1985;35:422–424. [Google Scholar]
- 31.Jannasch H W, Wirsen C O, Molyneaux S J. Chemoautotrophic sulfur-oxidizing bacteria from the Black Sea. Deep-Sea Res. 1991;38(Suppl. 2):S1105–S1120. [Google Scholar]
- 32.Jørgensen B B, Bak F. Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark) Appl Environ Microbiol. 1991;57:847–856. doi: 10.1128/aem.57.3.847-856.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato C, Li L, Tamaoka J, Horikoshi K. Molecular analyses of the sediment of the 11000-m deep Mariana Trench. Extremophiles. 1997;1:117–123. doi: 10.1007/s007920050024. [DOI] [PubMed] [Google Scholar]
- 34.Kersters K. The genus Deleya. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer Verlag; 1991. pp. 3189–3197. [Google Scholar]
- 35.Lane D J, Harrison A P, Stahl D, Pace B, Giovannoni S J, Olsen G J, Pace N R. Evolutionary relationships among sulfur- and iron-oxidizing bacteria. J Bacteriol. 1992;174:269–278. doi: 10.1128/jb.174.1.269-278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ledyard K M, DeLong E F, Dacey J W H. Characterization of a DMSP-degrading bacterial isolate from the Sargasso Sea. Arch Microbiol. 1993;160:312–318. [Google Scholar]
- 37.Llobet-Brossa E, Rossello-Mora R, Amann R I. Microbial community composition of Wadden sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malin G, Kirst G O. Algal production of dimethyl sulfide and its atmospheric role. J Phycol. 1997;33:889–896. [Google Scholar]
- 39.Müller P J. C/N ratios in pacific deep-sea sediments: effect of inorganic ammonium and organic nitrogen compounds sorbed by clays. Geochim Cosmochim Acta. 1977;41:765–776. [Google Scholar]
- 40.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 41.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3rd ed. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. [Google Scholar]
- 42.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 43.Noble P A, Dabinett P E, Gow J A. A numerical taxonomic study of pelagic and benthic surface-layer bacteria in seasonally-cold coastal waters. Syst Appl Microbiol. 1990;13:77–85. [Google Scholar]
- 44.Novitzky J A. Evidence for sedimenting particles as the origin of the microbial community in coastal marine sediment. Mar Ecol Prog Ser. 1990;60:161–167. [Google Scholar]
- 45.Pakulski J D, Kiene R P. Foliar Release of dimethyl-sulfoniopropionate from Spartina alterniflora. Mar Ecol Prog Ser. 1992;81:277–287. [Google Scholar]
- 46.Parkes R J, Cragg B A, Bale S J, Getliff J M, Goodman K, Rochelle P A, Fry J C, Weightman A J, Harvey S M. Deep bacterial biosphere in Pacific Ocean sediments. Nature. 1994;371:410–413. [Google Scholar]
- 47.Podgorsek L, Imhoff J F. Tetrathionate production by sulfur-oxidizing bacteria and the role of tetrathionate in the sulfur cycle of Baltic Sea sediments. Aquat Microb Ecol. 1999;17:255–265. [Google Scholar]
- 48.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 49.Rath J, Wu K Y, Herndl G J, DeLong E F. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol. 1998;14:261–269. [Google Scholar]
- 50.Reichenbach H. The order Cytophagales. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer Verlag; 1991. pp. 3631–3675. [Google Scholar]
- 51.Rethmeier J, Rabenstein A, Langer M, Fischer U. Detection of traces of oxidized and reduced sulfur compounds in small samples by combination of different high-performance liquid chromatography methods. J Chromatogr A. 1997;760:295–302. [Google Scholar]
- 52.Revsbech N P, Jørgensen B B. Microelectrodes: their use in microbial ecology. Adv Microb Ecol. 1985;9:293–352. [Google Scholar]
- 53.Robbins J A, Gustinis J. A squeezer for efficient extraction of pore water from small volumes of anoxic sediment. Limnol Oceanogr. 1976;21:905–909. [Google Scholar]
- 54.Ruby E G, Wirsen C O, Jannasch H W. Chemolithotrophic sulfur-oxidizing bacteria from the Galapagos Rift hydrothermal vents. Appl Environ Microbiol. 1981;42:317–324. doi: 10.1128/aem.42.2.317-324.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rüger H-J, Höfle M G. Marine star-shaped-aggregate-forming bacteria: Agrobacterium atlanticum sp. nov.; Agrobacterium meteori sp. nov.; Agrobacterium ferrugineum sp. nov., nom. rev.; Agrobacterium gelatinovorum sp. nov., nom. rev.; and Agrobacterium stellulatum sp. nov., nom. rev. Int J Syst Bacteriol. 1992;42:133–143. doi: 10.1099/00207713-42-1-133. [DOI] [PubMed] [Google Scholar]
- 56.Schippers A, Sand W. Bacterial leaching of metal sulfides proceeds by two indirect mechanisms via thiosulfate or via polysulfides and sulfur. Appl Environ Microbiol. 1999;65:319–321. doi: 10.1128/aem.65.1.319-321.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shiba T. Roseobacter litoralis gen. nov., sp. nov., and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain bacteriochlorophyll a. Syst Appl Microbiol. 1991;14:140–145. [Google Scholar]
- 58.Smith K L, Jr, Hinga K R. Sediment community respiration in the deep sea. In: Rowe G T, editor. Deep-sea biology. 8. The sea: ideas and observations on progress in the study of the seas. New York, N.Y: John Wiley & Sons; 1983. pp. 331–370. [Google Scholar]
- 59.Sorokin D. Oxidation of thiosulfate to tetrathionate by heterotrophic bacteria from aquatic environments. Mikrobiologiya. 1992;61:756–773. [Google Scholar]
- 60.Sorokin D Y, Lysenko A M. Heterotrophic bacteria from the Black Sea oxidizing reduced sulfur compounds to sulfate. Microbiology. 1993;62:594–602. [Google Scholar]
- 61.Sorokin D Y, Lysenko A M, Mityushina L L. Isolation and characterization of alkaliphilic chemoorganoheterotrophic bacteria oxidizing reduced inorganic sulfur compounds to tetrathionate. Microbiology. 1996;65:326–338. [Google Scholar]
- 62.Sorokin D Y. Sulfitobacter pontiacus gen. nov., sp. nov.: a new heterotrophic bacterium from the Black Sea, specialized on sulfite oxidation. Microbiology. 1995;64:295–305. [Google Scholar]
- 63.Sorokin D Y, Teske A, Robertson L A, Kuenen J G. Anaerobic oxidation of thiosulfate to tetrathionate by obligately heterotrophic bacteria. FEMS Microbiol Ecol. 1999;30:113–123. doi: 10.1111/j.1574-6941.1999.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki M T, Rappe M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swofford D A. PAUP, version 3.1.1. Washington, D.C.: Laboratory of Molecular Systematics, Smithsonian Institution; 1993. [Google Scholar]
- 66.Takami H, Inoue A, Fuji F, Horikoshi K. Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol Lett. 1997;152:279–285. doi: 10.1111/j.1574-6968.1997.tb10440.x. [DOI] [PubMed] [Google Scholar]
- 67.Takami H, Kobata K, Nagahama T, Kobayashi H, Inoue A, Horikoshi K. Biodiversity in deep-sea sites located near the south part of Japan. Extremophiles. 1999;3:97–102. doi: 10.1007/s007920050104. [DOI] [PubMed] [Google Scholar]
- 68.Tuttle J H. Organic carbon utilization by resting cells of thiosulfate-oxidizing marine heterotrophs. Appl Environ Microbiol. 1980;40:516–521. doi: 10.1128/aem.40.3.516-521.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tuttle J H, Jannasch H W. Occurrence and types of Thiobacillus-like bacteria in the sea. Limnol Oceanogr. 1972;17:532–543. [Google Scholar]
- 70.Tuttle J H, Jannasch H W. Sulfide and thiosulfate-oxidizing bacteria in anoxic marine basins. Mar Biol. 1973;20:64–70. [Google Scholar]
- 71.Tuttle J H, Holmes P E, Jannasch H W. Growth rate stimulation of marine Pseudomonads by thiosulfate. Arch Microbiol. 1974;99:1–14. doi: 10.1007/BF00696218. [DOI] [PubMed] [Google Scholar]
- 72.Uchino Y, Hirata A, Yokata A, Sugiyama J. Reclassification of marine Agrobacterium species: proposals of Stappia stellulata gen nov., comb. nov., Stappia aggregata gen. nov., nom. rev., Ruegeria atlantica gen. nov., comb. nov., Ruegeria gelatinovora comb. nov., Ruegeria algicola comb. nov., and Ahrensia kieliense gen. nov., sp. nov., nom. rev. J Gen Appl Microbiol. 1998;44:201–210. doi: 10.2323/jgam.44.201. [DOI] [PubMed] [Google Scholar]
- 73.van den Ende F, van Germerden H. Sulfide oxidation under oxygen limitation by a Thiobacillus thioparus isolated from a marine microbial mat. FEMS Microbiol Ecol. 1993;13:69–78. [Google Scholar]
- 74.Visscher P T, Quist P, van Gemerden H. Methylated sulfur compounds in microbial mats: in situ concentrations and metabolism by a colorless sulfur bacterium. Appl Environ Microbiol. 1991;57:1758–1763. doi: 10.1128/aem.57.6.1758-1763.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vreeland R H. The family Halomonadaceae. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer Verlag; 1991. pp. 3181–3188. [Google Scholar]
- 76.Wakeham S G, Lee C, Hedges J I, Hernes P J, Peterson M L. Molecular indicators of diagenetic status in marine organic matter. Geochim Cosmochim Acta. 1997;61:5363–5369. [Google Scholar]
- 77.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer Verlag; 1991. pp. 3352–3378. [Google Scholar]
- 78.Wirsen C O, Brinkhoff T, Kuever J, Muyzer G, Molyneaux S, Jannasch H W. Comparison of a new Thiomicrospira strain from the Mid-Atlantic Ridge with known hydrothermal vent isolates. Appl Environ Microbiol. 1998;64:4057–4059. doi: 10.1128/aem.64.10.4057-4059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]