Abstract
Monitoring the communal incidence of COVID-19 is important for both government and residents of an area to make informed decisions. However, continuous reliance on one means of monitoring might not be accurate because of biases introduced by government policies or behaviours of residents. Wastewater surveillance was employed to monitor concentrations of SARS-CoV-2 RNA in raw influent wastewater from wastewater treatment plants serving three Canadian Prairie cities with different population sizes. Data obtained from wastewater are not directly influenced by government regulations or behaviours of individuals. The means of three weekly samples collected using 24 h composite auto-samplers were determined. Viral loads were determined by RT-qPCR, and whole-genome sequencing was used to charaterize variants of concern (VOC). The dominant VOCs in the three cities were the same but with different proportions of sub-lineages. Sub-lineages of Delta were AY.12, AY.25, AY.27 and AY.93 in 2021, while the major sub-lineage of Omicron was BA.1 in January 2022, and BA.2 subsequently became a trace-level sub-variant then the predominant VOC. When each VOC was first detected varied among cities; However, Saskatoon, with the largest population, was always the first to present new VOCs. Viral loads varied among cities, but there was no direct correlation with population size, possibly because of differences in flow regimes. Population is one of the factors that affects trends in onset and development of local outbreaks during the pandemic. This might be due to demography or the fact that larger populations had greater potential for inter- and intra-country migration. Hence, wastewater surveillance data from larger cities can typically be used to indicate what to expect in smaller communities.
Keywords: Wastewater, Raw influent, Variant of concerns, RT-qPCR, SARS-CoV-2 RNA, Population size, Chemical tracer, BA.1, BA.2
Graphical abstract
1. Introduction
Wastewater surveillance has been a useful tool to inform public health officials and the general public within a geographical region about the state of the pandemic prevalence in their location (Peccia et al., 2020). Over the past year, wastewater surveillance developed for SARS-CoV-2 has evolved to monitor for several variants of concern (VOCs), including Alpha, Beta, Gamma, Delta and Omicron (Graber et al., 2021; Yaniv et al., 2021; Mishra et al., 2021; Ahmed et al., 2021; Izquierdo-Lara et al., 2021; Peterson et al., 2022; Johnson et al., 2022) and sub-lineages of the Delta and Omicron VOCs.
Differences in transmissibility have resulted in increased rates of infection compared to the ancestral strain and other more recent VOCs (Callaway, 2021; Li et al., 2022; Papanikolaou et al., 2022). The Delta VOC is characterized by multiple mutations, which sparked concern about the possibility of partial escape of immune defences from previous infections or the effectiveness of vaccination (Mishra et al., 2021; Ahmed et al., 2022a; Eyre et al., 2022). Nonetheless, frequencies of severe outcomes and mortalities in fully vaccinated and boosted populations have been less compared to unvaccinated or partially vaccinated populations (Papanikolaou et al., 2022). However, these variants have still resulted in increased numbers of symptomatic individuals, who in some cases have required hospitalization and therapeutic interventions. For instance, a study of Corona Virus disease (COVID-19) case data for Ontario, Canada, from February to June 2021, showed that the emergence of the Delta VOC resulted in greater rates of hospitalization, admission to intensive care (ICU), and death (Fisman and Tuite, 2021).
Clinical data from polymerase chain reaction (PCR) and whole-genome sequencing of individual patients was previously the main metric for monitoring case numbers and emerging variants. However, it is difficult to obtain reliable information on community transmission due to policy changes across Canada, including decreased availability of clinical testing of individuals and a move away from PCR testing to rapid antigen tests. Results of rapid antigen tests are not centrally collected. Also, there has been an increased number of asymptomatic COVID-positive patients due to increased vaccination rates (Karim and Karim, 2021). Therefore, efficient and accurate monitoring for emerging variants and overall viral loads in wastewater are useful for understanding the prevalence of SARS-CoV-2 in communities. This is particularly true because access to clinical testing and analytical facilities are prioritized for certain populations at greater risk for severe outcomes due to infection with COVID-19 (SHA, 2022; Arts et al., 2022). Hence, accessibility to clinical testing is limited to certain segments of the population (Covantes-Rosales et al., 2022). Therefore, to accurately identify and quantify prevalence of SARS-CoV-2 in a population, including variants of concern, wastewater surveillance appears to be the most promising public health alternative to individual mass testing. It provides a rapid, efficient, cost-effective and non-invasive integrative measure of the entire population, and there are no issues of privacy when samples are collected from municipal wastewater plants. Previous work has shown that clinical and wastewater data follow the same trends for Saskatoon, the largest urban center in Saskatchewan (Xie et al., 2022). Similarly, the major consensus SARS-CoV-2 genotypes detected in the wastewater were earlier identical to clinical genomes from the same area (Crits-Christoph et al., 2021). Therefore, sequencing of VOCs in wastewater is important where complete sequencing of all clinical cases is not possible.
Here we present the results of monitoring total SARS-CoV-2 RNA in wastewaters of three cold-region Prairie cities in Saskatchewan of various sizes, including Saskatoon, Prince Albert, and North Battleford, during a time when clinical testing became increasingly unavailable to the general population. A time lag between the arrival of the Omicron VOC among the three cities provided reliable monitoring to describe progressions of the fourth and fifth waves of SARS-COV-2 in Saskatchewan that were driven by the Delta and Omicron VOCs, respectively.
2. Material and methods
2.1. Study area overviews
Three cities in Saskatchewan, Canada, were studied, including Saskatoon (the most populated city with ≈300,000 people), Prince Albert (≈43,000 people), and North Battleford (≈19,300 people). These are the first, third, and seventh largest cities in the Province of Saskatchewan, with a population of approximately 1,181,000. Municipal wastewaters are received and treated by local municipal wastewater treatment plants (WWTPs) before being released to the South Saskatchewan (Saskatoon) or North Saskatchewan (Prince Albert and North Battleford) Rivers. The Saskatoon, Prince Albert, and North Battleford WWTPs received an average flow of 78, 12, and 4 million litres per day, respectively, during the study period (Table S1). One-litre composite wastewater samples were collected from the primary clarifier influent using an auto-sampler (60 mL every 15 min over 24 h) and maintained at 4 °C. The collected samples were transported on ice to the Toxicology Laboratory at the University of Saskatchewan to be heat-inactivated at 65 °C for 30 min and RNA extracted for qPCR analysis within 24 h. Overall, samples were received three times a week from each WWTP between August 2021 and January 31, 2022.
2.2. Sample pre-processing, viral enrichment, and wastewater environmental RNA (weRNA) extraction
Samples were enriched, concentrated, and RNA extracted as described previously (Xie et al., 2022) with slight modifications. Briefly, to assess recovery during the whole process, synthetic armoured viral particles (AQHRP; Armoured RNA Quant RNase P, Asuragen, TX, USA) were used as an internal spiking control for the whole process (Xie et al., 2022). A freshly diluted 10-μL aliquot (1.0 × 106 gene copies, gc) of the synthetic viral particles was mixed with 70 mL of raw influent sample. Virus particles were enriched using the PEG-8000 precipitation method by adding 7 g PEG-8000 and 1.6 g NaCl to the 70 mL influent sample (Ahmed et al., 2020a, Ahmed et al., 2020b). Enriched samples were agitated overnight (12–14 h) at 4 °C on an orbital shaker, with a speed of 10 rpm and angle of 360°) and then centrifuged at 12,000 ×g at 4 °C for 1 h to pellet the virus. After 1 h centrifuge, the supernatants were removed as much as possible. After pre-processing, weRNA was immediately extracted from pellets using RNeasy Power microbiome kits following the manufacturer's protocol (Qiagen, USA). The RNA was eluted with 100 μL AVE buffer (Xie et al., 2022). The recovery ratio (RR, Eq. (1)) was calculated to assess the practical performance of each batch (Eq. (1)).
| (1) |
where i and j represent sample and batch IDs, respectively; C AQHRP,i,j is the concentration of synthethic viral particles of sample i for batch j; C AQHRP,EPC,j is the concentration of external positive control (EPC) of batch j. The RR for this study was approximately 9.60 ± 8.04 %, consistent with RR reported earlier (Xie et al., 2022).
2.3. RT-qPCR assays for detection of viruses and variants of concern (VOCs)
Concentrations of SARS-CoV-2 and AQHRP were quantified by TaqMan RT-qPCR assays. Quantitative VOC assays were adopted based on Twist® Synthetic (Twist Bioscience, CA, USA) SARS-CoV-2 RNA standards for the detection of Omicron via N200 assays (Fuzzen et al., 2022) and Delta lineages via P681R assay (Thermofisher, CA, USA) (Xie et al., 2022). Synthetic quantitative RNA standards were confirmed by digital droplet PCR in eight replicates following the manufacturer's protocol (ddPCR, Bio-Rad Laboratories, CA, USA). Sequences of primers and probes are shown in Table S2; similarly, the operation of the RT-qPCR is shown in Table S3 and S4, as reported earlier (Xie et al., 2022; Fuzzen et al., 2022).
2.4. Next-generation sequencing
Whole-genome sequencing (WGS) of wastewater samples were done at the Division of Enteric Diseases, National Microbiology Laboratory, Public Health Agency of Canada (Winnipeg, MB, Canada). The method for sequencing was described in https://www.medrxiv.org/content/10.1101/2021.03.11.21253409v1.full.pdf (Landgraff et al., 2021). cDNA was synthesized using the SuperScript IV First-Strand Synthesis System (Invitrogen, USA). Tiled amplicons were amplified according to the ArticV3 protocol. Tiled amplicons were sequenced with MiSeq 300PE V3 chemistry (Illunima, USA). Mutations were identified on mapping files generated by SAMtools v 1.7 against a SARS-CoV-2 reference sequence (MN908947.3) (Danecek et al., 2021; Landgraff et al., 2021). SARS-CoV-2 lineage was assigned based on coverage of consensus mutations following the Pango Nomenclature proposal (Xie et al., 2022; Landgraff et al., 2021).
2.5. Statistical analyses
Plots were generated using Origin Pro 2021C (OriginLab Corporation USA). Viral loads were normalized to concentrations of the artificial sweetener, acesulfame (6-methyl-2,2-dioxooxathiazin-4-one; CAS 33665-90-6) by dividing numbers of copies of target genes by concentrations of acesulfame. Daily influent flow rates were used to normalize viral load by multiplying the gene copies by daily flow rates to obtain gene copies per day. A normality test was done using Shapiro-Wilk at 5 % decision level. Correlation coefficients were determined using Pearson correlation (r) or Spearman correlation depending on if the normality test was accepted or rejected.
3. Results and discussion
The load of SARS-CoV-2 viral RNA in wastewater can be used to follow the trajectory of COVID-19 infections in the cities studied (Fig. S1) (Crits-Christoph et al., 2021; Arts et al., 2022; Xie et al., 2022; Yu et al., 2022). Delta became the only VOC in the cities studied after it had replaced the Alpha VOC in June 2021 (Xie et al., 2022), which occurred before the current study. The Delta VOC was the only observed variant, with its sub-lineages detected throughout August until the first week of December 2021 (Fig. 1 ). Since several sub-lineages of Delta emerged, which were monitored through whole-genome sequencing (Table 1, Table 2, Table 3 ), it became clear that the specific sub-lineages circulating in various communities might be related to how frequently new people visited the community, which is a function of the total population and demography of the area. Migration has already been shown to play an important role in the spread of SARS-CoV-2 (Chen et al., 2020). Some studies also showed how new VOCs could enter communities through migration by detecting new VOCs in wastewaters of airplanes (Ahmed et al., 2022, Ahmed et al., 2022b). The emerging and the dominant, circulating VOCs in each community were monitored cost-effectively, with limited biases by wastewater surveillance (Ai et al., 2021). Wastewater surveillance has been a useful tool for monitoring overall SARS-CoV-2 RNA and understanding the specific VOCs dominating each community (Yu et al., 2022).
Fig. 1.
Pattern of viral load in three selected cities in Saskatchewan, including North Battleford (A), Prince Albert (B), and Saskatoon (C) over the study duration. The Delta variant (B.1.617 VOC (S_R681 gc/100 mL), S gene, is shown in red, while the Omicron variant (N_R203K.G204R, gc/100 mL), N gene is shown in green, and overall viral load, N gene, is shown in black.
Table 1.
Data obtained from whole genome sequencing of wastewater environmental RNA from August 2021 to January 2022 for Saskatoon.
| Date Collected | Nextclade (consensus) | Pangolin | % Breadth of coverage (≥ 5× depth) | Average depth of coverage | Median Depth of Coverage | VOC Detected (consensus) | Number of VOC mutations (consensus) | Frequencies of reads (> cov 30) with VOC mutation (consensus) | Number of mutations supporting presence of additional VOC (subconsensus) | Additional Delta sublineages or VOCs, VOIs detected in subconsensus sequences |
|---|---|---|---|---|---|---|---|---|---|---|
| 01/08/2021 | 21A (Delta) | AY.12 | 93.91 | 2903.88 | – | B.1.617.2 (Delta) | 13/13 | 0.9 | <3 (only common mutations were detected) | IE |
| 04/08/2021 | 21A (Delta) | B.1.617.2 | 93.42 | 2712.42 | 961 | B.1.617.2 (Delta) | 10/12 12/12 (cov <30) |
0.83 cov < 30 = 0.97 | P.1 = 4 (0.1) | Possible P.1 (Gamma) |
| 15/08/2021 | 21A (Delta) | AY.4 | 99.6 | 6365.48 | 10,240 | B.1.617.2 (Delta) sublineage AY.12 | 12/12 | 0.89 | <4 | IE |
| 22/08/2021 | 21A (Delta) | AY.4 | 99.55 | 5543.97 | 7060 | B.1.617.2 (Delta) sublineage AY.6 | 10/12 | 0.84 | <4 | IE |
| 02/09/2021 | 21A (Delta) | B.1.617.2 | 99.6 | 6385.45 | 6504 | B.1.617.2 (Delta) | 13/13 | 0.97 | AY.25 = 2 (~0.24) AY.15 = 1 (<0.1) |
AY.25 |
| 05/09/2021 | 21A (Delta) | B.1.617.2 | 99.6 | 5396.24 | 4696 | B.1.617.2 (Delta) | 13/13 | 0.99 | AY.25 = 2 (0.18) | AY.25 (weak signal) |
| 12/09/2021 | 21A (Delta) | B.1.617.2 | 99.6 | 6342.31 | 6567 | B.1.617.2 (Delta) | 13/13 | 0.99 | AY.25 = 4 (0.37) | AY.25 |
| 19/09/2021 | 21J (Delta) | B.1.617.2 | 99.6 | 6157.97 | 6342 | B.1.617.2 (Delta) | 13/13 | 0.99 | AY.25 = 4 (0.26) AY.27 = 3 (0.23) |
AY.25, AY.27 |
| 03/10/2021 | 21J (Delta) | B.1.617.2 | 99.6 | 5876.3 | 5910 | B.1.617.2 | 13/13 | 1 | AY.25 = 2 (0.32) AY.27 = 3 (0.27) |
AY.25, AY.27 |
| 13/10/2021 | 21J (Delta) | B.1.617.2 | 99.57 | 4742.91 | 5408 | B.1.617.2 | 13/13 | 1.0 | AY.25 = 4 (0.26) AY.27 = 3 (0.57) |
AY.25, AY.27 |
| 24/10/2021 | 21J (Delta) | AY.23 | 93.04 | 4125.28 | 4082 | Delta sublineage AY.23 | 11/13 | 0.84 | AY.25 = 2 (0.55) AY.27 = 3 (0.78) AY.33 = 2 (<0.1) |
AY.25, AY.27 |
| 31/10/2021 | 21J (Delta) | AY.9.2 | 95.31 | 2391.4 | 441 | AY.9.2 | 9/12 | 0.69 | AY.25 = 1 (0.61) AY.27 = 1 (0.38) |
AY.25, AY.27 |
| 14/11/2021 | 21I (Delta) | AY.27 | 99.39 | 3879.32 | 3450 | AY.27 | 11/13 | 0.84 | AY.25 = 3 (0.24) AY.93 = 4 (0.11) AY.103 = 1 (0.31) |
AY.25, AY.93, AY.103 |
| 24/11/2021 | 21I (Delta) | N_content:0.55 | 47.18 | 4140.94 | 0 | 21I (Delta) | 9/13 | 0.69 | AY.25 = 1 (0.25) AY.27 = 1 (0.12) |
AY.25, AY.27 |
| 08/12/2021 | 21J (Delta) | N_content:0.66 | 35.21 | 3692 | 0 | 21J (Delta) | 5/12 | 0.38 | AY.25 = 2 (0.38) | |
| 17/12/2021 | 21J (Delta) | N_content:0.85 | 16.81 | 548.38 | 0 | 21J (Delta) | 3/13 | 0.23 | <4 | IE |
| 26/12/2021 | 21K (Omicron) | BA.1 | 96.94 | 11,418.2 | 3739 | BA.1 (Omicron) | 36/51 | 0.58 | Delta = 10/13 (0.27) AY.53 = 1 (0.15) |
AY.53 |
| 02/01/2022 | 21K (Omicron) | BA.1 | 99.57 | 13,755.92 | 5132 | BA.1 (Omicron) | 48/51 | 0.64 | BA.2 = 3 (37/60)-0.37 Delta = 11 (0.23) AY.25/AY.25.1 = 5 (0.1) |
Moderate presence of 3 BA.2 mutations Low presence of Delta (AY.25/AY.25.1) |
| 09/01/2022 | 21K (Omicron) | BA.1 | 99.29 | 14,499.08 | 5575 | BA.1 (Omicron) | 51/51 | 0.96 | Delta = 4 (< 0.1) | Trace presence of 4 Delta mutations |
| 23/01/2022 | 21K (Omicron) | BA.1 | 98.65 | 98.65 | 98.65 | BA.1 (Omicron) | 51/51 | 0.94 | BA.2 = 14/28 (0.12) Delta = 4 (0.16) |
Low presence of BA.2 Low presence of Delta |
| 30/01/2022 | 21K (Omicron) | BA.1.1 | 98.41 | 9983.54 | 4365 | BA.1.1 (Omicron) | 50/51 | 0.95 | BA.2 = 10 (< 0.1) | Trace presence of BA.2 |
IE – Insufficient evidence; () - frequencies of reads for mutations supporting subconsensus sequences.
Table 2.
Data obtained from whole genome sequencing of wastewater environmental RNA from August 2021 to January 2022 for Prince Albert.
| Date Collected | Nextclade (consensus) | Pangolin | % Breadth of coverage (≥ 5× depth) | Average depth of coverage | Median Depth of Coverage | VOC Detected (consensus) | Number of VOC mutations (consensus) | Frequencies of reads (> cov 30) with VOC mutation (consensus) | Number of mutations supporting presence of additional VOC (subconsensus) | Additional Delta sublineages or VOCs, VOIs detected in subconsensus sequences |
|---|---|---|---|---|---|---|---|---|---|---|
| 04/08/2021 | 21A (Delta) | AY.4 | 98 | 3444.43 | 1289 | B.1.617.2 (Delta) sublineage AY.12 | 10/12 12/12 (cov <30) |
0.61 cov < 30 = 0.99 |
B.1.1.7 = 4 (<0.1) P.1 = 16 (0.17) |
Confirmed P.1 (Gamma)- weak Possible B.1.1.7 (Alpha) |
| 11/08/2021 | 21A (Delta) | B.1.617.2 | 98.61 | 5949.44 | 8666 | B.1.617.2 (Delta) | 11/12 | 0.89 | <4 | IE |
| 23/08/2021 | 21A (Delta) | B.1.617.2 | 99.6 | 4270.86 | 3396 | B.1.617.2 (Delta) | 10/12 | 0.84 | <4 | IE |
| 30/08/2021 | 21A (Delta) | B.1.617.2 | 98.61 | 5306.34 | 4909 | B.1.617.2 (Delta) | 13/13 | 0.99 | AY.25 = 2 (<0.1) | AY.25 (weak signal) |
| 06/09/2021 | 21A (Delta) | B.1.617.2 | 99.41 | 5755.65 | 5254 | B.1.617.2 (Delta) | 12/13 | 0.91 | AY.25 = 2 AY.25.1 = 1 (< 0.1) | AY.25 (weak signal) |
| 13/09/2021 | 21A (Delta) | N_content:0.64 | 38.17 | 1384.15 | 2 | 21A (Delta) | 7/13 | 0.54 | <3 | IE |
| 04/10/2021 | 21J (Delta) | B.1.617.2 | 98.61 | 5705.11 | 5627 | B.1.617.2 | 12/13 | 0.92 | AY.25 = 4 (0.16) | IE |
| 11/10/2021 | 21J (Delta) | AY.39 | 99.39 | 3966.28 | 3495 | AY.39 | 11/13 | 0.84 | AY.4 = 1 (0.14) | IE |
| 22/10/2021 | 21J (Delta) | AY.93 | 99.33 | 4396.12 | 4682 | AY.93 | 11/13 | 0.84 | AY.27 = 3 (0.36) AY.25 = 1 (< 0.1) |
AY.25 |
| 22/10/2021 | 21J (Delta) | B.1.617.2 | 99.57 | 3807 | 3345 | B.1.617.2 (Delta) | 13/13 | 1.0 | AY.15 = 1 (< 0.1) AY.25 = 4 (0.33) AY.27 = 2 (0.31) |
AY.25, AY.27 |
| 15/11/2021 | 21J (Delta) | N_content:0.41 | 74.48 | 1662.91 | 140 | 21J (Delta) | 9/12 | 0.69 | AY.27 = 2 (0.15) AY.93 = 2 (0.5) AY.5.2 = 1 (0.38) |
AY.27, AY.93, AY.5.2. |
| 29/11/2021 | 21I (Delta) | N_content:0.55 | 45.85 | 2864.19 | 0 | 21I (Delta) | 11/13 | 0.78 | AY.103 = 1 (0.65) | AY.103 |
| 10/12/2021 | 21I (Delta) | AY.27 | 99.58 | 17,582.41 | 5403 | AY.27 | 11/13 | 0.85 | <4 | |
| 04/01/2022 | 21K (Omicron) | B.1.1.529 | 98.81 | 13,699.9 | 4514 | BA.1 (Omicron) | 46/51 | 0.58 | BA.2 = 4 (34/60)-0.34 Delta = 13 (0.3) AY.27 = 6 (< 0.1) |
Moderate presence of 4 BA.2 mutations Moderate presence of Delta (trace presence of AY.27) |
| 24/01/2022 | 21K (Omicron) | BA.1.1 | 98.89 | 98.89 | 98.89 | BA.1.1 (Omicron) | 50/51 | 0.81 | BA.2 = 3/28 (<0.1) | Trace presence of BA.2 |
| 31/01/2022 | 21K (Omicron) | BA.1.1 | 98.78 | 17,763.47 | 9845 | BA.1.1 (Omicron) | 51/51 | 0.97 | <4 |
Table 3.
Data obtained from whole genome sequencing of wastewater environmental RNA from August 2021 to January 2022 for North Battleford.
| Date Collected | Nextclade (consensus) | Pangolin | % Breadth of coverage (≥ 5× depth) | Average depth of coverage | Median Depth of Coverage | VOC Detected (consensus) | Number of VOC mutations (consensus) | Frequencies of reads (> cov 30) with VOC mutation (consensus) | Number of mutations supporting presence of additional VOC (subconsensus) | Additional Delta sublineages or VOCs, VOIs detected in subconsensus sequences |
|---|---|---|---|---|---|---|---|---|---|---|
| 06/08/2021 | 20B | None | 49.54 | 1176.78 | 2 | None detected | n/a | n/a | B.1.1.7 = 4 (0.21) B.1.617.2 = 2 (<0.1) |
Possible B.1.1.7 (Alpha) |
| 20/08/2021 | 21A (Delta) | B.1.617.2 | 98.98 | 4986.72 | 5603 | B.1.617.2 (Delta) sublineage AY.6 | 10/12 | 0.84 | <4 | IE |
| 30/08/2021 | 21A (Delta) | B.1.617.2 | 99.6 | 6319.17 | 6650 | B.1.617.2 (Delta) | 13/13 | 0.99 | AY.25 = 2 (0.14) AY.21 = 1 (0.18) |
AY.25 (weak signal) |
| 03/09/2021 | 21A (Delta) | B.1.617.2 | 99.6 | 4648.12 | 3777 | B.1.617.2 (Delta) | 11/13 | 0.84 | AY.25 = 2 (<0.1) | AY.25 (weak signal) |
| 09/09/2021 | 21A (Delta) | AY.12 | 97.71 | 5627.37 | 5480 | Delta sublineage AY.12 | 11/13 | 0.84 | AY.25 = 4 (0.13) | AY.25 (weak signal) |
| 16/09/2021 | 21J (Delta) | B.1.617.2 | 98.72 | 5194.09 | 4595 | B.1.617.2 (Delta) | 11/13 | 0.84 | AY.25 = 2 (0.11) AY.27 = 1 < 0.1) |
AY.25 |
| 03/10/2021 | 21J (Delta) | B.1.617.2 | 99.6 | 6164.71 | 6232 | B.1.617.2 | 13/13 | 1 | AY.25 = 4 (0.16) AY.27 = 3 (0.15) |
IE |
| 09/10/2021 | 21J (Delta) | AY.39 | 99.52 | 5031.35 | 5945 | AY.39 | 11/13 | 0.85 | <4 | IE |
| 22/10/2021 | 21I (Delta) | B.1.617.2 | 99.57 | 4949.38 | 5961 | B.1.617.2 | 11/13 | 0.84 | AY.25 = 2 (0.19) AY.27 = 3 (0.68) |
AY.25, AY.27 |
| 24/10/2021 | 21J (Delta) | B.1.617.2 | 99.57 | 5234.73 | 6593 | B.1.617.2 (Delta) | 13/13 | 1.0 | AY.25 = 3 (0.18) AY.27 = 3 (0.22) |
AY.25, AY.27 |
| 12/11/2021 | 21A (Delta) | AY.70 | 85.12 | 3017.02 | 1311 | AY.70 | 7/13 | 0.54 | AY.25 = 1 (0.18) AY.27 = 1 (0.38) AY.93 = 1 (0.72) |
AY.25, AY.27, AY.93 |
| 26/11/2021 | 21I (Delta) | AY.74 | 91.33 | 6001.55 | 2523 | AY.74 | 11/13 | 0.85 | AY.4.1 = 1 (0.5) | AY.4.1 |
| 11/12/2021 | 21I (Delta) | AY.27 | 89.34 | 7322.47 | 1723 | AY.27 | 11/13 | 0.84 | AY.103 = 1 (0.35) AY.25 = 1 (0.21) |
AY.103 AY.25 |
| 03/01/2022 | 21I (Delta) | AY.27 | 99.6 | 16,478.08 | 7917 | AY.27 (Delta) | 13/13 | 0.73 | Moderate presence of BA.1 (Omicron) | |
| 21/01/2022 | 21K (Omicron) | BA.1 | 98.69 | 98.69 | 98.69 | BA.1 (Omicron) | 50/51 | 0.9 | BA.2 = 27/28 (0.41) Delta = 3 (0.16); 2 mutations overlap with Omicron |
Moderate presence of BA.2 Trace presence of Delta |
| 28/01/2022 | 21K (Omicron) | BA.1 | 98.65 | 12,386.05 | 7772 | BA.1 (Omicron) | 50/51 | 0.77 | BA.2 = 27 (0.42) | Moderate presence of BA.2 |
VOC sub-lineages (Table 1, Table 2, Table 3) demonstrated a pattern of occurrence of the virus lineages and the timing when new sub-lineages became apparent. Delta VOC and its AY lineages were the dominant VOC in wastewater, driving the fourth wave. The dominance could be because >75 % of the spike proteins in the Delta VOC are capable of invading human cells (Wang and Han, 2022). Delta has 15 mutations of the spike protein, while Omicron has 32 (CDC, 2022). Hence, it took, Omicron 7 days to replace Delta in Saskatoon, 15 days in Prince Albert, and it became the dominant VOC on the first day it was detected in North Battleford. Similarly, it has been shown that Omicron displaced Delta in Ontario province-wide analysis of both clinical and wastewater surveillance in only 2 weeks following its first detection in travellers through clinical genomic surveillance (Arts et al., 2022). Overall, Delta was displaced by Omicron VOC in less than a month because of the additional mutations and deletions. The additional mutations and deletions allow Omicron to have increased transmissibility (Table 1, Table 2, Table 3) (Karim and Karim, 2021).
The total viral load detected by RT-qPCR for the Omicron VOC was 2-fold greater than that observed for the Delta VOC. The maximum viral loads for Delta VOC were approximately 125,000, 110,000, and 182,000 gene copies/100 mL for Saskatoon, Prince Albert, and North Battleford, respectively, while the greatest viral loads for Omicron were 201,000, 340,000, and 190,000 gene copies/100 mL, for Saskatoon, Prince Albert, and North Battleford, respectively. Another study has shown that the viral load associated with Omicron was very high compared to the earlier viral loads from Alpha, Delta, and Beta (Covantes-Rosales et al., 2022). This large load of virus particles observed for Omicron could directly relate to its increased rate of infection because of its ability to infect vaccinated people. Interestingly, some studies have found that vaccination efficacy decreased with time (Levine-Tiefenbrun et al., 2021), hence increasing the possibility of re-infection (Neto et al., 2022). However, it is not clear if the rates of shedding viruses by people infected with Omicron are greater or lesser than that of Delta. Nevertheless, it has been reported that the doubling time for Omicron was 1 to 2 days, compared to 1 to 5 days for Delta (Karim and Karim, 2021). Furthermore, the proportion that was Omicron was also estimated to have an initial doubling time of 3.2 days (95 % CI = 3.1–3.4 days), faster than those of Delta (7.2 days; 95 % CI = 7.0–7.4 days) (Lambrou et al., 2022). Similarly, it was documented that Omicron can cause re-infection at the rate of 2.39 compared to Beta and Delta; therefore, the majority of people previously infected with other variants might be at risk of being infected again by the Omicron VOC (Ahmed et al., 2022a).
Consensus and sub-consensus genome matches from results of sequencing aligned well with the publicly available Omicron genomes, and therefore assignments could be made to either BA.1 or BA.2. In January 2021, the major subvariants of Omicron were BA.1 and BA.1.1, although trace amounts of BA.2 were detected infrequently and identified by sub-consensus sequence (Table 1, Table 2, Table 3). The mutations with coverage >30 % were considered consensus (Izquierdo-Lara et al., 2021). The consensus sequence represented the predominant lineages in the study areas but might not give the complete picture of other lineages in trace amounts, which might soon become the main lineages (Crits-Christoph et al., 2021). The breadth of coverage was generally >97 %, except on a few occasions (Table 1, Table 2, Table 3), but 100 % breadth of coverage was needed to have complete consensus viral genomes (Crits-Christoph et al., 2021). The sub-consensus sequence helped identify lineages present in trace amounts as they were too rare to be seen in the consensus sequence. Thus, the BA.2 sub-lineage and AY-lineages, which were not seen by consensus, were detected at trace, low, and moderate prevalence.
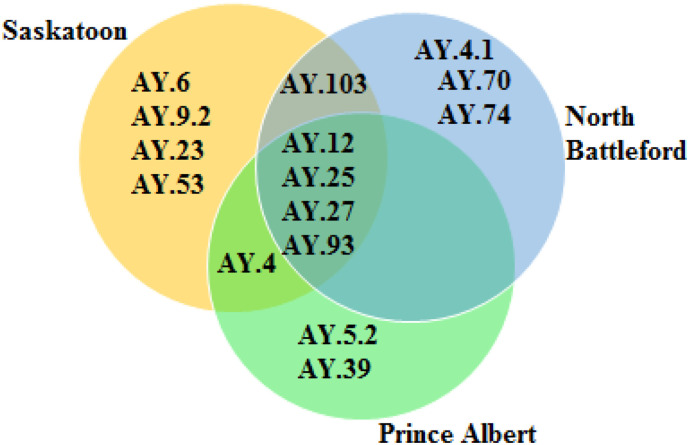
Monitoring the sub-lineage circulating is important because it might help understand the risk of hospitalization or mortality (Nyberg et al., 2022). Several sub-lineages of the Delta VOC were evident in the three cities (Table 1, Table 2, Table 3). >250 AY lineages have been detected worldwide (CDC, 2022); however, only 15 different types were sequenced in this study, out of which 10 were detected in consensus and 5 in sub-consensus sequences. The sub-lineages detected in the consensus sequence were AY.4, AY.12, AY.6, AY.23, AY.9.2, AY.39, AY.93, AY.27, AY.70, AY.74, and in sub-consensuses sequences were AY.25, AY.25.1, AY.103, AY.4.1, and AY.53. The sub-lineages found in the three cities were AY.12, AY.25, AY.27, and AY.93 (Fig. 2 ). These AY lineages are different from the characteristics AY lineages found in Brazil (AY.99.2, AY.43, AY.101, AY.34.1, AY.43.1, AY.43.2, AY.46.3, AY.100, AY.99.1, AY.36) (Gularte et al., 2022). Similarly, they are different from the signature AY-lineages found in Sir Lanka (AY.104 and AY.28) (Ranasinghe et al., 2022). In addition to the AY-lineages found in all three cities, some different sub-lineages were found in each city (Fig. 2). These included AY.4, AY.6, AY.23, AY.9.2, AY.53, and AY.103 in Saskatoon; AY.4, AY.5.2, and AY.39 in Prince Albert; and AY.103, AY.70, AY.4.1, and AY.74 in North Battleford. The mean depth of coverage, the number of mutations, and the read frequencies were all considered to confirm the assignment of consensus VOC or lineages. When the coverage was <30, lineage was only assigned when the number of mutations counted tallied the maximum number of mutations for the VOC. Overall, VOCs or sub-lineages with low read frequencies were not considered as possible VOCs or sub-lineages in either consensus or sub-consensus. For example, AY.33, AY.21, and AY.15 had 2, 1, and 1 number of mutations, respectively, but their number of reads was too low to be assigned.
Fig. 2.
Comparisons of AY lineages sequenced in the three study cities between August 2021 and December 2022.
The week and duration of the dominance of each of the AY-lineages are summarized in Table 1, Table 2, Table 3. Each of these AY-lineages is typical to a particular area or country; for example, AY.5.2 is called the Portugal lineage even though the percentage in most countries are as follows: Belgium 43.0 %, Portugal 38.0 %, United Kingdom 6.0 %, France 3.0 %, Netherlands 2.0 % (CDC, 2022). It is not surprising that AY.25 and AY.27 are present in the three cities and are the dominant AY-lineages given AY.25 is primarily a United States lineage at 97.0 % (Argentina 1.0 %), while AY.27 is a Canadian lineage with Canada 99.0 % (USA 1.0 %) (CDC, 2022). Sri Lanka was reported to be the origin of AY.28 and AY.104 found worldwide (Ranasinghe et al., 2022). Therefore analysis of lineages might be used to determine how infected people from different countries moved the virus into other countries (Nemudryi et al., 2020; Rambaut et al., 2021).
All the clades before the second week of December 2021 were characteristic of the Delta variant, with the predominant clades observed in the three cities being 21A, 21 J, and 21I (Table 1, Table 2, Table 3), with each of these occurring in different months (Table 1, Table 2, Table 3). However, in early August, the North Battleford WWTP still presented 20B associated with the Alpha variant. 21A was the major clade in August and the first three weeks in September before transitioning to 21J. The clade 21J has been described to have all the mutations of the 21A clade with an additional mutation in N gene (Gularte et al., 2022). This justifies its ability to replace 21A clade.
Omicron was first detected in Saskatoon wastewater before it was detected in wastewater of the other two smaller communities. It was detected on December 12, 2021, in Saskatoon, December 15 in Prince Albert, and December 29 in North Battleford. The discovery of Omicron in Saskatoon first before other cities might be because the city has a major airport, where most travellers arriving in the province first land before moving to the other areas. The city of Saskatoon also has a University, a Polytechnic, and many schools. Saskatoon is the commercial and industrial hub of the province. Closure of Universities and schools has been implicated to reduced the level of SARS-CoV-2 RNA in wastewater (Stephens et al., 2022). The presence of travellers and visitors in cities has been linked to the large Omicron viral load in Alberta wastewater (Hubert et al., 2022). Similarly, Omicron was reported in aircraft wastewater travelling between two countries (Ahmed et al., 2022a; Ahmed et al., 2022b). Therefore, cities with airports that receive travellers frequently have the potential to develop new VOCs first, and other new diseases compared to more remote communities. North Battleford was the last city where Omicron was detected because the population in the city was the least, and the possibility of community transmission in a smaller city is less than in more crowded cities with greater population densities. This observation is similar to an earlier report where two large Alberta cities, Calgary and Edmonton, exhibited a more rapid emergence of Omicron relative to the smaller and more remote Alberta municipalities of Brooks and Taber (Hubert et al., 2022). Therefore, larger cities with a greater influx of travellers and greater population densities have a greater risk of observing new lineages or variants (Arts et al., 2022).
When viral loads were normalized to concentrations of the artificial sweetener “acesulfame”, no changes were observed in the trend of the viral load (Fig. 3 ). The Spearman correlation coefficients (r) between unnormalized data and the gene copies normalized by acesulfame were r = 0.986 (p < 0.0001), 0.975 (p < 0.0001), and 0.978 (p < 0.0001) for Saskatoon, Prince Albert, and North Battleford, respectively. This lack of changes in trend might be because acesulfame was stable during the study period (Fig. S2). Acesulfame was better than pepper mild mottle virus or concentrations of creatinine or ammonia as indicators of populations contributing wastes to WWTPs and can be used to correct for effects of dilution (Xie et al., 2022). Hence, the population served by the WWTPs during the study period was relatively constant. Thus, normalization with acesulfame helped to ascertain that there were no variations in the numbers of persons contributing waste to the WWTPs. It also helps to correct the effects of dilution between sampling days, if any.
Fig. 3.
SARS-COV-2 viral load normalized to concentrations of the artificial sweetener, acesulfame (red) for three selected cities in Saskatchewan, including North Battleford (A), Prince Albert (B), and Saskatoon (C).
Like the gene copies per acesulfame, the gene copies per day correlated well with gene copies per 100 mL (Fig. 4 ). The Spearman correlation coefficients were 1.000 (p < 0.0001), 0.987 (p < 0.0001), and 0.999 (p < 0.0001) for Saskatoon, Prince Albert, and North Battleford, respectively, for inflow normalized data compared with unnormalized data. The high correlation between unnormalized data and the inflow normalized data confirmed that the population serving the WWTPs, and the inflow did not vary significantly between August 2021 and January 2022 (Table S1). This stability in the inflow could be because no rain or snow melted during the study period, especially during the Omicron wave (December 2021 and January 2022). Also, Saskatoon and North Battleford sewer systems were designed to ensure little or no contributions from runoff. Normalizing the viral load with actual water usage has been reported to reduce uncertainty in wastewater (Li et al., 2021). Thus, the huge difference between Omicron viral load and Delta viral load was not due to variation in population or snowmelt. Interestingly viral load normalized with acesulfame has positive significant correlation with inflow normalized data, r = 0.986 (p < 0.0001), r = 0.970 (p < 0.0001), and r = 0.98 (p < 0.0001), for Saskatoon, Prince Albert, and North Battleford, respectively. Hence, population variation, snow melting, or runoff did not affect the viral load presented by both Delta and Omicron VOC during the study period.
Fig. 4.
SARS-COV-2 viral load normalized to influent volume (red) of three selected cities in Saskatchewan, including North Battleford (A), Prince Albert (B), and Saskatoon (C).
4. Conclusions
The SARS-CoV-2 viral load of three cities with different population sizes is reported. The city with the largest population, Saskatoon, was the first to exhibit the Omicron VOC in wastewater and the first city to become Omicron VOC dominated. The AY-lineages associated with the USA (AY.25) and Canada (AY.27) were the major AY-lineages detected when Delta was the dominant VOC. Omicron displaced Delta variants within a week in Saskatoon as the major VOC because of its higher transmissibility due to having over 50 mutations. BA.1 and BA.1.1 were the dominant Omicron sub-lineages in January 2022, with only trace levels found for BA.2. The viral load in the fifth wave driven by Omicron was greater than that observed during the fourth wave, when Delta was the dominant VOC, likely due to the possibility of re-infection with Omicron by those already recovered from SARS-CoV-2 infections. There were strong correlations (>0.9) between normalized and unnormalized samples in the three cities because the population indicator was relatively stable during the study period. Also, inflow normalized viral load significantly correlated with acesulfame normalized viral load.
CRediT authorship contribution statement
JPG, MA, KNM, PDJ: provided funding, project conceptualization and management, method development; data collection and curation; wrote and edited the final manuscript.
YX, FFO: method development, data collection and curation, wrote the first draft, formatted and edited the final manuscript.
MA, JKC, JC, CM, CL: method development, chemical analyses, data collection, sequencing and curation.
KK, MK, MS: data collection.
MF, MRS: method development.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study is part of the project titled “Next generation solutions to ensure healthy water resources for future generations,” funded by the Global Water Futures program, Canada First Research Excellence Fund (#419205; Additional information is available at www.globalwaterfutures.ca) and the Public Health Agency of Canada. The authors acknowledge the support of the Saskatoon Wastewater Treatment Plant. The research was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (Project # 326415-07). The authors wish to acknowledge the support of an instrumentation grant from the Canada Foundation for Innovation. Prof. Giesy was supported by the Canada Research Chairs Program of the Natural Sciences and Engineering Research Council of Canada (NSERC) and a distinguished visiting professorship of Environmental Sciences from Baylor University in Waco, Texas, USA.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.156741.
Appendix A. Supplementary data
Supplementary data
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O'Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Smith W.J., Metcalfe S., Stephens M., Jennison A.V., Moore F.A., Bourke J., Schlebusch S., McMahon J., Hewitson G., Nguyen S., Barcelon J., Jackson G., Mueller J.F., Ehret J., Hosegood I., Tian W., Wang H., Yang L., Bertsch P.M., Tynan J., Thomas K.V., Bibby K., Graber T.E., Ziels R., Simpson S.L. Detection of the Omicron (B. 1.1. 529) variant of SARS-CoV-2 in aircraft wastewater. Sci. Total Environ. 2022;820:153171. doi: 10.1016/j.scitotenv.2022.153171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Simpson S.L., Bertsch P.M., Ehret J., Hosegood I., Metcalfe S.S., Smith W.J., Thomas K.V., Tynan J., Mueller J.F. Wastewater surveillance demonstrates high predictive value for COVID-19 infection on board repatriation flights to Australia. Environ. Int. 2022;158 doi: 10.1016/j.envint.2021.106938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y., Davis A., Jones D., Lemeshow S., Tu H., He F., Ru P., Pan X., Bohrerova Z., Lee J. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts E., Brown S., Bulir D., Charles T.C., DeGroot C.T., Delatolla R., Desaulniers J.P., Edwards E.A., Fuzzen M., Gilbride K., Gilchrist J., Goodridge L., Graber T.E., Habash M., Jüni P., Kirkwood A., Knockleby J., Kyle C., Landgraff C., Mangat C., Manuel D.G., McKay R.M., Mejia E., Mloszewska A., Ormeci B., Oswald C., Payne S.J., Peng H., Peterson S., Poon A.F.Y., Servos M.R., Simmons D., Sun J., Yang M., Ybazeta G. 2022. Community Surveillance of Omicron in Ontario: Wastewater-based Epidemiology Comes of Age. Preprint. Access May 2022. [DOI] [Google Scholar]
- Callaway E. The mutation that helps Delta spread like wildfire. Nature. 2021;596(7873):472–473. doi: 10.1038/d41586-021-02275-2. [DOI] [PubMed] [Google Scholar]
- CDC Centers for disease control and prevention. 2022. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html Access May 2022.
- Chen M., Li M., Hao Y., Liu Z., Hu L., Wang L. The introduction of population migration to SEIAR for COVID-19 epidemic modeling with an efficient intervention strategy. Information Fusion. 2020;64:252. doi: 10.1016/j.inffus.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covantes-Rosales C.E., Barajas-Carrillo V.W., Girón-Pérez D.A., Toledo-Ibarra G.A., Díaz-Reséndiz K.J.G., Navidad-Murrieta M.S., Ventura-Ramón G.H., Pulido-Muñoz M.E., Mercado-Salgado U., Ojeda-Durán A.J., Argüero-Fonseca A., Girón-Pérez M.I. Comparative analysis of age, sex, and viral load in outpatients during the four waves of SARS-CoV-2 in a Mexican medium-sized city. Int. J. Environ. Res. Public Health. 2022;19(9):5719. doi: 10.3390/ijerph19095719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph A., Kantor R.S., Olm M.R., Whitney O.N., Al-Shayeb B., Lou Y.C., Flamholz A., Kennedy L.C., Greenwald H., Hinkle A., Hetzel J. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. Clin. Sci. Epidemiol. MBio. 2021;12(1) doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O, Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre D.W., Taylor D., Purver M., Chapman D., Fowler T., Pouwels K.B., Walker A.S., Peto T.E. Effect of Covid-19 vaccination on transmission of alpha and Delta variants. N. Engl. J. Med. 2022;386:744. doi: 10.1056/NEJMoa2116597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisman D.N., Tuite A.R. Evaluation of the relative virulence of novel SARS-CoV-2 variants: a retrospective cohort study in Ontario, Canada. Cmaj. 2021;193()):E1619–E1625. doi: 10.1503/cmaj.211248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuzzen M.L., Harper N.B., Dhiyebi H.A., Srikanthan N., Hayat S., Peterson S.W., Yang I., Sun J.X., Edwards E.A., Giesy J.P., Mangat C.S., Graber T.E., Delatolla R., Servos M.R. medRxiv; 2022. Multiplex RT-qPCR Assay (N200) to Detect and Estimate Prevalence of Multiple SARS-CoV-2 Variants of Concern in Wastewater. Accessed in May 2022. [DOI] [Google Scholar]
- Graber T.E., Mercier É., Bhatnagar K., Fuzzen M., D'Aoust P.M., Hoang H.D., Tian X., Towhid S.T., Plaza-Diaz J., Eid W., Alain T., Butler A., Goodridge L., Servos M., Delatolla R. Near real-time determination of B. 1.1. 7 in proportion to total SARS-CoV-2 viral load in wastewater using an allele-specific primer extension PCR strategy. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gularte J.S., da Silva M.S., Mosena A.C.S., Demoliner M., Hansen A.W., Filippi M., Pereira V.M.D.A.G., Heldt F.H., Weber M.N., de Almeida P.R., Hoffmann A.T., Valim A.R.D.M., Possuelo L.G., Fleck J.D., Spilki F.R. Early introduction, dispersal and evolution of Delta SARS-CoV-2 in southern Brazil, late predominance of AY. 99.2 and AY. 101 related lineages. Virus Res. 2022;311 doi: 10.1016/j.virusres.2022.198702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert C.R., Acosta N., Waddell B.J., Hasing M.E., Qiu Y., Fuzzen M., Harper N.B., Bautista M.A., Gao T., Papparis C., Van Doorn J., Du K., Xiang K., Chan L., Vivas L., Pradham P., McCalder J., Low K., England W.E., Kuzma D., Conly J., Ryan M.C., Achari G., Hu J., Cabaj J.L., Sikora C., Svenson L., Zelyas N., Servos M., Meddings J., Hrudey S.E., Frankowski K., Parkins M.D., Pang X., Lee B.E. medRxiv; 2022. Emergence and Spread of the SARS-CoV-2 Omicron Variant in Alberta Communities Revealed by Wastewater Monitoring. Accessed in January 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Lara R., Elsinga G., Heijnen L., Munnink B.B.O., Schapendonk C.M., Nieuwenhuijse D., Kon M., Lu L., Aarestrup F.M., Lycett S., Medema G., Koopmans M.P.G., de Graff M. Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerg. Infect. Dis. 2021;27(5):1405. doi: 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R., Sharma J.R., Ramharack P., Mangwana N., Kinnear C., Viraragavan A., Glanzmann B., Louw J., Abdelatif N., Reddy T., Surujlal-Naicker S., Nkambule S., Mahlangeni N., Webster C., Mdhluli M., Gray G., Mathee A., Preiser W., Muller C., Street R. Tracking the circulating SARS-CoV-2 variant of concern in South Africa using wastewater-based epidemiology. Sci. Rep. 2022;12(1):1. doi: 10.1038/s41598-022-05110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398(10317):2126. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrou A.S., Shirk P., Steele M.K., Paul P., Paden C.R., Cadwell B., Reese H.E., Aoki Y., Hassell N., Zheng X.Y., Talarico S., Chen J.C., Oberste M.S., Batra D., McMullan L.K., Halpin A.L., Galloway S.E., MacCannell D.R., Kondor R., Barnes J., MacNeil A., Silk B.J., Dugan V.G., Scobie H.M., Wentworth D.E. Genomic surveillance for SARS-CoV-2 variants: predominance of the Delta (B. 1.617. 2) and omicron (B. 1.1. 529) variants—United States, June 2021–January 2022. MMWR Morb. Mortal. Wkly Rep. 2022;71(6):206. doi: 10.15585/mmwr.mm7106a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraff C., Wang L.Y.R., Buchanan C., Wells M., Schonfeld J., Bessonov K., Ali J., Robert E., Nadon C. medRxiv; 2021. Metagenomic Sequencing of Municipal Wastewater Provides a Near-complete SARS-CoV-2 Genome Sequence Identified as the B. 1.1. 7 Variant of Concern From a Canadian Municipality Concurrent With an Outbreak. Accessed in May 2022. [DOI] [Google Scholar]
- Levine-Tiefenbrun M., Yelin I., Alapi H., Katz R., Herzel E., Kuint J., Chodick G., Gazit S., Patalon T., Kishony R. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat. Med. 2021;27(12):2108. doi: 10.1038/s41591-021-01575-4. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang S., Shi J., Luby S.P., Jiang G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021;415 doi: 10.1016/j.cej.2021.129039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Deng A., Li K., Hu Y., Li Z., Shi Y., Xiong Q., Liu Z., Guo Q., Zou L., Zhang H., Zhang M., Ouyang F., Su J., Su W., Xu J., Lin H., Sun J., Peng J., Jiang H., Zhou P., Hu T., Luo M., Zhang Y., Zheng H., Xiao J., Liu T., Tan M., Che R., Zeng H., Zheng Z., Huang Y., Yu J., Yi L., Wu J., Chen J., Zhong H., Deng X., Kang M., Pybus O.G., Hall M., Lythgoe K.A., Li Y., Yuan J., He J., Lu J. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat. Commun. 2022;13(1):1. doi: 10.1038/s41467-022-28089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Mindermann S., Sharma M., Whittaker C., Mellan T.A., Wilton T., Klapsa D., Mate R., Fritzsche M., Zambon M., Ahuja J., Howes A., Miscouridou X., Nason G.P., Ratmann O., Semenova E., Leech G., Sandkühler J.F., Rogers-Smith C., Vollmer M., Unwin H.J.T., Gal Y., Chand M., Gandy A., Martin J., Volz E., Ferguson N.M., Bhatt S., Brauner J.M., Flaxman S. Changing composition of SARS-CoV-2 lineages and rise of Delta variant in England. EClinicalMedicine. 2021;39 doi: 10.1016/j.eclinm.2021.101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto F.P., Teixeira D.G., da Cunha D.C., Morais I.C., Tavares C.P., Gurgel G.P., do Nascimento S.D., dos Santos D.C., Sales A.D.O., Jerônimo S.M. medRxiv; 2022. SARS-CoV-2 Reinfections With BA. 1 (Omicron) Variant Among Fully Vaccinated Individuals in the Northeast of Brazil. Accessed in May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg T., Harman K., Zaidi A., Seaman S., Andrews N., Nash S.G., Charlett A., Lopez Bernal J., Myers R., Groves N., Gallagher E., Gharbia S., Chand M., Thelwall S., De Angelis D., Dabrera G., Presanis A. Hospitalisation and mortality risk for COVID-19 cases with SARS-CoV-2 AY. 4.2 (VUI-21OCT-01) compared to non-AY. 4.2 Delta variant sub-lineages. J. Infect. Dis. 2022 doi: 10.1093/infdis/jiac063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikolaou V., Chrysovergis A., Ragos V., Tsiambas E., Katsinis S., Manoli A., Papouliakos S., Roukas D., Mastronikolis S., Peschos D., Batistatou A., Kyrodimos E., Mastronikolis N. From Delta to omicron: S1-RBD/S2 mutation/deletion equilibrium in SARS-CoV-2 defined variants. Gene. 2022;814 doi: 10.1016/j.gene.2021.146134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W., Lidder R., Daigle J., Wonitowy Q., Dueck C., Nagasawa A., Mulvey M.R., Mangat C.S. RT-qPCR detection of SARS-CoV-2 mutations S 69–70 del, S N501Y and N D3L associated with variants of concern in Canadian wastewater samples. Sci. Total Environ. 2022;810 doi: 10.1016/j.scitotenv.2021.151283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. Addendum: a dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2021;6(3):415. doi: 10.1038/s41564-021-00872-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranasinghe D., Jayathilaka D., Jeewandara C., Gunasinghe D., Ariyaratne D., Jayadasa T., Kuruppu H., Wijesinghe A., Bary F., Madushanka D., Darshana P., Guruge D., Wijayamuni R., Ogg G.S., Malavige G.S. medRxiv; 2022. Molecular Epidemiology of AY. 28 and AY. 104 Delta Sub-lineages in Sri Lanka. Accessed in May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saskatchewan Health Authority Dash Board, n.d.Saskatchewan Health Authority Dash Board (n.d.). https://www.saskatchewan.ca/government/health-care-administration-and-provider-resources/treatment-procedures-and-guidelines/emerging-public-health-issues/2019-novel-coronavirus/testing-information/where. Accessed in May 2022.
- Stephens N., Béen F., Savic D. An analysis of SARS-CoV-2 in wastewater to evaluate the effectiveness of nonpharmaceutical interventions against COVID-19 in the Netherlands. ACS ES&T Water Article ASAP. 2022 doi: 10.1021/acsestwater.2c0007. [DOI] [PubMed] [Google Scholar]
- Wang C., Han J. Will the COVID-19 pandemic end with the Delta and omicron variants? Environ. Chem. Lett. 2022;p11 doi: 10.1007/s10311-021-01369-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Challis J.K., Oloye F.F., Asadi M., Cantin J., Brinkmann M., McPhedran K.N., Hogan N., Sadowski M., Jones P.D., Landgraff C., Mangat C., Servos M.R., Giesy J.P. RNA in municipal wastewater reveals magnitudes of COVID-19 outbreaks across four waves driven by SARS-CoV-2 variants of concern. ACS ES&T Water. 2022 doi: 10.1021/acsestwater.1c00349. [DOI] [PubMed] [Google Scholar]
- Yaniv K., Ozer E., Shagan M., Lakkakula S., Plotkin N., Bhandarkar N.S., Kushmaro A. Direct RT-qPCR assay for SARS-CoV-2 variants of concern (Alpha, B. 1.1. 7 and Beta, B. 1.351) detection and quantification in wastewater. Environmental Research. 2021;201:111653. doi: 10.1016/j.envres.2021.111653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A.T., Hughes B., Wolfe M.K., Leon T., Duong D., Rabe A., Kennedy L.C., Ravuri S., White B.J., Wigginton K.R., Boehm A.B., Vugia D.J. Estimating relative abundance of 2 SARS-CoV-2 variants through wastewater surveillance at 2 large metropolitan sites, United States. Emerg. Infect. Dis. 2022;28(5):940. doi: 10.3201/eid2805.212488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data