Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to pose serious threats to global health. We previously reported that AAK1, BIKE and GAK, members of the Numb-associated kinase family, control intracellular trafficking of multiple RNA viruses during viral entry and assembly/egress. Here, using both genetic and pharmacological approaches, we probe the functional relevance of NAKs for SARS-CoV-2 infection. siRNA-mediated depletion of AAK1, BIKE, GAK, and STK16, the fourth member of the NAK family, suppressed SARS-CoV-2 infection in human lung epithelial cells. Both known and novel small molecules with potent AAK1/BIKE, GAK or STK16 activity suppressed SARS-CoV-2 infection. Moreover, combination treatment with the approved anti-cancer drugs, sunitinib and erlotinib, with potent anti-AAK1/BIKE and GAK activity, respectively, demonstrated synergistic effect against SARS-CoV-2 infection in vitro. Time-of-addition experiments revealed that pharmacological inhibition of AAK1 and BIKE suppressed viral entry as well as late stages of the SARS-CoV-2 life cycle. Lastly, suppression of NAKs expression by siRNAs inhibited entry of both wild type and SARS-CoV-2 pseudovirus. These findings provide insight into the roles of NAKs in SARS-CoV-2 infection and establish a proof-of-principle that pharmacological inhibition of NAKs can be potentially used as a host-targeted approach to treat SARS-CoV-2 with potential implications to other coronaviruses.
Keywords: SARS-CoV-2, Numb-associated kinases, Kinase inhibitors, Host-targeted antivirals
1. Introduction
SARS-CoV-2 responsible for the COVID-19 pandemic continues to pose a major global health challenge (Zhou et al., 2020; Mallah et al., 2021). While effective vaccines are available to reduce COVID-19 severity, limited access to vaccines in various parts of the world, vaccine hesitancy, and continuous emergence of new viral variants are among the ongoing challenges. A few direct-acting antivirals (DAAs), including the protease inhibitor nirmatrelvir (Owen et al., 2021) and the polymerase inhibitor molnupiravir (Jayk Bernal et al., 2022), have demonstrated promising reduction of hospitalization and death rates. Nevertheless, widespread use of these DAAs may lead to the emergence of resistant viral variants. Indeed, viral escape mutants were identified in cultured cells infected with various coronaviruses under treatment with the polymerase inhibitors molnupiravir (Agostini et al., 2019) and remdesivir (Stevens et al., 2022). There is thus a need for host-targeted antiviral drugs that will effectively suppress viral replication with a high barrier to resistance.
SARS-CoV-2 entry into target cells is mediated by the angiotensin-converting enzyme 2 (ACE2) receptor and is enhanced by neuropilin-1 (NRP-1) (Cantuti-Castelvetri et al., 2020). Fusion of the SARS-CoV-2 envelope is thought to occur primarily at the plasma membrane where the cellular transmembrane protease serine 2 (TMPRSS2) cleaves the viral spike (S) protein at the S2’ site following its pre-cleavage at the S1/S2 site (Hoffmann et al., 2020). Nevertheless, fusion with endosomal membranes following clathrin-mediated endocytosis (CME) and cleavage of the S protein by cathepsin L has also been reported, particularly in cells expressing low level of TMPRSS2 (Bayati et al., 2021). Regardless of the entry mechanism, the replicase complex is then translated from the RNA genome followed by viral RNA replication. Virions are thought to assemble in the endoplasmic reticulum-golgi intermediate compartment (ERGIC) and egress via the secretory pathway (Klein et al., 2020).
The Numb-associated kinases family of Ser/Thr kinases is composed of adaptor-associated kinase 1 (AAK1), BMP-2 inducible kinase (BIKE/BMP2K), cyclin G-associated kinase (GAK), and serine/threonine kinase 16 (STK16). These kinases share only limited homology in their kinase domain and low homology in other protein regions (Sorrell et al., 2016). NAKs have been shown to regulate intracellular membrane trafficking (Sorensen and Conner, 2008; Sato et al., 2009). AAK1 and GAK phosphorylate the endocytic adaptor protein complex 2 (AP2M1) and the secretory AP complex 1 (AP1M1), thereby stimulating their binding to cellular cargo (Conner and Schmid, 2002). BIKE was identified as an accessory protein on a subset of clathrin-coated vesicles that binds the endocytic adaptor NUMB and phosphorylates AP2M1 (Kearns et al., 2001; Krieger et al., 2013; Sorrell et al., 2016; Pu et al., 2020). STK16, the most distantly related member of the NAK family, regulates various physiological activities including Golgi assembly, trans-Golgi network (TGN) protein secretion and sorting, as well as cell cycle and TGF-β signaling (In et al., 2014; López-Coral et al., 2018).
We have previously discovered that AAK1 and GAK mediate intracellular trafficking of hepatitis C virus (HCV), dengue virus (DENV) and Ebola virus (EBOV) during viral entry and assembly/egress (Neveu et al., 2012, 2015; Bekerman et al., 2017; Xiao et al., 2018). We have recently shown a requirement for BIKE in DENV infection (Pu et al., 2020), beyond its reported role in human immunodeficiency virus-1 (HIV-1) infection (Zhou et al., 2008). Additionally, we have demonstrated that pharmacological inhibition of AAK1/BIKE and/or GAK activity by known or novel small molecules has a broad-spectrum antiviral potential both in vitro and in vivo (Bekerman et al., 2017; Pu et al., 2020).
While AAK1 was proposed as a cellular target of SARS-CoV-2 based on in silico analysis (Richardson et al., 2020; Stebbing et al., 2020), the roles of NAKs in SARS-CoV-2 infection and as candidate targets for anti-SARS-CoV-2 therapy remain largely uncharacterized. Here, we sought to probe the functional relevance of the four members of the NAK family in SARS-CoV-2 infection and determine their role as candidate anti-SARS-CoV-2 targets.
2. Materials and methods
2.1. Cells
Calu-3 cells (ATCC) were grown in medium (DMEM; Gibco) supplemented with 10% fetal calf serum (FCS; Omega Scientific Inc.), 1% Pen-strep (Gibco) and 1% nonessential amino acids (NEAA, Gibco). The African green monkey kidney cell line (Vero E6, ATCC) was maintained in DMEM supplemented with 10% FCS, 1% l-glutamine 200 mM, 1% Pen-strep, 1% NEAA, 1% HEPES (Gibco), 1% Sodium pyruvate (Thermo Fisher Scientific). Vero E6-TMPRSS2 (JCRB cell bank, #cat JCRB1819) were maintained in DMEM supplemented with 10% FCS, 1% Pen-strep and 1 mg/ml G418 (Gibco, cat. #10131035). HEK-293T (ATCC) cells were grown in DMEM supplemented with 10% FCS, 1% l-glutamine, and 1% Pen-strep. Cells were maintained in a humidified incubator with 5% CO2 at 37 °C and were tested negative for mycoplasma.
2.2. Compounds
The following reagents were used: (5Z)-7-oxozeaenol (Cayman Chemical), sunitinib malate, (Selleckchem), erlotinib (LC Laboratories), gefitinib (Selleckchem), baricitinib (a gift from Dr. Chris Liang), SGC-GAK-1 and STK16-IN-1 (MedChemExpress). RMC-76 (compound 21b) and RMC-242 (compound 7b) were prepared as previously described (Verdonck et al., 2019; Martinez-Gualda et al., 2021).
2.3. Viral stocks preparation and sequencing
The 2019-nCoV/USA-WA1/2020 SARS-CoV-2 isolate (NR-52281) (BEI Resources) was passaged 3 times in Vero E6-TMPRSS2 cells. The rSARS-CoV-2/WT and rSARS-CoV-2 expressing Nluc-reporter genes (rSARS-CoV-2/Nluc) were generated as previously described (Chiem et al., 2020; Ye et al., 2020). Briefly, Vero E6 cells were transfected using Lipofectamine 2000 (Thermo Fisher) with 4 mg/well of pBeloBAC11-SARS-CoV-2/WT or – Nluc-2A. At 12 hours (h) post-transfection, medium was replaced by DMEM with 2% FBS. At 72 h, P0 virus-containing tissue culture supernatants were collected and stored at −80 °C. Following titration, P0 virus stock was used to generate a P1 stock by infecting Vero E6 monolayers with multiplicity of infection (MOI) of 0.0001 for 72 h. P1 virus was passaged twice in Vero E6-TMPRSS2 cells. Viral titers were determined by standard plaque assay on Vero E6 cells.
Work involving WT SARS-CoV-2 was conducted at the BSL3 facilities of Stanford University, George Mason University, and Texas Biomedical Research Institute according to CDC and institutional guidelines. SARS-CoV-2 stocks were deep sequenced on a MiSeq platform (Illumina). SARS-CoV-2 whole-genome amplicon-based sequencing was conducted by adapting an existing whole genome sequencing pipeline for poliovirus genotyping as described (Wang et al., 2021). All the experiments were performed using a P3 SARS-CoV-2 USA-WA1/2020, rSARS-CoV-2/WT or rSARS-CoV-2/Nluc containing 100% WT population with no deletion in the Spike multi-basic cleavage site.
2.4. rVSV-SARS-CoV-2-S pseudovirus production
As described in (Saul et al., 2021b), HEK-293T cells were transfected with 30 μg of a Spike expression plasmid. After 24 h, the medium was replaced and cells were treated with valproic acid (VPA). Following a 4 h incubation, the cells were infected with VSV-G pseudo typed ΔG-luciferase VSV virus (MOI = 3). At 6 h post-infection (hpi), cells were washed with PBS, and fresh medium containing anti-VSV-G hybridoma was added to neutralize the residual VSV-G pseudovirus. Twenty-four hours later, culture supernatant was harvested, clarified by centrifugation, filtered (0.22 μm) and stored at −80 °C. rVSV-SARS-CoV-2-S was titrated on Vero cells via luciferase assay, and TCID50 was determined. Positive wells were defined as having luminescence values at least 10-fold higher than the cell background.
2.5. Plaque assay and nanoluciferase (Nluc) assay
For plaque assays, monolayers of Vero E6 cells were infected with WT SARS-CoV-2 or rSARS-CoV-2/Nluc for 1 h at 37 °C. Cells were then overlaid with MEM and carboxymethyl cellulose and incubated at 37 °C. At 72 hpi, the overlay was removed, and the cells were submerged in 70% ethanol for fixation and viral inactivation followed by crystal violet staining. Nluc expression was determined on a plate reader GloMax Discover Microplate Reader (Promega) at 24 hpi in the infected cell supernatants using a luciferin solution obtained from the hydrolysis of its O-acetylated precursor, hikarazine-103 (prepared by Dr. Yves Janin, Pasteur Institute, France) as a substrate (Coutant et al., 2019, 2020).
2.6. Time-of-addition experiment
Calu-3 cells were infected with rSARS-CoV-2/WT (MOI = 1). Two hpi, the virus was removed and cells were washed twice with PBS. At 2, 5 and 8 hpi, 5 μM of RMC-76 or 0.1% DMSO were added. At 10 hpi, cell culture supernatants were collected for infectious viral titer measurement by plaque assay and cell lysates were used for measurement of intracellular SARS-CoV-2 nucleocapsid (N) copy number by RT-qPCR as described below.
2.7. RNA interference
The following siRNAs (10 pmole) were transfected into Calu-3 cells using Dharmafect-4 transfection reagent (#cat: T-2004-02, Dharmacon) 48 h prior to infection with SARS-CoV-2: siAAK1 (ID: s22494, Ambion), siBIKE (M-005071-01, Dharmacon), siGAK (ID: s5529, Ambion), siSTK16 (AM51331, Life Technology) and siNT (D-001206-13-05, Dharmacon).
2.8. Western blotting and antibodies
Calu-3 cells were lysed in RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, #cat 89900) supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Clarified protein lysates were run on 4%–12% Bis-Tris gels (Invitrogen) and transferred onto PVDF membranes using dry transfer (iBlot2, Invitrogen). Membranes were blocked and blotted with anti-AAK1 (Abcam, #cat ab134971), anti-BIKE (Santa Cruz Biotechnology, #cat sc-134284), anti-GAK (Abcam, #cat ab186120), anti-STK16 (Santa Cruz Biotechnology, #cat sc-374356) and anti–β-actin (Sigma-Aldrich, #cat A3854) antibodies. Signal was detected with secondary antibodies conjugated with HRP, and with ECL2 substrate (Pierce). The images were quantified using ImageJ software.
2.9. Infection assays
Vero E6 cells and Calu-3 were infected with SARS-CoV-2 (USA-WA1/2020) or rSARS-CoV-2/Nluc in triplicates (MOI = 0.05) in DMEM containing 2% FCS. After 1-h incubation at 37 °C, the viral inoculum was removed, cells were washed and new medium was added. Culture supernatants were harvested at 24 hpi and the viral titer was measured via plaque or Nluc assays.
2.10. Entry assays
Calu-3 cells were infected with rSARS-CoV-2/WT (MOI = 1) or a high inoculum of rVSV-SARS-CoV-2-S pseudovirus (MOI = 3). Following 2-h incubation, the viral inoculum was removed, cells were washed with PBS and lysed in TRIzol LS, and viral RNA level measured by RT-qPCR.
2.11. RT-qPCR
RNA was extracted from Calu-3 lysates using Direct-zol RNA Miniprep Plus Kit (Zymo Research) and reverse transcribed using High-Capacity cDNA RT kit (Applied Biosystems) according to the manufacturer's instructions. Primers and PowerUp SYBR Green Master Mix (Applied Biosystems) were added to the samples, and PCR reactions were performed with QuantStudio3 (Applied Biosystems) in triplicates. Target genes were normalized to the housekeeping gene (GAPDH). Sequences of primers used for RT-qPCR are available upon request.
2.12. Pharmacological inhibition
The inhibitors or DMSO were administered to cells 1 h prior to viral inoculation and maintained for the duration of the experiment. Viral infection was measured via Nluc (rSARS-CoV-2/Nluc) or plaque (SARS-CoV-2) assays.
2.13. Viability assays
Cell viability was assessed using alamarBlue reagent (Invitrogen) according to the manufacturer's protocol. Fluorescence was detected at 560 nm on an InfiniteM1000 plate reader (Tecan) and GloMax Discover Microplate Reader (Promega).
2.14. NanoBRET assay
NanoBRET for RMC-242 was performed at Carna Biosciences. Briefly, HEK293 cells were transiently transfected with the NanoLuc® Fusion DNA and incubated at 37 °C. 20 h post-transfection, NanoBRET™ tracer reagent and RMC-242 were added to the cells and incubated at 37 °C for 2 h. Nanoluciferase-based bioluminescence resonance energy transfer (BRET) was measured using NanoBRET™ Nano-Glo® Substrate on a GloMax® Discover Multimode Microplate Reader (Promega).
2.15. In vitro kinase assays
In vitro kinase assays to determine IC50 of gefitinib for AAK1 and BIKE were performed busing the LabChip platform (Nanosyn).
2.16. Multiplexed Inhibitor Bead (MIB) affinity chromatography/MS analysis
Kinome profiling was performed as previously described (Duncan et al., 2012; Asquith et al., 2018). Briefly, SUM159 cell lysates were incubated with DMSO or the indicated concentration of RMC-76 for 30 min on ice. Kinase fragments were then detected and analyzed by mass spectrometry. Kinase abundance was quantified label-free using MaxQuant software.
2.17. Data analysis of combination drug treatment
The MacSynergy II program was used to perform synergy/antagonism analysis, as previously described (Prichard and Shipman, 1996; Neveu et al., 2015; Bekerman et al., 2017). The combination's effect is determined by subtracting the theoretical additive values from the experimental values. A synergy peak is depicted by the three-dimensional differential surface plot above a theoretical additive plane and antagonism as depressions below it (Prichard and Shipman, 1990). Matrix data sets in three replicates were assessed at the 95% confidence level for each experiment. Synergy and log volume were calculated.
2.18. Statistical analysis
All data were analyzed with GraphPad Prism software. Half-maximal effective concentrations (EC50) and half-maximal cytotoxic concentrations (CC50) were measured by fitting of data to a 3-parameter logistic curve. P values were calculated by ordinary 1-way ANOVA with Dunnett's multiple comparisons tests as specified in each figure legend.
3. Results
3.1. NAKs are required for wild type (WT) SARS-CoV-2 infection
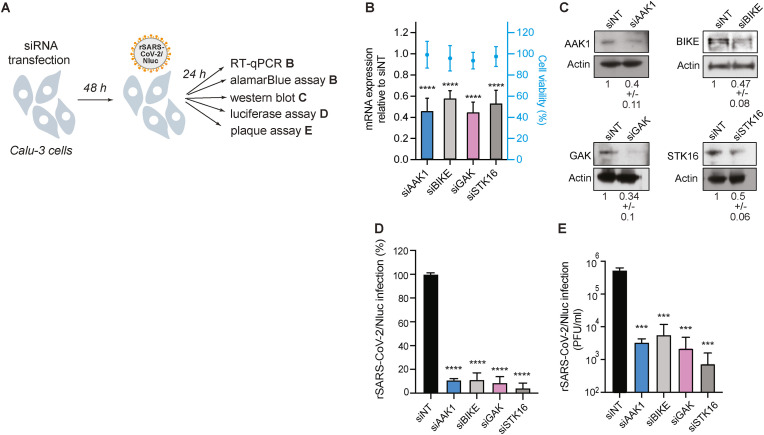
To probe the functional relevance of NAKs to WT SARS-CoV-2 infection, we first used an RNAi approach. Calu-3 (human lung epithelial) cells were depleted of AAK1, BIKE, GAK and STK16 individually via siRNAs (Fig. 1 A), and knockdown efficiency was confirmed via RT-qPCR (Fig. 1B) and western blot analysis (Fig. 1C). NAKs depletion had no apparent effect on Calu-3 cell viability, as measured by an alamarBlue assay (Fig. 1B). Nevertheless, it reduced SARS-CoV-2 infection by 89% ± 1.1% (siAAK1), 91% ± 5.7% (siBIKE), 91% ± 5.0% (siGAK), and 96% ± 4.3% (siSTK16) relative to the non-targeting (siNT) control, as measured via Nluc assay at 24 h post-infection (hpi) with rSARS-CoV-2/Nluc virus (SARS-CoV-2 USA-WA1/2020 isolate (NR-52281) expressing a nanoluciferase reporter gene)) (Chiem et al., 2020; Ye et al., 2020) (Fig. 1D). Moreover, a 2–3 log reduction in SARS-CoV-2 titer was measured in the infected cells relative to the siNT control via plaque assay at 24 hpi (Fig. 1E). These loss-of-function experiments provide genetic evidence that the four NAKs are required for effective SARS-CoV-2 infection.
Fig. 1.
NAKs are required for SARS-CoV-2 infection.
(A) Schematic of the experiments shown in panels B–E. (B) Confirmation of siRNA-mediated gene expression knockdown and cell viability in Calu-3 cells transfected with the indicated siRNAs. Shown is gene expression normalized to GAPDH and expressed relative to the respective gene level in the non-targeting (siNT) control at 48 h post-transfection measured by RT-qPCR and cell viability measured by alamarBlue assay. (C) Confirmation of siRNA-mediated protein expression knockdown efficiency by western blot. Representative membranes and quantitative data from two experiments are shown. (D) WT SARS-CoV-2 infection measured at 24 h post-infection (hpi) of the indicated NAK-depleted Calu-3 cells with rSARS-CoV-2/Nluc (USA-WA1/2020 strain; MOI = 0.05) by Nluc assays. (E) Viral titers measured at 24 hpi of the indicated NAK-depleted Calu-3 cells with rSARS-CoV-2/Nluc (USA-WA1/2020 strain; MOI = 0.05) by plaque assays. Data in all panels are representative of two or more independent experiments. Individual experiments had three biological replicates, means ± standard deviation (SD) are shown. Data are relative to siNT (B, D and E). ***P < 0.001; ****P < 0.0001 by 1-way ANOVA followed by Dunnett's multiple comparisons test. PFU, plaque-forming units.
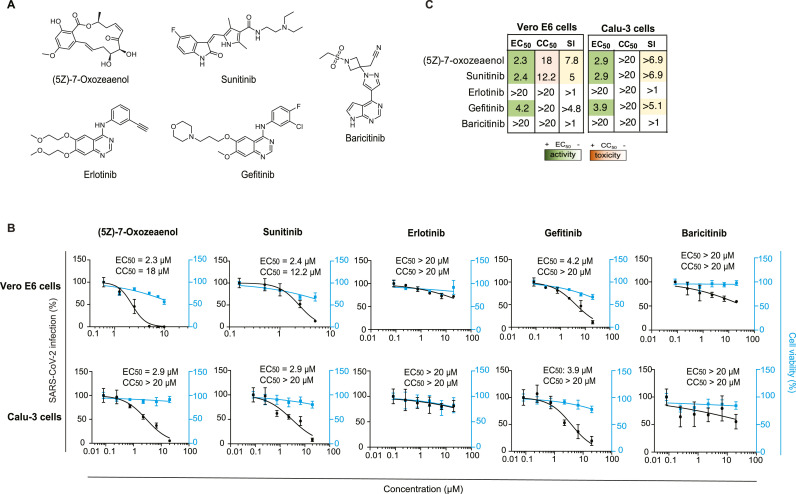
3.2. Known pharmacological inhibitors with anti-NAK activity suppress SARS-CoV-2 infection
To determine whether a similar effect on SARS-CoV-2 infection to that achieved genetically can be achieved pharmacologically, we determined the anti-SARS-CoV-2 activity of a set of 5 compounds: (5Z)-7-oxozeaenol, sunitinib, erlotinib, gefitinib and baricitinib (Table 1 and Fig. 2 A), via two orthogonal assays in two distinct cell lines. Vero E6 and Calu-3 cells were pretreated for 1 h with the compounds followed by infection with WT SARS-CoV-2 (2019-nCoV/USA-WA1/2020) or rSARS-CoV-2/Nluc (Chiem et al., 2021), respectively, both at an MOI of 0.05. The antiviral effect of the compounds was measured at 24 hpi via plaque (Vero E6 cells) or Nluc (Calu-3 cells) assays. Their effect on cellular viability was measured both in the infected (Fig. 2) and uninfected cells (Supplementary Fig. S1) via alamarBlue assays.
Table 1.
Kinase inhibitors with anti-NAK activity used in this study. Dissociation constant (Kd), enzymatic activity (IC50), percent binding of control (% of control) values measured in vitro on the four NAKs and NanoBRET GAK IC50 values measured in cells of the indicated kinase inhibitors. The source of KINOMEscan data (if available) and other targets of these compounds are also shown. Kd, IC50, and % of control are shown as *, ** and *** respectively. ^ represents data obtained in this study.
| Compound | Status |
|
NanoBRET GAK IC50 (nM) | Reference | Kinome | Other targets | |||
|---|---|---|---|---|---|---|---|---|---|
| AAK1 | BIKE | GAK | STK16 | ||||||
| (5Z)-7- Oxozeaenol | Investigational | 13%*** | 3.8%*** | 10%*** | 100%*** | NA | Pu et al.,2020 | ID: 20211 | TAK1 (8 nM), VEGFR2 (52 nM), ERK2 (80 nM) |
| Sunitinib | Approved (cancer) | 11 nM* | 5.5 nM* | 20 nM* | 250 nM* | NA | Davis et al.,2011 | Davis et al.,2011 | VEGFR2 (80 nM), PDGFRβ (2 nM), FLT3, KIT, PDGFRα |
| Erlotinib | Approved (cancer) | 1200 nM* | 1200 nM* | 3 nM* | >9000 nM* | 910 nM | Asquith et al., 2019a, Asquith et al., 2019b | Davis et al.,2011 | ErbB1 (0.7 nM), STK10 (19 nM), YSK4 (25 nM), SLK (26 nM) |
| Gefitinib | Approved (cancer) | >10,000 nM**a | >10,000 nM**a | 13 nM* | NA | 420 nM | Asquith et al., 2019a, Asquith et al., 2019b | Davis et al.,2011 | EGFR |
| Baricitinib | Approved (RA) | 17 nM* | 40 nM* | 134 nM∗ | 69 nM* | NA | Stebbing et al.,2021 | Klaeger et al.,2017 | JAK1 (5.9 nM), JAK2 (5.7 nM), ROCK1/2, TYK2 (53 nM), CAMK2A, MAP3K2, PRPF4B |
| RMC-76 | Experimental | 4 nM** | 3.8 nM*a | 280 nM*a | NA | NA | Figure S1 | NA | CDC42BPB, PRKD3, CLK1, CLK4, ULK3, MINK1 |
| RMC-242 | Experimental | 4100 nM*a | >15000 n M*a | 12 nM** | NA | 78 nMa | Martinez-Gualda et al.,2021 | NA | NA |
| SGC-GAK-1 | Experimental | NA 86%*** |
NA 92%*** |
2 nM* 9.7%*** |
NA 84%*** |
48 nM | Asquith et al., 2019a, Asquith et al., 2019b | Asquith et al., 2019a, Asquith et al., 2019b | RIPK2 (110 nM), ADCK3 (190 nM), NLK (520 nM) |
| STK-16-1N-1 | Experimental | NA 13%*** |
NA 9.5%*** |
NA 46%*** |
295 nM** 0.65%*** |
NA | Liu et al., 2016 | Liu et al.,2016 | PI3Kδ (856 nM), PI3Kγ (867 nM) |
Data obtained in this study, RA= Rheumatoid Arthritis
Fig. 2.
Known compounds with anti-NAK activity suppress SARS-CoV-2 infection.
(A) Chemical structures of known compounds with anti-NAK activity. (B) Dose response of WT SARS-CoV-2 infection to the indicated compounds in Vero E6 cells infected with USA-WA1/2020 strain and in Calu-3 cells infected with rSARS-CoV-2/Nluc (MOIs = 0.05) (black) measured via plaque and Nluc assays at 24 hpi, respectively. Dose response of cellular viability to the indicated compounds measured via alamarBlue assays are shown in blue. (C) Heat maps of the EC50 and CC50 values of the inhibitors color-coded based on the antiviral activity and cell viability measured in the indicated cell lines. Selectivity indices (SI, CC50 to EC50 ratios) greater than 5 are depicted in yellow. Data in all panels are representative of two or more independent experiments. Individual experiments had three or greater biological replicates. Shown are means ± SD.
(5Z)-7-oxozeaenol, an investigational anticancer natural product, is an ATP-competitive inhibitor (Ninomiya-Tsuji et al., 2003; Wu et al., 2013) that potently binds BIKE, GAK and AAK1 (3.8%, 10% and 13% binding of control, respectively, at 10 μM) (KINOMEscan, ID:20211) (Table 1), previously demonstrating a broad-spectrum antiviral activity against DENV, Venezuelan equine encephalitis virus (VEEV/TC-83), and EBOV (Pu et al., 2020). We measured a dose-dependent inhibition of SARS-CoV-2 infection in Vero E6 and Calu-3 cells upon drug treatment with (5Z)-7-oxozeaenol with EC50 values of 2.3 μM, and 2.9 μM, respectively (Fig. 2B and C). While mild toxicity was observed in Vero E6 cells with a CC50 of 18 μM, no apparent toxicity was measured at the tested concentrations in Calu-3 cells (Fig. 2B and C).
Sunitinib, an approved multi-kinase inhibitor with potent binding to AAK1 (dissociation constant (K d = 11 nM)), BIKE (K d = 5.5 nM), and GAK (K d = 20 nM) (Table 1) (Davis et al., 2011) demonstrated a moderate anti-SARS-CoV-2 effect in Vero E6 (EC50 = 2.4 μM) and Calu-3 (EC50 = 2.9 μM) cells (Fig. 2B and C). Like (5Z)-7-oxozeaenol, sunitinib reduced cellular viability in Vero E6 cells (CC50 = 12.2 μM), yet had no apparent toxicity in Calu-3 cells (CC50 > 20 μM).
Erlotinib, an approved anticancer EGFR inhibitor (K d = 0.7 nM) (KINOMEscan, ID:20160), showed no anti-SARS-CoV-2 activity and no cellular toxicity in both cell lines (EC50 > 20 μM, CC50 > 20 μM) (Fig. 2B and C). Notably, while erlotinib potently binds GAK in vitro (K d = 3 nM) (Davis et al., 2011), its intracellular anti-GAK activity is low with an IC50 of 910 nM, as measured by a nanoluciferase-based bioluminescence resonance energy transfer (NanoBRET) assay (Table 1) (Asquith et al., 2019b, 2020). Gefitinib, another approved drug with potent GAK binding activity (K d = 13 nM) (Davis et al., 2011) and moderate anti-GAK activity via the NanoBRET assay (IC50 = 420 nM) (Table 1), moderately suppressed SARS-CoV-2 infection in both cell lines (EC50 = 3.9–4.2 μM), with some toxicity in Vero E6 (CC50 = 13 μM) but not in Calu-3 cells (CC50 > 20 μM) (Fig. 2B and C).
Baricitinib, a janus kinase (JAK) inhibitor approved for inflammatory diseases and used for COVID-19 treatment for its predicted potential effect on AAK1-mediated SARS-CoV-2 entry, beyond JAK-mediated inflammation (Richardson et al., 2020; Stebbing et al., 2020), showed weak activity against SARS-CoV-2 in Vero E6 (EC50 > 20 μM) and largely no antiviral activity in Calu-3 cells (Fig. 2B and C), in line with its weaker anti-NAK activity (AAK1, K d = 17 nM; BIKE, K d = 40 nM; and GAK, K d = 134 nM) (Stebbing et al., 2021) (Table 1).
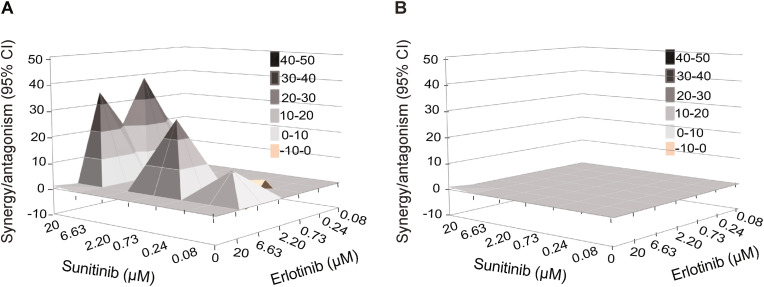
3.3. Sunitinib and erlotinib have a synergistic anti-SARS-CoV-2 effect
To determine whether a synergistic effect could be achieved with a combination treatment of compounds with anti-AAK1/BIKE and GAK activity, we measured the anti-SARS-CoV-2 activity of combination treatment with sunitinib and erlotinib. Calu-3 cells were pretreated for 1 h with the combination treatment followed by infection with rSARS-CoV-2/Nluc (MOI = 0.05). The effects of treatment on viral infection and cellular viability were measured at 24 hpi via Nluc and alamarBlue assays, respectively. Treatment with combinations of sunitinib and erlotinib exhibited synergistic inhibition of SARS-CoV-2 infection with a synergy volume of 38 μM2% at the 95% CI and calculated antagonism of −3.41 μM2% (Fig. 3 A) with no synergistic toxicity (Fig. 3B). Based on Prichard et al. (Prichard and Shipman, 1996), this level of synergy is considered minor but significant. These results are in agreement with our reported in vitro and in vivo HCV, DENV and EBOV data (Neveu et al., 2015; Bekerman et al., 2017), and point to combinations of anti-NAK inhibitors as a potential anti-coronaviral strategy.
Fig. 3.
Combination treatment with sunitinib/erlotinib has synergistic effect against SARS-CoV-2.
(A and B) Synergy/antagonism of sunitinib/erlotinib combination treatment on antiviral effect measured in Calu-3 cells infected with rSARS-CoV-2/Nluc (USA-WA1/2020 strain; MOI = 0.05) at 24 hpi via Nluc assays (A) and on cellular viability measured at 24 hpi in the same samples via alamarBlue assays (B). Data represents the differential surface analysis at the 95% confidence interval (CI), analyzed via the MacSynergy II program. Synergy and antagonism are indicated by the peaks above and below the theoretical additive plane, respectively. The level of synergy or antagonism is depicted by the color code on the figure. Data are representative of two independent experiments with three replicates each.
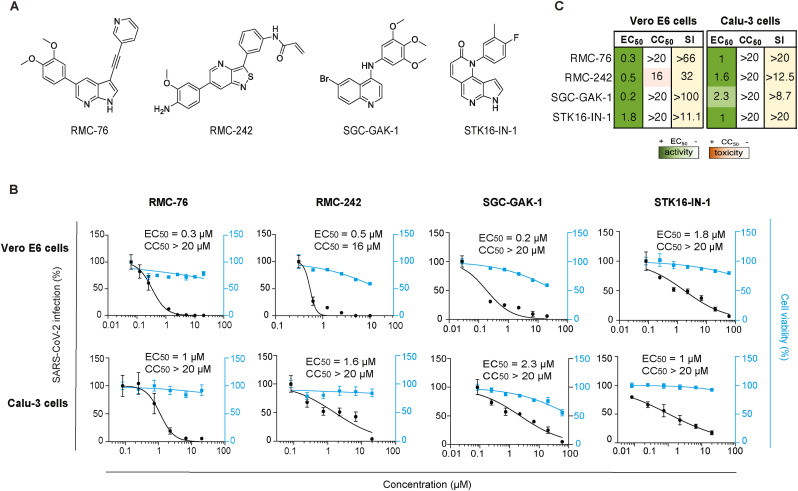
3.4. Novel, chemically distinct NAK inhibitors potently inhibit SARS-CoV-2 infection in vitro
Next, we determined whether chemically distinct, novel small molecules with anti-NAK activity (Table 1 and Fig. 4 A) can suppress SARS-COV-2 infection. RMC-76 is a pyrrolo [2,3-b]pyridine (compound 21b in (Verdonck et al., 2019)) with potent anti-AAK1 (IC50 = 4.0 nM) and anti-BIKE (K d = 3.8 nM) (Table 1) activity and reported activity against DENV and EBOV. RMC-76 effectively suppressed SARS-CoV-2 infection in both Vero E6 (EC50 = 0.3 μM) and Calu-3 (EC50 = 1 μM) cells (Fig. 4B and C), with no apparent toxicity in both cell lines (CC50 > 20 μM). We further evaluated the selectivity of RMC-76 via kinome profiling in cell lysates using multiplexed inhibitor beads kinome profiling coupled with mass spectrometry (MIB/MS) (Duncan et al., 2012; Asquith et al., 2018, 2019b). RMC-76 showed highest selectivity towards AAK1 and BIKE, with lower affinity for the off-target kinases CDC42BPB, PRKD3, CLK1, CLK4, ULK3, and MINK1 (Table 1 and Supplementary Fig. S2).
Fig. 4.
Novel compounds with anti-NAK activity suppress SARS-CoV-2 infection.
(A) Chemical structures of RMC-76, RMC-242, SCG-GAK-1, and STK16-IN-1. (B) Dose response of WT SARS-CoV-2 infection to the indicated compounds in Vero E6 cells infected with USA-WA1/2020 strain and in Calu-3 cells infected with rSARS-CoV-2/Nluc (MOIs = 0.05) (black) measured via plaque and Nluc assays at 24 hpi, respectively. Dose response of cellular viability to the indicated compounds measured via alamarBlue assays are shown in blue. (C) Heat maps of the EC50 and CC50 values of the inhibitors color-coded based on the antiviral activity and cell viability measured in the indicated cell lines. Selectivity indices (SI, CC50 to EC50 ratios) greater than 5 are depicted in yellow. Data in all panels are representative of two or more independent experiments. Individual experiments had three or greater biological replicates. Shown are means ± SD.
RMC-242, an N-(3-(6-(4-amino-3-methoxyphenyl) isothiazolo [4,3-b]pyridin-3-yl)phenyl) acrylamide (compound 7b in (Martinez-Gualda et al., 2021)) with anti-GAK (IC50 = 12 nM) activity (Table 1) has previously shown anti-DENV activity. RMC-242 dose-dependently suppressed SARS-CoV-2 infection in Vero E6 (EC50 = 0.5 μM) and Calu-3 (EC50 = 1.6 μM) cells. While RMC-242 exhibited mild toxicity in Vero cells (CC50 = 16 μM), no apparent toxicity was measured in Calu-3 cells (CC50 > 20 μM) (Fig. 4B and C, Supplementary Fig. S1). SGC-GAK-1, a 4-anilinoquinoline, (compound 11 in (Asquith et al., 2019a)) with potent anti-GAK activity (K d = 2 nM), has previously shown potent anti-DENV activity (Saul et al., 2020). SGC-GAK-1 demonstrated high or moderate anti-SARS-CoV-2 effect in Vero E6 (EC50 = 0.2 μM) and Calu-3 (EC50 = 2.3 μM) cells, respectively, with no apparent cellular toxicity (CC50 > 20 μM) (Fig. 4B and C, Supplementary Fig. S1).
Lastly, STK16-IN-1, a pyrrolonaphthyridinone compound with high selectivity against STK16 (IC50 = 295 nM) (Liu et al., 2016) effectively suppressed SARS-CoV-2 in Vero E6 (EC50 = 1.8 μM) and Calu-3 (EC50 = 1 μM) cells with no apparent toxicity in both cell lines (CC50 > 20 μM) (Fig. 4B and C, Supplementary Fig. S1).
These results validate NAKs as druggable antiviral targets and point to their pharmacological inhibition as a potential anti-SARS-CoV-2 strategy.
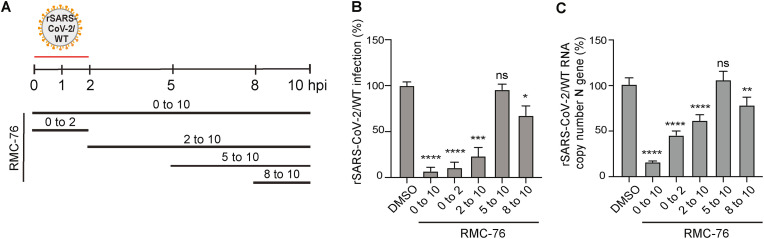
3.5. RMC-76 suppresses early and late stages of the SARS-CoV-2 life cycle
To pinpoint the stage of the viral life cycle that is inhibited by compounds with anti-NAK activity, we next conducted time-of-addition experiments in Calu-3 cells infected with rSARS-CoV-2/WT (MOI = 1). RMC-76 (with potent anti-AAK1/BIKE activity) was added at 2, 5 or 8 hpi (Fig. 5 A). Cell culture supernatants were harvested at 10 hpi, which represents a single cycle of SARS-CoV-2 replication, and infectious viral titers were measured by plaque assay. Treatment with RMC-76 initiated upon infection onset and maintained throughout the 10 h experiment (0–10) suppressed viral infection by 93% ± 4.1% relative to the DMSO control. RMC-76 treatment during the first 2 h of infection only (0–2) suppressed viral infection by +89% ± 5.9% relative to DMSO, indicating that RMC-76 suppresses viral entry (Fig. 5B). Nevertheless, addition of RMC-76 at 2 hpi (2–10) reduced viral infection by 77% ± 9.4% revealing a potential effect in a post-entry stage (Fig. 5B). Whereas treatment with RMC-76 initiated 5 hpi (5–10) had no effect on viral infection, treatment initiated at 8 hpi (8–10) reduced SARS-CoV-2 infection by 33% ± 10.6% (Fig. 5B). RMC-76 had a similar effect on intracellular SARS-CoV-2 nucleocapsid (N) copy number measured by RT-qPCR in cell lysates harvested at the same time points (Fig. 5C). It remains to be determined which property of RMC-76 accounts for the inability to capture its effect when added at 8–10 h in the 5–10 h time window. While RMC-76 is stable in plasma (T1/2 of >240 min) and thus unlikely to be unstable in cells, it is possible that it is being pumped out of the cells via a transporter.
Fig. 5.
RMC-76 inhibits temporally distinct stages of SARS-CoV-2 life cycle.
(A) Schematic of the time-of-addition experiments shown in panel B and C. Calu-3 cells were infected with rSARS-CoV-2/WT at an MOI of 1. At the indicated time points, 5 μM RMC-76 or DMSO were added to the infected cells. (B) Infectious virus titers measured by plaque assays in culture supernatants harvested at 10 hpi. (C) Intracellular SARS-CoV-2 nucleocapsid (N) copy numbers measured by RT-qPCR assays in cell lysates harvested at 10 hpi. Data are relative to DMSO and are representative of two independent experiments, each with three replicates, means ± SD are shown. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA followed by Dunnett's multiple comparisons test. Ns, non-significant.
These results provide evidence that compounds with anti-AAK1/BIKE activity suppress viral entry and a later stage of the SARS-CoV-2 life cycle (assembly and/or egress), but likely not viral RNA replication.
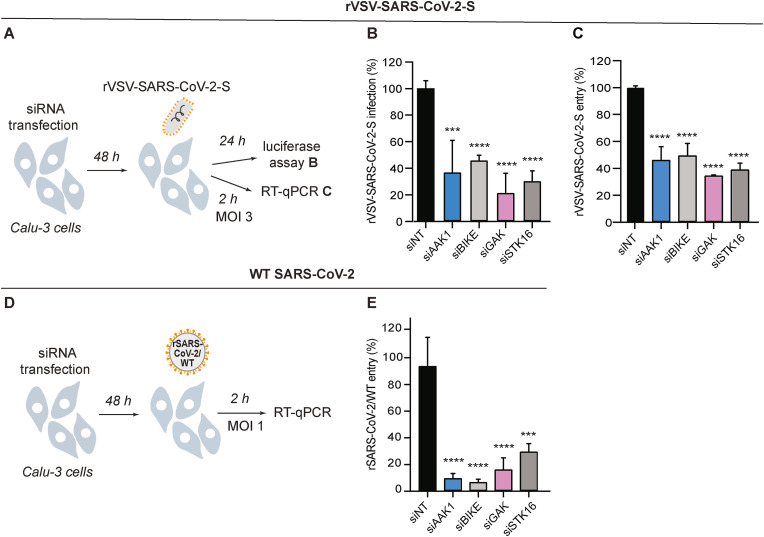
3.6. NAKs are required for SARS-CoV-2 pseudovirus entry
Since RMC-76 demonstrated an effect on viral entry and SARS-CoV-2 is thought to enter target cells in part via CME (Bayati et al., 2021; Koch et al., 2021), we tested the hypothesis that NAKs are involved in mediating SARS-CoV-2 entry. To this end, we first used vesicular stomatitis virus encapsidated RNA pseudotyped with the SARS-CoV-2 spike glycoprotein expressing a luciferase reporter gene (rVSV-SARS-CoV-2-S) (Khanna et al., 2020; Saul et al., 2021b). Calu-3 cells depleted of the individual NAKs (Fig. 1B and C) were infected with rVSV-SARS-CoV-2-S followed by luciferase assays at 24 hpi (Fig. 6 A). Silencing of NAKs expression suppressed rVSV-SARS-CoV-2-S infection by 50–70% relative to siNT (Fig. 6B).
Fig. 6.
NAKs are required for the entry of pseudovirus and WT SARS-CoV-2.
(A) Schematic of the experiments shown in panels B and C. (B) rVSV-SARS-CoV-2-S infection at 24 hpi of Calu-3 cells depleted of the indicated NAKs-by siRNAs (Fig. 1B and C) measured via luciferase assays. (C) rVSV-SARS-CoV-2-S entry at 2 hpi of Calu-3 cells (MOI 3) depleted of the indicated NAKs by siRNAs measured via RT-qPCR. (D) Schematic of the experiment shown in panel E. (E) rSARS-CoV-2/WT entry at 2 hpi of Calu-3 cells (MOI = 1) depleted of the indicated NAKs by siRNAs (Fig. 1B and C) measured via RT-qPCR. Data in all panels are representative of two or more independent experiments. Individual experiments had three biological replicates, means ± SD are shown. Data are relative to siNT (B, C, E). ***P < 0.001; ****P < 0.0001 by 1-way ANOVA followed by Dunnett's multiple comparisons test.
To confirm that the observed defect was indeed in viral entry, we next quantified the level of intracellular viral RNA at 2 hpi of Calu-3 cells depleted of the individual NAKs (Fig. 1, Fig. 6A) with a high-inoculum of rVSV-SARS-CoV-2-S (MOI 3). NAKs depletion reduced the intracellular viral copy number by 50–65% relative to siNT as measured via RT-qPCR, supporting a role for NAKs in SARS-CoV-2 entry (Fig. 6C).
3.7. NAKs are required for WT SARS-CoV-2 entry
To define the role of NAKs in the entry of WT SARS-CoV-2, we conducted entry experiments in Calu-3 cells infected with rSARS-CoV-2/WT virus (MOI = 1) (Fig. 6D). Suppression of NAKs expression (Fig. 1B and C) reduced the level of intracellular viral RNA measured via RT-qPCR at 2 hpi by 90% ± 3.3% (siAAK1), 93% ± 2.0% (siBIKE), 83% ± 8.5% (siGAK), and 70% ± 5.8% (siSTK16) relative to siNT, respectively (Fig. 6E).
Together, these findings provide genetic evidence that the four NAKs are involved in mediating SARS-CoV-2 infection and entry.
4. Discussion
We have previously demonstrated that AAK1 and GAK regulate intracellular trafficking of multiple RNA viruses via phosphorylation of the AP1 and AP2 complexes and represent targets for broad-spectrum antivirals (Neveu et al., 2015; Bekerman et al., 2017; Pu et al., 2018, 2020; Xiao et al., 2018; Verdonck et al., 2019). We have also reported the requirement for BIKE in DENV and unrelated RNA viral infections in part via AP2M1 phosphorylation (Pu et al., 2020). Nevertheless, the roles of NAKs in coronaviral infection remained largely unknown. Moreover, to our knowledge, the role of STK16 in viral infection has not been reported to date. Here, by integrating virology, genetic and pharmacological approaches, we demonstrate a requirement for all four NAKs in SARS-CoV-2 infection. Moreover, we provide a proof of concept for the potential utility of NAK inhibition as a strategy for treating SARS-CoV-2 infection.
AAK1 was previously identified as a candidate target for SARS-CoV-2 antiviral development via an artificial intelligence screen (Richardson et al., 2020; Stebbing et al., 2020). Using an RNAi approach, we provide evidence that AAK1 is required for SARS-CoV-2 infection in a biologically relevant cell culture model, in agreement with a recent study demonstrating a requirement for AAK1 in other human lung cells (Puray-Chavez et al., 2021). Beyond AAK1, we provide genetic evidence that BIKE, GAK and STK16, NAKs whose functional relevance in SARS-CoV-2 has remained unknown, are also proviral factors. To our knowledge, this is the first evidence that STK16 is required for any viral infection.
We provide evidence that NAKs are involved in regulating SARS-CoV-2 entry. Depletion of NAKs reduced the entry of pseudo- and WT SARS-CoV-2 via both Nluc assays and entry-specific assays measuring viral RNA at 2 hpi. Moreover, time-of-addition experiments revealed that pharmacological inhibition of AAK1/BIKE potently inhibited viral infection when added in the first two hpi. While the precise roles of NAKs in viral entry remains to be elucidated, phosphorylation of AP2M1, NAK substrate involved in CME, was reported to be induced upon SARS-CoV-2 infection in lung cells (Puray-Chavez et al., 2021) and to be required for SARS-CoV-2 pseudovirus entry (Wang et al., 2020). Notably, a conserved tyrosine motif in the cytoplasmic tail of ACE2 was shown to mediate its interaction with AP2M1 and to be essential for SARS-CoV-2 entry into cells with low TMPRSS2 expression (Wang et al., 2020). Sorting into late endosomes is the main penetration route of SARS-CoV-2 into cells with low or no TMPRSS2 expression. In TMPRSS2 expressing cells, beyond fusion-mediated penetration at the plasma membrane, SARS-CoV-2 sorting into the endocytic pathway also appears to play a role (Bayati et al., 2021; Koch et al., 2021). Our findings that NAKs, regulators in the endocytic pathway, are required for effective entry of WT SARS-CoV-2 into Calu-3 (high TMPRSS2 expressing) cells support the latter.
Beyond viral entry, the time-of-addition experiments provide evidence that NAKs are also involved in regulating later stage(s) of the viral life cycle, likely assembly and/or egress, but not viral RNA replication. This is in agreement with our prior findings involving their roles in unrelated RNA viral infections such as DENV, EBOV, and VEEV (Neveu et al., 2015; Bekerman et al., 2017; Pu et al., 2020; Saul et al., 2020, 2021a, 2021b; Huang et al., 2021). Together, these findings provide evidence that NAKs regulate temporally distinct stages of the SARS-CoV-2 life cycle and represent candidate targets for the development of anti-SARS-CoV-2 approaches.
Using compounds with potent anti-NAK activity as pharmacological tools we provide further support for NAKs requirement in SARS-CoV-2 infection and validate them as antiviral targets in lung epithelial cells. RMC-76 and sunitinib, novel and approved compounds with potent anti-AAK1/BIKE activity demonstrated antiviral effect against SARS-CoV-2. This is in line with a previous study reporting suppression of SARS-CoV-2 infection with a selective AAK1 inhibitor (SGC-AAK1-1) (Puray-Chavez et al., 2021). The role of AAK1 and BIKE in the entry of SARS-CoV-2, a stage of the viral life cycle that is inhibited by RMC-76, supports a hypothesis that inhibition of NAKs, at least in part, mediates the antiviral effect of this compound.
Compounds with anti-GAK activity have demonstrated variable antiviral effect. Erlotinib demonstrated no antiviral effect when used individually, gefitinib showed moderate antiviral activity, RMC-242 and SGC-GAK-1 showed potent or moderate antiviral activity against SARS-CoV-2, respectively. All four compounds potently bind GAK in an in vitro binding assay (K d values of: 3 nM (erlotinib), 13 nM (gefitinib), 12 nM (RMC-242), and 2 nM (SGC-GAK-1)), yet these K d values do not correlate with their antiviral activity. In contrast, the anti-GAK activity of these compounds in the cell-based target engagement NanoBRET assay (the most relevant biochemical measurement for predicting a biological effect in cells that takes into account compound properties, such as cell permeability) is variable (IC50 values of: 910 nM (erlotinib), 420 nM (gefitinib), 78 nM (RMC-242), 48 nM (SGC-GAK-1)) and largely correlates with their anti-SARS-CoV-2 effects. The STK16 inhibitor, STK16-IN-1 demonstrated anti-SARS-CoV-2 effect. Combined with the siRNA data, this finding validates the requirement for STK16 in SARS-CoV-2 infection.
Baricitinib, an approved anti-inflammatory drug, only weakly suppressed SARS-CoV-2 infection in both Vero E6 and Calu-3 cells, in agreement with previous reports (Wang et al., 2020; Stebbing et al., 2021). Baricitinib shows moderate binding to AAK1 (K d = 17 nM) and BIKE (K d = 40 nM) and low binding to GAK (K d = 134 nM) and STK16 (K d = 69 nM). Nevertheless, its anti-NAKs activity in a cell-based target engagement assay is not known, and it was shown to inhibit AP2M1 phosphorylation only at a high concentration (10 μM) (Wang et al., 2020). Additionally, the kinome of baricitinib reveals very potent binding to diverse kinases, with ROCK1/2, TYK2, CAMK2A, MAP3K2, and PRPF4B, being just several examples (Klaeger et al., 2017). As with other multi-kinase inhibitors, it is difficult to predict the antiviral effect of baricitinib, as it is driven by its net effect on both proviral and antiviral targets, as we have previously shown with erlotinib and sunitinib (Bekerman et al., 2017). Baricitinib is studied in COVID-19 patients (Kalil et al., 2021; Marconi et al., 2021) for its anti-inflammatory effect mediated by its known target, JAK, and its NAK-mediated antiviral effect predicted by an artificial intelligent screen (Richardson et al., 2020; Stebbing et al., 2020). Our findings suggest that the benefit demonstrated with baricitinib clinically likely does not result from its predicted anti-NAK effect and point out a limitation of the artificial intelligence approach for drug discovery in predicting antiviral activity.
Lastly, we show that combining compounds with anti-AAK1/BIKE and anti-GAK activities may provide a synergistic antiviral effect against SARS-CoV-2 in vitro. This finding is in agreement with our prior data with sunitinib/erlotinib combinations in HCV, DENV, and EBOV in vitro (Neveu et al., 2015; Bekerman et al., 2017) and the finding that their combination protected 85% and 50% of mice from DENV and EBOV challenges, respectively (Bekerman et al., 2017; Pu et al., 2018). AAK1/BIKE and GAK have partially overlapping functions (Zhang et al., 2005; Neveu et al., 2015; Pu et al., 2020), which may explain moderate or no antiviral effect with one drug alone, yet synergistic activity upon treatment with sunitinib/erlotinib combinations. The observed synergistic effect may also result from inhibition of additional targets by these drugs.
In summary, these findings validate NAKs as candidate druggable targets for antiviral strategies against SARS-CoV-2 infection and provide a proof of concept that developing anti-NAKs approaches may have a utility for treating SARS-CoV-2 and possibly other coronavirus infections.
Author contributions
M.K., and S.E., wrote the first version of the manuscript. M.K., S.S., L.G., M.K.S., N.B., C.Y.L., M.P.E., and G.J., designed and performed the experiments and conducted data analysis. C.Y., J.G.P., B.M.G., and J.J., provided reagents and guidance. S.E., B.A.P., L.M.S., C.R.M.A., A.N., and S.D., provided scientific oversight and guidance. S.E., provided the funding for this study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements and funding
This work was supported by awards number W81XWH-16-1-0691 and W81XWH2110456 from the Department of Defense, Congressionally Directed Medical Research Programs (CDMRP), award number HDTRA11810039 from The Defense Threat Reduction Agency (DTRA) Fundamental Research to Counter Weapons of Mass Destruction, and award number RO1-AI158569–01 from the National Institute of Allergy and Infectious Diseases (NIAID) to S.E. This work was partly supported by the NIH Common Fund Illuminating the Druggable Genome (IDG) program (NIH Grant U24DK116204). S.E. is a Chan Zuckerberg Biohub investigator. M.K. was supported by a Postdoctoral Fellowship in Translational Medicine by the PhRMA Foundation. We thank the Stanford in vitro BSL3 service Center and its Director Dr. Jaishree Garhyan for assistance in the BSL3. We also thank the staff of Stanford Clinical Virology Laboratory for their help sequencing the SARS-CoV-2 USA-WA1/2020 isolate used in this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105367.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Agostini M.L., et al. Small-molecule antiviral β-d-N 4-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019 doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asquith C.R., et al. SGC-GAK-1: a chemical probe for cyclin G associated kinase (GAK) J. Med. Chem. 2019 doi: 10.1021/acs.jmedchem.8b01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asquith C.R., et al. Identification and optimization of 4‐anilinoquinolines as inhibitors of cyclin G associated kinase. ChemMedChem. 2018 doi: 10.1002/cmdc.201700663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asquith C.R., et al. Design and analysis of the 4‐anilinoquin (az) oline kinase inhibition profiles of GAK/SLK/STK10 using quantitative structure‐activity relationships. ChemMedChem. 2020 doi: 10.1002/cmdc.201900521. [DOI] [PubMed] [Google Scholar]
- Asquith C.R., et al. Design of a cyclin G associated kinase (GAK)/epidermal growth factor receptor (EGFR) inhibitor set to interrogate the relationship of EGFR and GAK in chordoma. J. Med. Chem. 2019 doi: 10.1021/acs.jmedchem.9b00350. [DOI] [PubMed] [Google Scholar]
- Bayati A., et al. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021 doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekerman E., et al. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Invest. 2017 doi: 10.1172/JCI89857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020 doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiem K., et al. Generation and Characterization of recombinant SARS-CoV-2 expressing reporter genes. J. Virol. 2021 doi: 10.1128/JVI.02209-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiem K., et al. Generation of recombinant SARS‐CoV‐2 using a bacterial artificial chromosome. Curr Protoc Microbiol. 2020 doi: 10.1002/cpmc.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner S.D., Schmid S.L. Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J. Cell Biol. 2002 doi: 10.1083/jcb.200108123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutant E.P., et al. Bioluminescence profiling of NanoKAZ/NanoLuc luciferase using a chemical library of coelenterazine analogues. Chem. Eur J. 2020 doi: 10.1002/chem.201904844. [DOI] [PubMed] [Google Scholar]
- Coutant E.P., et al. Gram-scale synthesis of luciferins derived from coelenterazine and original insights into their bioluminescence properties. Org. Biomol. Chem. 2019 doi: 10.1039/c9ob00459a. [DOI] [PubMed] [Google Scholar]
- Davis M.I., et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011 doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- Duncan J.S., et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012 doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.T., et al. Optimization of 4-anilinoquinolines as dengue virus inhibitors. Molecules. 2021 doi: 10.3390/molecules26237338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- In J.G., et al. Serine/threonine kinase 16 and MAL2 regulate constitutive secretion of soluble cargo in hepatic cells. Biochem. J. 2014 doi: 10.1042/BJ20140468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayk Bernal A., et al. Molnupiravir for oral treatment of covid-19 in nonhospitalized patients. N. Engl. J. Med. 2022 doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil A.C., et al. Baricitinib plus remdesivir for hospitalized adults with covid-19. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns A.E., et al. Cloning and characterization of a novel protein kinase that impairs osteoblast differentiation in vitro. J. Biol. Chem. 2001 doi: 10.1074/jbc.M106163200. [DOI] [PubMed] [Google Scholar]
- Khanna K., et al. Thiol drugs decrease SARS-CoV-2 lung injury in vivo and disrupt SARS-CoV-2 spike complex binding to ACE2 in vitro. bioRxiv. 2020 doi: 10.1101/2020.12.08.415505. [DOI] [Google Scholar]
- Klaeger S., et al. The target landscape of clinical kinase drugs. Science. 2017 doi: 10.1126/science.aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020 doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J., et al. TMPRSS2 expression dictates the entry route used by SARS‐CoV‐2 to infect host cells. EMBO J. 2021 doi: 10.15252/embj.2021107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J.R., et al. Identification and selected reaction monitoring (SRM) quantification of endocytosis factors associated with Numb. Mol. Cell. Proteomics. 2013 doi: 10.1074/mcp.M112.020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., et al. Discovery of a highly selective STK16 kinase inhibitor. ACS Chem. Biol. 2016 doi: 10.1021/acschembio.6b00250. [DOI] [PubMed] [Google Scholar]
- López-Coral A., et al. A serine/threonine kinase 16-based phospho-proteomics screen identifies WD repeat protein-1 as a regulator of constitutive secretion. Sci. Rep. 2018 doi: 10.1038/s41598-018-31426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallah S.I., et al. COVID-19: breaking down a global health crisis. Ann. Clin. Microbiol. Antimicrob. 2021 doi: 10.1186/s12941-021-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi V.C., et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): a randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 2021;(21) doi: 10.1016/S2213-2600. 00331-00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gualda B., et al. Discovery of 3-phenyl- and 3-N-piperidinyl-isothiazolo[4,3-b] pyridines as highly potent inhibitors of cyclin G-associated kinase. Eur. J. Med. Chem. 2021 doi: 10.1016/j.ejmech.2021.113158. [DOI] [PubMed] [Google Scholar]
- Neveu G., et al. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012 doi: 10.1371/journal.ppat.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu G., et al. AP-2-associated protein kinase 1 and cyclin G-associated kinase regulate hepatitis C virus entry and are potential drug targets. J. Virol. 2015 doi: 10.1128/JVI.02705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J., et al. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 2003 doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- Owen D.R., et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021 doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Prichard M.N., Shipman C., Jr. A three-dimensional model to analyze drug-drug interactions. Antivir. Res. 1990 doi: 10.1016/0166-3542(90)90001-n. [DOI] [PubMed] [Google Scholar]
- Prichard M.N., Shipman C., Jr. Analysis of combinations of antiviral drugs and design of effective multidrug therapies. Antivir. Ther. 1996;1 [PubMed] [Google Scholar]
- Pu S.Y., et al. BIKE regulates dengue virus infection and is a cellular target for broad-spectrum antivirals. Antivir. Res. 2020 doi: 10.1016/j.antiviral.2020.104966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu S.Y., et al. Feasibility and biological rationale of repurposing sunitinib and erlotinib for dengue treatment. Antivir. Res. 2018 doi: 10.1016/j.antiviral.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puray-Chavez M., et al. Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell. Cell Rep. 2021 doi: 10.1016/j.celrep.2021.109364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P., et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;(20) doi: 10.1016/S0140-6736. 30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato J., et al. GAK, a regulator of clathrin‐mediated membrane trafficking, localizes not only in the cytoplasm but also in the nucleus. Gene Cell. 2009 doi: 10.1111/j.1365-2443.2009.01296.x. [DOI] [PubMed] [Google Scholar]
- Saul S., et al. Identification and evaluation of 4-anilinoquin(az)olines as potent inhibitors of both dengue virus (DENV) and Venezuelan equine encephalitis virus (VEEV) Bioorg. Med. Chem. Lett. 2021 doi: 10.1016/j.bmcl.2021.128407. [DOI] [PubMed] [Google Scholar]
- Saul S., et al. Discovery of pan-ErbB inhibitors protecting from SARS-CoV-2 replication, inflammation, and lung injury by a drug repurposing screen. bioRxiv. 2021 doi: 10.1101/2021.05.15.444128. [DOI] [Google Scholar]
- Saul S., et al. Potent antiviral activity of novel multi-substituted 4-anilinoquin(az)olines. Bioorg. Med. Chem. Lett. 2020 doi: 10.1016/j.bmcl.2020.127284. [DOI] [PubMed] [Google Scholar]
- Sorensen E.B., Conner S.D. AAK1 regulates Numb function at an early step in clathrin‐mediated endocytosis. Traffic. 2008 doi: 10.1111/j.1600-0854.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- Sorrell F.J., et al. Family-wide structural analysis of human numb-associated protein kinases. Structure. 2016 doi: 10.1016/j.str.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J., et al. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020;(20):30132–30138. doi: 10.1016/S1473-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J., et al. JAK inhibition reduces SARS-CoV-2 liver infectivity and modulates inflammatory responses to reduce morbidity and mortality. Sci. Adv. 2021 doi: 10.1126/sciadv.abe4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L.J., et al. Distinct genetic determinants and mechanisms of SARS-CoV-2 resistance to remdesivir. bioRxiv. 2022 doi: 10.1101/2022.01.25.477724. [DOI] [Google Scholar]
- Verdonck S., et al. Synthesis and structure-activity relationships of 3,5-Disubstituted-pyrrolo[2,3-b]pyridines as inhibitors of adaptor-associated kinase 1 with antiviral activity. J. Med. Chem. 2019 doi: 10.1021/acs.jmedchem.9b00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., et al. Multiplex SARS-CoV-2 genotyping reverse transcriptase PCR for population-level variant screening and epidemiologic surveillance. J. Clin. Microbiol. 2021 doi: 10.1128/JCM.00859-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.G., et al. Sunitinib reduces the infection of SARS-CoV, MERS-CoV and SARS-CoV-2 partially by inhibiting AP2M1 phosphorylation. Cell Discov. 2020 doi: 10.1038/s41421-020-00217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., et al. Mechanism and in vitro pharmacology of TAK1 inhibition by (5Z)-7-Oxozeaenol. ACS Chem. Biol. 2013 doi: 10.1021/cb3005897. [DOI] [PubMed] [Google Scholar]
- Xiao F., et al. 2018. Interactions between the Hepatitis C Virus Nonstructural 2 Protein and Host Adaptor Proteins 1 and 4 Orchestrate Virus Release. Mbio. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C.J., et al. Rescue of SARS-CoV-2 from a single bacterial artificial chromosome. mBio. 2020 doi: 10.1128/mBio.02168-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.X., et al. Multiple roles for cyclin G-associated kinase in clathrin-mediated sorting events. Traffic. 2005 doi: 10.1111/j.1600-0854.2005.00346.x. [DOI] [PubMed] [Google Scholar]
- Zhou H., et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008 doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.