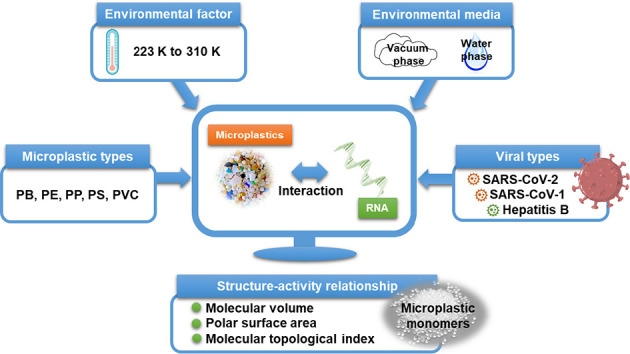
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the coronavirus disease-19 (COVID-19) pandemic spread across the world and remains difficult to control. Environmental pollution and habitat conditions do facilitate SARS-CoV-2 transmission as well as increase the risk of exposure to SARS-CoV-2. The coexistence of microplastics (MPs) with SARS-CoV-2 affects the viral behavior in the indoor and outdoor environment, and it is essential to study the interactions between MPs and SARS-CoV-2 because they both are ubiquitously present in our environment. To determine the mechanisms underlying the impact of MPs on SARS-CoV-2, we used molecular dynamic simulations to investigate the molecular interactions between five MPs and a SARS-CoV-2 RNA fragment at temperatures ranging from 223 to 310 K in vacuum and in water. We furthermore compared the interactions of MPs and SARS-CoV-2 RNA fragment to the performance of SARS-CoV-1 and Hepatitis B virus (HBV) RNA fragments in interacting with the MPs. The interaction affinity between the MPs and the SARS-CoV-2 RNA fragment was found to be greater than the affinity between the MPs and the SARS-CoV-1 or HBV RNA fragments, independent of the environmental media, temperature, and type of MPs. The mechanisms of the interaction between the MPs and the SARS-CoV-2 RNA fragment involved electrostatic and hydrophobic processes, and the interaction affinity was associated with the inherent structural parameters (i.e., molecular volume, polar surface area, and molecular topological index) of the MPs monomers. Although the evidence on the infectious potential of SARS-CoV-2 RNA is not fully understood, humans are exposed to MPs via their lungs, and the strong interaction with the gene materials of SARS-CoV-2 likely affects the exposure of humans to SARS-CoV-2.
Keywords: Microplastic pollution, SARS-CoV-2, Nucleic acid material, Behavior and fate, Environmental conditions
Abbreviations: COVID-19, coronavirus disease-19; DNA, deoxyribonucleic acid; Ee, interaction energy derived from electrostatic energy; Eint, interaction energy; EMP, energies of the isolated microplastics; EMP-virus, energies of the complex of microplastics and viral RNA fragment or nucleocapsid protein; Ep, interaction energy derived from potential energy; Et, interaction energy derived from total energy; Ev, interaction energy derived from ‘van der Waals’ energy; Evirus, energies of the viral RNA fragment or nucleocapsid protein; FSE, frameshift stimulation element; HBV, Hepatitis B virus; MD, molecular dynamics; MPs, microplastics; MTI, molecular topological index; PB, polybutene; PE, polyethylene; PP, polypropylene; PS, polystyrene; PSA, polar surface area; PVC, polyvinyl chloride; RNA, ribonucleic acid; SARS-CoV-1, severe acute respiratory syndrome coronavirus in 2002/2003; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VM, volume of molecule
Graphical abstract
1. Introduction
The global pandemic of the coronavirus disease-19 (COVID-19) has suddenly made us realize that viruses have become important biological pollutants. The outbreak of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) not only seriously threatens human health (Topol, 2020; Turner et al., 2021), but also greatly increases environmental stress (Adelodun et al., 2021a, Adelodun et al., 2021b; Bedrosian et al., 2021). It is thus essential to understand the environmental fate and the behavioral dynamics of the coronavirus. The SARS-CoV-2 can travel in all environmental compartments like water (Navarro et al., 2021; Sala-Comorera et al., 2021), air (Dubey et al., 2021; Razzini et al., 2020), and soil (Anand et al., 2021; Steffan et al., 2020). A nucleic acid material (DNA or RNA) enclosed in a nucleocapsid protein is referred to as the non-enveloped structure of a virus particle (Müller et al., 2019). This is in contrast to the enveloped structure of a virus particle which contains a biological membrane. An envelope increases viral sensitivity to external physical stressors (pH, heat, dryness, etc.) as biological membranes are relatively fragile structures. Consequently, the SARS-CoV-2 as an enveloped virus is more sensitive to environmental factors than non-enveloped viruses (Achak et al., 2021). Thus, it is reasonable to believe that the non-enveloped structural materials of the SARS-CoV-2 could be more resistant to these inactivation factors and are likely to maintain their stability for a long time. Furthermore, studies on the nucleic acid material of SARS-CoV-2 are used for its detection and control in the environment and even for the implementation of personal health prevention measures.
Microplastics (MPs, i.e. particle sizes <5 mm) are one of the most common and persistent emerging human-made pollutants. MPs are widespread in a ubiquitous fashion (Sheng et al., 2021), like they are detected in coastal waters (Roscher et al., 2021), freshwater and sediment (Zhang et al., 2021a), influents and effluents of sewage treatments (Nakao et al., 2021), agricultural soils (Boughattas et al., 2021), the atmosphere (Amato-Lourenço et al., 2020), and biosphere (Patil et al., 2022; Rezania et al., 2018).
The plethora of sources that can contribute to the release of MPs into air have been summarized in (Catarino et al., 2018; UNEP, 2016). In addition, the main sources of indoor and outdoor plastic debris released into the air and subject to human inhalation are illustrated by Amato-Lourenço et al. (2020). The indoor concentrations ranged between 1.0 and 60.0 fibers/m3 whereas outdoor concentrations were significantly lower as they range between 0.3 and 1.5 fibers/m3 (Dris et al., 2017). This is important to quantify and realize, because MPs have been reported as carriers or vectors for concurrent pollutants, e.g. metals (Li et al., 2021), organic pollutants (Yu et al., 2021), and they exhibit diverse interactive effects (Bhagat et al., 2021; Kim et al., 2017; Sun et al., 2021). In addition, MPs are becoming a novel ecological habitat termed the plastisphere (Zettler et al., 2013), and could facilitate the survival and dissemination of bacterial and fungal pathogens (Moresco et al., 2021), and antibiotic resistance genes (Li et al., 2021). Importantly, plastic pollution could be a secondary pathway for the transmission of human pathogenic viruses (Moresco et al., 2021) via the respiratory exposure route. We focus here on the MPs–SARS-CoV-2 interactions because both the virus as well as the sources of MPs (like fibers from clothes, building materials, household objects, polymer fragments in urban dust) are closely correlated to the presence of human. It was also reported that SARS-CoV-2 remains more stable on plastic surfaces than on stainless steel, glass, and ceramics (Gidari et al., 2021) which has its consequences for the oral and hand contact exposure routes for humans. Amato-Lourenço et al. (2022) found that SARS-CoV-2 aerosols may bind to total suspended particles, such as MPs, and facilitate virus entry into the human body. Moreover, SARS-CoV-2 virus particles have the ability to sorb to the surface of MPs released during washing processes (Belišová et al., 2022). Hence, there is an urgent need to further explore the interactions and mechanisms of MPs and SARS-CoV-2.
Virus stability in the environment is strongly influenced by the size and structure of the virus particle (including the presence or absence of an envelope), the type of genome (DNA or RNA), a transmission route such as faecal-oral and air droplets, the presence of vectors or carriers like the MPs, and the viral concentration of the contamination source. As known, the intrinsic properties such as polymer type of MPs dictate their interaction affinity with other co-contaminants (Fred-Ahmadu et al., 2020; Menéndez-Pedriza and Jaumot, 2020). Besides, many environmental factors can affect the stability of viruses in the environment (Aboubakr et al., 2021; Achak et al., 2021; Paul et al., 2021), in humans (Matson et al., 2020), and on common touch surfaces (Aboubakr et al., 2021). Notably, temperature (Paul et al., 2021) and relative humidity (Zhao et al., 2020) are the two critical factors that determine the fate and transport of coronaviruses given certain environmental conditions. Therefore, searching for some key characteristics that may affect the interaction of the MPs and SARS-CoV-2 is a noteworthy issue.
In silico methods are a promising approach and play a significant role in elucidating the mechanisms of the interactions of pollutants and biomacromolecules (Chen et al., 2019; Ge et al., 2011). In particular, the molecular simulation method such as molecular dynamics (MD) simulation is a practical in silico method in environmental applications (Chen et al., 2021; Feng et al., 2022; Sun et al., 2013). In addition, the molecular simulation method has shown to be an effective tool in exploring the interactions between MPs and SARS-CoV-2, and offered theoretical insights into the adsorption/separation and inactivation of carbon nanoparticles with a SARS-CoV-2 RNA fragments (Zhang et al., 2021b). This way in silico methods can not only contribute to minimizing the challenge of time-consuming and labor-intensive virus experiments under high risks of infection, but also to meeting our precautionary demand for options to handle any new versions of the coronavirus that might emerge in the future.
In light of the demands from the exploration of the interaction and mechanism between MPs and SARS-CoV-2, this knowledge gap needs to be addressed. Hence, in this work for the first time MPs were studied theoretically by MD simulation to characterize their interactions with the non-enveloped structural materials of SARS-CoV-2 including a nucleocapsid protein and a SARS-CoV-2 RNA fragment in the water phase and in the vacuum phase (as a reference for the water phase and as an approximation to the gas phase). Two reference viruses, namely SARS-CoV-1 (homologous coronavirus similar to SARS-CoV-2) and Hepatitis B virus (HBV, non-coronavirus dissimilar to SARS-CoV-2) were selected to compare the performance in interacting with MPs. The influence of five different MP types and the temperature as an environmental factor is considered. The objectives of this study were divided in several parts: 1) Comparison of the interactions of the MPs with the nucleocapsid protein and with the viral RNA fragments; 2) Interaction mechanisms between the MPs and viral RNA fragments; and 3) Correlation of the interaction affinity and molecular parameters of MP monomers.
2. Computational methods
2.1. MD simulation
The selected three-dimensional structure models of the SARS-CoV-2 RNA fragment determined by Zhang et al. (2021c), the SARS-CoV-1 RNA fragment determined by Robertson et al. (2005), and the HBV RNA fragment determined by LeBlanc et al. (2021) were used as model compounds for the simulation of the interactions between MPs and the viral RNA fragments. It should be noted that the SARS-CoV-2 RNA fragment is a model molecule of a frameshift stimulation element (FSE) from the SARS-CoV-2 RNA genome (Zhang et al., 2021c). The FSE plays an important role in the virus replication cycle and has emerged as a major drug target (Lan et al., 2022). The selected three-dimensional structure models of the SARS-CoV-2 nucleocapsid protein determined by Kang et al. (2020), the SARS-CoV-1 nucleocapsid protein determined by Huang et al. (2004), and the HBV nucleocapsid protein determined by Böttcher and Nassal (2018) were used as model compounds for the simulation of the interactions between MPs and the viral nucleocapsid protein. The structures of the RNA fragments [PDB ID: 6XRZ (SARS-CoV-2), 1XJR (SARS-CoV-1), 6VAR (HBV)] and nucleocapsid proteins [(PDB ID: 6M3M (SARS-CoV-2), 1SSK (SARS-CoV-1), 6HU7 (HBV)] were obtained from the RCSB Protein Data Bank (Burley et al., 2019).
The polymer chains derived from five plastic monomers were built as model compounds for MPs including polybutene (PB), polyethylene (PE), polypropylene (PP), polystyrene (PS), and polyvinyl chloride (PVC) within the simulation. All the simulations were carried out in a box with three-dimensional boundary conditions. The dimensions of the simulation boxes were a = b = c = 85 Å, a = b = c = 90°. The length of the simulation box in each direction was large enough to enable the interactions between the MP polymer chain and the materials of the viruses. The process of building the MP models refers to the simulation methods developed by Guo et al. (2019) with slight modifications. The MP polymer chains were built and energy minimized using the smart geometry optimization algorithm, which is the combination of steepest descent, conjugate gradient, and quasi-Newton geometry optimization algorithms. Then the optimized polymer chain was randomly packed in rectangular boxes with three-dimensional periodic boundary conditions by Amorphous Cell Construction. For each box, only one polymer chain was added. The amount of PB, PE, PP, PS, and PVC monomer molecules were 200, 600, 500, 200, and 600. The MP-virus systems included one polymer chain, one RNA fragment or one nucleocapsid protein, and either a vacuum layer (83 Å) or a water layer (83 Å). For the water system, 1000 water molecules were incorporated in each unit cell. The smart geometry optimization algorithm was used to minimize the energy of the simulation systems. Then the MD calculations were performed in the canonical ensemble NVT system in which the number of molecules [N], volume [V], and temperature [T] of the system are kept constant at 223, 263, 273, 298, and 310 K. These temperatures represent the range from a low-temperature environment to the temperature of the human body. The universal force field was adopted in the simulation framework. The van der Waals interaction cut-off was 12.5 Å, and the Ewald method (accuracy 0.001 kcal/mol) was used. The simulation was performed for 100 ps which allowed the studied system to reach equilibrium, and each step was 1.0 fs. A Nose thermostat was adopted. All the simulations were performed with the Materials Studio software package (ver. 8.0).
2.2. Interaction energy
For the interaction systems, the magnitude of the interaction energy (E int) is an indication of the magnitude of the driving force towards complexation. A negative value reflects stable adsorption on the plastisphere. E int was calculated by
| (1) |
where E MP-virus, E MP, and E virus represent the energies of the complex, the isolated MPs, and the viral RNA fragment or nucleocapsid protein, respectively.
2.3. Molecular parameters and linear correlation models
The MP monomers' molecular parameters (Table S1) such as volume of molecule (V M), polar surface area (PSA), and molecular topological index (MTI) were selected to correlate with E int so as to develop a quantitative relationship between the inherent properties of MPs and E int. The molecular parameters were calculated using Multiwfn 3.8 software (Lu and Chen, 2012a, Lu and Chen, 2012b). Correlation of interaction affinity and molecular parameters of the MP monomers was described using a polynomial relationship by performing linear regression models in Sigma Plot, ver. 14.0 (Systat Software Inc., San Jose, CA).
2.4. Statistical analysis
Statistically significant differences between test groups were determined by independent t-test and one-way analysis of variance with the Waller-Duncan test post hoc, at a significance level of p < 0.05 (IBM SPSS Statistics for Windows, ver. 23.0, IBM Corp., Armonk, NY). Linear regression analysis at the significant level of p < 0.05 was carried out using the SPSS.
3. Results
3.1. Comparison of interactions of MPs with viral nucleocapsid protein and RNA fragments
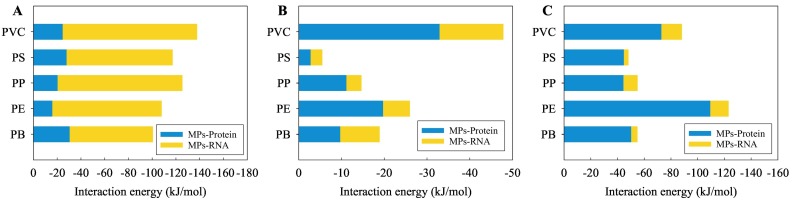
To fully understand the interactions between the MPs and the non-enveloped structures of the virus, the interactions of the MPs with the nucleocapsid protein and with the viral RNA fragments were compared after geometry optimization (Fig. 1 ). As shown in Fig. 1A, for the SARS-CoV-2, the absolute E int values between the MPs and the nucleocapsid protein were significantly lower (p < 0.05) than those between the MPs and the RNA fragment. In contrast, for the HBV (Fig. 1C), the absolute E int values between the MPs and the nucleocapsid proteins were significantly higher (p < 0.05) than those between the MPs and the RNA fragments. For the SARS-CoV-1 (Fig. 1B), the absolute E int values between the MPs and the nucleocapsid proteins were higher than the corresponding values between the MPs and the RNA fragments, but the two groups showed no significant difference (p > 0.05). Moreover, there was no significant difference in the absolute E int values between the interactions of the MPs with the nucleocapsid proteins of the SARS-CoV-2 and the SARS-CoV-1. However, the absolute E int values between the MPs and the nucleocapsid proteins of the HBV were significantly higher than those between the MPs and the nucleocapsid proteins of the SARS-CoV-2 or the SARS-CoV-1 (p < 0.05). In addition, the absolute E int values between the MPs and the RNA fragments of the SARS-CoV-2 were significantly higher than those between the MPs and the nucleocapsid proteins of the SARS-CoV-1 or the HBV (p < 0.05). Moreover, no significant difference in the absolute E int values between the interactions of the MPs with the RNA fragments of the SARS-CoV-1 and the HBV was found.
Fig. 1.
Interaction energies of five types of MPs with the SARS-CoV-2 RNA fragment and the nucleocapsid protein (A), the SARS-CoV-1 RNA fragment and the nucleocapsid protein (B), as well as the HBV RNA fragment and the nucleocapsid protein (C), as obtained by geometry optimization.
Generalizing, when comparing the nucleocapsid protein and the RNA fragment, then the MPs exhibited a stronger interaction with the RNA fragment for the SARS-CoV-2, while the MPs exhibited a stronger interaction with the nucleocapsid protein for the HBV. Furthermore, this difference in the interactions was not affected by the type of MP. The plastic types were a bit more discriminative for SARS-CoV-1 and HBV compared to the SARS-CoV-2 that had interactions energies all similar for each type of MPs.
3.2. Interaction mechanisms between MPs and viral RNA fragments
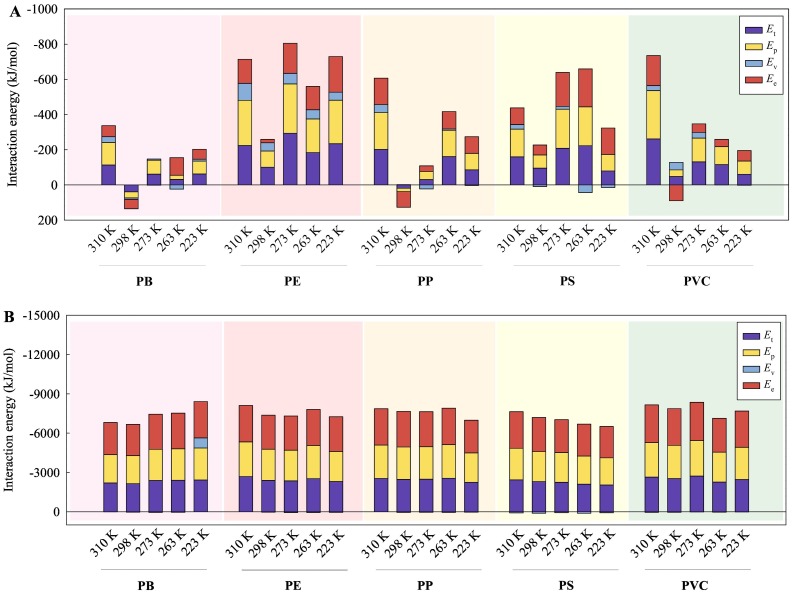
To reveal the mechanisms of the interactions of the MPs with the viral RNA fragments, the values of E int as derived from the total energy (E t), the potential energy (E p), the ‘van der Waals’ energy (E v), and the electrostatic energy (E e) are summarized in Figs. 2 , S1, and S2. As shown in Fig. 2A and B, the computed E int values were negative across most of the temperature range in vacuum and the full temperature range in water. This indicates that the MPs can form stable complexes with the SARS-CoV-2 RNA fragment. Furthermore, the computed E int derived from the E e between the MPs and the SARS-CoV-2 RNA fragment were generally closer to the E int values derived from the E t/E p than the E int values derived from the E v in both vacuum and water. Moreover, there were no significant differences between the E int values derived from the E e and E t/E p (p > 0.05) in vacuum, but significant differences between the E int values derived from the E v and E t/E p (p < 0.05). This implies that the electrostatic interaction contributed mainly to the mechanism of interaction between the MPs and SARS-CoV-2 RNA fragment. The genetic material of the SARS-CoV-2 is positive single-stranded RNA (Zhang et al., 2021c), whereas the studied MPs are neutral and the electrostatic interactions are mainly ion-induced dipole interactions.
Fig. 2.
Interaction energies of the five types of MPs with the SARS-CoV-2 RNA fragment in vacuum (A) and in water (B) at different temperatures. Et: interaction energy derived from total energy, Ep: interaction energy derived from potential energy, Ev: interaction energy derived from ‘van der Waals’ energy, and Ee: interaction energy derived from electrostatic energy.
Moreover, the absolute E int values derived from the E t, E p, or E e for the interactions between the MPs and the SARS-CoV-2 RNA fragment (Fig. 2B) in water were significantly greater than those in vacuum (p < 0.05) (Fig. 2A), implying that the interaction affinity of the MPs with the SARS-CoV-2 RNA fragment in water was stronger compared with the affinity in vacuum. This may be caused by the hydrophobicity of MPs (Ding et al., 2020; Zhang et al., 2020), which can provide stronger interactions with the viral RNA fragment in water.
As depicted in Figs. S1 and S2, the E int values derived from the E t and E p for the interaction between the MPs and the SARS-CoV-1 RNA or HBV RNA fragments in vacuum and water phases were significantly lower than those for the interaction between the MPs and SARS-CoV-2 RNA fragment (p < 0.05). This means that the MPs exhibited stronger interaction with the SARS-CoV-2 RNA fragment than with the SARS-CoV-1 RNA and the HBV RNA fragments. Moreover, most of the E int values for the interaction between the MPs and the SARS-CoV-1 RNA fragment or the HBV RNA fragment tended to be positive. This implies that the complexes of the MPs with SARS-CoV-1 RNA fragment or HBV RNA fragment were instable. As a result, it is difficult to analysis the interaction mechanisms of the MPs and the SARS-CoV-1 RNA fragment or the HBV RNA fragment.
3.3. Correlation of interaction affinity and temperatures
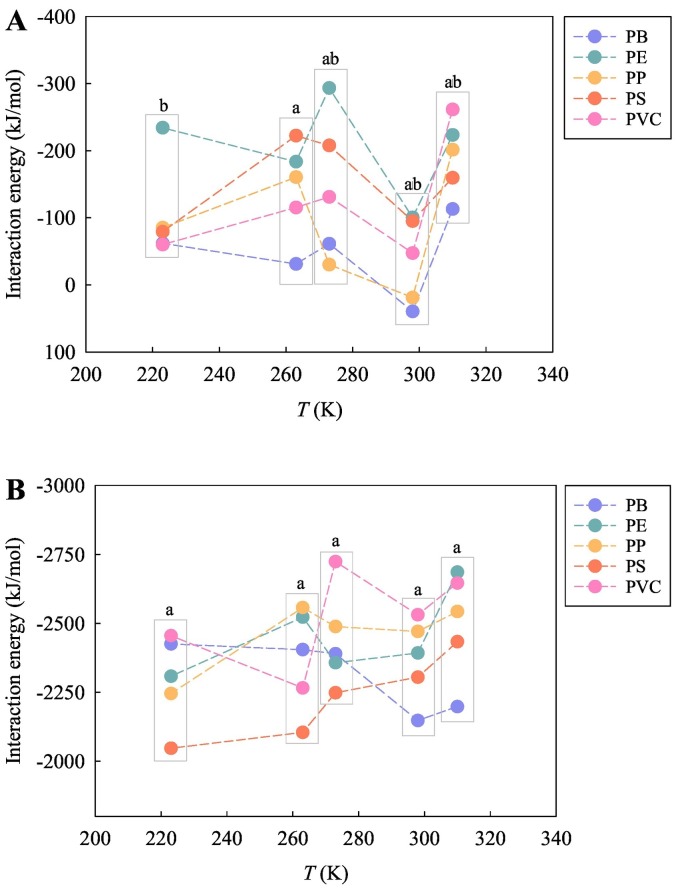
To test the impact of the studied temperature on the interactions of the MPs with the viral RNA fragments, the variation of the interaction affinity with the temperatures was plotted (Figs. 3 , S3, and S4). In general, for each of the MPs, the E int values derived from the total energies fluctuated with the temperature. In particular, the E int values between the MPs and SARS-CoV-2 RNA fragment tended to reach the highest value at 298 K in vacuum (Fig. 3A), implying that the interaction affinity between the MPs and SARS-CoV-2 RNA fragment was lowest at 298 K. In water, the E int values between the PS MPs and SARS-CoV-2 RNA fragment decreased with an increase of the temperature (Fig. 3B). A similar phenomenon occurs in the interaction between the PS MPs and SARS-CoV-1 RNA fragment in water (Fig. S3B). In terms of considering the various types of MPs as a whole, the E int values were not significantly different between the temperatures (Figs. 3, S3, and S4). This also means that temperature was not a determinative factor affecting the interaction affinity between the MPs and viral RNA fragments in the present simulation study.
Fig. 3.
Variation of the interaction energies derived from the total energies of the five types of MPs with the SARS-CoV-2 RNA fragment in vacuum (A) and in water (B) with the studied temperatures (223, 263, 273, 298, and 310 K). Different letters represent statistically significant differences between the treatments (p < 0.05).
3.4. Correlation of interaction affinity and molecular parameters of MP monomers
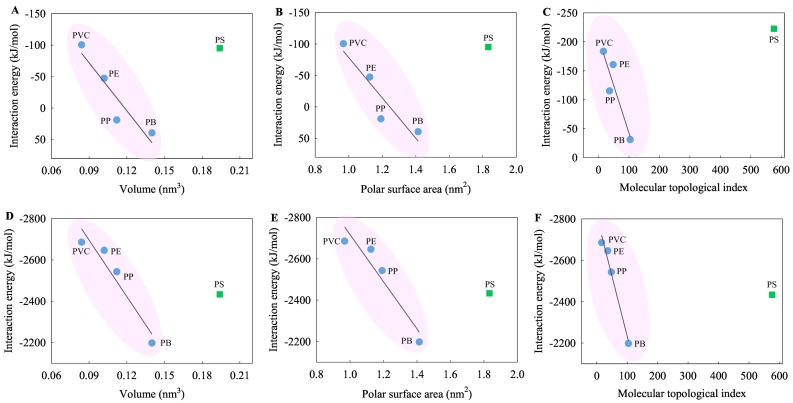
To explore the impact of the inherent properties of MPs on their interactions with the viral RNA fragments, a correlation was conducted between the interaction affinity and molecular parameters of MP monomers (Tables 1 , S2, S3, and Fig. 4 ). As shown in Table 1, the E int values derived from the total energies for the interaction of each of the MPs with the SARS-CoV-2 RNA fragment in vacuum and water phases correlated with the molecular parameters V M, PSA, and MTI of the MP monomers to varying degree. The degree of correlation tended to be higher in vacuum and at 310 and 298 K in water except for the PS MPs with aromatic hydrocarbons. In particular, the E int values correlated highly (Fig. 4A–C) and significantly (Fig. 4D–F) with the molecular parameters except for the PS MPs. On the whole, the greater the V M, PSA, and MTI values, the stronger the interactions between the MPs and the SARS-CoV-2 RNA fragment (Fig. 4).
Table 1.
Correlation coefficients between the Eint values derived from the total energies between the MPs and SARS-CoV-2 RNA fragment and the molecular parameters of the MP monomers.a
| Correlation model | Temperature (K) |
Volume (nm3) |
Polar surface area (nm2) |
Molecular topological index |
|||
|---|---|---|---|---|---|---|---|
| n = 5 | n = 4 | n = 5 | n = 4 | n = 5 | n = 4 | ||
| Eint in vacuum | 310 | 0.652 | 0.839 | 0.648 | 0.821 | 0.427 | 0.892 |
| 298 | 0.068 | 0.929 | 0.063 | 0.927 | 0.399 | 0.866 | |
| 273 | 0.065 | 0.791 | 0.073 | 0.797 | 0.235 | 0.683 | |
| 263 | 0.203 | 0.888 | 0.194 | 0.896 | 0.502 | 0.917 | |
| 223 | 0.510 | 0.739 | 0.523 | 0.760 | 0.274 | 0.645 | |
| Eint in water | 310 | 0.615 | 0.959 | 0.616 | 0.959 | 0.326 | 0.989 |
| 298 | 0.535 | 0.704 | 0.530 | 0.704 | 0.346 | 0.803 | |
| 273 | 0.577 | 0.123 | 0.563 | 0.123 | 0.621 | 0.223 | |
| 263 | 0.749 | 0.171 | 0.756 | 0.171 | 0.800 | 0.168 | |
| 223 | 0.669 | 0.334 | 0.659 | 0.334 | 0.817 | 0.376 | |
The correlation was tested for five types (n = 5) of MPs (PB, PE, PP, PS, and PVC)/four types (n = 4) of MPs (PB, PE, PP, and PVC) and the SARS-CoV-2 RNA fragment; the magnitude of correlation coefficient (R) reflects the degree of correlation between the Eint and molecular parameter values; the bold numbers indicate high values of the correlation coefficients (R > 0.800); the numbers marked in both bold and italic indicate a significant correlation at the 0.05 level (p < 0.05).
Fig. 4.
Variation of the interaction energies derived from the total energies of the five types of MPs with the SARS-CoV-2 RNA fragment at 298 K (A and B) and 263 K (C) in vacuum and at 310 K (D, E, and F) in water with the molecular parameters of the MP monomers.
Generally, the E int values derived from the total energies for the interaction of the SARS-CoV-1 (Table S2) or the HBV (Table S3) RNA fragment with the MPs in vacuum and water phases correlated moderately or weakly with the molecular parameters V M, PSA, and MTI of the MP monomers. It can be also found that there was a higher correlation between the E int values for the interaction of the SARS-CoV-1 RNA fragment with the MPs and the molecular parameters of the MP monomers at 310 K in vacuum except for the PS MPs. In addition, for the interaction of the MPs with the SARS-CoV-1 and the HBV RNA fragment, no significant correlation was found between the E int values and the molecular parameters of the MP monomers.
4. Discussion
Owing to the high prevalence of both enteric and respiratory viruses in the population and the environment, there is significant potential for human viruses to become associated with the plastisphere (Moresco et al., 2021). There are many sources of MPs in the environment and potential pathways for the interaction, colonisation, and dissemination of viruses. We have studied the interaction between three different viruses and five different MPs in water and vacuum air. For these exposure routes we have taken different conditions; being different temperatures, and different coating of the virus. These coatings have been modelled theoretically how the genetic material such as the RNA of a virus is released into cells after the virus undergoes fusion. There, the RNA segments are covered with the nucleocapsid protein enabling to travel to specific organelles such as the ribosome.
The first pathway described is via the respiratory path: MPs can enter the human body through breathing, mainly due to the presence of MP pollution in the air (Amato-Lourenço et al., 2020); indoor dust as well as air in cities were shown to be large contributors. So not only the virus and MPs dose will be higher indoors, also interaction effectivity is large. It makes it a large potential exposure route for humans. It has been proven that face masks can release large numbers of MPs, which were detected in nasal mucus of mask wearers and can be inhaled by human beings (Ma et al., 2021). In a way face masks are preventing inhalation of virus for human not infected, but those infected may even breathe the virus out. It is speculated that the virus can bind to MPs from the mask and human beings inhale them again as an agglomerate.
Second, the SARS-CoV-2 is transmitted primarily through respiratory droplets (Stadnytskyi et al., 2020) and/or aerosols (Liu et al., 2020a). Airborne dust is another transmission route linked to infectious diseases (Maestre et al., 2021; Moreno et al., 2021). More severe weather phenomena such as sandstorms may exacerbate the migration of the virus (Meo et al., 2021). The adsorption of the SARS-CoV-2 on these airborne media can contribute to the long-range transport of the virus. Note that the airborne transmission route refers to the presence of particles with diameter < 5 μm, who can remain in the air for long periods (Morawska and Cao, 2020). The particle sizes of the MPs are also in this scale range. Thus, MPs dispersed in air can be inhaled by humans (Amato-Lourenço et al., 2020). The MPs can be released into the atmospheric air via several sources, e.g., synthetic textiles (Chen et al., 2020), tire wear particles (Lee et al., 2020), domestic laundry dryers (O'Brien et al., 2020), etc. Hence, there is a high probability that the MPs and the SARS-CoV-2 will meet in the atmospheric environment. It has been reported that SARS-CoV-2 aerosols may bind to MPs and facilitate virus entry into the human body (Amato-Lourenço et al., 2022). Our results show that the MPs stabilized the SARS-CoV-2 RNA fragment in both vacuum and water. This also means that the MPs could act as a carrier capable of carrying the gene materials of the SARS-CoV-2 and become a new airborne media for the transport of the virus.
Third, a non-droplet transmission is also possible, as the infectious SARS-CoV-2 particles are also present in human excretions (Wiktorczyk-Kapischke et al., 2021). The fragment of the SARS-CoV-2 RNA has been frequently detected in various countries in wastewater (Kumar et al., 2020; La Rosa et al., 2020; Randazzo et al., 2020), particularly hospital effluent (Gonçalves et al., 2021). The transmission of SARS-CoV-2 via the fecal-oral route highlights the presence and persistence of SARS-CoV-2 in the aquatic environment (Arslan et al., 2020). Moreover, the SARS-CoV-2 RNA is relatively stable in sewage and non-chlorinated drinking water (Ahmed et al., 2020). The viral RNA was also found to be relatively stable in contrast to the rapid inactivation of infectious SARS-CoV-2 in river and in sea water (Sala-Comorera et al., 2021). The COVID-19 pandemic has a huge impact on the plastic waste management in many countries, in large due to the sudden surge of medical waste which has led to a potential significant release of MPs (Khoo et al., 2021). Recent studies indicated that MPs have a significant abundance in sewage. Therefore, the sewage treatment system may be an important site for the interaction between the MPs and the gene materials of SARS-CoV-2. Belišová et al. (2022) also confirmed the ability of SARS-CoV-2 virus particles to sorb to the surface of MPs, specifically microfibers in wastewater. The present results implied that the MPs stabilized the SARS-CoV-2 RNA fragment in the water phase, regardless of temperature and MP types. Additionally, the persistence of the SARS-CoV-2 RNA fragment when present on the MPs was different from the persistence of the SARS-CoV-1 and HBV RNA fragments. In comparison, the SARS-CoV-2 RNA fragment preferred to maintain on the MPs, which may cause the gene materials of the SARS-CoV-2 to be long lasting on the MPs.
The fourth path is via the oral route such as food and water. The results in Fig. 2 also indicated that the interaction affinity of the MPs with the SARS-CoV-2 RNA fragment in water was stronger compared with the affinity in vacuum by a factor of 10 at least. This means the MPs and viral genetic material may co-present in dairy products we eat. If the MPs are entered through the food chain (Bouwmeester et al., 2015; Mercogliano et al., 2020), the MPs enter cells via endocytosis and then are released into the cytoplasm. Particularly, the intestinal tract is the main place where MPs exist and is the channel into the circulatory system (Fournier et al., 2021; Visalli et al., 2021). In the meanwhile, it is confirmed that the SARS-CoV-2 can effectively infect intestinal epithelial cells and their precursors (Lamers et al., 2020), which reveals the fact that the intestinal tract is the potential infection site of the SARS-CoV-2 in humans. Taken together, an intercellular environment provides an opportunity for interaction between the MPs and the viral RNA segments/nucleocapsid protein. In our study, we revealed that the MPs showed stronger interaction with the SARS-CoV-2 RNA fragment than with its nucleocapsid protein. Comparison and analysis on the E int also supported the finding that the MPs interacted with the SARS-CoV-2 RNA fragment more strongly than with the SARS-CoV-1 or HBV RNA fragments. This also means that the MPs are more apt to stabilize the genetic materials of the SARS-CoV-2 in the intercellular environment, whereas this interaction may limit the transcription and replication of the viral RNA genomes.
The fifth potential route is via inanimate surfaces such as plastic, stainless steel, and glass has been established (Corpet, 2021; Gidari et al., 2021) on which the persistence of the SARS-CoV-2 is detected. For instance, Gidari et al. (2021) showed the ability of SARS-CoV-2 to persist on most common materials such as glass, stainless steel, and plastic with half-lives of 4.2, 4.4, and 5.3 h respectively. The SARS-CoV-2 is thus more stable on plastics than on steel or on glass. With the global outbreak and spread of COVID-19, disposable surgical masks as effective and cheap protective medical equipment have been widely used by the public. The random disposal of masks may result in new and greater MP pollution, because masks made of polymer materials would release MPs after entering the environment. More importantly, potential co-release of the MPs and the SARS-CoV-2 into the environment will be ineluctable. This might be expected as the result of the unreasonable disposal of the masks, especially the masks contaminated with the virus. MPs have been detected in the air. Thus, MPs can deposit upon the surface of various materials. Thus, there may be an opportunity for the interaction of MPs and the virus RNA. There is evidence that the SARS-CoV-2 RNA fragment has been detected on frozen food packaging (Han et al., 2021; Liu et al., 2020b), and aquatic products can be a route of transmission of COVID-19. Positive detection of COVID-19 nucleic acid in the samples of frozen food packaging is still occurring. Our theoretical investigation also indicated that the MPs stabilized the SARS-CoV-2 RNA fragment at very low temperatures ranging from 273 to 223 K. The presence of the genetic material of SARS-CoV-2 on the surfaces is not the same as the presence of the infectious virus, but indicates the transit and contact of infected individuals (Casabianca et al., 2022). Therefore, theoretical evidence of interactions between the MPs and the SARS-CoV-2 RNA fragment could support practices (e.g. strict sanitization of medical equipment, supplies, fabrics, environmental surfaces, and air contaminated with pathogens) that reduce the risk of SARS-CoV-2 infection and cut off its transmission route.
The plastisphere is a diverse microbial community of heterotrophs, autotrophs, predators, and symbionts (Zettler et al., 2013). Several studies demonstrated that the gene materials of microorganisms can be extracted from MPs and subsequently identified (Debeljak et al., 2017; Zettler et al., 2013). Regardless of environmental media and temperature, a stable binding between the MPs and the SARS-CoV-2 fragment was proven theoretically. After such a binding, the SARS-CoV-2 fragment is more difficult to degrade in the natural environment. This also means that entering the plastisphere appears to be an important process that significantly affects the global environmental fate of SARS-CoV-2.
SARS-CoV-2 belongs to the family of enveloped, single-strand RNA viruses (Mei and Tan, 2021). The viral membrane of SARS-CoV-2 surrounds a helical nucleocapsid in which the viral genome is encapsulated by the nucleocapsid protein (Savastano et al., 2020). The biological membrane, known as an envelope, contains lipids and proteins. An envelope may increase the viral sensitivity to physical influencing factors (pH, heat, dryness, etc.) as biological membranes are relatively fragile structures. The nucleocapsid protein of SARS-CoV-2 is produced at high levels within infected cells, enhances the efficiency of viral RNA transcription, and is essential for viral replication (Savastano et al., 2020). It is reported that the SARS-CoV-2 RNA is likely to persist for a long time in untreated wastewater (Ahmed et al., 2020). Consequently, it is essential to elucidate the interactions of the MPs with the nucleocapsid protein and SARS-CoV-2 RNA fragment. Further studies are warranted to evaluate the interaction of the MPs with other structural proteins of SARS-CoV-2, e.g. spike, membrane, and envelope. Furthermore, the interactions as addressed in this study are the first stepping stone to meet our precautionary demand for options to handle any new versions of the coronavirus that might emerge in the future.
It was also found that there are differences in the interaction affinity between the MPs with different compositions and SARS-CoV-2 RNA fragment (Fig. 1, Fig. 2). Notably, the molecular parameters of the PS monomer performed very different in affecting the interaction affinity as compared to the other MP monomers (Fig. 4). The benzene ring contained in PS allowed it to form π-π interactions with the SARS-CoV-2 RNA fragment that might modulate the interaction affinity. The differences in the composition of MPs are most directly reflected in the functional groups contained in their polymeric structural units. The properties of the MP monomer compounds can determine the mechanism of interaction of MPs with organic pollutants, which in turn exhibit a different interaction affinity for organic pollutants (Lee et al., 2014). In addition, changes in environmental conditions such as temperature can modulate the interaction between the MPs and SARS-CoV-2 RNA fragment (Fig. 3). Other factors such as pH, salinity, and dissolved organic matter which may result in differences in the interaction can also not be neglected. Accordingly, the single and combined effects of different environmental factors on the interaction of the MPs and SARS-CoV-2 will need to be considered in subsequent studies.
It is undeniable that in silico methods still have limitations in both space and time scales, which weakens their correlation with experimental observations and available experimental data. Moreover, quality assurance is required to minimize uncertainty in the calculation of toxicological data. In spite of this, in the face of the urgency of the COVID-19 pandemic, in silico methods are a useful tool to investigate the interaction of environmental pollutants such as MPs with the novel coronavirus, particularly the proposed methodologies that rely upon alternatives to biological testing with high risk of infection. Furthermore, in silico methods have the advantages of preliminary screening of high-risk combinations of multiple co-existing pollutants (e.g. SARS-CoV-2 and MPs) in the environment, and it will save valuable research time and efforts (e.g. model validation) as well as prevent infection during experimental testing.
5. Conclusions
In this work, we carried out MD simulations to investigate the interactions between five MPs and RNA fragments of three viruses including, SARS-CoV-2, SARS-CoV-1, and HBV at temperatures ranging from 223 to 310 K, in vacuum and in water phases. The estimated E int implied that the interactions of the MPs with the SARS-CoV-2 RNA fragment were stronger than those with the SARS-CoV-1 and HBV RNA fragments, regardless of the environmental media, temperature, and MP types. Furthermore, the electrostatic and hydrophobic processes were the predominant mechanisms for the interactions between the MPs and the SARS-CoV-2 RNA fragment, and the interaction affinity was associated with the inherent structural parameters (i.e., V M, PSA, and MTI) of the MP monomers. Our theoretical results suggest that MPs are capable of regulating the behavior and fate of the SARS-CoV-2 RNA fragment in the environment. While MPs are within air, food and water, this plastic pollution could be a secondary pathway for the transmission of human pathogenic virus and hence have consequences for the exposure of humans to SARS-CoV-2, both by the respiratory pathway (enhancing potential exposure) and the touch pathway where the plastic surface binds the SARS-CoV-2 RNA fragment and thus lowers potential exposure and infectious risks for human. It should be noted that the SARS-CoV-2 RNA fragment can be immobilized by MPs which are ubiquitous in the human environments and thus their persistence and circulation would prolong the presence of virus RNA in the environment. This in silico work serves to minimize the challenges of conducting time-consuming and labor-intensive virus experiments with a high risk of infection, while meeting our precautionary need for options to deal with any new versions of coronaviruses that may emerge in the future.
CRediT authorship contribution statement
Fan Zhang: Conceptualization, Methodology, Investigation, Data curation, Formal analysis, Writing – original draft, Visualization, Funding acquisition. Zhuang Wang: Conceptualization, Formal analysis, Resources, Writing – review & editing, Funding acquisition, Project administration. Martina G. Vijver: Conceptualization, Formal analysis, Writing – review & editing, Supervision, Funding acquisition, Project administration. Willie J.G.M. Peijnenburg: Conceptualization, Formal analysis, Writing – review & editing, Supervision, Funding acquisition, Project administration.
Declaration of competing interest
The authors declare no competing financial interest.
Acknowledgements
This article pays tribute to those who are fighting COVID-19. This work was supported by the European Union's Horizon 2020 research and innovation program “NanoinformaTIX” (814426) that supported W.J.G.M.P. and M.G.V. and the National Natural Science Foundation of China (31971522) to Z.W. F.Z. greatly acknowledges the support from the China Scholarship Council (202008320308). We also thank the reviewers for their valuable comments on the manuscript.
Editor: Thomas Kevin V
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.156812.
Appendix A. Supplementary data
Supplementary material
References
- Aboubakr H.A., Sharafeldin T.A., Goyal S.M. Stability of SARS-CoV-2 and other coronaviruses in the environment and on common touch surfaces and the influence of climatic conditions: a review. Transbound. Emerg. Dis. 2021;68:296–312. doi: 10.1111/tbed.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achak M., Alaoui Bakri S., Chhiti Y., M’hamdi Alaoui F.E., Barka N., Boumya W. SARS-CoV-2 in hospital wastewater during outbreak of COVID-19: a review on detection, survival and disinfection technologies. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.143192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelodun B., Kareem K.Y., Kumar P., Kumar V., Choi K.S., Yadav K.K., Yadav A., El-Denglawey A., Cabral-Pinto M., Son C.T., Krishnan S., Khan N.A. Understanding the impacts of the COVID-19 pandemic on sustainable Agri-food system and agroecosystem decarbonization nexus: a review. J. Clean. Prod. 2021;318 doi: 10.1016/j.jclepro.2021.128451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelodun B., Ajibade F.O., Tiamiyu A.O., Nwogwu N.A., Ibrahim R.G., Kumar P., Kumar V., Odey G., Yadav K.K., Khan A.H., Cabral-Pinto M.M.S., Kareem K.Y., Bakare H.O., Ajibade T.F., Naveed Q.N., Islam S., Fadare O.O., Choi K.S. Monitoring the presence and persistence of SARS-CoV-2 in water-food-environmental compartments: state of the knowledge and research needs. Environ. Res. 2021;200 doi: 10.1016/j.envres.2021.111373. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato-Lourenço L.F., de Souza Xavier Costa N., Dantas K.C., dos Santos Galvão L., Moralles F.N., Lombardi S.C.F.S., Júnior A.M., Lindoso J.A.L., Ando R.A., Lima F.G., Carvalho-Oliveira R., Mauad T. Airborne microplastics and SARS-CoV-2 in total suspended particles in the area surrounding the largest medical centre in Latin America. Environ. Pollut. 2022;292:118299. doi: 10.1016/j.envpol.2021.118299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato-Lourenço L.F., dos Santos Galvão L., de Weger L.A., Hiemstra P.S., Vijver M.G., Mauad T. An emerging class of air pollutants: potential effects of microplastics to respiratory human health? Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand U., Bianco F., Suresh S., Tripathi V., Núñez-Delgado A., Race M. SARS-CoV-2 and other viruses in soil: an environmental outlook. Environ. Res. 2021;198 doi: 10.1016/j.envres.2021.111297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan M., Xu B., Gamal El-Din M. Transmission of SARS-CoV-2 via fecal-oral and aerosols-borne routes: environmental dynamics and implications for wastewater management in underprivileged societies. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedrosian N., Mitchell E., Rohm E., Rothe M., Kelly C., String G., Lantagne D. A systematic review of surface contamination, stability, and disinfection data on SARS-CoV-2 (Through july 10, 2020) Environ. Sci. Technol. 2021;55:4162–4173. doi: 10.1021/acs.est.0c05651. [DOI] [PubMed] [Google Scholar]
- Belišová N., Konečná B., Bachratá N., Ryba J., Potočárová A., Tamáš M., Phuong A.L., Púček O., Kopáček J., Mackul'ak T. Sorption of SARS-CoV-2 virus particles to the surface of microplastics released during washing processes. Int. J. Environ. Res. Public Health. 2022;19:281. doi: 10.3390/ijerph19010281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat J., Nishimura N., Shimada Y. Toxicological interactions of microplastics/nanoplastics and environmental contaminants: current knowledge and future perspectives. J. Hazard. Mater. 2021;405 doi: 10.1016/j.jhazmat.2020.123913. [DOI] [PubMed] [Google Scholar]
- Böttcher B., Nassal M. Structure of mutant hepatitis B core protein capsids with premature secretion phenotype. J. Mol. Biol. 2018;430:4941–4954. doi: 10.1016/j.jmb.2018.10.018. [DOI] [PubMed] [Google Scholar]
- Boughattas I., Hattab S., Zitouni N., Mkhinini M., Missawi O., Bousserrhine N., Banni M. Assessing the presence of microplastic particles in tunisian agriculture soils and their potential toxicity effects using Eisenia andrei as bioindicator. Sci. Total Environ. 2021;796 doi: 10.1016/j.scitotenv.2021.148959. [DOI] [PubMed] [Google Scholar]
- Bouwmeester H., Hollman P.C.H., Peters R.J.B. Potential health impact of environmentally released micro- and nanoplastics in the human food production chain: experiences from nanotoxicology. Environ. Sci. Technol. 2015;49:8932–8947. doi: 10.1021/acs.est.5b01090. [DOI] [PubMed] [Google Scholar]
- Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S., Feng Z., Ghosh S., Goodsell D.S., Green R.K., Guranović V., Guzenko D., Hudson B.P., Kalro T., Liang Y., Lowe R., Namkoong H., Peisach E., Periskova I., Prlić A., Randle C., Rose A., Rose P., Sala R., Sekharan M., Shao C., Tan L., Tao Y.-P., Valasatava Y., Voigt M., Westbrook J., Woo J., Yang H., Young J., Zhuravleva M., Zardecki C. RCSB protein data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabianca A., Orlandi C., Amagliani G., Magnani M., Brandi G., Schiavano G.F. SARS-CoV-2 RNA detection on environmental surfaces in a university setting of Central Italy. Int. J. Environ. Res. Public Health. 2022;19:5560. doi: 10.3390/ijerph19095560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarino A.I., Macchia V., Sanderson W.G., Thompson R.C., Henry T.B. Low levels of microplastics (MP) in wild mussels indicate that MP ingestion by humans is minimal compared to exposure via household fibres fallout during a meal. Environ. Pollut. 2018;237:675–684. doi: 10.1016/j.envpol.2018.02.069. [DOI] [PubMed] [Google Scholar]
- Chen G., Feng Q., Wang J. Mini-review of microplastics in the atmosphere and their risks to humans. Sci. Total Environ. 2020;703 doi: 10.1016/j.scitotenv.2019.135504. [DOI] [PubMed] [Google Scholar]
- Chen X., Li X., Li Y. Toxicity inhibition strategy of microplastics to aquatic organisms through molecular docking, molecular dynamics simulation and molecular modification. Ecotoxicol. Environ. Saf. 2021;226 doi: 10.1016/j.ecoenv.2021.112870. [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang M., Fu H., Qu X., Zhang Z., Kang F., Zhu D. Spectroscopic and molecular modeling investigation on inhibition effect of nitroaromatic compounds on acetylcholinesterase activity. Chemosphere. 2019;236 doi: 10.1016/j.chemosphere.2019.124365. [DOI] [PubMed] [Google Scholar]
- Corpet D.E. Why does SARS-CoV-2 survive longer on plastic than on paper? Med. Hypotheses. 2021;146 doi: 10.1016/j.mehy.2020.110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeljak P., Pinto M., Proietti M., Reisser J., Ferrari F.F., Abbas B., van Loosdrecht M.C.M., Slat B., Herndl G.J. Extracting DNA from ocean microplastics: a method comparison study. Anal. Methods. 2017;9:1521–1526. doi: 10.1039/C6AY03119F. [DOI] [Google Scholar]
- Ding L., Mao R., Ma S., Guo X., Zhu L. High temperature depended on the ageing mechanism of microplastics under different environmental conditions and its effect on the distribution of organic pollutants. Water Res. 2020;174 doi: 10.1016/j.watres.2020.115634. [DOI] [PubMed] [Google Scholar]
- Dris R., Gasperi J., Mirande C., Mandin C., Guerrouache M., Langlois V., Tassin B. A first overview of textile fibers, including microplastics, in indoor and outdoor environments. Environ. Pollut. 2017;221:453–458. doi: 10.1016/j.envpol.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Dubey A., Kotnala G., Mandal T.K., Sonkar S.C., Singh V.K., Guru S.A., Bansal A., Irungbam M., Husain F., Goswami B., Kotnala R.K., Saxena S., Sharma S.K., Saxena K.N., Sharma C., Kumar S., Aswal D.K., Manchanda V., Koner B.C. Evidence of the presence of SARS-CoV-2 virus in atmospheric air and surfaces of a dedicated COVID hospital. J. Med. Virol. 2021;93:5339–5349. doi: 10.1002/jmv.27029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Liu Y., Xu Y., Li S., Liu X., Dai Y., Zhao J., Yue T. Benzo[a]pyrene and heavy metal ion adsorption on nanoplastics regulated by humic acid: cooperation/competition mechanisms revealed by molecular dynamics simulations. J. Hazard. Mater. 2022;424 doi: 10.1016/j.jhazmat.2021.127431. [DOI] [PubMed] [Google Scholar]
- Fournier E., Etienne-Mesmin L., Grootaert C., Jelsbak L., Syberg K., Blanquet-Diot S., Mercier-Bonin M. Microplastics in the human digestive environment: a focus on the potential and challenges facing in vitro gut model development. J. Hazard. Mater. 2021;415 doi: 10.1016/j.jhazmat.2021.125632. [DOI] [PubMed] [Google Scholar]
- Fred-Ahmadu O.H., Bhagwat G., Oluyoye I., Benson N.U., Ayejuyo O.O., Palanisami T. Interaction of chemical contaminants with microplastics: principles and perspectives. Sci. Total Environ. 2020;706 doi: 10.1016/j.scitotenv.2019.135978. [DOI] [PubMed] [Google Scholar]
- Ge C., Du J., Zhao L., Wang L., Liu Y., Li D., Yang Y., Zhou R., Zhao Y., Chai Z., Chen C. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc. Natl. Acad. Sci. 2011;108:16968–16973. doi: 10.1073/pnas.1105270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidari A., Sabbatini S., Bastianelli S., Pierucci S., Busti C., Bartolini D., Stabile A.M., Monari C., Galli F., Rende M., Cruciani G., Francisci D. SARS-CoV-2 survival on surfaces and the effect of UV-C light. Viruses. 2021;13:408. doi: 10.3390/v13030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J., Koritnik T., Mioč V., Trkov M., Bolješič M., Berginc N., Prosenc K., Kotar T., Paragi M. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.143226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Liu Y., Wang J. Sorption of sulfamethazine onto different types of microplastics: a combined experimental and molecular dynamics simulation study. Mar. Pollut. Bull. 2019;145:547–554. doi: 10.1016/j.marpolbul.2019.06.063. [DOI] [PubMed] [Google Scholar]
- Han J., Zhang X., He S., Jia P. Can the coronavirus disease be transmitted from food? A review of evidence, risks, policies and knowledge gaps. Environ. Chem. Lett. 2021;19:5–16. doi: 10.1007/s10311-020-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo K.S., Ho L.Y., Lim H.R., Leong H.Y., Chew K.W. Plastic waste associated with the COVID-19 pandemic: crisis or opportunity? J. Hazard. Mater. 2021;417 doi: 10.1016/j.jhazmat.2021.126108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Chae Y., An Y.-J. Mixture toxicity of nickel and microplastics with different functional groups on Daphnia magna. Environ. Sci. Technol. 2017;51:12852–12858. doi: 10.1021/acs.est.7b03732. [DOI] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T.C.T., Allan M.F., Malsick L.E., Woo J.Z., Zhu C., Zhang F., Khandwala S., Nyeo S.S.Y., Sun Y., Guo J.U., Bathe M., Näär A., Griffiths A., Rouskin S. Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells. Nat. Commun. 2022;13:1128. doi: 10.1038/s41467-022-28603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc R.M., Kasprzak W.K., Longhini A.P., Olenginski L.T., Abulwerdi F., Ginocchio S., Shields B., Nyman J., Svirydava M., Del Vecchio C., Ivanic J., Schneekloth J.S., Jr., Shapiro B.A., Dayie T.K., Le Grice S.F.J. Structural insights of the conserved "priming loop" of hepatitis B virus pre-genomic RNA. J. Biomol. Struct. Dyn. 2021 doi: 10.1080/07391102.2021.1934544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Ju M., Kim Y. Estimation of emission of tire wear particles (TWPs) in Korea. Waste Manag. 2020;108:154–159. doi: 10.1016/j.wasman.2020.04.037. [DOI] [PubMed] [Google Scholar]
- Lee H., Shim W.J., Kwon J.-H. Sorption capacity of plastic debris for hydrophobic organic chemicals. Sci. Total Environ. 2014;470–471:1545–1552. doi: 10.1016/j.scitotenv.2013.08.023. [DOI] [PubMed] [Google Scholar]
- Li R., Zhu L., Yang K., Li H., Zhu Y.-G., Cui L. Impact of urbanization on antibiotic resistome in different microplastics: evidence from a large-scale whole river analysis. Environ. Sci. Technol. 2021;55:8760–8770. doi: 10.1021/acs.est.1c01395. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., Liu X., Xu K., Ho K.-F., Kan H., Fu Q., Lan K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Liu P., Yang M., Zhao X., Guo Y., Wang L., Zhang J., Lei W., Han W., Jiang F., Liu W.J., Gao G.F., Wu G. Cold-chain transportation in the frozen food industry may have caused a recurrence of COVID-19 cases in destination: successful isolation of SARS-CoV-2 virus from the imported frozen cod package surface. Biosaf. Health. 2020;2:199–201. doi: 10.1016/j.bsheal.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Chen F. Multiwfn: a multifunctional wavefunction analyzer. J. Comput. Chem. 2012;33:580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- Lu T., Chen F. Quantitative analysis of molecular surface based on improved marching tetrahedra algorithm. J. Mol. Graph. Model. 2012;38:314–323. doi: 10.1016/j.jmgm.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Ma J., Chen F., Xu H., Jiang H., Liu J., Li P., Chen C.C., Pan K. Face masks as a source of nanoplastics and microplastics in the environment: quantification, characterization, and potential for bioaccumulation. Environ. Pollut. 2021;288 doi: 10.1016/j.envpol.2021.117748. [DOI] [PubMed] [Google Scholar]
- Maestre J.P., Jarma D., Yu J.-R.F., Siegel J.A., Horner S.D., Kinney K.A. Distribution of SARS-CoV-2 RNA signal in a home with COVID-19 positive occupants. Sci. Total Environ. 2021;778 doi: 10.1016/j.scitotenv.2021.146201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson M.J., Yinda C.K., Seifert S.N., Bushmaker T., Fischer R.J., van Doremalen N., Lloyd-Smith J.O., Munster V.J. Effect of environmental conditions on SARS-CoV-2 stability in human nasal mucus and sputum. Emerg. Infect. Dis. 2020;26:2276–2278. doi: 10.3201/eid2609.202267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei M., Tan X. Current strategies of antiviral drug discovery for COVID-19. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.671263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez-Pedriza A., Jaumot J. Interaction of environmental pollutants with microplastics: a critical review of sorption factors, bioaccumulation and ecotoxicological effects. Toxics. 2020;8:40. doi: 10.3390/toxics8020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meo S.A., Almutairi F.J., Abukhalaf A.A., Alessa O.M., Al-Khlaiwi T., Meo A.S. Sandstorm and its effect on particulate matter PM 2.5, carbon monoxide, nitrogen dioxide, ozone pollutants and SARS-CoV-2 cases and deaths. Sci. Total Environ. 2021;795 doi: 10.1016/j.scitotenv.2021.148764. [DOI] [PubMed] [Google Scholar]
- Mercogliano R., Avio C.G., Regoli F., Anastasio A., Colavita G., Santonicola S. Occurrence of microplastics in commercial seafood under the perspective of the human food chain. A review. J. Agric. Food Chem. 2020;68:5296–5301. doi: 10.1021/acs.jafc.0c01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno T., Pintó R.M., Bosch A., Moreno N., Alastuey A., Minguillón M.C., Anfruns-Estrada E., Guix S., Fuentes C., Buonanno G., Stabile L., Morawska L., Querol X. Tracing surface and airborne SARS-CoV-2 RNA inside public buses and subway trains. Environ. Int. 2021;147 doi: 10.1016/j.envint.2020.106326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco V., Oliver D.M., Weidmann M., Matallana-Surget S., Quilliam R.S. Survival of human enteric and respiratory viruses on plastics in soil, freshwater, and marine environments. Environ. Res. 2021;199 doi: 10.1016/j.envres.2021.111367. [DOI] [PubMed] [Google Scholar]
- Müller T.G., Sakin V., Müller B. A spotlight on viruses—application of click chemistry to visualize virus-cell interactions. Molecules. 2019;24:481. doi: 10.3390/molecules24030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao S., Akita K., Ozaki A., Masumoto K., Okuda T. Circulation of fibrous microplastic (microfiber) in sewage and sewage sludge treatment processes. Sci. Total Environ. 2021;795 doi: 10.1016/j.scitotenv.2021.148873. [DOI] [PubMed] [Google Scholar]
- Navarro A., Gómez L., Sanseverino I., Niegowska M., Roka E., Pedraccini R., Vargha M., Lettieri T. SARS-CoV-2 detection in wastewater using multiplex quantitative PCR. Sci. Total Environ. 2021;797 doi: 10.1016/j.scitotenv.2021.148890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien S., Okoffo E.D., O'Brien J.W., Ribeiro F., Wang X., Wright S.L., Samanipour S., Rauert C., Toapanta T.Y.A., Albarracin R., Thomas K.V. Airborne emissions of microplastic fibres from domestic laundry dryers. Sci. Total Environ. 2020;747 doi: 10.1016/j.scitotenv.2020.141175. [DOI] [PubMed] [Google Scholar]
- Patil S.M., Rane N.R., Bankole P.O., Krishnaiah P., Ahn Y., Park Y.-K., Yadav K.K., Amin M.A., Jeon B.-H. An assessment of micro- and nanoplastics in the biosphere: a review of detection, monitoring, and remediation technology. Chem. Eng. J. 2022;430 doi: 10.1016/j.cej.2021.132913. [DOI] [Google Scholar]
- Paul D., Kolar P., Hall S.G. A review of the impact of environmental factors on the fate and transport of coronaviruses in aqueous environments. npj Clean Water. 2021;4:7. doi: 10.1038/s41545-020-00096-w. [DOI] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzini K., Castrica M., Menchetti L., Maggi L., Negroni L., Orfeo N.V., Pizzoccheri A., Stocco M., Muttini S., Balzaretti C.M. SARS-CoV-2 RNA detection in the air and on surfaces in the COVID-19 ward of a hospital in Milan, Italy. Sci. Total Environ. 2020;742:140540. doi: 10.1016/j.scitotenv.2020.140540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania S., Park J., Md Din M.F., Mat Taib S., Talaiekhozani A., Kumar Yadav K., Kamyab H. Microplastics pollution in different aquatic environments and biota: a review of recent studies. Mar. Pollut. Bull. 2018;133:191–208. doi: 10.1016/j.marpolbul.2018.05.022. [DOI] [PubMed] [Google Scholar]
- Robertson M.P., Igel H., Baertsch R., Haussler D., Ares M., Scott W.G. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005;3:86–94. doi: 10.1371/journal.pbio.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscher L., Fehres A., Reisel L., Halbach M., Scholz-Böttcher B., Gerriets M., Badewien T.H., Shiravani G., Wurpts A., Primpke S., Gerdts G. Microplastic pollution in the weser estuary and the german North Sea. Environ. Pollut. 2021;288 doi: 10.1016/j.envpol.2021.117681. [DOI] [PubMed] [Google Scholar]
- Sala-Comorera L., Reynolds L.J., Martin N.A., O'Sullivan J.J., Meijer W.G., Fletcher N.F. Decay of infectious SARS-CoV-2 and surrogates in aquatic environments. Water Res. 2021;201 doi: 10.1016/j.watres.2021.117090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savastano A., Ibáñez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Liu Y., Wang K., Cizdziel J.V., Wu Y., Zhou Y. Ecotoxicological effects of micronized car tire wear particles and their heavy metals on the earthworm (Eisenia fetida) in soil. Sci. Total Environ. 2021;793 doi: 10.1016/j.scitotenv.2021.148613. [DOI] [PubMed] [Google Scholar]
- Stadnytskyi V., Bax C.E., Bax A., Anfinrud P. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc. Natl. Acad. Sci. 2020;117:11875–11877. doi: 10.1073/pnas.2006874117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.J., Derby J.A., Brevik E.C. Soil pathogens that may potentially cause pandemics, including severe acute respiratory syndrome (SARS) coronaviruses. Curr. Opin. Environ. Sci. Health. 2020;17:35–40. doi: 10.1016/j.coesh.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Xie H.-B., Chen J., Li X., Wang Z., Sheng L. Molecular dynamics simulations on the interactions of low molecular weight natural organic acids with C60. Chemosphere. 2013;92:429–434. doi: 10.1016/j.chemosphere.2013.01.039. [DOI] [PubMed] [Google Scholar]
- Sun S., Shi W., Tang Y., Han Y., Du X., Zhou W., Zhang W., Sun C., Liu G. The toxic impacts of microplastics (MPs) and polycyclic aromatic hydrocarbons (PAHs) on haematic parameters in a marine bivalve species and their potential mechanisms of action. Sci. Total Environ. 2021;783 doi: 10.1016/j.scitotenv.2021.147003. [DOI] [PubMed] [Google Scholar]
- Topol E.J. COVID-19 can affect the heart. Science. 2020;370:408–409. doi: 10.1126/science.abe2813. [DOI] [PubMed] [Google Scholar]
- Turner J.S., Kim W., Kalaidina E., Goss C.W., Rauseo A.M., Schmitz A.J., Hansen L., Haile A., Klebert M.K., Pusic I., O'Halloran J.A., Presti R.M., Ellebedy A.H. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature. 2021;595:421–425. doi: 10.1038/s41586-021-03647-4. [DOI] [PubMed] [Google Scholar]
- UNEP . United Nations Environment Programme, Nairobi. 2016. Marine plastic debris and microplastics—global lessons and research to inspire action and guide policy change. [DOI] [Google Scholar]
- Visalli G., Facciolà A., Pruiti Ciarello M., De Marco G., Maisano M., Di Pietro A. Acute and sub-chronic effects of microplastics (3 and 10 μm) on the human intestinal cells HT-29. Int. J. Environ. Res. Public Health. 2021;18:5833. doi: 10.3390/ijerph18115833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiktorczyk-Kapischke N., Grudlewska-Buda K., Wałecka-Zacharska E., Kwiecińska-Piróg J., Radtke L., Gospodarek-Komkowska E., Skowron K. SARS-CoV-2 in the environment—non-droplet spreading routes. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Mo W.Y., Luukkonen T. Adsorption behaviour and interaction of organic micropollutants with nano and microplastics—a review. Sci. Total Environ. 2021;797 doi: 10.1016/j.scitotenv.2021.149140. [DOI] [PubMed] [Google Scholar]
- Zettler E.R., Mincer T.J., Amaral-Zettler L.A. Life in the "plastisphere": microbial communities on plastic marine debris. Environ. Sci. Technol. 2013;47:7137–7146. doi: 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen H., He H., Cheng X., Ma T., Hu J., Yang S., Li S., Zhang L. Adsorption behavior and mechanism of 9-nitroanthracene on typical microplastics in aqueous solutions. Chemosphere. 2020;245 doi: 10.1016/j.chemosphere.2019.125628. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Liu T., Liu L., Fan Y., Rao W., Zheng J., Qian X. Distribution and sedimentation of microplastics in taihu Lake. Sci. Total Environ. 2021;795 doi: 10.1016/j.scitotenv.2021.148745. [DOI] [PubMed] [Google Scholar]
- Zhang F., Wang Z., Vijver M.G., Peijnenburg W.J.G.M. Probing nano-QSAR to assess the interactions between carbon nanoparticles and a SARS-CoV-2 RNA fragment. Ecotoxicol. Environ. Saf. 2021;219 doi: 10.1016/j.ecoenv.2021.112357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Zheludev I.N., Hagey R.J., Haslecker R., Hou Y.J., Kretsch R., Pintilie G.D., Rangan R., Kladwang W., Li S., Wu M.T.-P., Pham E.A., Bernardin-Souibgui C., Baric R.S., Sheahan T.P., Glenn J.S., Chiu W., Das R., D′Souza V. Cryo-EM and antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. Nat. Struct. Mol. Biol. 2021;28:747–754. doi: 10.1038/s41594-021-00653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Qi Y., Luzzatto-Fegiz P., Cui Y., Zhu Y. COVID-19: effects of environmental conditions on the propagation of respiratory droplets. Nano Lett. 2020;20:7744–7750. doi: 10.1021/acs.nanolett.0c03331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material