Abstract
Background
Comorbidities are risk factors for development of severe coronavirus disease 2019 (COVID-19). However, the extent to which an underlying comorbidity influences the immune response to severe acute respiratory syndrome coronavirus 2 remains unknown.
Objective
Our aim was to investigate the complex interrelations of comorbidities, the immune response, and patient outcome in COVID-19.
Methods
We used high-throughput, high-dimensional, single-cell mapping of peripheral blood leukocytes and algorithm-guided analysis.
Results
We discovered characteristic immune signatures associated not only with severe COVID-19 but also with the underlying medical condition. Different factors of the metabolic syndrome (obesity, hypertension, and diabetes) affected distinct immune populations, thereby additively increasing the immunodysregulatory effect when present in a single patient. Patients with disorders affecting the lung or heart, together with factors of metabolic syndrome, were clustered together, whereas immune disorder and chronic kidney disease displayed a distinct immune profile in COVID-19. In particular, severe acute respiratory syndrome coronavirus 2–infected patients with preexisting chronic kidney disease were characterized by the highest number of altered immune signatures of both lymphoid and myeloid immune branches. This overall major immune dysregulation could be the underlying mechanism for the estimated odds ratio of 16.3 for development of severe COVID-19 in this burdened cohort.
Conclusion
The combinatorial systematic analysis of the immune signatures, comorbidities, and outcomes of patients with COVID-19 has provided the mechanistic immunologic underpinnings of comorbidity-driven patient risk and uncovered comorbidity-driven immune signatures.
Key words: COVID-19, SARS-CoV-2, immune response, spectral flow cytometry, comorbidity, metabolic syndrome, chronic kidney disease, heart disease, immune disorder, algorithm-guided analysis
Abbreviations used: ACE2, Angiotensin-converting enzyme 2; CKD, Chronic kidney disease; CM, Central memory; COVID-19, Coronavirus disease 2019; DC, Dendritic cell; EM, Effector memory; HD, Heart disease; ID, Immune disorder; LAD, Lung-associated disorder; NK, Natural killer; OR, Odds ratio; PCA, Principal component analysis; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; TEMRA, T effector memory CD45RA+; T2DM, Type 2 diabetes mellitus; WHO, World Health Organization
Graphical abstract
The coronavirus disease 2019 (COVID-19) pandemic is continuously unfolding, with more than 5 million deaths to date following infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1 The majority of people infected with SARS-CoV-2 show mild symptoms, and some are even asymptomatic.2 Nevertheless, a relevant share of patients with COVID-19 not only develop fever, cough, and shortness of breath but also experience complications of pneumonia, respiratory failure, and severe acute respiratory distress syndrome.3, 4, 5 As seriously ill patients with COVID-19 regularly experience multiorgan failure affecting areas such as the cardiovascular system and kidneys next to the lungs, the medical history—and specifically, a history of comorbidities—plays a pivotal role in COVID-19 disease severity.
It is vital to identify patients at higher risk of development of severe COVID-19. This is important for prevention of SARS-CoV-2 infections, for therapeutic interventions once COVID-19 develops, and for COVID-19 vaccination strategy. Several comorbidities and old age are established risk factors for severe COVID-19.6 , 7 The most common comorbidities in hospitalized patients with COVID-19 are hypertension, diabetes, and obesity,7 followed by cardiovascular and respiratory system disease.6 Chronic kidney disease (CKD), cancer, and different immune disorders (IDs) have also been associated with worse disease outcomes.8, 9, 10, 11 However, the data as to whether and the extent to which a specific comorbidity increases the risk for severe COVID-19 are contradictory.
Severe cases of COVID-19 are mediated by immunopathology12—specifically, emergency myelopoiesis and a dysregulated, exhausted T-cell and natural killer (NK) cell compartment.13, 14, 15 Moreover, the uncontrolled production of cytokines—the so-called cytokine storm—is a main driver of the immunopathogenesis observed in severe COVID-19 cases.16 The extent to which an underlying comorbidity, and therewith the preceding immune status of an individual, influences the immune response to SARS-CoV-2 is unknown. As new therapeutics for patients with COVID-19 enter the market, understanding the underlying comorbidity-driven immunopathologic mechanisms in the immune response to SARS-CoV-2 is crucial for correct patient selection and individualized and optimized therapy.
Here, we have analyzed our previously generated high-dimensional single-cell spectral flow data set13 for the role of specific comorbidities in the immune response of patients affected by COVID-19. This has enabled us to link the single-cell immune signature of patients with COVID-19 to specific comorbidities. The combinatorial systematic analysis of patient immune signatures, comorbidities, and patient outcomes sheds light onto the mechanistic underpinnings of comorbidity-driven patient risk.
Methods
Samples from patients with COVID-19 and healthy donors
Clinical routine data and blood samples for PBMC isolation and cryopreservation were collected at the University Hospital Tuebingen in Germany, the Toulouse University Hospital in France (within the framework of the COVID-BioToul biobank [ClinicalTrials.gov identifier NCT04385108]), and the Nantes University Hospital in France (see Table E1, A in the Online Repository at www.jacionline.org), as previously reported.13 In brief, longitudinal collection of PBMC samples of patients with COVID-19 was performed (see Tables E1, A and E1, B in the Online Repository at www.jacionline.org). The patients included in this study were recruited between April 2020 and November 2020 (first and second waves; Wuhan and Alpha clades); in other words, they were recruited before the vaccine and mAb era. The grading of COVID-19 patients was executed according to the maximum severity of disease during the study based on the World Health Organization (WHO) ordinal scale17; the appropriate severity grade was then allocated to all related samples from each individual patient. Blood samples from healthy controls were collected at the blood transfusion center (Etablissement Français du Sang, Nantes, France) (see Table E1, A). Obesity was defined by a body mass index higher than 30 kg/m2. CKD included chronic renal insufficiency, history of kidney transplant, and dialysis. Hypertension was based on prior diagnosis (systolic [≥140 mmHg] and/or diastolic [≥90 mmHg] blood pressure at rest, measured on ≥2 days) and assigned to the patient independently of current blood pressure and antihypertensive therapy. The category heart disease (HD) included chronic cardiac disease, coronary artery disease, (ischemic) cardiomyopathy, cardiac arrhythmia, Fallot cardiopathy, and history of myocardial infarction. Lung-associated disorders (LAD) included bronchial asthma, lung cancer, chronic bronchitis, and smoking. The classification diabetes was assigned for patients diagnosed with type 2 diabetes mellitus (T2DM). The classification ID was assigned for patients diagnosed with a rheumatologic disorder (such as rheumatoid arthritis), polymyalgia rheumatica, or SLE or history of receiving immunosuppressive therapy after organ transplant. The category cancer included patients diagnosed with prostate cancer, melanoma, non–small cell lung cancer, and gastric cancer before development of COVID-19, independent of current anticancer therapy.
Spectral flow cytometry data
Spectral flow cytometry data used for this study are available from Mendeley Data at https://doi.org/10.17632/ffkvft27ds.2.13 In brief, PBMC samples were acquired on a Cytek Aurora spectral flow cytometer (Cytek Biosciences, Fremont, Calif) by using a surface panel, a panel focused on cytokine production in lymphoid cells (phorbol myristate acetate/ionomycin–stimulated), and a panel for cytokines in myeloid cells (R848-stimulated). Quality control, transformation, normalization, and downstream analysis were performed by using FlowJo (TreeStar) and R environment, as previously reported.13
Statistical analysis and visualization
Effect sizes were determined by using a nonparametric Wilcoxon signed rank test combined with a rank biserial correlation. These tests were performed by using the wilcox.effsize function from the rstatix package in R. Effect sizes greater than 0.3 (for severe vs mild COVID-19 comparison) or 0.2 (for comorbidity vs other samples) were considered relevant for further analysis. The value 0.3 is considered to be a medium strength effect. Frequencies of immune populations and median expression were compared by using the nonparametric Mann-Whitney-Wilcoxon test from the ggpubr package in R, including Benjamini-Hochberg correction using the stats package. The upset plots were made by using the Complex UpSet package in R. The principal component analysis (PCA) plots were made by using the fviz_pca function from the factoextra package, and the forest plots were created by using the forest plot package in R. For the correlogram, Pearson's r correlation coefficients were computed by using the Hmisc package, and the resulting correlation matrix was visualized by using the corrplot package. For correlation measurements, we used a linear regression model by applying the lm() and summary() functions. All other plots were drawn by using ggplot2. The significance of the statistical tests between groups is represented by P values of less than .05, .01, .001, or .0001.
Results
Cohort characteristics
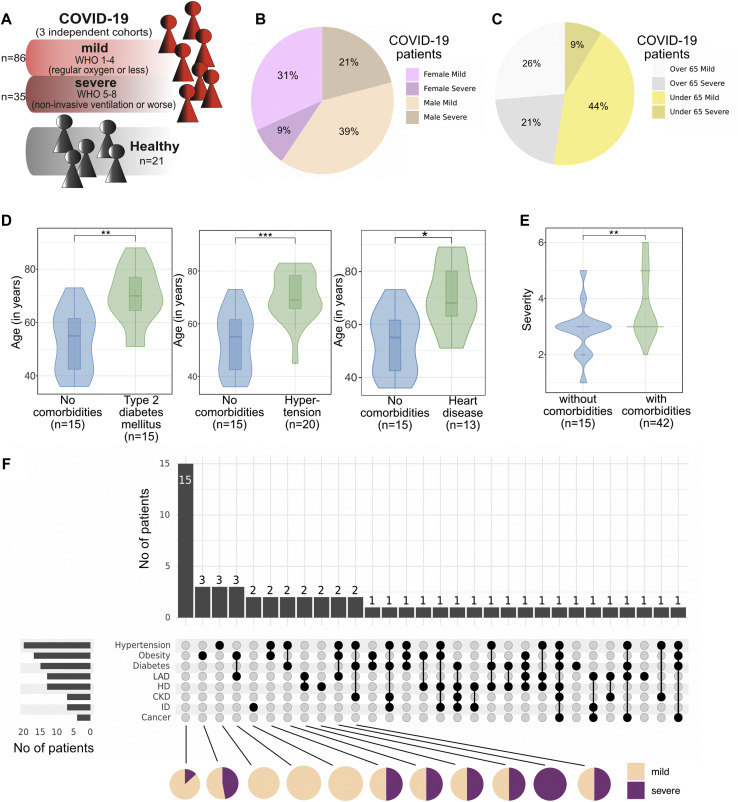
Three independent cohorts comprising a total of 57 patients with COVID-19 (121 PBMC samples) across Germany (Tuebingen) and France (Toulouse and Nantes) were analyzed in a longitudinal manner together with samples from 21 healthy controls (see Tables E1, A and E1, B). Although we initially anticipated center-specific batch effects in our multicenter study, this was not the case.13 Patients with COVID-19 were categorized on the basis of the WHO ordinal scale (World Health Organization, 2020), subdividing mild (WHO scale score of 1-4) and severe (WHO scale score of 5-8) disease (Fig 1 , A and see Tables E1, A and E1, C in the Online Repository at www.jacionline.org). The cohort of patients with COVID-19 consisted of 40% females and 60% males, with the latter presenting a higher rate of severe COVID-19 (Fig 1, B). Apart from male sex and in line with previous reports, patients older than 65 years were more likely to develop severe COVID-19 (Fig 1, C). Older age is associated with accumulation of comorbidities, as also represented by our study cohort (see Fig E1, A in the Online Repository at www.jacionline.org): the patients with T2DM, hypertension, or HD were significantly older than the patients without any comorbidity (Fig 1, D), whereas this was not the case for obesity, LAD (smoking, chronic bronchitis, lung cancer, or bronchial asthma), CKD, ID, or cancer (see Fig E1, B). When patients with and without comorbidities were compared, the severity of COVID-19 was significantly higher in patients burdened with comorbidity (Fig 1, E and see Table E1, C in the Online Repository at www.jacionline.org). Notably, 30 patients (52.6% of the cohort) presented with more than 1 type of comorbidity. Co-occurrence of specific comorbidities is depicted in Fig 1, F: the most frequent combinations involve hypertension, obesity, and T2DM—factors of the so-called metabolic syndrome (Fig 1, F and see Fig E2, A-G in the Online Repository at www.jacionline.org).
Fig 1.
A, Schematic of the study cohort. B, Proportion of female and male patients developing mild or severe COVID-19. C, Proportion of patients younger and older than 65 years who develop mild or severe COVID-19. D, Violin plot showing the age of patients with the indicated comorbidity versus that of patients without the comorbidity. Box plots depicting the median and 25th and 75th age percentiles. E, Violin plot showing the severity of COVID-19 in patients with at least 1 comorbidity versus in patients without comorbidity. F, UpSet plot to visualize co-occurrence of comorbidities in individual patients with COVID-19. Black dots connected by black lines represent the different comorbidity combinations. Bar plots on top display the number of patients per comorbidity combination. Bar plots on the left display the number of patients per comorbidity. Pie charts below show the proportion of patients with severe and mild COVID-19 in each comorbidity combination (shown for all combinations with more than 2 patients). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; Mann-Whitney test and Benjamini-Hochberg correction.
Patients with CKD demonstrate an increased risk for development of severe COVID-19
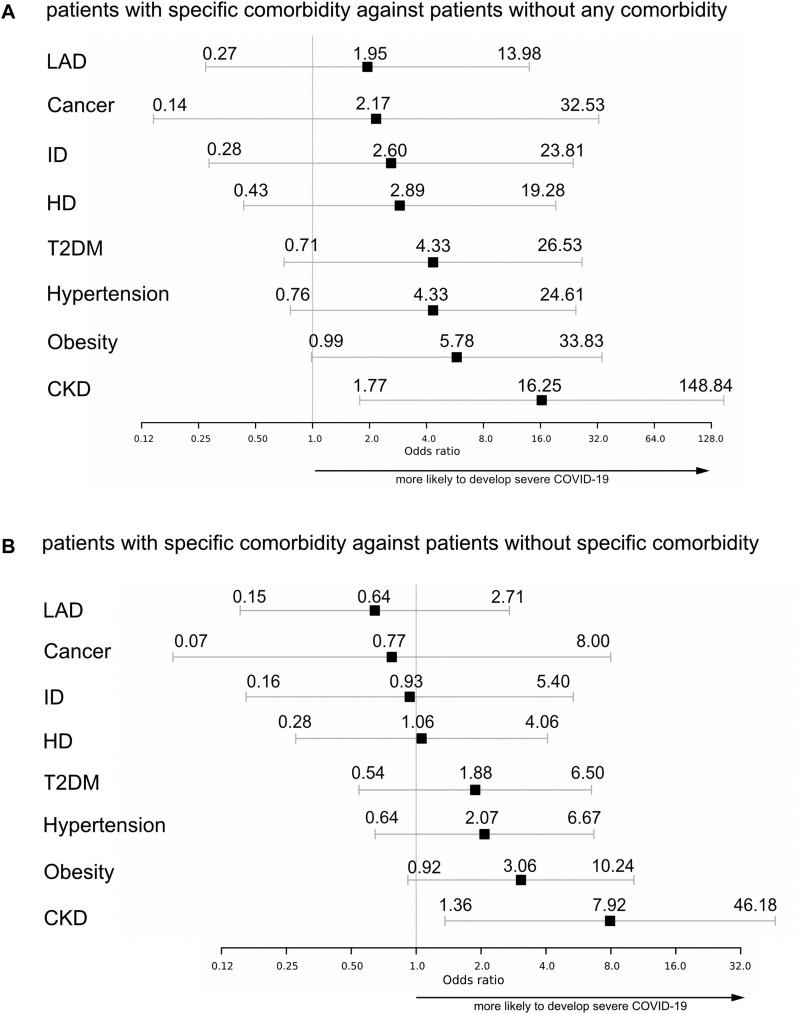
To investigate the influence of specific preexisting medical conditions on the development of severe COVID-19, we compared all patients with a specific comorbidity against patients without any comorbidity and estimated the odds ratios (ORs) for development of severe COVID-19 (Fig 2 , A). All comorbidity groups demonstrated a tendency toward more severe COVID-19, with CKD presenting significantly increased risk (OR = 16.25 [95% CI = 1.77-148.84). To minimize age being a confounding factor, we further compared patients with each comorbidity against all other patients with COVID-19 and, once again, the estimated ORs for severe COVID-19 for each comorbidity (Fig 2, B). CKD still revealed a significant co-occurrence, with an OR of 7.92 (95% CI = 1.36-46.18) for severe COVID-19, whereas obese patients showed a strong trend in the same direction. Notably, in both analyses, patients with LAD had the lowest estimated OR for severe COVID-19 of all comorbidities in our cohort.
Fig 2.
A, Forest plot of estimated ORs for development of severe COVID-19. Comparison of patients with specific comorbidity against patients without any comorbidity. B, Forest plot of estimated ORs for development of severe COVID-19. Comparison of patients with a specific comorbidity against patients without the specific comorbidity.
Patients with COVID-19 display severity- and comorbidity-specific immune profiles
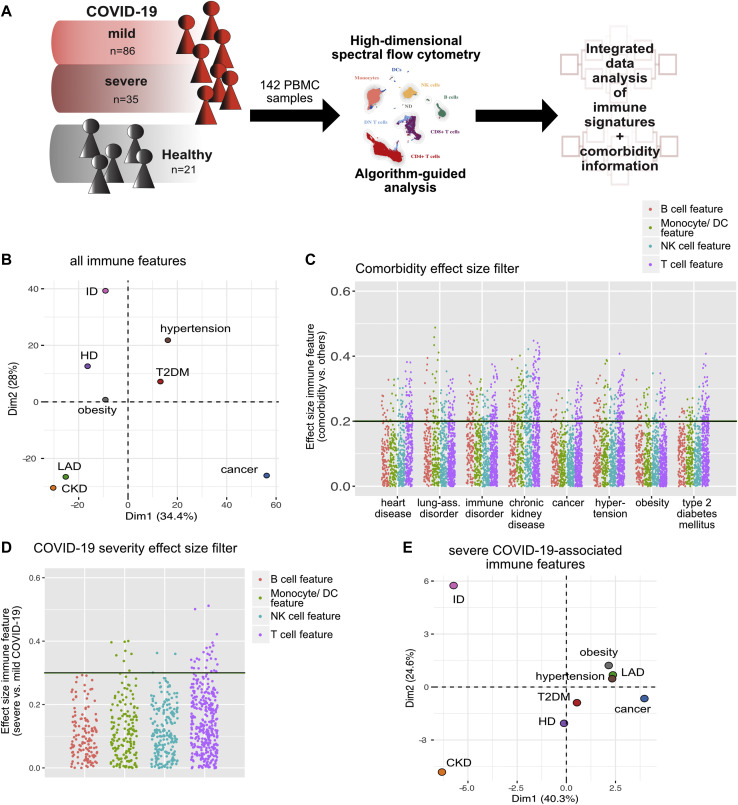
Because several comorbidities are associated with dysregulation of the immune system,7 , 18, 19, 20, 21, 22, 23, 24, 25 we questioned whether the immune dysregulation seen in comorbidities influences the immune response to SARS-CoV-2 infection and whether severity of COVID-19 can be linked to the immune signature of preexisting medical conditions. To do so, we made use of the detailed immune profiles that we had generated from PBMC samples (collected on day 0-96 after hospital admission due to COVID-19) by using high-parametric single-cell spectral flow cytometry.13 The data set provides information about the differentiation and activation status of lymphocyte and myeloid subpopulations (T cells, NK cells, B cells, monocytes, and dendritic cells [DCs]), as well as their cytokine polarization profiles. The generated data set resulted in hundreds of immune features, such as frequency of plasmacytoid DCs, mean fluorescence intensity of PD-1 in CD4+ effector memory (EM) T cells, and percentage of IFN-γ–positive cells among CD4+ central memory (CM) T cells. To take immune alteration at different time points into account, we treated all samples as individual and combined all time points of the longitudinal data set. We further enriched our data with clinical information (specifically, information on comorbidities) to link the immune response in COVID-19 not only to severity but also to a preexisting medical condition (Fig 3 , A). Of note, we did not find any significant changes in lymphocyte or granulocyte counts specific to a comorbidity group (see Fig E1, C and D).
Fig 3.
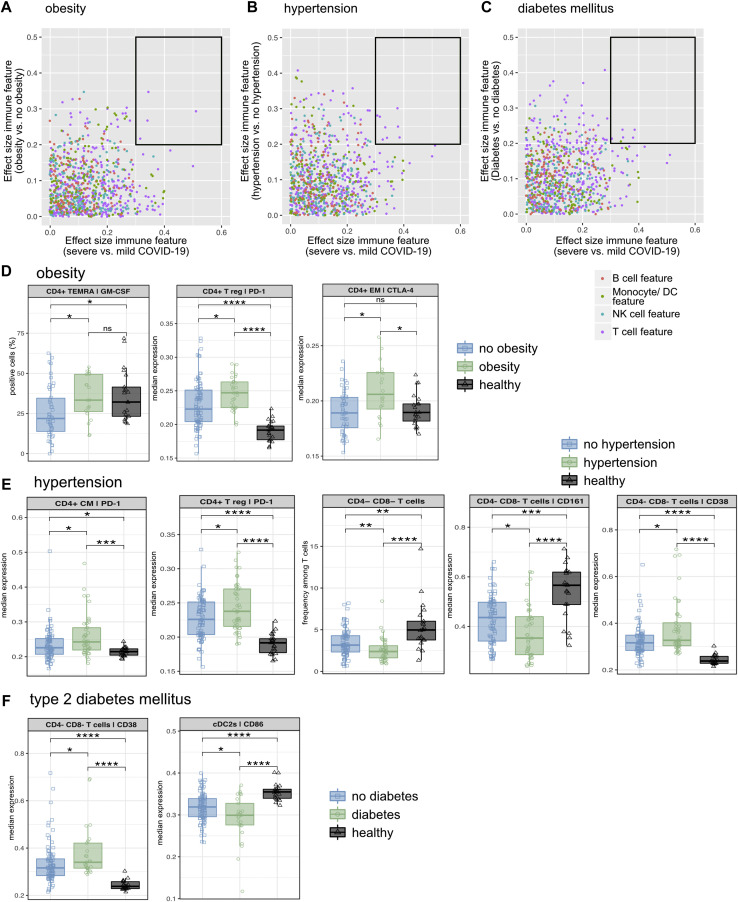
A, Schematic of the experimental approach. B, PCA of the total immune compartment on the basis of all extracted immune features grouped by comorbidity. C, Comorbidity effect size (ES) filter. Dot plot displaying the ES of every immune feature calculated for the indicated comorbidity versus without the indicated comorbidity. Each dot represents 1 immunologic feature, colored for the affected immune compartment. An ES of 0.2 represents the filter cutoff for further in-depth analysis of immune features. D, COVID-19 severity ES filter. Dot plot displaying the ES of every immune feature calculated for severe versus mild COVID-19. Each dot represents 1 immunologic feature, colored for the affected immune compartment. An ES of 0.3 represents the filter cutoff for further in-depth analysis of immune features. E, PCA of the total immune compartment based on all severe COVID-19–associated (ES severe vs mild COVID-19 > 0.3) immune features grouped by comorbidity.
To gain an initial overview of the global immune profiles of patients with COVID-19 with specific comorbidities, we performed PCA. The median values for each immune feature per sample were integrated; the resulting data set was then grouped by comorbidity (Fig 3, B and see Table E1, D in the Online Repository at www.jacionline.org). Because patients belong to multiple comorbidity groups, similar values in the PCA can also stem from an overlap of comorbidities in patients. Interestingly, patients with COVID-19 and preexisting CKD, LAD, and cancer were separated from the other comorbidities (Fig 3, B), suggesting a more distinct immune profile of patients having COVID-19 along with these comorbidities.
To identify immune signatures that are specifically relevant to severe COVID-19 and also associated with a specific comorbidity, the list of immune features was prefiltered by Wilcoxon effect size, which is a statistical measure combining fold change and statistical significance (with a value of 0.1-0.3 indicating a small effect and a value of >0.3 indicating an intermediate or large effect).26 , 27 First, we computed the effect size of immune signatures from samples from patients with a specific comorbidity versus from all other COVID-19 samples, thereby extracting all immune alterations associated with a specific preexisting medical condition (Fig 3, C). Second, the effect size per immune feature of samples from patients with severe versus mild COVID-19 was estimated (Fig 3, D), after which only severe COVID-19–associated immune features were used for PCA analysis (Fig 3, E and see Table E1, D). Interestingly, CKD and ID were now clearly separated, whereas all of the other comorbidities were clustered together (Fig 3, E). Hypertension, T2DM, and obesity, all of which are factors of the metabolic syndrome, as well as HD and LAD, appeared closely together, suggesting a more similar severe COVID-19–associated immune profile in those patients.
To further investigate the effect of age on all immune features associated with severe COVID-19, we performed a detailed correlation analysis. To gain an overall overview, for each feature, computed Pearson r coefficients values were visualized in a heatmap plot (see Fig E2, H). Furthermore, candidate correlating features were linearly correlated with age (Fig E2, I). Interestingly, although significant, all R 2 values turned out to be less than 0.1, highlighting the fact that there is no biologically relevant correlation between age and the expression value/frequency of the analyzed immune features. Of note, only 3 of the severe COVID-19–associated immune features showed differences that were sex dependent (see Fig E3, A and B in the Online Repository at www.jacionline.org). Although the samples from male patients in most comorbidity groups demonstrated a higher severity grade than the samples from female patients with COVID-19 (see Fig E3, C), we cannot exclude sex as a potential confounding factor.
In conclusion, ID and CKD appear to have the most distinct effects on the SARS-CoV-2 immune response, whereas comorbidities of the metabolic syndrome, cancer, LAD, and HD, show a similar pattern.
The metabolic syndrome combines immune alterations of both innate and adaptive immune branches
Prefiltered immune features, which exhibited a “comorbidity effect size” greater than 0.2 (Fig 3, C) and a “COVID-19 severity effect size” greater than 0.3 (Fig 3, D), were selected for further in-depth analysis (Fig 4, Fig 5, Fig 6 and see Figs E4-E7 and Table E1, E in the Online Repository at www.jacionline.org). The lower comorbidity effect size filter cutoff was used to include all potentially relevant immune features for next step analysis. To calculate the exact association of severe COVID-19–associated immune features with a specific comorbidity, we introduced a Mann-Whitney U-test with Benjamini-Hochberg correction comparing prefiltered immune features of patients with COVID-19 with and without individual comorbidity.
Fig 4.
A-C, Dot plots displaying the effect size (ES) of severe versus mild COVID-19 (x-axis) against the ES of the indicated comorbidity versus without the indicated comorbidity (y-axis). Each dot represents 1 immunologic feature, colored for the affected immune compartment. The black box highlights immune features for further in-depth analysis. D-F, The median frequency or median expression and 25th and 75th percentiles of the indicated parameters in the indicated FlowSOM-generated immune cell clusters. Comparison of obese patients, nonobese patients, and healthy controls (D), patients with hypertension, patients without hypertension, and healthy controls (E), patients with T2DM, patients with no T2DM, and healthy controls (F). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; Mann-Whitney test and Benjamini-Hochberg correction.
Fig 5.
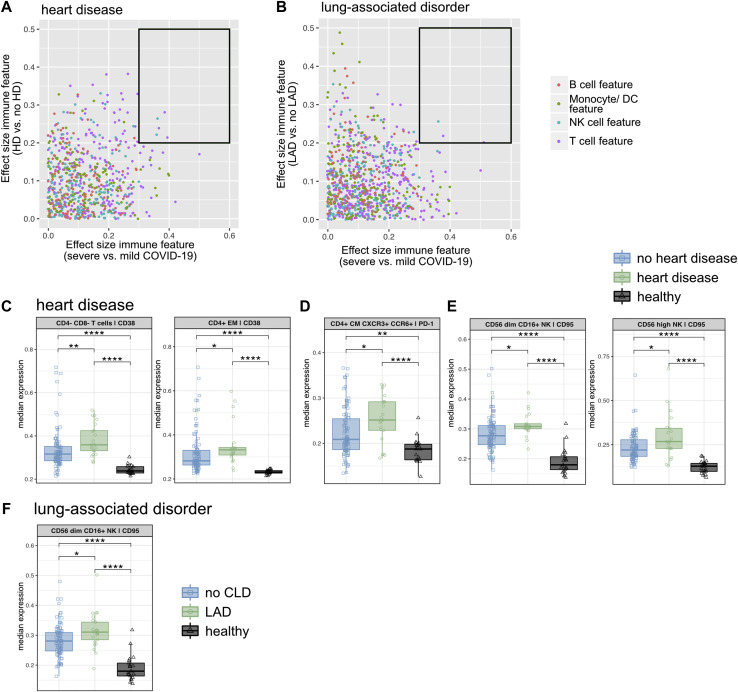
A and B, Dot plot displaying the effect size (ES) of severe versus mild COVID-19 (x-axis) against the ES of the indicated comorbidity versus without the indicated comorbidity (y-axis). Each dot represents 1 immunologic feature, colored for the affected immune compartment. The black box highlights immune features for further in-depth analysis. C-F, The median expression and 25th and 75th percentiles of the indicated parameters in the indicated FlowSOM-generated immune cell clusters. Comparison of patients with HD, patients with no HD, and healthy controls (C-E), and patients with an LAD, patients with no LAD, and healthy controls (F). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; Mann-Whitney test and Benjamini-Hochberg correction.
Fig 6.
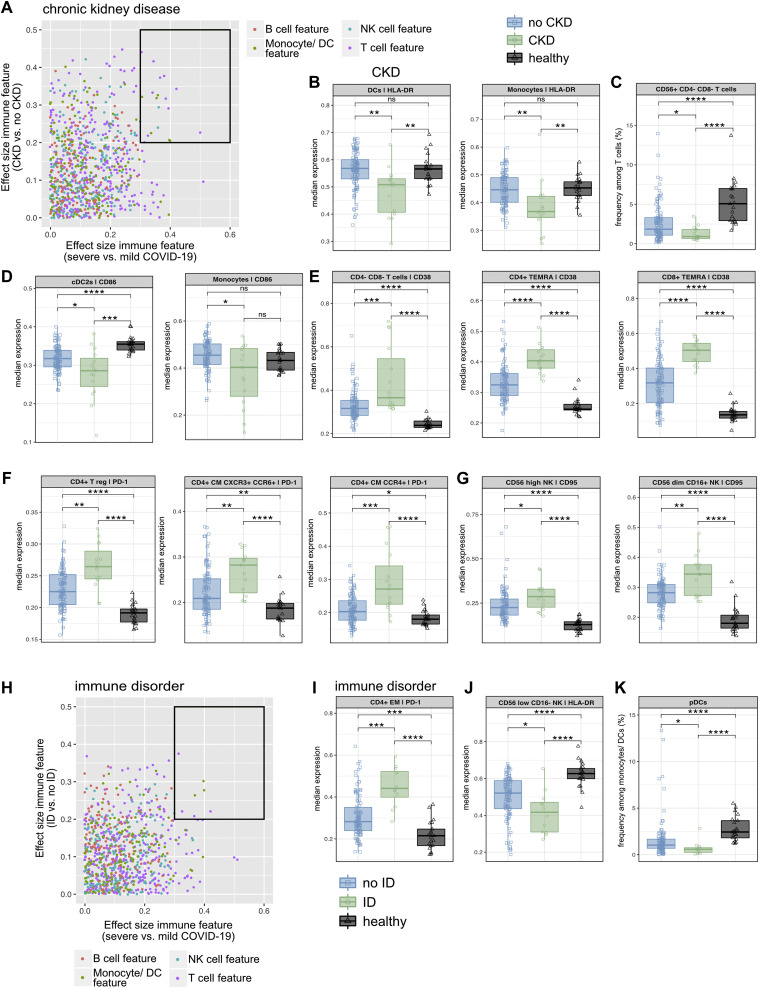
A, Dot plot displaying the effect size (ES) of severe versus mild COVID-19 (x-axis) against the ES of the indicated comorbidity versus without the indicated comorbidity (y-axis). Each dot represents 1 immunologic feature, colored for the affected immune compartment. The black box highlights immune features for further in-depth analysis. B-G, The median frequency or median expression and 25th and 75th percentiles of the indicated parameters in the indicated FlowSOM-generated immune cell clusters. Comparison of patients with CKD, patients without CKD, and healthy controls. H, Dot plot displaying the ES of severe versus mild COVID-19 (x-axis) against the ES of the indicated comorbidity versus without the indicated comorbidity (y-axis). Each dot represents 1 immunologic feature, colored for the affected immune compartment. The black box highlights immune features for further in-depth analysis. I-K, The median frequency or median expression and 25th and 75th percentiles of the indicated parameters in the indicated FlowSOM-generated immune cell clusters. Comparison of patients with an ID, patients with no ID, and healthy controls. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; Mann-Whitney test and Benjamini-Hochberg correction.
Within our study cohort, only 4 patients with COVID-19 had cancer co-occurring with other comorbidities (see Fig E4, A). An in-depth analysis of prefiltered immune features (see Fig E4, B) did not show severe COVID-19–associated signatures that were specific for cancer (see Fig E4, C), most likely owing to limited power.
In the case of obese patients with COVID-19 (Fig 4, D and see Fig E5, A), GM-CSF expression in CD4+ T EM CD45RA+ (TEMRA) cells, PD-1 expression in CD4+ regulatory T cells, and cytotoxic T-lymphocyte antigen-4 expression in CD4+ EM T cells were significantly higher than in nonobese patients with COVID-19 (Fig 4, D). Patients with COVID-19 and hypertension likewise demonstrated an impaired CD4+ T-cell compartment, with higher PD-1 expression in CD4+ CM and regulatory T cells (Fig 4, E). Furthermore, hypertensive patients with COVID-19 displayed reduced frequencies of CD4–CD8– (TCRγδ cell–enriched) T cells. Interestingly, this subset shows higher expression of CD38 as a sign of activation and reduced expression of CD161, resembling impaired innate activity and pointing to a dysregulated unconventional T-cell profile associated with hypertension (Fig 4, E). In contrast, patients with COVID-19 and T2DM were not only characterized by a T-cell signature (higher expression of CD38 in CD4–CD8– [TCRγδ cell–enriched] T cells) but also showed lower expression of the costimulatory signal CD86 in conventional DCs type 2 (Fig 4, F).
To summarize, severe COVID-19–associated immune alterations were concentrated in the unconventional and CD4+ T-cell compartment in patients with obesity and hypertension, pointing to increased activation/exhaustion and impaired innate activity. In contrast, T2DM was linked to impaired myeloid costimulation.
HD and LAD are associated with hyperactivated and exhausted T cells and NK cells
The prefiltered immune signatures associated with severe COVID-19 as well as with either HD or LAD were concentrated in the T-cell and NK cell compartment (Fig 5 and see Fig E6). Expression of the activation marker CD38 in CD4–CD8– (TCRγδ cell–enriched) and CD4+ EM T cells was found to be higher in patients with COVID-19 and HD than in healthy controls and patients with COVID-19 without HD (Fig 5, C). Expression of the immune checkpoint PD-1 was significantly higher in CD4+CXCR3+CCR6+ (TH 1/17 cell–enriched) T cells (Fig 5, D), and higher expression of CD95 was found in CD56high (cytokine-producing) and CD56dim CD16+ (cytotoxic) NK cells (Fig 5, E). The latter NK cell immune signature was also found to be characteristic for patients with LAD (Fig 5, F). Of note, all these immune features characteristic for HD and LAD were shared across patients with CKD (see Fig E2, D-F and Table E1, E).
In short, HD and LAD are associated with hyperactivated and exhausted T-cell and NK cell subsets.
The immune response in patients with COVID-19 with CKD and ID displays major dysregulations affecting all immune branches
When compared with the immune response in all other study patients with COVID-19, the immune response in individuals with CKD was characterized by the highest number of immune alterations associated with severe disease (Fig 6, A-G and see Fig E7, A-F and Table E1, E). Although no patient with COVID-19 had only CKD and none of the other comorbidities (Figs E2, F and E7, A), several immune signatures appeared to be unique in CKD, namely, lower expression of HLA-DR in DCs and monocytes (Fig 6, B and see Fig E7, B) and a reduced frequency of CD56+CD4–CD8– (NK T cell–like) T cells among all T cells (Fig 6, C). These specific features did not appear significantly in any of the other comorbidity groups. Further supporting such an overall altered immune response in patients with COVID-19 with CKD, we also identified reduced expression of CD86 in myeloid subsets (Fig 6, D and see Fig E7, C), significantly higher expression of the activation and exhaustion marker CD38 and PD-1 on several unconventional and conventional T-cell subsets (Fig 6, E and F and see Figs E7, D and E), higher expression of CD95 in cytotoxic and cytokine-producing NK cell subpopulations (Fig 6, G), and reduced expression of HLA-DR in CD56lowCD16– NK cells (see Fig E7, F), with all of these features pointing toward a massive deregulation affecting both the innate and adaptive immune compartments.
Similar to patients with COVID-19 and CKD, patients with COVID-19 and preexisting ID presented significant immune features affecting both the myeloid and lymphoid compartments, albeit limited in number (Fig 6, H-K and see Fig E7, G-I). Next to signs of hyperactivation and exhaustion (higher expression of PD-1 and CD38 in CD4+ T-cell subsets [Fig 6, I and see Fig E7, G and H]), impaired innate immunity seems to play a role in the SARS-CoV-2 immune response in patients with ID, represented by reduced HLA-DR in CD56lowCD16– NK cells (Fig 6, J). Although these features were also seen in patients with CKD, reduced frequencies of plasmacytoid DCs among all monocytes and DCs turned out to be unique to the cohort of patients with ID (Fig 6, K).
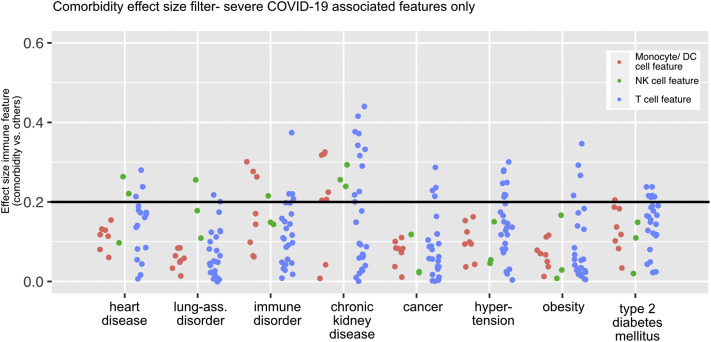
In summary, the immune profile associated with both severe COVID-19 and CKD as well as ID was characterized by an overall dysregulation of all immune branches (Fig 7 )—specifically, a hyperactivated, exhausted T-cell compartment and compromised innate immunity. These results were distinct from the comorbidity-driven immune signatures of HD and LAD associated with T-cell and NK cell hyperactivation/exhaustion and that of cancer, hypertension, and obesity, which solely affect the T-cell compartment (Fig 7).
Fig 7.
Dot plot displaying the effect size (ES) of every severe COVID-19–associated immune feature calculated for the indicated comorbidity versus without the indicated comorbidity. Each dot represents 1 immunologic feature, colored for the affected immune compartment. An ES of 0.2 represents the filter cutoff for further in-depth analysis of immune features.
Discussion
Several large studies of COVID-19 and comorbidity are out there and reporting risk calculations between comorbidities and COVID-19 severity. However, none of them take into account the immune profile as a potential underlying mechanism for the variable outcomes. Our study is not geared primarily to reveal which comorbidities dictate COVID-19 outcomes. Instead, our combinatorial systematic analysis of patients’ immune signatures, comorbidities, and outcomes sheds light onto the mechanistic underpinnings of comorbidity-driven patient risk. In general, the identified features comprised increased T-cell and NK cell activation and exhaustion as well as signs of impaired antigen presentation—with a comorbidity-specific immune profile for each subgroup, respectively.
Obesity has been confirmed as a risk factor for severity and death in COVID-19,28 , 29 which is in line with our results demonstrating that obese patients with COVID-19 were 3.1 times more likely than nonobese patients to experience a severe disease course. Overall, obesity could lead to increased risk of severe COVID-19 through immune cell and hormone deregulation, which then leads to decreased host defense and a higher chance for hyperinflammation.30 , 31 Insulin signaling has immunostimulatory effects on T cells, inducing growth, proliferation, and production of cytokines in T cells. All of these pathways are critical to T-cell function and thus host defense, and their dysregulation leads, in the case of obesity, to a proinflammatory condition.29 In line with this, we detected higher GM-CSF expression in CD4+ CM T cells as a characteristic signature in our obese cohort. This feature was distinct to nonobese patients, but it did not differ from that in healthy controls. Thus, in this cohort, it might not play a major role as an early pathogenic driver of the hyperinflammation seen in severe COVID-19.13 , 32 Higher expression of cytotoxic T-lymphocyte antigen-4 on CD4+ EM T cells could be part of the chronic inflammatory state found in obese individuals as a potential negative feedback loop.29
Our analysis showed hypertension demonstrating an OR of 2.1 for severe COVID-19, which is consistent with the findings of previous studies presenting hypertension as being significantly associated with adverse outcomes in COVID-19.33 A recent study using single-cell RNA sequencing showed a distinct inflammatory predisposition of immune cells from airway samples of patients with COVID-19 and hypertension, which was correlated with COVID-19 severity.20 Our data add results of the circulating immune system comprising decreased frequencies of altered CD4–CD8– (TCRγδ cell–enriched) T cells (most likely owing to their migration to the lung34) characterizing impaired early response to viral infection.35
Next to obesity and hypertension, another factor of the metabolic syndrome, namely, T2DM, has been associated with worse outcomes in COVID-19.31 , 36 Our study estimates an OR of 1.9 of patients with diabetes developing severe COVID-19. Hyperglycemia and insulin resistance promote synthesis of proinflammatory cytokines, oxidative stress, and production of adhesion molecules that mediate tissue inflammation. Poorly controlled diabetes has been linked to an inhibited lymphocyte proliferative response and impaired monocyte, macrophage, and neutrophil functions.37 , 38 Interestingly, although being related to obesity and hypertension through the metabolic syndrome, in our study focusing on severe COVID-19–associated immune signatures, it was characterized not only by unconventional T-cell dysregulation but also by reduced expression of the costimulatory receptor CD86 in conventional DCs type 2. One could hypothesize that patients with COVID-19 and several comorbidities that are part of the metabolic syndrome might display dysregulation of several immune branches, thus explaining these patients’ highly increased need for mechanical ventilation, intensive care unit admissions, and COVID-19 mortality.39
Whereas previous reports have described patients with HD as well as with chronic obstructive pulmonary disease and smoking being more likely to develop severe COVID-19,7 , 40 , 41 our analysis was not able to confirm significance for this assumption when such patients were compared with all patients without this specific comorbidity. Removing all samples collected from patients having asthma, which has been hypothesized to even have a protective effect through an increase in TH2 cell counts and decreased expression of the SARS-CoV-2 entry receptor angiotensin-converting enzyme 2 (ACE2) in lung tissue,42, 43, 44 did not change these results for the edited LAD group (data not shown). Of note, increased ACE2 expression was observed in the bronchial epithelium of obese patients with COVID-19 and chronic obstructive pulmonary disease.45
Both HD and LAD in our study cohort were characterized by alterations of the T-cell and NK cell compartment. Specifically, patients with HD demonstrated signs of T-cell hyperinflammation and exhaustion, consistent with reported inflammatory signatures associated with HD in patients without SARS-CoV-2 infection.46 All of the immune features described in patients with HD were shared with those with CKD. Interestingly, the PCA analysis showed HD as being the comorbidity located closest to CKD, which supports the similarity between these 2 comorbidities in terms of effects on the immune response to SARS-CoV-2.
CKDs are the most prominent risk factors for COVID-19 mortality.8 , 47 We confirmed an increased risk for severe COVID-19 in patients with CKD in our study cohort, with an OR of 7.9 (95% CI = 1.4-46.2). Normal kidney function contributes to immune homeostasis by filtering circulating cytokines and immunogenic pathogen components and thereby limiting inflammation, which are important functions in COVID-19 in particular, as the cytokine storm plays a key role in disease progression. In advanced kidney disease, reduced filtering capacity leads to increased innate immune cell activation, which in turn leads to increased production of cytokine and reactive oxygen species, further increasing inflammation.24 Additionally, lymphocyte number and function are decreased in patients with CKD, making them immunocompromised.24 , 48 Together with a deregulated renin-angiotensin system including the coronavirus entry receptor ACE2,49 the effects of CKD on the immune system lead to increased inflammation and host susceptibility to infections, especially SARS-CoV-2.24 , 50 , 51 Our data resemble those for a similar condition after SARS-CoV-2 infection: increased expression of PD-1, CD38, and CD95 on several T-cell and NK cell subsets points to a hyperactivated and exhausted T-cell and NK cell compartment.13 , 52 , 53 Furthermore, reduced expression of HLA-DR and CD86 in monocytes and DCs points to impaired antigen presentation in these individuals. Of note, the amount of significantly changed and CKD-specific immune features was higher than in any other comorbidity cohort, thereby supporting an immune profile of patients with COVID-19 and CKD that is characterized by an overall dysregulation in both the lymphoid and myeloid compartments leading to such a severe disease. Next to CKD, alterations in both immune branches were also associated with preexisting ID, but not to this extent.
Several risk assessments of comorbidities and severe COVID-19 have been performed, but there is a striking lack of investigative studies examining the underlying immunologic mechanisms. Our study uncovers novel associations between specific comorbidities and the severe COVID-19–associated immune response, presenting characteristic immune landscapes depending on the underlying medical condition of a patient. These results enrich the database of COVID-19 immunopathology and provide a source for more individualized treatments of patients with COVID-19 based on their comorbidities. Using therapies targeting the immunologic changes specific to severe COVID-19 and even a patient’s comorbidities could be a promising tool to alleviate the severity of this disease.
Limitations of the study
We used a “real-life” cohort of patients with COVID-19; thus, the sample numbers for patients with COVID-19 and rare comorbidities was low, and it reduced the power of the analysis for these cohorts. Furthermore, as patients regularly had multiple comorbidities, the effects could be biased. To take immune alteration at different time points into account, we treated all samples as individual and combined all time points of the longitudinal data set, resulting in different numbers of samples per patient within the data set. Because of the retrospective design of this study, we cannot completely exclude further confounding factors such as sex. Despite these challenges, this study is the first of its kind, analyzing comorbidities of patients with COVID-19 and their association with specific immune signatures and thereby providing novel insights into the underlying immunopathology in patients with COVID-19 with different medical histories.
Key messages.
-
•
The immune response associated with severe COVID-19 displayed comorbidity-specific characteristics.
-
•
The metabolic syndrome combined impaired innate immunity and adaptive immune dysregulation as possible underlying mechanisms for a worse COVID-19 outcome.
-
•
CKD before SARS-CoV-2 infection was associated with increased risk of development of severe COVID-19 and was linked to an overall dysregulated lymphoid and myeloid compartment.
-
•
The mechanistic underpinnings of comorbidity-driven patient risk serve as a basis for precision medicine.
Acknowledgments
We thank the patients and blood donors who contributed to this study, as well as the hospitals in Nantes, Toulouse, and Tübingen for sample and data collection. Regarding the COVID-BioToul biobank (ClinicalTrials.gov identifier NCT04385108) specifically, we thank the Centre de Ressources Biologiques “Toulouse – Bio Ressources” (CRB TBR), the Clinical Research Center 1436, and the Delegation for Clinical Research and Innovation of the Toulouse University Hospital for their highly valuable implication. We thank the Biological Resource Centre for Biobanking (CHU Nantes, Nantes Université, Centre de ressources biologiques (BB-0033-00040), F-44000 Nantes, France). The graphical abstract was created with Biorender.com.
Footnotes
Supported by the Swiss National Science Foundation, Switzerland (grants 733 310030_170320, 310030_188450, and CRSII5_183478 to B.B, 31CA30_195883 [to S.K., M.C., M.B., and B.B.]), The LOOP Zurich, the Vontobel Foundation (to B.B.), the European Union’s Horizon 2020 Research and Innovation Program (grant 847782 [to B.B. and A.R.]), and the European Research Council (grant 882424 [to B.B.]), the Agence National de la Recherche and Region Pays de la Loire - Flash COVID-19:COVARDS project (to A.R.), the INSPIRE regional initiative (to R.L.), and the French Ministry of Health with the participation of the Groupement Interrégional de Recherche Clinique et d’Innovation Sud-Ouest Outre-Mer (PHRCI 2020 IMMUNOMARK-COV [to R.L. and G.M.-B.]). S. Kreutmair is the recipient of a postdoctoral fellowship of the Deutsche Forschungsgemeinschaft. N.G.N. is the recipient of a University Research Priority Program postdoctoral fellowship. F.I. received a PhD fellowship from the Studienstiftung des deutschen Volkes.
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Supplementary data
References
- 1.World Health Organization Weekly epidemiological update on COVID-19. 2020;1:4. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---8-june-2022 Available at: [Google Scholar]
- 2.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price-Haywood E.G., Burton J., Fort D., Seoane L. Hospitalization and mortality among Black patients and White patients with Covid-19. N Engl J Med. 2020;382:2534–2543. doi: 10.1056/NEJMsa2011686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arentz M., Yim E., Klaff L., Lokhandwala S., Riedo F.X., Chon M., et al. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington state. JAMA. 2020;323:1612–1614. doi: 10.1001/jama.2020.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velavan T.P., Meyer C.G. The COVID-19 epidemic. Trop Med Int Health. 2020;25:278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 7.Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gok M., Cetinkaya H., Kandemir T., Karahan E., Tuncer İ.B., Bukrek C., et al. Chronic kidney disease predicts poor outcomes of COVID-19 patients. Int Urol Nephrol. 2021;53:1891–1898. doi: 10.1007/s11255-020-02758-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favalli E.G., Gerosa M., Murgo A., Caporali R. Are patients with systemic lupus erythematosus at increased risk for COVID-19? Ann Rheum Dis. 2021;80:e25. doi: 10.1136/annrheumdis-2020-217787. [DOI] [PubMed] [Google Scholar]
- 10.Arleo T., Tong D., Shabto J., O’Keefe G., Khosroshahi A. Clinical course and outcomes of COVID-19 in rheumatic disease patients: a case cohort study with a diverse population. Clin Rheumatol. 2021;40:2633–2642. doi: 10.1007/s10067-021-05578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai A., Sachdeva S., Parekh T., Desai R. COVID-19 and cancer: lessons from a pooled meta-analysis. JCO Glob Oncol. 2020;6:557–559. doi: 10.1200/GO.20.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soy M., Keser G., Atagündüz P., Tabak F., Atagündüz I., Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39:2085–2094. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreutmair S., Unger S., Núñez N.G., Ingelfinger F., Alberti C., De Feo D., et al. Distinct immunological signatures discriminate severe COVID-19 from non-SARS-CoV-2-driven critical pneumonia. Immunity. 2021;54:1578–1593.e5. doi: 10.1016/j.immuni.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur S., Bansal R., Kollimuttathuillam S., Gowda A.M., Singh B., Mehta D., et al. The looming storm: blood and cytokines in COVID-19. Blood Rev. 2021;46 doi: 10.1016/j.blre.2020.100743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization WHO R&D blueprint novel coronavirus COVID-19 therapeutic trial synopsis. World Health Organ. 2020;1-9 https://www.who.int/publications/i/item/covid-19-therapeutic-trial-synopsis Available at: [Google Scholar]
- 18.Milner J.J., Beck M.A. The impact of obesity on the immune response to infection. Proc Nutr Soc. 2012;71:298–306. doi: 10.1017/S0029665112000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinh Q.N., Drummond G.R., Sobey C.G., Chrissobolis S. Roles of inflammation, oxidative stress, and vascular dysfunction in hypertension. Biomed Res Int. 2014;2014 doi: 10.1155/2014/406960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trump S., Lukassen S., Anker M.S., Chua R.L., Liebig J., Thürmann L., et al. Hypertension delays viral clearance and exacerbates airway hyperinflammation in patients with COVID-19. Nat Biotechnol. 2021;39:705–716. doi: 10.1038/s41587-020-00796-1. [DOI] [PubMed] [Google Scholar]
- 21.Berbudi A., Rahmadika N., Tjahjadi A.I., RuslamiR Type 2 diabetes and its impact on the immune system. Curr Diabetes Rev. 2020;16:442–449. doi: 10.2174/1573399815666191024085838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sopori M. Effects of cigarette smoke on the immune system. Nat Rev Immunol. 2002;2:372–377. doi: 10.1038/nri803. [DOI] [PubMed] [Google Scholar]
- 23.Marsland B.J., Königshoff M., Saglani S., Eickelberg O. Immune system dysregulation in chronic lung disease. Eur Respir J. 2011;38:500–501. doi: 10.1183/09031936.00103211. [DOI] [PubMed] [Google Scholar]
- 24.Tecklenborg J., Clayton D., Siebert S., Coley S.M. The role of the immune system in kidney disease. Clin Exp Immunol. 2018;192:142–150. doi: 10.1111/cei.13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mari D., Di Berardino F., Cugno M. Chronic heart failure and the immune system. Clin Rev Allergy Immunol. 2002;23:325–340. doi: 10.1385/CRIAI:23:3:325. [DOI] [PubMed] [Google Scholar]
- 26.McGough J.J., Faraone S.V. Estimating the size of treatment effects: moving beyond p values. Psychiatry (Edgmont) 2009;6:21–29. [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan G.M., Feinn R. Using effect size-or why the P value is not enough. J Grad Med Educ. 2012;4:279–282. doi: 10.4300/JGME-D-12-00156.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J., Hu J., Zhu C. Obesity aggravates COVID-19: a systematic review and meta-analysis. J Med Virol. 2021;93:257–261. doi: 10.1002/jmv.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammad S., Aziz R., Al Mahri S., Malik S.S., Haji E., Khan A.H., et al. Obesity and COVID-19: what makes obese host so vulnerable? Immun. Ageing. 2021;18:1. doi: 10.1186/s12979-020-00212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai S.-H., Liao W., Chen S.-W., Liu L.-L., Liu S.-Y., Zheng Z.-D. Association between obesity and clinical prognosis in patients infected with SARS-CoV-2. Infect Dis Poverty. 2020;9:80. doi: 10.1186/s40249-020-00703-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren H., Yang Y., Wang F., Yan Y., Shi X., Dong K., et al. Association of the insulin resistance marker TyG index with the severity and mortality of COVID-19. Cardiovasc Diabetol. 2020;19:58. doi: 10.1186/s12933-020-01035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thwaites R.S., Sanchez Sevilla Uruchurtu A., Siggins M.K., Liew F., Russell C.D., Moore S.C., et al. Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abg9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang X., Shi L., Wang Y., Xiao W., Duan G., Yang H., et al. The association of hypertension with the severity and mortality of COVID-19 patients: evidence based on adjusted effect estimates. J Infect. 2020;81:e44–e47. doi: 10.1016/j.jinf.2020.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saris A., Reijnders T.D.Y., Nossent E.J., Schuurman A.R., Verhoeff J., van Asten S., et al. Distinct cellular immune profiles in the airways and blood of critically ill patients with COVID-19. Thorax. 2021;76:1010–1019. doi: 10.1136/thoraxjnl-2020-216256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Acquisto F., Crompton T. CD3+CD4-CD8- (double negative) T cells: saviours or villains of the immune response? Biochem Pharmacol. 2011;82:333–340. doi: 10.1016/j.bcp.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 36.Guo W, Li M, Dong Y, Zhou H, Zhang Z, Tian, C, et al. Diabetes is a risk factor for the progression and prognosis of COVID-19 [e-pub ahead of print]. Diabetes Metab Res Rev https://doi.org/10.1002/dmrr.3319. [DOI] [PMC free article] [PubMed]
- 37.Knapp S. Diabetes and infection: is there a link?--a mini-review. Gerontology. 2013;59:99–104. doi: 10.1159/000345107. [DOI] [PubMed] [Google Scholar]
- 38.Melvin W.J., Audu C.O., Davis F.M., Sharma S.B., Joshi A., DenDekker A., et al. Coronavirus induces diabetic macrophage-mediated inflammation via SETDB2. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2101071118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohia P, Kapur S, Benjaram S, Pandey A, Mir T, Seyoum B. Metabolic syndrome and clinical outcomes in patients infected with COVID-19: does age, sex, and race of the patient with metabolic syndrome matter [e-pub ahead of print]? J Diabetes https://doi.org/10.1111/1753-0407.13157. [DOI] [PMC free article] [PubMed]
- 40.Rabbani G., Shariful Islam S.M., Rahman M.A., Amin N., Marzan B., Robin R.C., et al. Pre-existing COPD is associated with an increased risk of mortality and severity in COVID-19: a rapid systematic review and meta-analysis. Expert Rev Respir Med. 2021;15:705–716. doi: 10.1080/17476348.2021.1866547. [DOI] [PubMed] [Google Scholar]
- 41.Patanavanich R., Glantz S.A. Smoking is associated with COVID-19 progression: a meta-analysis. Nicotine Tob Res. 2020;22:1653–1656. doi: 10.1093/ntr/ntaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman-Cribbin W., Rapp J., Alpert N., Tuminello S., Taioli E. The impact of asthma on mortality in patients with COVID-19. Chest. 2020;158:2290–2291. doi: 10.1016/j.chest.2020.05.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carli G., Cecchi L., Stebbing J., Parronchi P., Farsi A. Is asthma protective against COVID-19? Allergy. 2021;76:866–868. doi: 10.1111/all.14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson D.J., Busse W.W., Bacharier L.B., Kattan M., O’Connor G.T., Wood R.A., et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J Allergy Clin Immunol. 2020;146:203–206.e3. doi: 10.1016/j.jaci.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higham A., Singh D. Increased ACE2 expression in bronchial epithelium of COPD patients who are overweight. Obesity (Silver Spring) 2020;28:1586–1589. doi: 10.1002/oby.22907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodondi N., Marques-Vidal P., Butler J., Sutton-Tyrrell K., Cornuz J., Satterfield S., et al. Markers of atherosclerosis and inflammation for prediction of coronary heart disease in older adults. Am J Epidemiol. 2010;171:540–549. doi: 10.1093/aje/kwp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ERA-EDTA-Council, ERACODA-Working Group Chronic kidney disease is a key risk factor for severe COVID-19: a call to action by the ERA-EDTA. Nephrol Dial Transplant. 2021;36:87–94. doi: 10.1093/ndt/gfaa314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hartzell S., Bin S., Cantarelli C., Haverly M., Manrique J., Angeletti A., et al. Kidney failure associates with T cell exhaustion and imbalanced follicular helper T cells. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.583702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silhol F., Sarlon G., Deharo J.-C., Vaïsse B. Downregulation of ACE2 induces overstimulation of the renin-angiotensin system in COVID-19: should we block the renin-angiotensin system? Hypertension. 2020;43:854–856. doi: 10.1038/s41440-020-0476-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Girndt M., Sester M., Sester U., Kaul H., Köhler H. Defective expression of B7-2 (CD86) on monocytes of dialysis patients correlates to the uremia-associated immune defect. Kidney Int. 2001;59:1382–1389. doi: 10.1046/j.1523-1755.2001.0590041382.x. [DOI] [PubMed] [Google Scholar]
- 51.Dounousi E., Duni A., Naka K.K., Vartholomatos G., Zoccali C. The innate immune system and cardiovascular disease in ESKD: monocytes and natural killer cells. Curr. Vasc Pharmacol. 2021;19:63–76. doi: 10.2174/1570161118666200628024027. [DOI] [PubMed] [Google Scholar]
- 52.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.