Abstract
Acetic acid plays a crucial role in the organoleptic balance of many fermented products. We have investigated the factors controlling the production of acetate by Saccharomyces cerevisiae during alcoholic fermentation by metabolic engineering of the enzymatic steps involved in its formation and its utilization. The impact of reduced pyruvate decarboxylase (PDC), limited acetaldehyde dehydrogenase (ACDH), or increased acetoacetyl coenzyme A synthetase (ACS) levels in a strain derived from a wine yeast strain was studied during alcoholic fermentation. In the strain with the PDC1 gene deleted exhibiting 25% of the PDC activity of the wild type, no significant differences were observed in the acetate yield or in the amounts of secondary metabolites formed. A strain overexpressing ACS2 and displaying a four- to sevenfold increase in ACS activity did not produce reduced acetate levels. In contrast, strains with one or two disrupted copies of ALD6, encoding the cytosolic Mg2+-activated NADP-dependent ACDH and exhibiting 60 and 30% of wild-type ACDH activity, showed a substantial decrease in acetate yield (the acetate production was 75 and 40% of wild-type production, respectively). This decrease was associated with a rerouting of carbon flux towards the formation of glycerol, succinate, and butanediol. The deletion of ALD4, encoding the mitochondrial K+-activated NAD(P)-linked ACDH, had no effect on the amount of acetate formed. In contrast, a strain lacking both Ald6p and Ald4p exhibited a long delay in growth and acetate production, suggesting that Ald4p can partially replace the Ald6p isoform. Moreover, the ald6 ald4 double mutant was still able to ferment large amounts of sugar and to produce acetate, suggesting the contribution of another member(s) of the ALD family.
Acetate, a by-product of yeast alcoholic fermentation, plays a significant role in the organoleptic balance of many fermented products as the main component of volatile acidity. In beer, the level of acetic acid is usually around 57 to 145 mg/liter and must remain below the taste threshold of 0.4 g/liter (14). In wine, acetic acid is highly undesirable above 0.8 g/liter (8). However, higher levels are sometimes produced, depending on the strain, on must composition, and on the winemaking process, substantially affecting wine quality. Strains with reduced acetate production would therefore be of high value in enology. In addition, the reduction of acetate production is also of great interest for adjusting the by-product formation of yeast strains overproducing glycerol. We have previously constructed wine yeast strains overproducing glycerol that could be of great value in enology to improve the quality of wines lacking body (24, 34). The only critical side effect of glycerol overproduction was the high level of acetate formed (34).
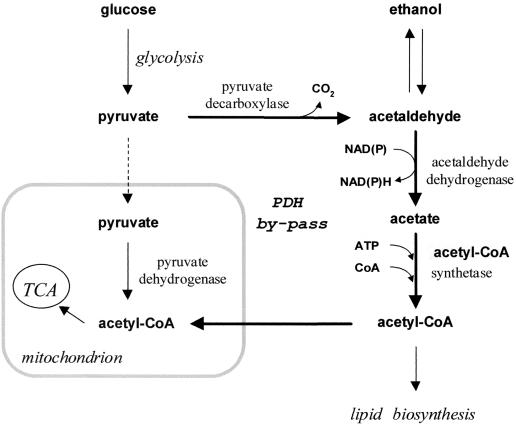
The aim of this study was to better understand how the amount of acetate produced by yeast is controlled during fermentative metabolism. During alcoholic fermentation, acetate is formed by Saccharomyces cerevisiae via the cytosolic pyruvate dehydrogenase (PDH) bypass, which is an alternative route to the PDH reaction for the conversion of pyruvate to acetyl-coenzyme A (CoA). This so-called PDH bypass involves the enzymes pyruvate decarboxylase (PDC; EC 4.1.1.1), acetaldehyde dehydrogenase (ACDH; EC 1.2.1.5 and EC 1.2.1.4), and acetyl-CoA synthetase (ACS; EC 6.2.1.1) (Fig. 1). Acetyl-CoA can be formed in the mitochondria by oxidative decarboxylation of pyruvate catalyzed by the PDH complex. However, due to the inability of S. cerevisiae to transport acetyl-CoA out of the mitochondria, the PDH bypass has an essential role in providing acetyl-CoA in the cytosolic compartment (9, 32, 44). Acetyl-CoA is further processed in the cytosol, in particular for lipid synthesis. In addition to being a substrate for acetyl-CoA synthetase, a physiological role of acetate formation may be the regeneration of reducing equivalents (NADH and NADPH) for maintaining the redox balance.
FIG. 1.
Enzymes involved in the PDH bypass. TCA, tricarboxylic acid cycle.
Pyruvate decarboxylase catalyzes the thiamine PPi- and Mg2+-dependent decarboxylation of pyruvate to acetaldehyde and carbon dioxide. Three structural genes, PDC1, PDC5 (40), and PDC6 (12), potentially encode this enzyme. The role of PDC6 remains unclear. During growth on glucose, 90% of the PDC activity is due to Pdc1p in wild-type cells (11, 12). On the other hand, PDC5 expression is strongly enhanced in a mutant with PDC1 deleted (11, 38, 40). The full expression of PDC1 and PDC5 requires the presence of a functional PDC2 gene, which encodes a positive transcriptional regulator (13).
ACDH, the second enzyme of the PDH bypass, oxidizes acetaldehyde to acetic acid. Two enzymes located in different compartments have been isolated from S. cerevisiae. A mitochondrial enzyme, activated by K+ and thiols, that can use both NAD+ and NADP+ (15) and a cytosolic enzyme that is Mg2+ activated and specific for NADP+ (6, 41) were reported. It has been proposed that the cytosolic enzyme is responsible for the formation of acetate on glucose and that the mitochondrial enzyme plays a major role during growth on ethanol (22, 36). These enzymes belong to the family of S. cerevisiae aldehyde dehydrogenases (ALDH), which contains five members (S. cerevisiae Genome Database [http://genome-www.stanford.edu/Saccharomyces/]). The nomenclature used to designate these genes in the last few years has been very confused and has only very recently been clarified (28). ALD2 (YMR170c), ALD3 (YMR169c), and ALD6 (YPL061w) correspond to the cytosolic isoforms, while ALD4 (YOR374w) and ALD5 (YER073w) encode the mitochondrial enzyme. The two tandem-repeat genes ALD2 and ALD3 have been characterized as the cytosolic stress-inducible aldehyde dehydrogenases using NAD as a cofactor, and they are glucose repressed (27, 28, 30). The main cytosolic Mg2+-activated ACDH isoform is encoded by ALD6 and preferentially uses NADP (23). This isoform plays a role both on ethanol (deletion of this gene led to the inability to use ethanol as a carbon source) and on glucose. Since the deletion mutant was viable on glucose, it was suggested that the enzyme encoded by ALD6 was not solely responsible for the production of cytosolic acetyl-CoA (23). ALD5 (YER073w) was shown to encode a minor form of the mitochondrial NAD(P) ACDH (49), which might play a role in regulation or biosynthesis of electron transport chain components (19). Recently, the gene ALD4 (YOR374w) was shown to encode the mitochondrial K+-activated acetaldehyde dehydrogenase that is active during growth on ethanol (named ALD7) (21, 42). Based on the observation that the ability of a mutant with ALD4 deleted to grow on glucose was not affected while its growth on ethanol was severely impaired, it was concluded that only Ald6p plays a role during growth on glucose (42). In contrast, it was recently reported that a strain in which ALD6 (denoted ALD1) and ALD4 (denoted ALD2) had been disrupted failed to grow on glucose alone unless acetate was added (49). In all these studies, the contribution of each isoform to acetate production under fermentative metabolism has never been examined.
The formation of acetyl-CoA from acetate is catalyzed by ACS and involves hydrolysis of ATP. It has been suggested that when respiring cells are exposed to excess glucose, acetate production might be due to a limited in vivo activity of ACS (46). Two structural genes encoding ACS, ACS1 and ACS2, have been described. ACS1 is subject to glucose repression (17, 44), and its product is therefore only present during respiratory and respirofermentative growth (5). In contrast, ACS2 is constitutively expressed (44). A mutant with ACS2 disrupted can no longer grow on glucose, since the cytosolic compartment cannot be supplied with acetyl-CoA (43).
The aim of this study was to investigate how acetate excretion can be reduced during alcoholic fermentation. We have examined the roles of PDC, ACS, and the major cytosolic and mitochondrial forms of ACDH in controlling acetate production by constructing strains with reduced or increased levels of the corresponding activity. The consequences for acetate and end-product formation, growth, and the fermentation rate were investigated during fermentation under enological conditions.
MATERIALS AND METHODS
Yeast and bacterial strains.
Escherichia coli DH5α was used for cloning experiments. E. coli cultivation and media were as described previously (37). The strain V5 (MATa ura3) derived from the Champagne strain was previously described (24). Under enological conditions, this strain exhibits fermentation performance close to that of industrial strains and superior to that of laboratory strains. Yeast strains were maintained and grown on YPD medium (1% Bacto Yeast Extract, 2% Bacto Peptone, and 2% glucose), or on minimal medium SD (2% glucose, 0.67% yeast nitrogen base without amino acids) with uracil (20 mg/liter).
Fermentation media and conditions.
Batch fermentation experiments were carried out under conditions previously defined to simulate enological fermentation (1). The synthetic medium MS simulating a standard grape juice containing 20% glucose was as described previously (1) but without proline. The total nitrogen amount was 300 mg/liter (80 mg of ammoniacal nitrogen/liter). Cells were precultured on MS medium at 28°C in 50-ml flasks without agitation for 36 h. Fermentation was performed by inoculation of the precultured cells at a density of 106 per ml in fermentors with a working volume of 200 ml or 1.1 liters equipped with fermentation locks and was carried out at 28°C with continuous stirring (500 rpm). CO2 release was determined by automatic measurement of fermentor weight loss every 20 min (35). A linear correlation has been established between ethanol and sugar concentrations and the volume of CO2 released (7).
DNA manipulation, cloning techniques, and transformation methods.
Restriction and modification enzymes were used according to the manufacturer's instructions. E. coli plasmid DNA was prepared using standard protocols (37). Purified oligonucleotides were synthesized by Eurogentec. E. coli transformation was carried out by the CaCl2-RbCl2 method (10). Transformation of S. cerevisiae was performed using the lithium acetate procedure (39).
PDC1 deletion.
To delete PDC1, the PCR-based gene disruption method previously described (48) was used. A PCR fragment was amplified from pFA6-kanMX4. The forward primer PDC1-F1 (5′-TATCTTCTACTCATAACCTCACGCAAAATAACACAGTCAACGTACGCTGCAGGTCGAC-3′) has 18-nucleotide homology with the pFA6-kanMX4 multiple cloning sites (MCS) and a 40-nucleotide extension (underlined sequence) corresponding to the region −50 to −11 upstream of the start codon of the PDC1 open reading frame (ORF). The reverse primer PDC1-R1 (5′-ATGCTTATAAAACTTTAACTAATAATTAGAGATTAAATCGATCGATGAATTCGAGCTCG-3′) has 20-nucleotide homology with the pFA6-kanMX4 MCS and 39 nucleotides (underlined sequence) corresponding to the region immediately downstream of the stop codon of the PDC1 ORF. The replacement of the PDC1 ORF by the amplified module was verified by PCR analysis of total DNA isolated from Kanr transformants, using as the forward primer PDC1a (5′-CTCTCCTTGGAATCAGA-3′), corresponding to the region −126 to −109 upstream of the start codon of the PDC1 ORF, and as the reverse primer PDC1b (5′-GGTAAGTGACAGTGCAG-3′), corresponding to the region +74 to +95 downstream of the stop codon of the PDC1 ORF. The deletion was confirmed by Southern blot analysis using KpnI-digested DNA and a pFA6-kanMX4 or KpnI internal fragment as probes.
ACS2 overexpression.
The ACS2 gene was PCR amplified from total DNA isolated from the V5 strain, using as the forward primer 5′-CCGCGGCCGCGGTTAGTGATTGTTATAC-3′ and as the reverse primer 5′-CCGCGGCCGCTTTCCTAGCTGACCAG-3′, in which NotI sites (in italics) were introduced. The underlined sequences correspond to the region −64 to −43 upstream of the start codon of the ACS2 gene and to the region +133 to +151 downstream of the stop codon of the ACS2 ORF. The PCR fragment was digested by NotI and cloned into the NotI site of the pFL60 plasmid (26) to give the pFL-ACS2 vector.
Deletion of ALD6 and ALD4.
The genes ALD6 and ALD4 were deleted by the PCR-based gene disruption method (48). To delete ALD6 in the strain V5, two primers were used to amplify a PCR fragment from pFA6-kanMX4 corresponding to the Kanr module and to nucleotide extensions to direct the integration in the ALD6 gene. The forward primer ALD6KanF (5′-AAACATCAAGAAACATCTTTAACATACACAAACACATCGTACGCTGCAGGTCGAC-3′) has 18-nucleotide homology with the pFA6-kanMX4 MCS and a 37-nucleotide extension (underlined sequence) corresponding to the region −51 to −15 upstream of the start codon of the ALD6 ORF. The reverse primer, ALD6Kanr (5′-TTTGTGTATATGACGGAAAGAAATGCAGGTTGGTACATTAATCGATGAATTCGAGCTCG-3′), has 19-nucleotide homology with the pFA6-kanMX4 MCS and 40 nucleotides (underlined sequence) corresponding to the region immediately downstream of the stop codon of the ALD6 ORF (+1501 to +1540).
The deletion was verified by PCR analysis of total DNA isolated from Kanr transformants, using as the forward primer ALD6V1 (5′-GGGCGCGCCGCGGA-3′), corresponding to the region −421 to −408 upstream of the start codon of the ALD6 ORF, and as the reverse primer ALD6V2 (5′-GCAGTAAGACCAAGTAAGT-3′), corresponding to the region +1555 to +1573. The disrupted phenotype was confirmed by Southern blot analysis of genomic DNA and of chromosome blots. Total DNA was digested by NsiI and hybridized with a probe corresponding to a ClaI-digested PCR fragment generated using primers ALD6V1 and ALD6V2. A HindIII-ClaI internal ALD6 fragment obtained by digestion of the PCR product was used for hybridization on chromosome blots.
The gene ALD4 was inactivated in the strains V5 and V5 ald6 using a similar strategy but with the URA3 gene as a marker. The forward primer, 5′-ATGTT CAGTAGATCTACGCTCTGCTTAAAGACGTCTGCATCCGGAGATGAT CAGATCTGGC-3′, has 19-nucleotide homology with the URA3 gene of the plasmid pVT100-U (47) and a 42-nucleotide extension (underlined sequence) corresponding to the region +1 to +42 downstream of the start codon of the ALD4 ORF. The reverse primer, 5′-CACCAGGCTTATTGATGACCTTACTCGTCCAATTTGGCACCGTCATTATAAAAATCATTACGAC-3′, has 24-nucleotide homology with the URA3 gene of pVT100-U and a 40-base extension corresponding to the region +1542 to +1580 of ALD4.
The strains V5 and V5 ald6 were transformed by the PCR products. The correctness of the deletion was checked by PCR analysis using the primers ALD4V1 (5′-GCGGGACTTCCGTCCA-3′) and ALD4V2 (5′-CATCAAGGTCTCTGATGC-3′), corresponding to the regions −247 to −233 and +1867 to +1883 of ALD4, respectively. The deletion was confirmed by Southern blot analysis of genomic DNA and of chromosome blots. Total DNA was digested by ScaI and hybridized with a PCR ALD4 fragment generated using the primers ALD4V1 and ALD4V2. An XhoI-NsiI internal ALD4 fragment obtained by digestion of the PCR product was used for hybridization on chromosome blots.
ALD6 overexpression.
The ALD6 gene was PCR amplified from total DNA isolated from the strain V5, using as the forward primer 5′-AAGGATCCATGACTAAGCTACACTTTG-3′ and as the reverse primer 5′-AAGGATCCTTACAACTTAATTCTGACA-3′, with homology with the regions immediately downstream of the start codon and surrounding the stop codon, respectively. BamHI sites (in italics) were introduced. The 15-kb PCR fragment was digested by BamHI and cloned into the BamHI site of the pVT100-U plasmid (47) to give the pVT-ALD6 vector.
Analytical methods.
Yeast cells were counted using an electronic particle counter (ZM; Coultronics). Acetic, pyruvic, and succinic acids, glycerol, and ethanol were analyzed by high-pressure liquid chromatography and by enzymatic assays (Boehringer detection kit), 2,3-butanediol was analyzed by gas chromatography, and acetaldehyde was analyzed by the enzymatic method as previously described (24). Reducing sugar was measured by a colorimetric method using 3.5-dinitrosalicylic acid as previously described (25).
Cell extracts and enzyme assays.
Samples of cultures were harvested at 2,000 × g and washed twice in 10 mM phosphate buffer (pH 7.5). The cells were suspended in 100 mM phosphate buffer containing 2 mM MgCl2 and 1 mM dithiothreitol and broken using 0.5-mm glass beads. Debris was removed by centrifugation at 15,000 × g. The supernatant was used as a cell extract. Enzyme activities were assayed immediately after the preparation of cell extracts. The protein concentration was determined by the Bradford method (3) using bovine serum albumin as the standard.
The PDC and ACS activities were determined as previously described (9, 31). The ACS activity was assayed fluorometrically at excitation and emission wavelengths of 340 and 460 nm, respectively. The solutions of the Boehringer acetate kit were used. The reaction mixture consisted of solution 1 (600 μl), solution 2 (2.4 μl), solution 3 (malate dehydrogenase and citrate synthetase; 4.2 μl), and 120 μl of a solution containing 3.35 mM CoA and 41.3 mM ATP in a final volume of 2 ml. The reaction was started with 10 mM potassium acetate. The ACDH activity was determined as previously described (31) except that MgCl2 (10 mM) was used instead of KCl.
RESULTS
Impact of reduced pyruvate decarboxylase activity.
The strain V5 was transformed with the PCR product obtained by amplification of the KanMX module of the plasmid pFA6-kanMX4 with the primers PDC1-F1 and PDC1-R1. Total DNA isolated from three Kanr transformants was subjected to PCR analysis using two primers, PDC1a and PDC1b. A 1.5-kb fragment was detected in all three transformants (instead of the 1.9-kb fragment in the wild-type strain), which was consistent with the deletion of PDC1 as predicted from the yeast genome sequence. This was further confirmed by Southern blot analysis.
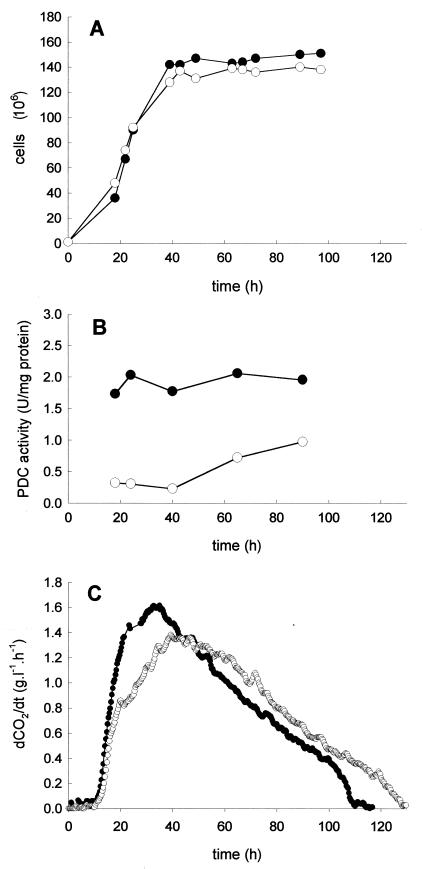
Figure 2 presents the growth on synthetic medium MS simulating the composition of a grape must, the specific PDC activity, and the fermentation rate of V5 pdc1 with respect to the wild-type strain. The growth of the mutant was unaffected (Fig. 2A), while a slight decrease in the CO2 production rate (Fig. 2C) was observed during the growth phase for the deletion mutant, resulting in an increased fermentation time. The pyruvate decarboxylase activity, measured in cell extracts over different time points of the fermentation, was reduced about fourfold in the mutant strain V5 pdc1 compared to that in the parental strain, V5 (Fig. 2B).
FIG. 2.
Consequences of PDC1 deletion for growth (A), PDC activity (B), and fermentation rate (C). Fermentations were performed in 1-liter fermentors filled with MS medium as described in Materials and Methods. ●, V5; ○, V5 pdc1. The experiment was repeated three times with similar results. dCO2/dT, CO2 production rate.
To determine the consequences of a reduced PDC activity for metabolic flux, the concentrations of pyruvate, acetaldehyde, ethanol, acetate, and glycerol produced by the deletion and the wild-type strains were compared. Although the pyruvate production of the pdc1 strain was 2.4 times greater than that of the wild-type strain, no significant difference in the amounts of other metabolites between the two strains was observed (Table 1). Since acetaldehyde production varies greatly during fermentation, increasing during the growth phase and subsequently decreasing during the stationary phase, its concentration was monitored throughout the fermentation, but again no difference was detected between the mutant and the control strains. Finally, no significant difference was found between the amounts of ethanol formed by V5 and V5 pdc1 (data not shown).
TABLE 1.
Concentrations of metabolites for V5 and V5 pdc1 after fermentation on MS medium
| Metabolite | Metabolite concna (g/liter)
|
|
|---|---|---|
| V5 | V5 pdc1 | |
| Pyruvate | 0.17 | 0.41 |
| Acetaldehyde | 33b | 23b |
| Acetate | 0.91 | 0.89 |
| Glycerol | 6.6 | 6.4 |
The concentrations of metabolites were determined after complete glucose exhaustion.
Milligrams per liter.
Impact of overexpression of ACS2.
To investigate whether acetate excretion can be reduced by overproduction of ACS, we introduced the gene ACS2 on the multicopy plasmid pFL61 under the control of the PGK1 promoter. The transformed strains formed small colonies on SD medium (minimal medium containing 2% glucose) and exhibited very slow growth on this medium (data not shown). The growth defect was partially alleviated during growth on MS medium, but the final population was significantly reduced (Fig. 3A). The acetyl-CoA synthetase activity, measured at different times during fermentation (Table 2), was four- to sevenfold greater in the strain overexpressing ACS2 than in the control strain. During growth on MS medium, V5/pFLACS2 exhibited delayed fermentation activity but was able to complete the degradation of sugars (Fig. 3B). Despite this delay, the CO2 production rate remained higher than that of the control strain during the stationary phase, resulting in a reduction of the fermentation time. Overproduction of Acs2p had no significant effect on acetate production during fermentation. The final concentrations of acetate, acetaldehyde, glycerol, and pyruvate were similar in strain V5/pFLACS2 and the wild-type strain (Table 3).
FIG. 3.
Consequences of ACS2 overexpression for growth (A) and fermentation rate (B) on MS medium. ●, V5/pFL61 (control); ○, V5/pFLACS2. Fermentation conditions were as described in the legend to Fig. 2. The experiment was repeated three times with similar results. dCO2/dt, CO2 production rate.
TABLE 2.
Acetoacetyl-CoA synthetase specific activities of strains during fermentation on MS medium
| Strain | ACS activitya (mU/mg of protein) for fermentation time (h) of:
|
||||
|---|---|---|---|---|---|
| 22 | 27 | 31 | 35 | 46 | |
| V5/pFL61 | 3.3 ± 0.8 | 31 ± 3.4 | 11.8 ± 1.9 | 12.4 ± 1.3 | 33.9 ± 1.9 |
| V5/pFLACS2 | 25.3 ± 6.5 | 146.3 ± 2.6 | 81.5 ± 7 | 85.7 ± 17.5 | 133.4 ± 3.9 |
Mean and standard deviation for three determinations.
TABLE 3.
Concentrations of metabolites for V5 and V5/pFLACS2 after fermentation on MS mediuma
| Metabolite | Metabolite concn (g/liter)
|
|
|---|---|---|
| V5/pFL61 | V5/pFLACS2 | |
| Pyruvate | 0.18 | 0.22 |
| Acetaldehyde | 35b | 50b |
| Acetate | 0.85 | 0.85 |
| Glycerol | 7.5 | 7.5 |
Fermentations were performed in 1-liter fermentors at 28°C with agitation. The concentrations of metabolites were determined after complete glucose exhaustion.
Milligrams per liter.
Impact of limited acetaldehyde dehydrogenase.
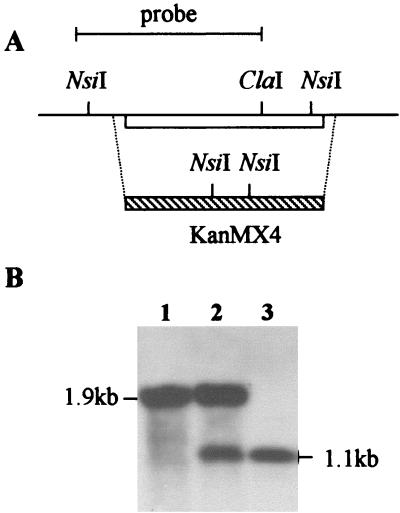
To limit acetaldehyde dehydrogenase, we deleted ALD6, the structural gene coding for a cytosolic, Mg2+-activated acetaldehyde dehydrogenase (23). PCR analysis of the total DNA of three Kanr transformants strongly suggested that two copies of ALD6 were present in the wild-type strain, V5. In two of the three transformants, one ALD6 copy was replaced by the KanMX4 module, while the two ALD6 copies were deleted in the third transformant (data not shown). This was further confirmed by Southern blot analysis of total DNA digested by NsiI. The detection of two NsiI fragments of 1.9 and 1.1 kb in two transformants was consistent with the existence of both an intact and a deleted ALD6 copy in these strains (Fig. 4, lane 2). In contrast, the two ALD6 copies were disrupted in a third transformant, as indicated by the detection of only the 1.1-kb NsiI fragment (Fig. 4, lane 3).
FIG. 4.
Southern blot analysis of mutants with one or two ALD6 genes deleted. (A) Restriction map of the ALD6 region and of the KanMX4 fragment used for disruption and position of the fragment used as a probe. (B) Total DNA from the wild-type strain, V5 (lane 1), V5 ALD6 ald6 (lane 2), and V5 ald6 (lane 3) was digested by NsiI and hybridized with the probe covering the ALD6 coding region plus the 5′ flanking region.
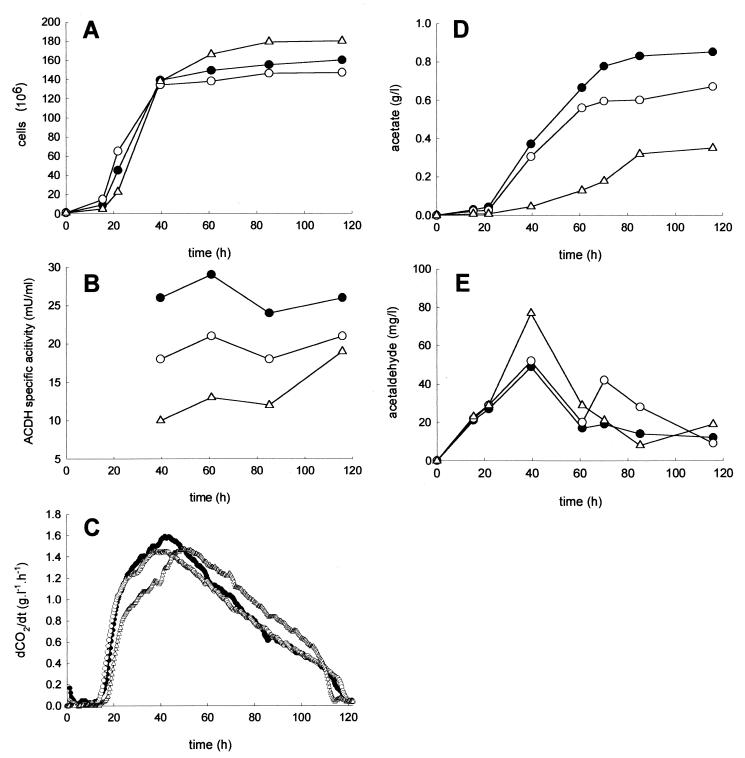
The effects of the disruption of ALD6 were investigated in cell extracts of cultures during fermentation on the synthetic medium MS simulating the composition of a grape must. As shown in Fig. 5B, the single- and double-disrupted mutants displayed reductions in NADP-dependent ACDH activity of about 30 and 60%, respectively, compared to the control strain. The mutants displayed growth similar to that of the wild-type strain (Fig. 5A). The fermentation rate was unaffected for the single-disrupted mutant and slightly modified for the strain with the two ALD6 genes deleted (Fig. 5C). As shown in Fig. 5D, the limitation of the cytosolic ALDH encoded by ALD6 results in a marked decrease in acetate production compared to the wild-type strain. The residual acetate amount produced by the strains in which one or two copies of ALD6 had been deleted represented 75 and 40% of the production of the wild-type strain, respectively. The reduction in the amount of acetate was therefore proportional to the decrease in NADP-dependent acetaldehyde activity, indicating that this enzyme is limiting for acetate production. Moreover, the strain devoid of Ald6p was still able to produce acetate, suggesting the contribution of other ALD genes. A transient increase in acetaldehyde concentration, particularly marked for the double-disrupted strain, was observed during fermentation (Fig. 5E). However, the final concentration was not significantly different from that of the V5 strain.
FIG. 5.
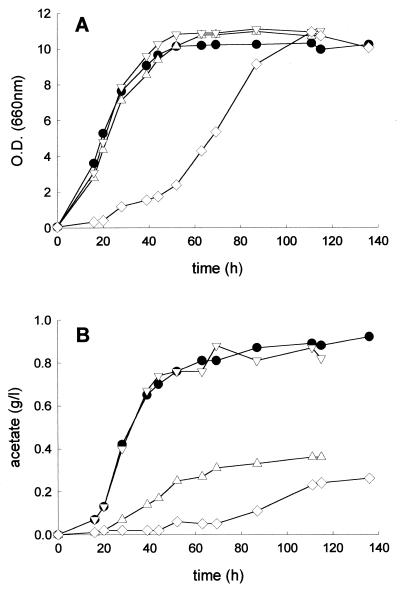
Consequences of ALD6 disruption for growth (A), NADP-dependent ACDH activity (B), fermentation rate (C), acetate (D), and acetaldehyde (E) production. Fermentation conditions were as described in the legend to Fig. 2. The specific NADP-dependent acetaldehyde dehydrogenase activity was determined on crude extracts immediately after they had been prepared, as described in Materials and Methods. ●, V5; ○, V5 with one ALD6 copy deleted (V5 ALD6 ald6); Δ, V5 with the two ALD6 copies deleted (V5 ald6). dCO2/dt, CO2 production rate.
To confirm that the level of Ald6p controls acetate production, ALD6 was cloned on the multicopy pVT100-U vector under the control of the ADHI promoter and terminator. This vector was introduced into the V5 strain to give the strain V5-ALD6, which was further tested for acetate production during alcoholic fermentation. As expected, the amount of acetate produced after fermentation on MS medium was higher for the recombinant strain (1.5 g/liter) than for the control strain (0.85 g/liter).
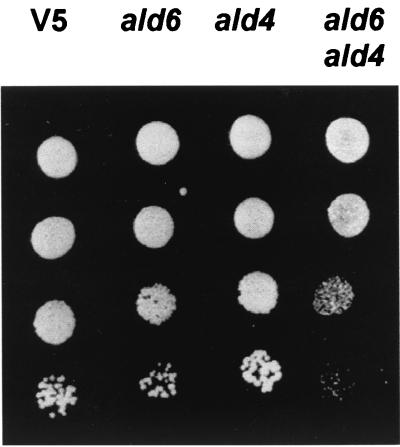
To identify other genes responsible for the acetate formed by a strain in which the two ALD6 gene copies have been deleted, we disrupted the gene ALD4, encoding the mitochondrial K+-activated ACDH, in the wild-type and ald6 strains. The verification of the disruption by PCR and Southern blotting revealed, as observed for ALD6, the presence of two ALD4 copies. Ura+ transformants with one ALD4 copy or both copies deleted were obtained for the two strains V5 and V5 ald6, as demonstrated by PCR and Southern blot analysis (data not shown). While a V5 strain lacking the ALD4 gene(s) exhibited normal growth and produced the same amount of acetate as the wild type, the growth and acetate production of the strain lacking both Ald6p and Ald4p were markedly affected compared to those of the wild-type and the ald6 strains (Fig. 6). The ald6 ald4 mutant was still able to form acetate. Low levels were detected during half of the fermentation, and then the production increased to reach a concentration slightly below that of the ald6 strain (Fig. 6B). The ald6 ald4 mutant was also able to complete the fermentation on a rich sugar medium, although sugar exhaustion was achieved 40 h later than for the other strains (data not shown). It was recently reported (49) that a mutant devoid of Ald6p and Ald4p was unable to grow on glucose. The possibility that the increased acetate production around midfermentation was due to a recombination between an inactivated ALD allele(s) and other putative ALD genes was ruled out by PCR analysis of cells taken at the end of the fermentation (data not shown). Another possibility might be the selection in the growth medium of cells bearing additional genomic mutations. To examine this hypothesis, cells collected at the end of fermentation were plated on yeast-peptone-dextrose (YPD) medium and submitted to a second fermentation experiment. The growth and acetate production patterns were identical to those of the initial ald6 ald4 mutant (data not shown), excluding the possibility of additional mutation. To investigate if the specific growth conditions used in this study (a sugar-rich medium) could explain the differences in the phenotype of the mutant, the ald6 ald4 mutant was grown on SD medium plus uracil (Fig. 7). The mutant was able to grow, although to a lesser extent than the wild type or the ald6 and ald4 mutants. Similarly, the double mutant was able to form colonies on the same medium smaller than those of the other three strains.
FIG. 6.
Comparison of growth (A) and acetate production (B) of a mutant lacking Ald6p, Ald4p, or both Ald6p and Ald4p. ●, V5; Δ, V5 ald6; ▿, V5 ald4; ◊, V5 ald6 ald4. Fermentation conditions were as described in the legend to Fig. 2. Complete sugar exhaustion was achieved after 110 h of fermentation for all strains except the ald6 ald4 strain, which finished fermentation at 140 h. The experiment is representative of three independent experiments. OD, optical density.
FIG. 7.
Growth of ald6, ald4, and ald6 ald4 mutants on SD medium plus uracil after 4 days at 28°C. The cells were pregrown in YPD medium. Equal amounts of cells from a dilution series in 1:10 steps were spotted onto SD medium plus uracil.
As shown in Table 4, reduction of acetate formation by the mutants V5 ald6 and V5 ald6 ald4 led to marked shifts in the yields of fermentation end products. The concentration of acetaldehyde was increased, and that of pyruvate was proportionally decreased. A marked increase in the concentrations of glycerol and 2,3-butanediol was also observed in the two mutants, while that of succinate was only slightly enhanced in the ald6 ald4 mutant. Although a decrease in acetate formation was expected to trigger large changes in metabolite formation to counterbalance the deficit in reduced cofactors, the increase in glycerol production is unexpected in the context of NAD(P)H shortage.
TABLE 4.
Concentrations of metabolites for V5, V5 ald6, and V5 ald6 ald4 after fermentation on MS mediuma
| Metabolite | Metabolite concn (g/liter)
|
||
|---|---|---|---|
| V5 | V5 ald6 | V5 ald6 ald4 | |
| Pyruvate | 0.12 | 0.07 | 0.07 |
| Acetaldehyde | 31b | 41b | 77b |
| Acetate | 0.87 | 0.36 | 0.28 |
| Glycerol | 6.6 | 7.8 | 9.5 |
| Succinate | 0.58 | 0.60 | 0.70 |
| 2,3-Butanediol | 0.76 | 1.1 | 1.8 |
Fermentations were performed in 1-liter fermentors at 28°C with agitation. The concentrations of metabolites were determined after complete glucose exhaustion.
Milligrams per liter.
DISCUSSION
Although acetic acid plays an important role as (off) flavor in many fermented products, little is known about the factors involved in controlling its production by S. cerevisiae. Since pyruvate decarboxylase catalyzes the first step of the ethanol-specific pathway, strong reduction of PDC activity was expected to restrict alcoholic fermentation and acetate production. Due to its important role as a pivotal enzyme in NADH reoxidation, this might have resulted in the adjustment of the redox balance to supply oxidized cofactors (i.e., by increasing glycerol or decreasing acetate production). In this work we report that a pdc1 mutant exhibiting a markedly decreased pyruvate decarboxylase activity during batch fermentation on MS medium produced the same amount of acetate as the wild-type strain, while the flux to ethanol was only very slightly limited. Despite a twofold increase in pyruvate production, the fermentation rate was slightly decreased and the ethanol production was unaffected. As a consequence, we were unable to detect changes in the production of glycerol and succinate, indicating that the redox balance is not significantly modified in the mutant. Furthermore, the glycerol yields of strains overexpressing GPD1, encoding glycerol-3-phosphate dehydrogenase, were found to be similar in both V5 pdc1 and wild-type backgrounds (F. Remize and S. Dequin, unpublished data). These data suggest that PDC is not a major factor in controlling metabolic flux during fermentation in the sugar-rich and low-nitrogen MS medium. This is consistent with the observation that a fourfold overexpression of PDC did not enhance alcoholic fermentation or growth rate (38) in growing shake flask cultures. In contrast, it has been previously reported that a YSH 306 strain with PDC2, which encodes a positive transcription regulator of PDC1 and PDC5 and exhibits 19% of PDC residual activity, deleted displays a decrease of 30% in ethanol yield, resulting in increased glycerol formation in batch culture on YPD medium (29). The limitation of the ethanol branch in this mutant is probably due to PDC activities of both the wild-type YSH306 and the corresponding pdc2 mutant (29) lower than those of V5 and V5 pdc1 (this study). In any case, our results argue against a major role for PDC in controlling ethanol and acetate flux.
Amplification of ACS2 did not result in enhanced acetate utilization, despite a four- to sevenfold increase in ACS activity. A high expression level of this enzyme might trigger perturbations of metabolic flux, since the growth of the ACS2-overexpressed strain was affected, depending on the growth conditions. The amplification of this enzymatic step could lead to increased ATP consumption and to a modification of the intracellular pools of acetyl-CoA and CoA, which are potent effectors of key enzymes in carbon metabolism. Such modifications might explain both the reduction in biomass formation and the stimulation of the fermentation rate during the stationary phase. Interestingly, a marked increase in acetate production was observed in a strain overexpressing ACS1 and exhibiting a 6- to 12-fold increase in ACS activity in glucose-limited chemostat cultures (4), which might reflect such modifications.
In contrast to PDC and ACS, the cytosolic acetaldehyde dehydrogenase was shown to be a key enzyme for the control of acetate formation. A requirement for Ald6p for optimal growth on glucose, suggesting a role for Ald6p in acetate formation, was previously observed (23), while others reported a wild-type phenotype for ald6 (49). In this study, the reduction and increase of acetate formation by strains with ALD6 deleted or overexpressing ALD6, respectively (and exhibiting growth similar to that of the wild-type strain) clearly demonstrate that Ald6p has a major role in acetate formation during sugar fermentation and that the level of this enzyme controls the amount of acetate formed. This is consistent with the observation of decreased acetate production by a mutant exhibiting reduced NADP-dependent ACDH activity (18).
An interesting finding is the observation that the mitochondrial isoform Aldp4p might also be involved in the production of acetic acid under certain circumstances. While ALD4 was shown to be involved in growth on ethanol, its role on glucose was previously ruled out on the basis of the observation that an ald4 mutant exhibited normal growth on this substrate (42). In contrast, it was reported that a double mutant (ald6 ald4) was no longer able to grow on glucose, suggesting that both genes are involved in acetate formation during fermentation (49). In this paper, we show that the deletion of ALD4 alleles did not affect growth on glucose or acetate production, strongly supporting the view that Ald4p does not play a role during fermentative metabolism. In contrast, the altered growth and acetate production of the ald6 ald4 mutant suggest that Ald4p could partially replace the main isoform, ALD6. In the absence of Ald6p, acetaldehyde produced by decarboxylation of pyruvate would be transported from the cytosol to the mitochondria to generate acetate and then acetyl-CoA in the cytosol. The existence of such a pathway, called the mitochondrial pyruvate dehydrogenase bypass, operative during respiratory metabolism, was recently proposed (2). The fact that the synthesis of mitochondrial NAD(P)-dependent acetaldehyde dehydrogenase is repressed in the presence of glucose (15) and the observation of a wild-type phenotype for ald4 (this study) suggest that Ald4p could be deregulated in the ald6 mutant. Furthermore, the observation that the double mutant can grow, albeit slowly, and produce acetate strongly suggests the contribution of one or more of the other members of the ACDH family. Whether this additional gene(s) is functional in a wild-type strain or induced to compensate for the loss of Ald6p and Ald4p will need to be elucidated. These results are in disagreement with the quasiabsence of growth of an ald6 ald4 mutant (referred as ald1 ald2) previously reported (49). Although the delayed growth might have escaped attention under the test conditions (the sizes of colonies formed on SD plates after 6 days of growth), the discrepancies observed might be due to differences in growth conditions or in the genetic backgrounds of the strains. However, the observation that the double mutant V5 ald6 ald4 can grow on SD medium as well as on YPD medium in the absence of acetate disproves the hypothesis of a growth medium effect. On the other hand, we cannot exclude the possibility that the strain V5 contains additional or differently regulated ALD alleles. It was shown in this study (and by other unpublished data) that this strain, a meiotic segregant of an industrial wine yeast strain, is at least partially diploid. Moreover, wine yeast strains are known to display structural chromosomal divergences from laboratory strains, which may influence gene expression (33). A careful study of ald6 ald4 mutants in different genetic backgrounds will be necessary to specify the role of ALD4 and of the other ALD genes coding for minor isoforms of the ACDH family.
The reduction of acetate production results in transient increased formation of acetaldehyde. Since this compound is toxic to the cells (16), the increased formation of 2,3-butanediol might reflect a detoxication mechanism. On the other hand, the marked increase in glycerol production observed is unexpected, since this leads to a more pronounced deficit in reduced cofactors. Since the Km of Ald6p for NAD is 170 times higher than that of NADP (49), the deletion of ALD6 must result in a decrease in NADPH formation. However, the NADP/NAD ratio might also be affected in these mutants, depending on the contribution of other ALDHs that can use NAD (Ald2p and Ald3p [28]) or NAD and NADP (Ald4p). The increased glycerol production, therefore, could reflect deregulation mechanisms. Further characterization of these mutants is needed to understand how the cell will cope with a reduction of acetate production. In S. cerevisiae, the couples NAD-NADH and NADP-NADPH constitute distinct biochemical compartments due to the absence of transdehydrogenase activity (20, 45). However, the existence of systems which could serve as transdehydrogenase, producing NADPH from NADH, has recently been postulated (e.g., the coupling of glycerol production and degradation [30]). The existence of at least five ACDH isoforms with different cofactor specificities, one being able to suppress the loss of the other, as shown for Ald6p and Ald4p in this study, supports the idea of a role for these isoenzymes in managing the intracellular cytosolic and mitochondrial NADPH-NADH pools.
Genetic engineering strategies to minimize acetate formation are of considerable interest for industrial purposes. In wine and beer, the production of acetate in large amounts is undesirable. In glycerol-overproducing yeast, acetate formation is greatly increased. Furthermore, the reduction of acetate formation could also be of great interest for the biomass-directed applications of S. cerevisiae, since acetate production may have a detrimental effect on these applications. The results presented demonstrate that the level of Ald6p controls the amount of acetate formed by S. cerevisiae on glucose. Deletion of ALD6 has been shown to efficiently reduce acetate formation during wine fermentation. While the amounts of acetaldehyde, glycerol, succinate, and 2,3-butanediol produced by the engineered strain under enological conditions were slightly increased, they remained within the concentration ranges commonly found in wines. Inactivation of Ald6p is therefore a promising option for engineering industrial yeasts involved in these fermentation fields. However, the increased production of compounds that play, in particular, a role in maintaining the redox balance will have to be specifically addressed in relation to the characteristics of the product.
ACKNOWLEDGMENTS
This work was supported by the European Community in the framework of the Biotechnology-Cell Factory project BIO-CT95-0161.
We thank E. Baptista and C. Camarasa for assistance in fermentation experiments and high-pressure liquid chromatography analyses.
REFERENCES
- 1.Bely M, Sablayrolles J M, Barre P. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in enological conditions. J Ferm Bioeng. 1990;70:246–252. [Google Scholar]
- 2.Boubekeur S, Bunnoust O, Camougrand N, Castroviejo M, Rigoulet M, Guerin B. A mitochondrial pyruvate dehydrogenase bypass in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:21044–21048. doi: 10.1074/jbc.274.30.21044. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.De Jong-Gubbels P, van der Berg M A, Luttik M A H, Steensma H Y, van Dijken J P, Pronk J T. Overproduction of acetyl-coenzyme A synthetase isoenzymes in respiring Saccharomyces cerevisiae cells does not reduce acetate production after exposure to glucose excess. FEMS Microbiol Lett. 1998;165:15–20. doi: 10.1111/j.1574-6968.1998.tb13121.x. [DOI] [PubMed] [Google Scholar]
- 5.De Virgilio C, Bürckert N, Barth G, Neuhaus J M, Bollerand T, Wiemken A. Cloning and disruption of a gene required for growth on acetate but not on ethanol: the acetyl-coenzyme A synthetase gene of Saccharomyces cerevisiae. Yeast. 1992;8:1043–1051. doi: 10.1002/yea.320081207. [DOI] [PubMed] [Google Scholar]
- 6.Dickinson F M. The purification and some properties of the Mg2+-activated cytosolic aldehyde dehydrogenase of Saccharomyces cerevisiae. Biochem J. 1996;315:393–399. doi: 10.1042/bj3150393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Haloui N, Picque D, Corrieu G. Alcoholic fermentation in winemaking: on line measurement of density and carbon dioxide evolution. J Food Eng. 1988;8:17–30. [Google Scholar]
- 8.Fleet H, Heard G M. Yeast—growth during fermentation. In: Fleet G, editor. Wine microbiology and biotechnology. Chur, Switzerland: Harwood Academic Publishers; 1992. pp. 27–54. [Google Scholar]
- 9.Flikweert M T, van der Zanden L, Janssen W M T M, Steesma H Y, van Dijken J P, Pronk J T. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast. 1996;12:247–257. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C247::AID-YEA911%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 10.Hanhahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. I. Oxford, England: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 11.Hohmann S, Cederberg H. Autoregulation may control the expression of yeast pyruvate decarboxylase in Saccharomyces cerevisiae. Eur J Biochem. 1990;188:615–621. doi: 10.1111/j.1432-1033.1990.tb15442.x. [DOI] [PubMed] [Google Scholar]
- 12.Hohmann S. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol. 1991;173:7963–7969. doi: 10.1128/jb.173.24.7963-7969.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohmann S. Characterization of PDC2, a gene necessary for high level expression of pyruvate decarboxylase structural genes in Saccharomyces cerevisiae. Mol Gen Genet. 1993;241:657–666. doi: 10.1007/BF00279908. [DOI] [PubMed] [Google Scholar]
- 14.Hough J S, Briggs D E, Stevensand R, Young T W. Beer flavour and beer quality. In: Hough J S, editor. Malting and brewing science. II. London, United Kingdom: Chapman and Hall; 1982. pp. 839–876. [Google Scholar]
- 15.Jacobson M K, Bernofsky C. Mitochondrial aldehyde dehydrogenase from Saccharomyces cerevisiae. Biochem Biophys Acta. 1974;350:277–291. doi: 10.1016/0005-2744(74)90502-6. [DOI] [PubMed] [Google Scholar]
- 16.Jones R P. Biological principles for the effects of ethanol. Enzyme Microb Technol. 1989;11:130–153. [Google Scholar]
- 17.Kratzer S, Shuller H J. Carbon source-dependent regulation of the acetyl coenzyme A synthetase-encoding gene ACS1 from Saccharomyces cerevisiae. Gene. 1995;161:75–79. doi: 10.1016/0378-1119(95)00289-i. [DOI] [PubMed] [Google Scholar]
- 18.Kurita O, Ito H. Isolation and characterization of mutants partially deficient in aldehyde dehydrogenase in Saccharomyces cerevisiae. Biosci Biotech Biochem. 1994;58:609–615. [Google Scholar]
- 19.Kurita O, Nishida Y. Involvement of mitochondrial aldehyde dehydrogenase ALD5 in maintenance of the mitochondrial electron transport chain in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1999;187:281–287. doi: 10.1111/j.1574-6968.1999.tb08856.x. [DOI] [PubMed] [Google Scholar]
- 20.Lagunas R, Gancedo J M. Reduced pyridine nucleotide balance in glucose-growing Saccharomyces cerevisiae. Eur J Biochem. 1973;37:90–94. doi: 10.1111/j.1432-1033.1973.tb02961.x. [DOI] [PubMed] [Google Scholar]
- 21.Larsson N, Norbeck J, Karlsson H, Karlsson K A, Blomberg A. Identification of two-dimensional gel electrophoresis resolved yeast proteins by matrix-assisted laser desorption ionisation mass spectrometry. Electrophoresis. 1997;18:418–423. doi: 10.1002/elps.1150180316. [DOI] [PubMed] [Google Scholar]
- 22.Llorente N, de Castro I N. Physiological role of yeasts NAD(P)+ and NADP+-linked aldehyde dehydrogenases. Rev Esp Fisiol. 1977;33:135–142. [PubMed] [Google Scholar]
- 23.Meaden P G, Dickinson F M, Mifsud A, Teissier W, Westwater J, Bussey H, Midgley M. The ALD6 gene of Saccharomyces cerevisiae encodes a cytosolic, Mg2+-activated acetaldehyde dehydrogenase. Yeast. 1997;13:1319–1327. doi: 10.1002/(SICI)1097-0061(199711)13:14<1319::AID-YEA183>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 24.Michnick S, Roustan J L, Remize F, Barre P, Dequin S. Modulation of glycerol and ethanol yields during alcoholic fermentation in Saccharomyces cerevisiae strains overexpressed or disrupted for GPD1 encoding glycerol-3-phosphate dehydrogenase. Yeast. 1997;13:783–793. doi: 10.1002/(SICI)1097-0061(199707)13:9<783::AID-YEA128>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 25.Miller G L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- 26.Minet M, Dufour M E, Lacroute F. Complementation of Saccharomyces cerevisiae aoxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- 27.Miralles V J, Serrano R. A genomic locus in Saccharomyces cerevisiae with four genes up-regulated by osmotic stress. Mol Microbiol. 1995;17:653–662. doi: 10.1111/j.1365-2958.1995.mmi_17040653.x. [DOI] [PubMed] [Google Scholar]
- 28.Navarro-Avino J, Prasad R, Miralles V J, Benito R M, Serrano R. A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast. 1999;15:829–842. doi: 10.1002/(SICI)1097-0061(199907)15:10A<829::AID-YEA423>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Nevoigt E, Stahl U. Reduced pyruvate decarboxylase and increased glycerol-3-phosphate dehydrogenase [NAD+] levels enhance glycerol production in Saccharomyces cerevisiae. Yeast. 1996;12:1331–1337. doi: 10.1002/(SICI)1097-0061(199610)12:13%3C1331::AID-YEA28%3E3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 30.Norbeck J, Blomberg A. Metabolic and regulatory changes associated with growth of Saccharomyces cerevisiae in 1.4 M NaCl. J Biol Chem. 1997;272:5544–5554. doi: 10.1074/jbc.272.9.5544. [DOI] [PubMed] [Google Scholar]
- 31.Postma E, Verduyn C, Scheffers A, van Dijken J P. Enzymatic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol. 1989;55:468–477. doi: 10.1128/aem.55.2.468-477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pronk J T, Steesma H Y, van Dijken J P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Rachidi N, Barre P, Blondin B. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine yeast strain of Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:841–850. doi: 10.1007/s004380050028. [DOI] [PubMed] [Google Scholar]
- 34.Remize F, Roustan J L, Sablayrolles J M, Barre P, Dequin S. Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in by-product formation and to a stimulation of fermentation rate in stationary phase. Appl Environ Microbiol. 1999;65:143–149. doi: 10.1128/aem.65.1.143-149.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sablayrolles J M, Barre P, Grenier P. Design of laboratory automatic system for studying alcoholic fermentations in anisothermal enological conditions. Biotechnol Tech. 1987;1:181–184. [Google Scholar]
- 36.Saigal D, Cunningham S J, Farrès J, Weiner H. Molecular cloning of the mitochondrial dehydrogenase gene of Saccharomyces cerevisiae by genetic complementation. J Bacteriol. 1991;173:3199–3208. doi: 10.1128/jb.173.10.3199-3208.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schaaf I, Green J B A, Gozalbo D, Hohmann S. A deletion of the PDC1 gene for pyruvate decarboxylase of yeast causes a different phenotype than previously isolated point mutations. Curr Genet. 1989;15:75–81. doi: 10.1007/BF00435452. [DOI] [PubMed] [Google Scholar]
- 39.Schiestl R H, Gietz R D. High efficiency transformation of intact cells using single stranded nucleic acid as carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 40.Seeboth P G, Bohnsack K, Hollenberg C P. pdc10 mutants of Saccharomyces cerevisiae give evidence for an additional structural PDC gene: cloning of PDC5, a gene homologous to PDC1. J Bacteriol. 1990;172:678–685. doi: 10.1128/jb.172.2.678-685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seegmiller J E. TNP-linked aldehyde dehydrogenase from yeast. Methods Enzymol. 1955;1:511–514. [Google Scholar]
- 42.Teissier W D, Meaden P G, Dickinson F M, Midgley M. Identification and disruption of the gene encoding the K+-activated acetaldehyde dehydrogenase of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1998;164:29–34. doi: 10.1111/j.1574-6968.1998.tb13063.x. [DOI] [PubMed] [Google Scholar]
- 43.Van den Berg M A, Steesma H Y. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur J Biochem. 1995;231:704–713. doi: 10.1111/j.1432-1033.1995.tb20751.x. [DOI] [PubMed] [Google Scholar]
- 44.Van den Berg M A, de Jong-Gubbels P, Steesma H Y, van Dijken J P, Pronk J T. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- 45.van Dijken J P, Scheffers W A. Redox balances in the metabolism of sugar by yeasts. FEMS Microbiol Rev. 1986;32:199–224. [Google Scholar]
- 46.Van Urk H, Voll W S L, Scheffers W A, van Dijken J P. Transient-state analysis of metabolic fluxes in Crabtree-positive and Crabtree-negative yeasts. Appl Environ Microbiol. 1990;56:281–287. doi: 10.1128/aem.56.1.281-287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vernet T, Dignard D, Thomas D. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 48.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Mann C J, Bai Y, Ni L, Weiner H. Molecular cloning, characterization, and potential roles of cytosolic and mitochondrial aldehyde dehydrogenases in ethanol metabolism in Saccharomyces cerevisiae. J Bacteriol. 1998;180:822–830. doi: 10.1128/jb.180.4.822-830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]