Abstract
Little is known of hematopoietic stem (HSC) and progenitor (HPC) cell self-renewal. The role of Brahma (BRM), a chromatin remodeler, in HSC function is unknown. Bone marrow (BM) from Brm−/− mice manifested increased numbers of long- and short-term HSCs, GMPs, and increased numbers and cycling of functional HPCs. However, increased Brm−/− BM HSC numbers had decreased 2° and 3° engraftment, suggesting BRM enhances HSC self-renewal. Valine was elevated in lineage negative Brm−/− BM cells, linking intracellular valine with Brm expression. Valine enhanced HPC colony formation, replating of human cord blood (CB) HPC-derived colonies, mouse BM and human CB HPC survival in vitro, and ex-vivo expansion of normal mouse BM HSCs and HPCs. Valine increased oxygen consumption rates of WT cells. BRM through CD98 was linked to regulated import of branched chain amino acids, such as valine, in HPCs. Brm−/− LSK cells exhibited upregulated interferon response/cell cycle gene programs. Effects of BRM depletion are less apparent on isolated HSCs compared to HSCs in the presence of HPCs, suggesting cell extrinsic effects on HSCs. Thus, intracellular valine is regulated by BRM expression in HPCs, and the BRM/valine axis regulates HSC and HPC self-renewal, proliferation, and possibly differentiation fate decisions.
INTRODUCTION
Hematopoietic stem cells (HSCs) self-renew and differentiate to hematopoietic progenitor cells (HPCs) and more mature cells. Self-renewal controls HSC pool size1–3. HPCs have limited self-renewal activity in vitro4–6. Control of HSC/HPC function and self-renewal are being identified4–8, but underlying mechanisms still remain poorly defined.
Chromatin remodeling complexes unwind compact chromatin structures allowing gene expression. One remodeler, the switch/sucrose nonfermentable (SWI/SNF) complex is conserved from yeast to mammals9,10. Mammalian Brahma (BRM)-related gene-1 (BRG1)/BRM associated factor (BAF) is a functional equivalent of the yeast SWI/SNF complex11,12. Although other 15 subunits of BAF complexes are identical, ATPase subunits BRG1 and BRM form mutually exclusive protein complexes13,14. BRG1 or BRM is essential for BAF-mediated nucleosome remodeling; subunit mutations inactivate BAF activity15,16. Brg1 knockout mice are embryonic lethal. BRG1 is essential for embryonic stem cell self-renewal and proliferation17–21. However, Brm−/− mice are viable22. Silencing Brg1, but not Brm, activates p53-mediated senescence in cancer cells23. Biochemical studies suggest distinct roles for BRG1 and BRM complexes.24 Their roles in HSC/HPC regulation in vivo remain unknown.
Mitochondria play roles in bioenergetic and metabolic demands of HSCs, controlling HSC and HPC homeostasis25–27. Brief exposure of HSCs to ambient air oxygen stimulates mitochondrial metabolism and decreased long-term (LT)-HSCs, concomitant with differentiation to HPCs26. Impaired energy metabolism/ mitochondrial activity leads to HSC exhaustion28–31. Mitochondria are not only primary sites of fatty acid oxidation, but of branched chain amino acid (BCAA) metabolism30. BCAAs, essential amino acids in metazoans, are from dietary sources. Dietary valine restriction in mice results in HSC ablation in bone marrow (BM)32. However, valine’s role in HSC/HPC homeostasis/self-renewal lacked genetic and additional evidence. Administration of BCAA also induces interferon signaling in non-hematopoietic cell types33 though its effects on interferon response in HSCs/HPCs are not known. We show that lineage negative (lin−) cells from Brm−/−mice have elevated valine levels, compared to WT mouse cells. Brm−/−mice have increased HSC numbers, yet no competitive advantage in primary transplants, and greatly reduced engraftment in secondary (2°) and tertiary (3°) transplantation.26,34. Exogenous valine increased numbers, survival, and replating capacity in vitro of functional HPCs and ex-vivo expansion of phenotyped HSCs/HPCs. Brm and valine were linked to CD98 cell surface expression. HSCs were affected by cell extrinsic effects in absence of BRM, HPCs were affected by cell intrinsic effects in the absence of BRM, and both were affected by cell intrinsic effects of valine. Thus, intracellular valine levels, apparently controlled by Brm, regulate HSC/HPC functions
METHODS AND MATERIALS
Mice.
Male and female C57BL/6J, Boy/J, and B6 x Boy/J F1 mice (8–10 weeks old) were obtained from on-site breeding at Indiana University School of Medicine (IUSM). Brm −/− and Brm +/− mice (on a C57Bl/6 mouse strain background)22 were provided by Dr. Ching-Pin Chang, IUSM. Animals were maintained under temperature- and light-controlled conditions (21–24 °C, 12 hour light/12 hour dark cycle) and were housed according to age, sex and genotype. Mice were fed ad libitum, and matched by age and sex. Studies were IUSM IACUC approved.
Isolation of Mouse Lin− BM Cells and Human CD34+ CB Cells.
Mouse BM was isolated from femurs of mice prior to use by flushing in PBS (Lonza; Walkersville, MD, USA). Lineage depletion used a mouse lineage cell depletion kit (Miltenyi Biotec; Aurburn, CA, USA) using two sequential columns. Human CB, obtained from the Cleveland Cord Blood Bank and Cord:Use, was within 36 hours post-delivery. CB was washed in PBS prior to Ficoll-Paque™ PLUS (GE Healthcare Bio-Sciences AB; Pittsburgh, PA, USA) separation of mononuclear cells (LD cells). CD34+ CB cells were isolated using CD34 immunomagnetic selection kit (Miltenyi Biotec) using two sequential columns (purity ≥97% CD34+ cells). Mouse Lin− cell purity was ≥92%. IUSM IRB approved CB studies.
Flow Cytometry.
Mouse BM cells, collected at ~2.5×106 cells per tube, were washed in PBS, incubated in fluorescently-conjugated antibodies (S. Table 1) for 20 minutes at room temperature, washed in PBS, and fixed in 1% formaldehyde. Samples were analyzed by LSRII flow cytometer (BD Biosciences) for immunophenotypically defined cell populations according to S. Table 2. (Lin− Sca-1+ c-Kit+ (LSK); long-term (LT)-HSCs; short-term (ST)-HSCs; multipotent progenitors (MPPs); common myeloid progenitors (CMPs); granulocyte macrophage progenitors (GMPs); myeloid erythroid progenitors (MEPs); and common lymphoid progenitors (CLPs)).
HPC Assays.
CFU-GM, BFU-E and CFU-GEMM colonies were scored from whole mouse BM plated at 5×104 cells/mL, and CFU-GM and CFU-GEMM were scored from 5×104 low-density human (LD) CB or 500–1000 CD34+ cells in 1% methylcellulose/Iscoves Modified Dulbeccos Medium (IMDM) with 30% FBS (Hyclone, non-dialyzed or dialyzed) and assay dependent growth factors (see figure legends and supplemental methods). We were interested in HPC derived mouse BM colonies and did not score CFU-E. CFU-E detect precursor, not progenitor, cells. Growth factors for human HPC allows detection of CFU-GEMM, not BFU-E37.Cultures were plated with control medium, 10 mM L-Valine (Sigma), or 10 mM L-Leucine (Sigma), optimal levels of these amino acids. For HPC survival assays, growth factor addition was delayed, and colony counts compared to growth factors added at Day 06–8,36. Individual human CFU-GEMM were selected for secondary replating. Colonies were not pooled for replating.
% Functional HPC in S-Phase of the Cell Cycle.
High specific activity tritiated thymidine ([3H]Tdr) kill assays were utilized6,26,35. BM cells were pulse-treated with 50 μCi of high specific activity [3H]Tdr (20 Ci/mmol; DuPont NEN) at 37°C for 20 minutes, and washed twice prior to plating in HPC colony assays. Colonies in [3H]Tdr treated plates were compared to vehicle control treated plates, and percent CFU in S-phase calculated. This is the only means of assessing the percent functional HPC in S-phase of the cell cycle.
Engrafting Studies.
Competitive transplantations were performed. Donor BM cells from wildtype or Brm−/− mice (CD45.1− CD45.2+)26,35 were mixed (1:1 ratio at 1×105 cells) with competitor BM from Boy/J mice (CD45.1+ CD45.2−) and injected intravenously into lethally irradiated (950 cGy) B6 x Boy/J F1 mice (F1 mice; CD45.1+ CD45.2+). Percentage donor CD45.1− CD45.2+ cells in PB were determined by flow cytometry. At 4 months BM from B6 x Boy/J F1 mice was analyzed for percent donor cell engraftment, myeloid/lymphoid ratio, and numbers of donor-derived LT-HSCs. Secondary and tertiary transplants (2×106 BM cells from recipient mice) were transplanted into lethally irradiated B6xBoy/J F1 mice and analyzed similarly to that of primary transplants.
Ex Vivo HSC Expansion Assays.
WT C57BL/6 or Brm−/− Lin− BM cells were isolated and cultured at 50,000 cells/well in mouse HSC expansion media containing RPMI-1640 media (Lonza) with 10% FBS (Fisher Scientific), 100 ng/mL rmSCF (R&D Systems; cat. # 455-MC-010), 100 ng/mL rmTPO (R&D Systems; cat. # 488-TO-200/CF), and 100 ng/mL rmFLT3L (BioLegend; cat. # 550706) with 10 mM L-Valine (Sigma), 10 mM L-Leucine (Sigma) or vehicle control at 37°C at 5% CO2 and 5% O2. After 4 days in culture, flow cytometry determined LT-HSC frequency and HPC colony assays were performed. Absolute numbers of output populations were compared to number of input populations to determine expansion. For separation of cell extrinsic/intrinsic effects, LT-HSCs were separated from ST-HSCs and MPPs WT C57BL/6 or Brm−/− BM using FACSAria and SORP Aria flow cytometer sorters (BD Biosciences). 1000 sorted LT-HSCs (called ‘HSC’) or 1000 sorted LT-HSCs + 4000 sorted ST-HSC/MPPs (called ‘LSK’; 5000 cells total) were plated per well in mouse HSC expansion media and assayed as described above +/− 10mM L-Valine. Engrafting studies were performed (see supplemental methods for details).
Metabolite Profiling of lin− cells.
Mouse BM cells were collected from femurs and tibia by gentle grinding with a mortar and pestle in cold PBS. 5×106 lin− cells from WT or Brm−/− BM were snap frozen and sent to Human Metabolome Technologies, Yamagata, Japan for quantitative mass spectrometry. Cationic and anionic metabolites were measured using CE-TOFMS or CE-MS/MS 38–42. Absolute quantification was performed for 116 metabolites including glycolytic and TCA cycle intermediates, amino acids, and nucleic acids. Metabolite concentrations were calculated by normalizing peak areas of each metabolite with respect to area of internal standard and using standard curves (obtained by three-point calibrations).
Cellular Respirometry.
100,000 BM or lin− cells were plated in each well of a 96-well for OCR analysis on an XF96 Seahorse Extracellular Flux Analyzer, Agilent Technologies, Lexington, MA26,43–45. After three readings, oligomycin was injected, followed by FCCP and rotenone (Agilent Technologies).
RNA-sequencing
LSK cells were isolated from age and sex-matched Brm−/− or WT mice by flow sorting. RNA was harvested using Qiagen RNeasy Micro Plus Kit. cDNA Library preparation and paired end sequencing was performed by the IUSM Medical Genomics Core. Analysis was performed using FastQC, Cutadapt, STAR, HTSeq, and DESeq2. RNA-seq data will be deposited at Gene Expression Omnibus (GEO).
Statistics.
One-way ANOVA with post-hoc Tukey’s Multiple Comparison Test or Student’s t test was utilized unless otherwise indicated.
RESULTS
Phenotypic BM HSC/HPC, Functional HPC Numbers and Cycling Status in Brm−/− mice.
Brm−/− mouse BM contained significantly increased LT-and ST-HSCs as well as GMP (Fig. 1a–c), with mild trends to increased MPP and CMP progenitors (S. Fig. 1a,b), with no effects on MEP or CLP(S. Fig. 1c,d). Brm heterozygotes (−/+) showed little or no changes in phenotyped HSCs/HPCs numbers (Fig. 1a–c and S. Fig. 1a–d). Functional HPC populations, determined by in vitro colony assay and cycling of functional HPC (assessed by high specific activity tritiated thymidine kill assay) demonstrated increased numbers (Fig. 1d–f) and cycling status (Fig. 1g–i) of granulocyte macrophage CFU-GM, BFU-E, and CFU-GEMM progenitors.
Figure 1. Increased hematopoietic stem cell (HSC) numbers in Brm−/− mouse bone marrow (BM).
(a-c) Immunophenotyping of BM harvested from wildtype (WT; n=11), Brm+/− (n=8) and Brm−/− (n=11) mice was performed to examine long-term (LT)-HSC (a), short-term (ST)-HSC (b), and granulocyte macrophage progenitor cell (GMP; c) populations using flow cytometry. Data are mean ± SEM. (d-f) Colony-forming units (CFU) granulocyte-macrophage (GM; d), burst-forming units-erythrocyte (BFU-E; e), and CFU-granulocyte-erythrocyte-macrophage-megakaryocyte (GEMM; f) numbers per femur were determined by colony assay. Data are mean ± SEM of individually assessed mice per group plated in triplicate. n= 4–7 mice per group. (g-i) Percent cycling (CFUs in S-phase) CFU-GM (g), BFU-E (h), and CFU-GEMM (i) was determined by a high specific activity tritiated thymidine (3HTdr) kill assay. Data are mean ± SEM of individually assessed mice per group plated in triplicate. n= 4–7 mice per group. (a-i) * p<0.05, ** p<0.01, and *** p<0.001 when using one-way ANOVA with post-hoc Tukey’s Multiple Comparison Test.
Brm−/− HSCs Demonstrate Defective Self-Renewal Capacity.
An in vivo competitive repopulating assay of wildtype (WT) and Brm−/− BM cells analyzed engraftment into lethally-irradiated congenic recipient mice (CD45.1−CD45.2+ donor cells, CD45.1+CD45.2− competitor cells, and F1 (CD45.1+CD45.2+) recipient mice). Chimerism of WT and Brm−/− donor cells in primary mice were not significantly different in peripheral blood (PB) at months 2–4 (Fig. 2a,b), BM at month 4 (Fig. 2c), or phenotypically-defined donor HSCs in BM at month 4 (Fig. 2d), even though there were increased phenotyped Brm−/− HSCs in initial donor pools (Fig. 1a,b). No differences were noted in myeloid/lymphoid ratios of donor BM (month 4; S. Fig. 1e). Mice receiving primary Brm−/− donor recipient BM demonstrated significant decreases in engrafting capability compared to primary WT donor recipient BM at months 2–4 for PB (Fig. 2e,f) and at month 4 for BM (Fig. 2g) in secondary mice. Significant decreases in numbers of phenotypically-defined donor HSCs (Fig. 2h), without changes in the myeloid/lymphoid ratios in BM of secondary transplants at month 4 were noted (S. Fig. 1f). Significantly decreased engraftment of Brm−/− BM was seen at months 2 (PB) and 4 (PB and BM) following tertiary engraftment (Fig. 2i–k) with significantly reduced phenotypically-defined HSC numbers in BM 4 months after transplantation (Fig. 2l). Secondary and tertiary engraftment, established measures of HSC self-renewal activity, were deficient for Brm−/− BM cells, suggesting a critical role for BRM in mouse BM HSC self-renewal.
Figure 2. Diminished self-renewal capacity of HSCs from Brm−/− mice following serial transplantation into secondary and tertiary irradiated mouse recipients.
(a-d) Primary (1°) BM transplantation (BMT) was performed using WT or Brm−/− mice (CD45.1− CD45.2+) as donors, Boy/J mice (CD45.1+ CD45.2−) as competitors and lethally irradiated B6 x Boy/J F1 mice (CD45.1+ CD45.2+) as recipients. At 2 (a) and 4 (b) months, peripheral blood (PB) was examined for percent donor cells. At 4 months BM was examined for percent CD45.1− CD45.2+ (donor) cells (c) and for the number of donor LSK CD150+ CD41− CD48− HSC by flow cytometry (d). (e-h) Secondary (2°) BMT was performed using BM from 1° recipient mice injected into lethally irradiated B6 x Boy/J F1 recipients. Analysis was performed as in 1° BMT. (i-l) Tertiary (3°) BMT was performed using BM from 2° BMT recipient mice injected into lethally irradiation B6 x Boy/J F1 recipients. Analysis was performed as in 1° BMT. (a-l) n=7–8 mice per group. Data are mean ± SEM. Statistics was performed using Student’s T test.
Increased levels of valine in lin− cells from Brm−/− mice.
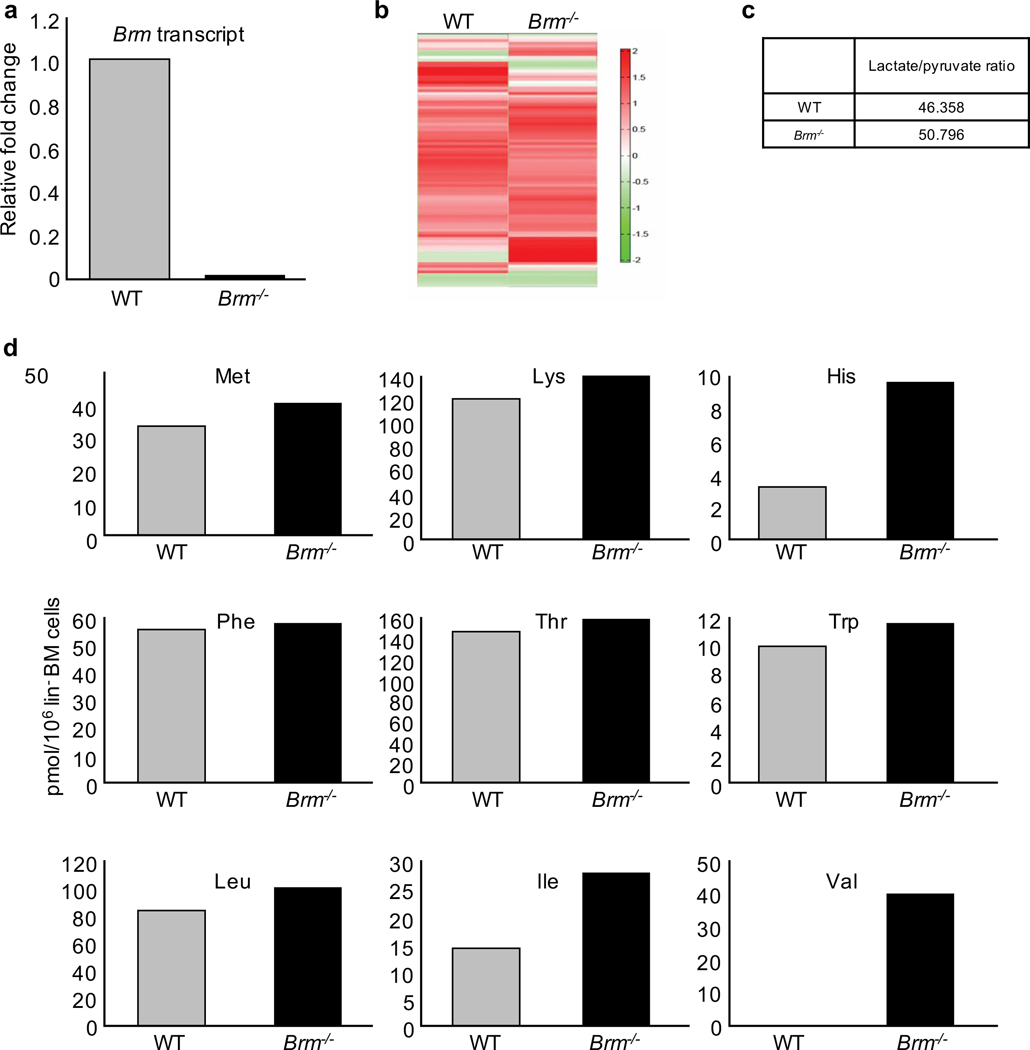
To evaluate metabolic pathways, we used targeted quantitative metabolic profiling of 116 metabolites by mass spectrometry. These 116 metabolites are involved in glycolysis, oxidative phosphorylation, pentose phosphate pathway and amino acid metabolism (S. Table 3). Metabolic analysis was performed on lin− cells (containing HSCs and HPCs). Knockout of Brm was confirmed in the lin− population (Fig. 3a). Hierarchical clustering analysis showed differences in 116 metabolites from WT and Brm−/− cells (Fig. 3b). Lactate/pyruvate ratio, a major metabolic indicator, revealed similar lactate/pyruvate ratios (Fig. 3c). Total non-essential amino acid quantification showed no significant changes in most amino acids in Brm−/− compared to WT cells. Conditional essential amino acids cystine and proline were below detectable levels (S. Table 3). Glutamine levels were undetectable in WT cells and low, but measurable (8 pmol/106cells) in Brm−/− cells (S. Table 1). Valine levels were not detected in WT, but highly elevated in Brm−/− cells (Fig. 3d). Other BCAA, leucine and isoleucine, exhibited increased intracellular levels in Brm−/− cells, though effects are more muted (Fig. 3d). Because valine is apparently essential for HSC survival and proliferation32, studies without mechanistic insight, our studies focused on valine, and functional HPCs. Quantitative serum metabolic analysis revealed no significant differences in all essential amino acids including valine in Brm−/− compared to WT mice serum (S. Table 2). This demonstrates that Brm−/− lin− BM cells manifest increased intracellular levels of valine, without changes in serum valine.
Figure 3. Metabolic profiling of lin− BM cells of WT and Brm−/− mice.
(a) Quantitative transcript levels of Brm transcripts in total RNA isolated from lin− cells. (b) Hierarchical clustering showing differences in 116 metabolites from lin− cells that are obtained from a pool of 6 male and 6 female WT or Brm−/− (KO) mice. (c) Ratios of lactate/pyruvate in WT and Brm−/− Lin− BM cells. (d) Graphs show absolute concentrations of essential amino acids (pmol/106 Cells) in lin− BM cells from WT and Brm−/− mice.
Valine enhances WT BM HPC colony numbers, replating capacity, survival, and ex-vivo expansion of HSCs and HPCs.
We assessed whether exogenously added valine to WT mouse BM cell cultures influenced HPC colony numbers. WT BM cells were cultured with either undialyzed FBS (containing valine: 279 uM) or dialyzed FBS where valine was greatly decreased (22 uM; S. Table 3 and S. Fig. 2). Valine, but not leucine, at 10 mM, a dose pre-determined to be effective, significantly enhanced colony formation by mouse BM CFU-GM, BFU-E, and CFU-GEMM stimulated by PWMSCM, SCF and EPO (Fig. 4a–c) when cultured with dialyzed FBS. In undialyzed FBS, only CFU-GM numbers were increased by valine addition. CFU-GM colonies stimulated by GM-CSF and SCF (S. Fig. 3a), and macrophage progenitors (CFU-M) stimulated by M-CSF and SCF (S. Fig. 3b) were significantly increased by valine with dialyzed FBS cultures. Valine (10mM), but not leucine (10mM), enhanced colony formation of CFU-GM (S. Fig. 3c) and CFU-GEMM (S. Fig. 3d) in low density (LD) human CB cells, and CFU-GM (Fig. 4d) and CFU-GEMM (Fig. 4e) in CD34+ purified human CB cells stimulated by EPO, SCF, GM-CSF, and IL-3. This cytokine combination does not allow detection of BFU-E37. Valine enhanced WT, but not Brm−/− HPCs (Fig. 4f; S. Fig. 3e).
Figure 4. Effects of control diluent, valine or leucine and non-dialyzed (normal) or dialyzed FBS on primary HPC colony formation of normal mouse BM and human CB CD34+ cells.
(a-c) C57BL/6 BM cells were cultured in presence of 30% normal or dialyzed FBS plated in the presence of control diluent, valine (10mM), or leucine (10mM) and indicated growth factors in a colony formation assay. Data are shown as number of CFU-GM (a), BFU-E (b), and CFU-GEMM (c) per 5×104 BM cells/plate. Data represents the combination of 3 separate experiments. (d & e) CD34+ human CB cells were cultured in presence of 30% normal or dialyzed FBS plated in the presence of diluent, valine (10mM), or leucine (10mM) and the indicated growth factors in a colony formation assay. Data are shown as number of CFU-GM (d) and CFU-GEMM (e) colonies per 500 cells/plated. Data represents the combination of 2 separate experiments utilizing two separate CB units. (f) Data are mean ± SD of 3 biological replicates. Statistics were performed using One-way ANOVA with post-hoc Tukey’s Multiple Comparison Test. ** p<0.01, *** p<0.001, and **** p<.0001 when comparing to vehicle of same mouse phenotype. ⱡ ⱡ p<0.01, ⱡ ⱡ ⱡ p<0.001, and ⱡ ⱡ ⱡ ⱡ p<0.0001 when comparing treatment between wildtype and Brm−/− mice.
HPCs manifest limited self-renewalcapacity6–8,36. CFU-GEMM colonies formed from human CD34+ CB cells cultured with EPO, SCF, GM-CSF and IL-3 in dialyzed FBS in primary plates, in presence of either control medium, 10 mM valine, or 10 mM leucine after 14 days (Fig. 5a–d) were individually replated into secondary plates. Single cell preparations of the CFU-GEMM colonies were verified microscopically. Secondary dishes contained dialyzed FBS plus EPO, SCF, GM-CSF and IL-3, but with no valine or leucine added. Valine, but not leucine added in primary plates, significantly enhanced numbers of secondary CFU-GM (Fig. 5b), BFU-E (Fig. 5c), and CFU-GEMM (Fig. 5d) colonies. Valine also enhanced percent of replates with at least 1 colony, and the ratio of numbers of secondary colonies per single colony replated for the HPCs (S. Table 4).
Figure 5. Human CB secondary colony formation, and mouse BM HPC survival.
(a-d) Primary human CB CFU-GEMM colonies were individually harvested from each group and replated with normal serum. Results are shown as number of total secondary colonies (a), and CFU-GM (b), BFU-E (c), and CFU-GEMM colonies (d). Total number of primary colonies replated, number of replated cultures with at least 1 colony, and other additional information are provided in S. Table 4. (a-d) Data are mean ± SEM. * p<0.05, ** p<0.01, and *** p<0.001 when using one-way ANOVA with post-hoc Tukey’s Multiple Comparison Test. C57BL/6 BM cells were cultured with 10% (e) or 30% (f-h) normal (non-dialyzed) or dialyzed FBS in the presence of control diluent, valine (10mM), or leucine (10mM) and the presence of the indicated growth factors. (e) CFU-GM were enumerated by adding GM-CSF and SCF to the culture dish at days 0 (DO), 1 (D1), or 2 (D2). (f-h) CFU-GM (f), BFU-E (g), and CFU-GEMM (h) colonies were enumerated by adding PWMSCM, SCF, and EPO to the culture dish at D0, D1 or D2. Data is shown as number of colonies per 5×104 cells/plated. (e-h) Data are mean ± SEM. a, p<0.05 when compared to DO of the same amino acid (AA) treatment and serum group. b, p<0.01 when compared to control of same day, same serum group, but different AA treatment. c, p<0.01 when compared to same day, same AA treatment, but different serum group. Data are mean ± SEM. * p<0.05, ** p<0.01, and *** p<0.001 when using one-way ANOVA with post-hoc Tukey’s Multiple Comparison Test.
Enhancing effects of valine on in vitro survival of HPCs cultured under delayed growth factor addition26 was assessed with WT mouse BM cells (Fig. 5e–h). BM cells were plated with 10 mM valine or leucine with either non-dialyzed or dialyzed FBS at day 0, and growth factors added days 0, 1, or 2. Valine, but not leucine, significantly enhanced colony survival when cultured in GM-CSF and SCF (Fig. 5e) and of CFU-GM (Fig. 5f), BFU-E (Fig. 5g), and CFU-GEMM (Fig. 5h) when cultured with PWMSCM, SCF and EPO (addition of growth factors delayed for 1 or 2 days). Effects were most notable with dialyzed FBS.
Valine, not leucine, enhanced ex-vivo expansion of phenotypically-defined HSCs/HPCs in mouse Lin− BM cells cultured 4 days in presence of SCF, FLT3L and TPO (S. Fig. 4a–d). Valine increased Brm−/− BM expansion ex-vivo of LT-HSCs, but not to extent of WT BM (S. Fig. 5a), but did not increase HPCs of Brm−/− BM (S. Fig. 5b–d). Differences in expansion in the Brm−/− HSCs compared to HPCs may reflect different accumulation of intracellular valine in different subpopulations of cells.
Valine stimulates mitochondrial respiration in mouse lin− BM.
We studied mitochondrial respiration using normal WT lin− mouse BM cells. Among BCAAs, valine is a unique amino acid as its metabolism in mitochondria yields 3 molecules of NADH and a FADH243. These reducing equivalents stimulate mitochondrial respiration coupled oxidative phosphorylation43. Because mitochondrial metabolism plays a critical role in HSC biology25–29,44–48 and Brm−/− mice manifest increased phenotypically-defined HSCs with decreased self-renewal capacity and increased numbers of phenotypically-defined and functional HPCs, while selectively increasing intracellular levels of valine in lin− cells, we hypothesized that valine stimulates mitochondrial metabolism. Total mouse BM and lin− cells, commonly used to assess mitochondrial functions, were incubated with valine (6mM, but as active as 10mM used in other studies) and mitochondrial respiration was measured on a Seahorse analyzer (Fig. 6). Valine did not alter basal or maximal respiration in BM cells from Brm−/− mice, but significantly increased both and maximal respiration in WT cells (Fig. 6a–d). Although basal oxygen consumption mirrored that of total BM cells, valine significantly stimulated maximal respiration in Brm−/− lin− cells (Fig. 6c,d). Valine stimulated basal oxygen consumption in WT BM and lin− cells but not in Brm−/− cells, suggesting that BRM plays an important role in regulating valine availability for mitochondrial respiration.
Figure 6. Valine stimulates respiration in lin− WT BM cells.
(a) Seahorse tracers showing oxygen consumption rate (OCR) in bone marrow cells exposed to 6 mM valine. B. Bar graphs show quantification of OCR from A. C. Tracers show OCR in lin− cells. B. Quantification of OCR from C. ** : p<0.001, * : p<0.05 : as assessed by one-way ANOVA with post-hoc Tukey’s Multiple Comparison Test for multiple groups.
Link between BRM and import of valine.
We sought to examine mechanisms by which intracellular valine is increased in lin- cells and to examine how this may cause the observed phenotypes in HSCs/HPCs. We analyzed effects of BRM expression on expression of amino acid transport protein CD98. CD98 regulates import of branched chain amino acids including valine, leucine, and isoleucine. We determined expression of CD98hc, one member of the CD98 heterodimer, by flow cytometry in WT or Brm−/− mice. CD98 expression is modestly but insignificantly increased in Brm−/− HSCs compared to WT (S. Fig. 6a–c), differences enriched specifically in LT-HSCs compared to ST-HSCs (S. Fig. 6d–e). Differences in cell surface CD98 is more apparent in MPPs, with increased CD98 (S. Fig. 6f–g) and a significantly different distribution of CD98-, CD98+, and CD98high expression, with Brm−/− cells exhibiting a higher proportion of CD98high cells (S. Fig. 6h). We performed chromatin immunoprecipitation followed by qPCR (ChIP-qPCR) in lin- BM, examining BRM binding to Slc3a2 locus (codes for CD98hc). BRM binds to promoter regions both at 1000bp upstream of transcription start site (TSS) (S Fig. 6i) and at TSS (S. Fig. 6k). Data are consistent with our mined ChIP-seq data in a human cell line: BRM binds at the SLC3A2 promoter for the transcript that encodes the CD98hc isoform that is orthologous to mouse CD98hc (S. Fig. 6k). Thus BRM localizes to the locus coding for CD98hc and may repress expression in lin- BM leading to increased CD98high populations of HPCs. These differences may account for some observed phenotypes in HPCs. These slight biological changes are not likely to account for observed changes in HSCs.
Brm−/− LSK exhibit transcriptomic changes associated with increased inflammation and cell cycle
Changes in CD98 are not likely to account for all observed phenotypic changes in the most primitive cells. We examined transcriptomic changes in Brm−/− compared to WT cells. RNA-sequencing (RNA-seq) of LSK cells revealed that 96 genes are upregulated and 57 genes are downregulated in Brm−/− LSK cells(Fig. 7a). Fast gene set enrichment analysis (FGSEA) revealed genes associated with amino acid transport and catabolic processes were not strongly enriched in differentially expressed genes (representative plots Fig. 7b–c). Slc3a2 was not significantly differentially expressed in Brm−/− LSK, possibly due to proportions of HSC in LSK cells that exhibit little to slight changes in CD98, thus potentially muting any effects seen in HPC populations of LSK cells. Gene programs associated with interferon signaling and those associated with cell cycle were highly upregulated in Brm−/− LSK compared to WT LSK (representative plots Fig. 7d–e). This suggests that amino acid transport may play indirect or more modest roles in Brm−/− LSK, while the primary effects on these cells may instead be responses to inflammation leading to increased cell cycling. Exposure to increased BCAA induces inflammatory responses33. We hypothesized that increases in intracellular valine in lin- Brm−/− cells may be attributed primarily to HPCs, while observed effects on HSCs may be due to cell extrinsic effects caused by signaling induced in HPCs.
Figure 7. Brm−/− LSK exhibit a transcriptomic profile showing enriched interferon response and cell cycle genes.
(a-e) Lin-Sca1+Kit+ cells were isolated using flow cytometry sorting from age and sex-matched WT or Brm−/− mice (n=3 each). RNA was extracted, cDNA libraries were prepared and sequenced. (a) Log-ratio/mean average (MA) plot showing overall transcriptomic differences of Brm−/− LSK vs WT LSK. Significantly differentially expressed (DE) genes as determined by DESeq2 are highlighted in red (upregulated) or blue (downregulated). Brm is indicated with an arrow confirming lack of expression in Brm−/− LSK. (b-e) Fast gene set enrichment (FGSEA) plots for the Brm−/− LSK vs WT LSK using the indicated a priori defined gene sets. Source of gene set is listed in parentheses. NES=Normalized enrichment score; Padj=p-value adjusted for multiple testing.
Brm−/− HSCs are affected by cell extrinsic effects
To examine if Brm−/− HSCs are affected by cell extrinsic effects, we isolated LT-HSCs and LSK (containing LT-HSC, ST-HSC, and MPP) separately from Brm−/− and WT mice and treated cells +/− valine (Fig. 8a). Brm−/− LT-HSCs cultured by themselves did not exhibit enhanced expansion of LT-HSC, CFU-GM, CFU-GEMM, or BFU-E compared to WT LT-HSCs (Fig. 8b, S. Fig. 7a,c,e). Brm−/− LSK cultures exhibited stronger expansion of LT-HSCs, CFU-GM, and CFU-GEMM (Fig. 8c, S. Fig. 7b,d,f). Interestingly, valine addition strongly enhanced expansion of Brm−/− LT-HSCs when cultured alone, but only slightly enhanced expansion of Brm−/− LT-HSCs when cultured with subpopulations present in LSK cells. This is likely the result of combinations of cell intrinsic effects on HSCs from added valine and cell extrinsic effects exerted by the Brm−/− HPCs. To examine whether expansion of functional LT-HSCs are affected by cell extrinsic effects, we performed competitive transplants using WT or Brm−/− LT-HSCs or LSK cells expanded +/− valine. LT-HSCs cultured alone exhibited no changes in functional HSC expansion, demonstrated by similar donor chimerism in recipient mice, regardless of BRM status or valine treatment (Fig. 8d, S. Fig. 8a,c). This suggests that cell intrinsic effects of added valine lead to expansion of phenotypically defined HSCs, but not necessarily functional HSCs. LSK cultures exhibited reduced donor chimerism in recipient mice after valine treatment of WT LSK and after valine treatment of Brm−/− LSK (Fig. 8e, S. Fig. 8b,d). This may be surprising given enhanced effects on LT-HSC expansion; we infer that addition of valine in ex vivo culture, which drove cells to expand more rapidly, may have pushed cells to earlier exhaustion in vivo.
Figure 8. Populations of Brm−/− HPC exert cell extrinsic effects on HSC while valine treatment induces cell intrinsic effects.
(a) Wildtype (WT) and Brm−/− HSC (1000 sorted LT-HSC) and LSK cells (1000 sorted LT-HSC + 4000 sorted ST-HSC/MPP; 5000 cells total) were cultured in expansion media with either vehicle control or 10mM valine. (b&c) Immunophenotyping to examine LT-HSC numbers was performed on day 4 from expansion assays using input HSC (b) and input LSK (c). (b&c) Data are mean ± SD of 3 biological replicates. Statistics were performed using One-way ANOVA with post-hoc Tukey’s Multiple Comparison Test. *** p<0.001 and **** p<.0001 when comparing to vehicle of same mouse phenotype. † p<0.05 and †† p<0.01 when comparing same treatment between WT and Brm−/− mice. Number above bar indicates average fold change increase from input (i.e. expansion). (d-e) Engrafting studies were performed using 1000 WT HSC, 1000 Brm−/− HSC, 5000 WT LSK, and 5000 Brm−/− LSK input cells and the progeny cells from the HSC and LSK expansions groups (WT ‘HSC’ + vehicle, WT ‘HSC’ + valine, WT ‘LSK’ + vehicle, WT ‘LSK’ + valine, Brm−/− ‘HSC’ + vehicle, Brm−/− ‘HSC’ + valine, Brm−/− ‘LSK’ + vehicle, and Brm−/− ‘LSK’ + valine). Data are the mean ± SD of 4–5 mice per group. Shown are bone marrow chimerism. Statistics were performed using One-way ANOVA with post-hoc Tukey’s Multiple Comparison Test. (f-g) Engrafting studies were performed using 100,000 total BM cells from mice that had been treated 24 hours prior with 10mg valine via IP injection. Percentages of CD45.1-CD45.2+ donor chimerism in the bone marrow (f) and total numbers of CD45.1-CD45.2+Lin-Sca1+Kit+CD48-CD41-CD150+ donor derived HSC per femur were calculated by cell surface staining and flow cytometry. Data from two independent experiments were pooled. Statistics were performed using unpaired t-tests. *=p<0.05.
Valine treatment of donor mice improves HSC engraftment
To examine whether valine addition by itself affects HSC expansion and function in vivo, we treated donor mice intraperitoneally with 10mg valine, harvested and used BM 24 hours later for transplantation of lethally irradiated recipients. When using valine treated donors, mice exhibited a trend toward higher engraftment of CD45.1-CD45.2+ donor cells at months 2 and 4 in the peripheral blood (S. Fig. 8e–f) and BM (Fig. 8f). There are significantly higher numbers of donor HSCs found in the treated group (Fig. 8g). Donor cell myeloid/lymphoid ratios remained unchanged (S. Fig. 8g). This indicates that increased availability of valine has direct and unique effects on HSCs compared to effects of loss of BRM and increased intracellular valine in lin- populations.
DISCUSSION
We identified chromatin remodeling factor BRM as a regulator of proliferation, survival, and replating (in vitro self-renewal) efficiency of BM HPCs that also affects HSC self-renewal and ex-vivo expansion of HSCs and HPCs. This is likely mediated by increased intracellular valine in HPCs, driving cell intrinsic enhancement of HPC growth/function while causing HPCs to exert cell extrinsic effects on HSCs, possibly in form of interferon signaling. Combinations of genetic, unbiased quantitative mass spectrometry, metabolic analysis, and transcriptomic profiling suggest that BRM plays a crucial role in these events by regulating, directly or indirectly, intracellular BCAA availability, mitochondrial metabolism, and interferon/cell cycle responses.
Mice with constitutive loss of BRM have significantly more BM LT-HSC, ST-HSC, and GMP compared to WT mice. They exhibit increased numbers and cycling status of CFU-GM, BFU-E, and CFU-GEMM, read outs for functional HPCs. However, despite increased cell numbers, Brm−/− HSCs have defective self-renewal capacity, evidenced by poor engraftment in secondary and tertiary transplantation. That loss of BRM results in decreased HSC self-renewal capability but increased numbers of functional HPCs suggests expression of BRM is involved in enhancing HSC self-renewal. BRM may serve as a functional HSC to HPC fate determinant, with loss of BRM stimulating proliferation of HSCs, leading to exhaustion.
Metabolite profiling revealed lin− Brm−/− cells have strongly increased levels of valine, and moderate increases in leucine and isoleucine. Among BCAAs, valine but not leucine or isoleucine plays roles in survival and proliferation of HSCs32. Quantitative analysis of serum metabolites revealed that valine levels are similar in WT and Brm−/− mouse serum, suggesting effects are due to intracellular, not extracellular, valine. We showed that treatment with valine but not leucine enhanced CFU-GM, BFU-E, and CFU-GEMM in mouse BM and CFU-GM and CFU-GEMM in human LD CB and purified human CD34+ cells. Valine treatment enhanced secondary replating of mouse CFU-GM, BFU-E, and CFU-GEMM and promoted survival of CFU-GM, BFU-E, and CFU-GEMM after delayed growth factor addition. Valine treatment of wildtype cells enhanced ex vivo expansion of HSCs and HPCs. This suggests that valine promotes proliferation of HSCs and HPC proliferation, survival, and function.
BCAAs including valine get metabolized in the mitochondria.30 We explored whether increased valine levels in lin- BM cells affected mitochondrial metabolism. Valine stimulates mitochondrial oxygen consumption rate (OCR) in total and lin− BM cells. Valine increased both basal and maximal OCR in BM cells from WT mice, whereas BM cells from Brm−/− mice did not respond to valine. It is likely that Brm−/− BM cells accumulate valine at a maximal or near maximal levels. Thus, additional availability of valine does not further increase OCR. This implies that valine stimulates increased mitochondrial metabolism in BM cells and BRM dependent accumulation of intracellular valine affects this stimulation. Mitochondrial metabolism plays critical roles in HSC/HPC proliferation and function45–48. This suggests that accumulated intracellular valine resulting from loss of BRM in HPCs leads to increased mitochondrial metabolism, thus providing potential mechanisms by which BRM loss in HSCs/HPCs drives increased proliferation.
We explored whether there are clear links between reduced BRM expression, increased intracellular valine, and phenotypic responses of HSCs/HPCs to valine and in cells lacking BRM. Intracellular valine levels of the heterogeneous lin- population increased in Brm−/− cells. Cell surface staining and flow cytometry demonstrated that this accumulation may be due in part to BRM regulation of cell surface expression of valine transport protein CD98 on the surface of HPCs. However, HSCs exhibit only minor changes in CD98 expression in Brm−/− compared to WT cells, suggesting that CD98 likely does not significantly account for increased HSC expansion ex vivo and reduced functionality in vivo. Indeed, RNA-seq analysis of Brm−/− LSK revealed that BRM does not regulate gene programs associated with BCAA transport or processing in these primitive hematopoietic cells. Brm−/− LSK cells have enhanced expression of interferon response and cell cycle gene programs. Thus, observed effects in HSCs lacking BRM may instead be due to cell extrinsic effects exerted on HSCs by the larger, more mature HPC population found within lin- BM.
To test whether cell extrinsic effects are involved, we isolated LT-HSCs and tested whether lack of BRM or valine treatment affects LT-HSCs in the same manner as it does when the LT-HSCs are a part of heterogeneous LSK populations. Interestingly, Brm−/− LT-HSCs did not have enhanced ex vivo expansion compared to WT when treated alone, but did have enhanced expansion when treated as a part of the LSK population. This strongly indicates that effects on LT-HSCs are due to cell extrinsic effects. Also, valine addition strongly enhanced expansion of Brm−/− LT-HSCs when treated alone, showing that Brm−/− LT-HSCs can consume additional valine, driving enhanced expansion. Valine addition caused a more modest increase in expansion in Brm−/− LT-HSCs when cultured as a part of the LSK population, suggesting that the expansion in these cells is mostly caused by cell extrinsic effects. Additional valine may have an additive effect on expansion because Brm−/− LT-HSCs can consume more valine. There is no additive effect on expansion of Brm−/− CFU-GM, BFU-E, or CFU-GEMM with addition of valine, likely because HPCs are already saturated with intracellular valine. Finally, valine treatment of donor mice prior to BM harvest and transplantation into lethally irradiated recipients shows a higher level of engraftment for donor derived HSCs compared to vehicle treatment. This indicates that valine treatment can have a direct enhancing effect on functional HSC and is an important corollary to previous studies showing that valine depletion reduces numbers and functions of HSC32.
Our data strongly suggests that BRM, a cell intrinsic factor, limits intracellular valine of HPCs, thus modulating proliferation, survival, functionality, and mitochondrial metabolism of HPCs, due in part to regulation of CD98 amino acid transport protein. BCAA are capable of inducing interferon responses in non-hematopoietic cell types33. We suggest that reduced BRM expression leads to enhanced interferon responses and cell cycling of HSC, likely due to external stimuli released by HPCs due to excessive accumulation of intracellular valine. We also show that valine treatment induces similar enhancing effects on HPC expansion and function as loss of BRM, and that valine treatment enhances expansion and function of HSCs in a different manner than loss of BRM. Thus, we identified a novel axis of regulation for self-renewal, survival, and expansion of HSCs/HPCs wherein BRM regulates intracellular valine content of HPC, which in turn affects elf-renewal of HSCs/HPCs and possibly HSC to HPC differentiation fate decisions.
Supplementary Material
ACKNOWLEDGMENTS
These studies were supported by the Public Health Service Grants from the NIH to HEB: R35 HL139599 (Outstanding Investigator Award), R01 DK109188, and U54 DK106846. JR was supported as a postdoctoral fellow by NIH T32 DK007519 (PI HEB). We thank Dr. Ching-Pin Chang for Brm−/− mice.
Footnotes
Competing interests statement: Dr. Broxmeyer is on the Scientific Advisory Board of Elixell Corp, a stem cell company. The other authors have no competing financial interests to disclose
COMPETING INTERESTS
HEB is on the Scientific Advisory Board of Elixell Corp, a stem cell company. None of the other authors have conflicts of interest to disclose.
REFERENCES
- 1.Miyamoto K, Araki KY, Naka K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1(1):101–112. [DOI] [PubMed] [Google Scholar]
- 2.Shaheen MB HE Cytokine/Receptor Families and Signal Transduction. In: Hematology: Basic Principles and Practice. (ed 7th Edition ). Philadelphia, PA.: Elsevier Saunders; 2018. [Google Scholar]
- 3.Shaheen MBHE. Principles of Cytokine Signaling. In: Hematology: Basic Principles and Practice, 6th Edition (ed 6th Edition ). Philadelphia, PA.: Elsevier Saunders; 2013. [Google Scholar]
- 4.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453(7193):306–313. [DOI] [PubMed] [Google Scholar]
- 6.Broxmeyer HE, Lee M-R, Hangoc G, et al. Hematopoietic stem/progenitor cells, generation of induced pluripotent stem cells, and isolation of endothelial progenitors from 21- to 23.5-year cryopreserved cord blood. Blood. 2011;117:4773–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carow CE, Hangoc G, Broxmeyer HE. Human multipotential progenitor cells (CFU-GEMM) have extensive replating capacity for secondary CFU-GEMM: an effect enhanced by cord blood plasma. Blood. 1993;81(4):942–949. [PubMed] [Google Scholar]
- 8.Carow CE, Hangoc G, Cooper SH, Williams DE, Broxmeyer HE. Mast cell growth factor (c-kit ligand) supports the growth of human multipotential progenitor cells with a high replating potential. Blood. 1991;78(9):2216–2221. [PubMed] [Google Scholar]
- 9.Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23(8):403–412. [DOI] [PubMed] [Google Scholar]
- 10.Muchardt C, Yaniv M. When the SWI/SNF complex remodels…the cell cycle. Oncogene. 2001;20(24):3067–3075. [DOI] [PubMed] [Google Scholar]
- 11.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21(3):396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol. 2009;19(2):120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raab JR, Runge JS, Spear CC, Magnuson T. Co-regulation of transcription by BRG1 and BRM, two mutually exclusive SWI/SNF ATPase subunits. Epigenetics Chromatin. 2017;10(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15(5):603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanton BZ, Hodges C, Calarco JP, et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet. 2017;49(2):282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong AK, Shanahan F, Chen Y, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60(21):6171–6177. [PubMed] [Google Scholar]
- 17.Bottardi S, Ross J, Pierre-Charles N, Blank V, Milot E. Lineage-specific activators affect beta-globin locus chromatin in multipotent hematopoietic progenitors. EMBO J. 2006;25(15):3586–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bultman S, Gebuhr T, Yee D, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6(6):1287–1295. [DOI] [PubMed] [Google Scholar]
- 19.Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135(3):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27(2):317–328. [DOI] [PubMed] [Google Scholar]
- 21.Singhal N, Esch D, Stehling M, Scholer HR. BRG1 Is Required to Maintain Pluripotency of Murine Embryonic Stem Cells. Biores Open Access. 2014;3(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 1998;17(23):6979–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene. 2009;28(27):2492–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11(2):377–389. [DOI] [PubMed] [Google Scholar]
- 25.Filippi MD, Ghaffari S. Mitochondria in the maintenance of hematopoietic stem cells: new perspectives and opportunities. Blood. 2019;133(18):1943–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantel CR, O’Leary HA, Chitteti BR, et al. Enhancing Hematopoietic Stem Cell Transplantation Efficacy by Mitigating Oxygen Shock. Cell. 2015;161(7):1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snoeck HW. Mitochondrial regulation of hematopoietic stem cells. Curr Opin Cell Biol. 2017;49:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gan B, Hu J, Jiang S, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468(7324):701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurumurthy S, Xie SZ, Alagesan B, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468(7324):659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annu Rev Nutr. 1984;4:409–454. [DOI] [PubMed] [Google Scholar]
- 31.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468(7324):653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taya Y, Ota Y, Wilkinson AC, et al. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science. 2016;354(6316):1152–1155. [DOI] [PubMed] [Google Scholar]
- 33.Honda M, Takehana K, Sakai A, et al. Malnutrition impairs interferon signaling through mTOR and FoxO pathways in patients with chronic Hepatitis C. Gastroenterology. 2011; 141(1):128–140.e2. [DOI] [PubMed] [Google Scholar]
- 34.Broxmeyer HE, Christopherson K, Hangoc G, et al. CD1d expression on and regulation of murine hematopoietic stem and progenitor cells. Blood. 2012;119(24):5731–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broxmeyer HE, Hoggatt J, O’Leary HA, et al. Dipeptidylpeptidase 4 negatively regulates colony-stimulating factor activity and stress hematopoiesis. Nat Med. 2012;18(12):1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu RY, Wang X, Pixley FJ, et al. BCL-6 negatively regulates macrophage proliferation by suppressing autocrine IL-6 production. Blood. 2005;105(4):1777–1784. [DOI] [PubMed] [Google Scholar]
- 37.Broxmeyer HE, Hangoc G, Cooper S, et al. Growth characteristics and expansion of human umbilical cord blood and estimation of its potential for transplantation in adults. PNAS. 1992; 89(9):4109–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanai Y, Segawa H, Miyamoto K, Uchino H, Takeda E, Endou H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J Biol Chem. 1998;273(37):23629–23632. [DOI] [PubMed] [Google Scholar]
- 39.Meier C, Ristic Z, Klauser S, Verrey F. Activation of system L heterodimeric amino acid exchangers by intracellular substrates. EMBO J. 2002;21(4):580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soga T, Heiger DN. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2000;72(6):1236–1241. [DOI] [PubMed] [Google Scholar]
- 41.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res. 2003;2(5):488–494. [DOI] [PubMed] [Google Scholar]
- 42.Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2002;74(10):2233–2239. [DOI] [PubMed] [Google Scholar]
- 43.Overmyer KA, Evans CR, Qi NR, et al. Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab. 2015;21(3):468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120(4):483–495. [DOI] [PubMed] [Google Scholar]
- 45.Anso E, Weinberg SE, Diebold LP, et al. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat Cell Biol. 2017;19(6):614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito K, Turcotte R, Cui J, et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science. 2016;354(6316):1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M, Snoeck HW. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature. 2016;529(7587):528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohrin M, Shin J, Liu Y, et al. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347(6228):1374–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.