Abstract
The green fluorescent protein (GFP) gene, gfp, of the jellyfish Aequorea victoria is being used as a reporter system for gene expression and as a marker for tracking prokaryotes and eukaryotes. Cells that have been genetically altered with the gfp gene produce a protein that fluoresces when it is excited by UV light. This unique phenotype allows gfp-tagged cells to be specifically monitored by nondestructive means. In this study we determined whether a gfp-tagged strain of Pseudomonas fluorescens continued to fluoresce under conditions under which the cells were starved, viable but nonculturable (VBNC), or dead. Epifluorescent microscopy, flow cytometry, and spectrofluorometry were used to measure fluorescence intensity in starved, VBNC, and dead or dying cells. Results obtained by using flow cytometry indicated that microcosms containing VBNC cells, which were obtained by incubation under stress conditions (starvation at 37.5°C), fluoresced at an intensity that was at least 80% of the intensity of nonstressed cultures. Similarly, microcosms containing starved cells incubated at 5 and 30°C had fluorescence intensities that were 90 to 110% of the intensity of nonstressed cells. VBNC cells remained fluorescent during the entire 6-month incubation period. In addition, cells starved at 5 or 30°C remained fluorescent for at least 11 months. Treatment of the cells with UV light or incubation at 39 or 50°C resulted in a loss of GFP from the cells. There was a strong correlation between cell death and leakage of GFP from the cells, although the extent of leakage varied depending on the treatment. Most dead cells were not GFP fluorescent, but a small proportion of the dead cells retained some GFP at a lower concentration than the concentration in live cells. Our results suggest that gfp-tagged cells remain fluorescent following starvation and entry into the VBNC state but that fluorescence is lost when the cells die, presumably because membrane integrity is lost.
Increasingly, genetically modified microorganisms (GMMs) are being engineered for specific environmental applications, such as plant growth promotion, insect or plant pathogen control, and bioremediation. The release of GMMs must be regulated to minimize potentially hazardous consequences. Environmental concerns include dispersal from the release site, the effect on the indigenous microbial population, and the potential for gene transfer (7). Due to the large number of microorganisms found in natural samples, it would be difficult to distinguish GMMs from the indigenous population by traditional identification methods. For these reasons molecular marker systems and detection methods have been developed (9, 10, 11, 15).
The ideal marker system should enable detection and quantification of specific organisms and provide an indication of their viability. It should also confer a unique characteristic that is both simple and inexpensive to detect. Some marker systems currently in use include antibiotic resistance genes, genes encoding luciferase enzymes (e.g., luxAB) that provide a bioluminescent phenotype, and genes encoding metabolic enzymes (e.g., lacZY) that allow marked cells to be distinguished after a substrate is converted to a colored or chemiluminescent product (9, 10). One disadvantage of some marker systems is the natural occurrence of the phenotype in the indigenous microbial population. For example, Lac+ bacterial cells are often found in habitats such as soil (6). Another problem with many markers is a requirement for a particular substrate. For example, when luxAB genes are used to mark cells, an aldehyde substrate is required for bioluminescence to be observed. It is possible to use the entire lux operon for marking cells to avoid this dependence on substrate addition, but the requirement for substrate synthesis places an energy burden on the cells (5). Luciferase markers are also dependent on oxygen and cellular energy reserves for the luciferase energy reaction. However, bacterial cells are often starved in nature when energy resources are limited and cannot be detected by bioluminescence assays. These limitations have prompted the development of new markers.
Currently, there is great interest in the gfp gene as a potential marker for tracking and visualizing bacteria in environmental samples. This gene, which is found naturally in the jellyfish Aequorea victoria, encodes the green fluorescent protein (GFP) (3). In A. victoria, GFP is activated by oxygen-dependent energy transfer between aequorin and GFP. In the absence of aequorin, GFP emits green light at 508 nm when it is excited with UV light at 396 nm (3). Several GFP mutants have been constructed with altered fluorescence properties; one of these is the P11 mutant, which has enhanced fluorescence and a red-shifted excitation wavelength (8).
Introduction of the gfp gene into a cell results in fluorescence that does not require any substrates, is not species specific, and can be detected by nondestructive means (3). The only thing required of the host cell is gene expression. Once GFP is synthesized and the posttranslational modifications (cyclization and oxidation) take place, the tagged cell is fluorescent as long as the protein is maintained. One copy of gfp inserted into the chromosome is sufficient to detect fluorescing cells by flow cytometry (23), but greater fluorescence intensity is achieved when two copies of gfp are present in the chromosome (24).
One concern with the use of GFP as a marker during release studies is the extreme stability of the GFP protein (21). A possible consequence of this stability is that tagged cells might remain fluorescent regardless of viability. If this is the case, the presence of the phenotype would not be an indication that viable cells are present. On the other hand, during a stress response (for example, a nutrient starvation response) cells synthesize proteases that are known to degrade some existing proteins in order to obtain energy and amino acids for essential protein synthesis (16). Thus, GFP may be degraded by proteases. For temporal studies of gene expression in environmental samples, unstable GFP variants have been constructed which are more susceptible to such cellular proteases (1). Altered stability has been obtained by adding a protease-specific peptide sequence to the GFP which results in degradation of GFP.
One situation that has a profound effect on bacterial cells is the viable-but-nonculturable (VBNC) state. This state occurs when a cell is not able to grow on media normally used for growth but remains viable (18). The VBNC state has been found largely in gram-negative organisms, and Vibrio vulnificus is the best-studied organism (18). The inducing conditions vary from organism to organism, but all of them appear to be normal environmental stresses. In the case of V. vulnificus, low temperature (<10°C) has been shown to induce the VBNC state (26). Other examples of stresses include high temperature and nutrient depletion.
In previous studies workers found that gfp-tagged pseudomonads remained fluorescent during long-term starvation (23, 25). However, the number of culturable bacteria declined during starvation (25). The question remains whether the higher number of GFP-containing cells enumerated by flow cytometry compared to colonies grown on agar medium is due to counting of GFP fluorescent VBNC cells or if dead cells are fluorescent and are enumerated by flow cytometry.
The aim of this study was to determine whether a gfp-tagged strain of Pseudomonas fluorescens is fluorescent during starvation and entry into a VBNC state. Also, we wanted to determine whether GFP fluorescence of cells is linked to cell viability. Such information is necessary in order to judge the usefulness of GFP as a marker for cells in environmental samples.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. fluorescens A506, wt (14), and A506::gfp2 (24) were used in the experiments described below. Strain A506::gfp2 was chromosomally marked with two copies of the mutant gfp gene, P11 (8), which results in fluorescence intensity that is greater than that of wild-type GFP (24). P11 emits light at 502 nm when it is excited at 471 nm (8). The pseudomonads were routinely grown in Luria broth (LB) at 30°C. Kanamycin (50 μg/ml) was added to the A506::gfp2 culture medium.
Microcosm preparation.
Log-phase cells were grown in LB, diluted in phosphate-buffered saline (PBS), and inoculated into PBS to obtain microcosms containing ca. 106 CFU/ml. One-hundred-milliliter microcosms were prepared in 125-ml plastic, screw-cap flasks and maintained under static conditions. The microcosms were incubated at 5, 30, or 37.5°C, and the cells were starved at all three temperatures. A temperature of 37.5°C was chosen since it been shown previously that this temperature places P. fluorescens cells in the VBNC state (19).
Culturability assays.
The culturability of cells was determined by plating dilutions of microcosm samples onto Luria agar (LA). A microcosm was said to be nonculturable when there was less than 0.1 CFU/ml, as determined by a lack of growth after a 10-ml microcosm sample was filtered through a 0.22-μm-pore-size filter (Poretics, Livermore, Calif.) and the filter was placed on LA and incubated at 30°C for 2 days. Cell viability was determined as described below to distinguish nonculturable cells from dead cells.
Viability assays.
Viability assays were performed by using 5-cyano-2,3-ditolyl tetrazolium chloride (CTC), which fluoresces red when it is reduced by an active electron transport chain (22). A 1-ml sample of cells from one of the microcosms described above was incubated overnight in 500 μl of a 8 mM CTC solution (final concentration, 3 mM). This was followed by incubation for at least 30 min in 50 μl of a 1-μg/ml 4,6-diamidino-2-phenylindole (DAPI) working solution, which was used as a counterstain for microscopic determinations of total numbers of cells. After staining, the cells were collected by filtration on black polycarbonate filters (pore size, 0.2 μm; diameter, 25 mm; Poretics). An epifluorescence microscope (series BH2; Olympus) was used to determine the viable cell counts and total cell counts. The percentage of viable cells in the population was then determined.
Flow cytometry.
For some experiments, samples were analyzed with a Becton Dickinson model FACStar Plus flow cytometer equipped with a 100-mW air-cooled argon ion laser (488 nm) by using LYSYS II software. Fluorescence compensation was performed by using Becton Dickinson CaliBRITE beads as directed by the manufacturer. Green fluorescence emission at wavelengths between 515 and 545 nm was collected with a photomultiplier tube (type FL1 PMT) by using a type DF530/30 bandpass filter. Forward angle light scatter (FSC) was collected by using a type BP488/10 bandpass filter. The FSC neutral-density filter was removed before the bacteria were analyzed. Stabilized isotonic saline was used as the sheath fluid, and a sterile 0.2-μm-pore-size filter was piggybacked onto the sheath line. All of the cultures that were analyzed were fixed with 50 μl of 37% formaldehyde, since this concentration of formaldehyde has been shown previously to have no adverse effect on GFP fluorescence (13). After fixation, the samples were washed once in PBS and then resuspended in 1 ml of PBS. The threshold trigger was set to FL1 (green) fluorescence, and the FSC and side angle light scatter (SSC) amp gains were set to LOG. The instrument voltage values for SSC, FL1, and FL2 (orange-red fluorescence) were initially set at 400, 400, and 600 V, respectively, and then were optimized for log-phase gfp-tagged P. fluorescens. For each cell sample run, data for 10,000 events were collected. Log-phase cells of gfp-tagged and nontagged strains were used as positive and negative controls, respectively. The fluorescence of the gfp-tagged stressed cells was compared to the maximum fluorescence and the baseline fluorescence by using the mean fluorescence channel number. It should be noted that only the fluorescence of GFP present in intact cells is detected by flow cytometry; any GFP lost to the surrounding medium is not detected.
P. fluorescens A506::gfp2 cells treated with heat (see below) were analyzed with a model FACScalibur flow cytometer equipped with a 15-mW air-cooled argon ion laser (488 nm) as the excitation source. For each measurement, data for 10,000 events were collected. Green fluorescence at wavelengths of 500 to 550 nm was detected with a fluorescence detector set at a photomultiplier tube voltage of 600 V with logarithmic gain. FSC was collected by using a diode with an amplification factor of 10 and was processed in log gain. SSC and FL3 (red fluorescence) were detected in log gain by using a photomultiplier tube set at 400 and 600 V, respectively. Propidium iodide (PI)- and CTC-stained cells were detected with the FL3 detector. The fluorescence intensity of the cells was expressed as the geometric mean (G-mean), which was defined as follows: G-mean = 10ΣlogX(i)/n, where X(i) is the channel or linear value for the ith event and n is the number of events.
UV light treatment.
P. fluorescens A506::gfp2 log-phase cells were grown in LB, washed once, and resuspended in 10 ml of PBS. A 1-ml sample was removed and fixed with 50 μl of 37% formaldehyde. The remaining 9 ml was transferred to a sterile petri dish and exposed to 245-nm UV light for 5 min. After exposure, cell death was confirmed by a lack of growth on LA. Furthermore, previous studies (unpublished data) performed in our laboratory indicated that cells treated with UV in this way exhibited no detectable response to CTC staining (i.e., were not viable). Samples (1 ml) were removed, fixed, and centrifuged, and the supernatants were removed. The dead cells were resuspended in PBS, and the fluorescence intensities of both the supernatant and the cells were determined by spectrofluorometry performed with a Hitachi model F-2500 fluorescence spectrophotometer. The parameters used were as follows: excitation wavelength, 471 nm; emission wavelength, 502 nm; 700 V; slit width, 10 nm. Log-phase cells of P. fluorescens A506 were incubated for 30 min at 25°C in the supernatant removed at each time point. The fluorescence intensity was measured by flow cytometry. Samples (1 ml) were prepared for flow cytometric analysis as described above.
In a separate experiment, the effect of UV treatment on P. fluorescens A506::gfp2 cell size, shape, and fluorescence properties was determined. The cells were washed, resuspended in 0.9% NaCl, treated with short-wavelength UV light for 60 min, and then left at room temperature for 70.5 h. Then 1-ml portions were removed, washed, and resuspended in a solution containing 1 ml of 0.9% NaCl and 2 μl of 20 mM PI before analysis with the FACScalibur flow cytometer. PI can be used as a stain for dead cells because viable cells exclude the stain (17).
Heat treatment.
P. fluorescens A506::gfp2 cells were grown to the early stationary phase in LB, washed once, and resuspended in 0.9% NaCl. The culture was divided into 4-ml portions and then heat treated at 50°C for 0 to 30 min or at 39°C for 0 to 5 h. After heat treatment the cell preparations were divided into 1-ml portions and centrifuged, and the supernatants were saved for analysis by spectrofluorometry. The cells were resuspended in either a solution containing 535 μl of 5 mM CTC and 465 μl of 0.9% NaCl, a solution containing 1 ml of 0.9% NaCl and 2 μl of 20 mM PI, or 1 ml of 0.9% NaCl (to count GFP fluorescent cells and CFU). For CFU enumeration the cells were diluted and plated onto LA supplemented with 50 μg of kanamycin per ml. GFP fluorescent cells and PI-stained cells were examined by flow cytometry. CTC-stained cells were incubated overnight at room temperature in the dark prior to flow cytometric analysis as this was found to result in optimum fluorescence. An analysis of the supernatant for leakage of GFP from the cells during heat treatment was performed by using a model LS50B spectrofluorometer (Perkin-Elmer, Beaconsfield, United Kingdom) set to excitation at a wavelength of 471 nm and emission at a wavelength of 502 nm. Before analysis, the supernatant was filtered through a 0.22-μm-pore-size sterile filter to remove any remaining cells.
RESULTS
Starvation and entry into the VBNC state.
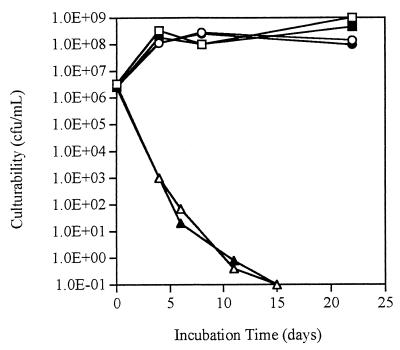
After incubation at 37.5°C under starvation conditions for 15 days, both P. fluorescens A506::gfp2 and wild-type strain A506 were nonculturable (Fig. 1). However, viability assays performed with these cells after 21 days indicated that at least 1% (gfp-tagged) or 3% (nontagged) of the total cell populations (as determined by DAPI staining) were viable. In contrast, both gfp-tagged and wild-type cells incubated in microcosms at 5 or 30°C remained culturable, and the cell concentrations were between 108 and 109 CFU/ml during the 21-day study period (Fig. 1). Since the results showed that the responses of wild-type and tagged cells were the same, there was no apparent adverse effect on survival due to the presence of two copies of the gfp gene in the chromosome.
FIG. 1.
Culturability of P. fluorescens A506 and A506::gfp2 under starvation conditions at different incubation temperatures. Symbols: ■, A506 at 5°C; ●, A506 at 30°C; □, A506::gfp2 at 5°C; ○, A506::gfp2 at 30°C; ▴, A506 at 37.5°C; ▵, A506::gfp2 at 37.5°C. The results shown are the results of single trials, but the same trends were observed in four replicate experiments.
Fluorescence intensity.
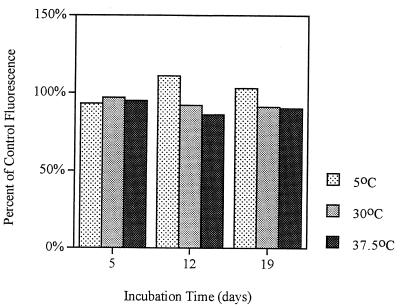
The fluorescence intensities of both starved and VBNC cells remained at levels that allowed detection of the cells by flow cytometry (Fig. 2). Throughout this study the fluorescence intensity of P. fluorescens A506::gfp2 at 37.5°C was more than 80% of the fluorescence intensity of the same strain in the log phase. The fluorescence intensity of starved cells incubated at 30°C never dropped below 90% of the fluorescence intensity of the tagged log-phase cells, while the fluorescence intensity of starved cells incubated at 5°C remained between 90 and 110% of the fluorescence intensity of the tagged log-phase cells.
FIG. 2.
GFP fluorescence intensity of starved and VBNC cells. Samples (1 ml) were removed from microcosms, fixed, and analyzed by flow cytometry. The results shown are the results of single trials, but the same trends were observed in four replicate experiments.
Fluorescence following UV light treatment.
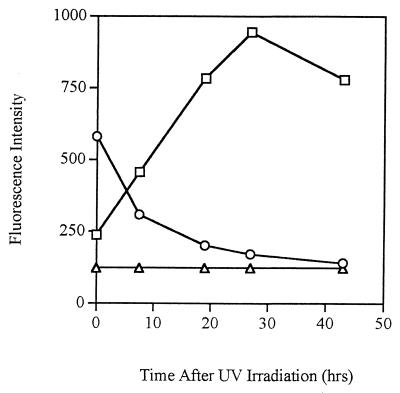
UV light was used to kill gfp-tagged log-phase cells. The fluorescence intensity of the population, as determined by spectrofluorometry, decreased when cells died (Fig. 3), while the fluorescence intensity of the supernatant increased. Within 10 h following exposure to UV light, the fluorescence intensity of the supernatant surpassed that of the cells. This indicates that rather than being degraded, intact GFP was released from the cells into the surrounding environment. Although the supernatant became increasingly fluorescent with time, incubation of nontagged P. fluorescens cells in the fluorescent supernatant did not result in the cells becoming fluorescent (results not shown).
FIG. 3.
GFP fluorescence intensity of P. fluorescens A506::gfp2 cells (○) and culture supernatant (□) following UV treatment (wavelength, 245 nm) for 5 min. The fluorescence of nontagged P. fluorescens A506 (▵) was used as the baseline fluorescence. This experiment was repeated five times, and the same trend was observed in each experiment. The fluorescence intensity of the cells shown is the average for the whole bacterial population.
Fluorescence following heat treatment.
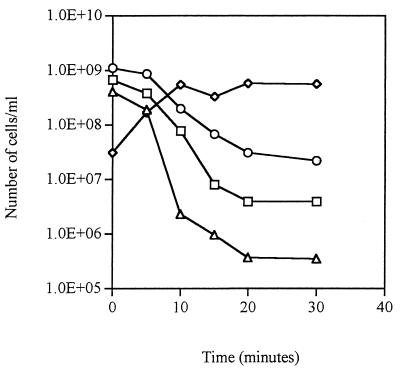
During incubation at 50°C, the number of GFP fluorescent P. fluorescens A506 cells (Fig. 4) decreased at a rate that paralleled the loss of culturability (as determined by CFU) and viability (as determined by CTC). A corresponding increase in the number of PI-stained (dead) cells was observed. Therefore, in this experiment there was a clear distinction between living cells that were still GFP fluorescent and dead cells that had lost the ability to fluoresce.
FIG. 4.
Effect of heat killing on GFP fluorescence following a 30-min exposure of P. fluorescens A506::gfp2 cells to 50°C. Symbols: ○, total number of GFP fluorescent cells; ▵, culturable cell count; □, number of viable cells as determined by using CTC; ◊, number of dead cells as determined by using PI. This experiment was performed in duplicate, and the results shown are averages.
In a separate experiment, release of GFP from the cells during heat treatment at 50°C was studied in more detail (Table 1). The number of PI-stained cells was highly negatively correlated with the number of GFP fluorescent cells (r = −0.99), indicating that most dead cells were no longer fluorescent. Also, there was a very high positive correlation between the number of PI-stained cells and the release of GFP from the cells (as determined by spectrofluorometry) (r = 0.99). During treatment a small proportion of the PI-stained cells retained GFP fluorescence, and this proportion decreased during the treatment (Table 1). The GFP fluorescence intensities (G-mean values) of both live cells and fluorescent PI-stained cells decreased during treatment, indicating that both types of cells leaked GFP (Table 1). However, the fluorescence intensity of PI-stained cells that retained GFP fluorescence was always lower than the fluorescence intensity of cells that were not stained with PI, indicating that more GFP was lost from dead or dying cells (Table 1).
TABLE 1.
Effect of heat treatment on P. fluorescens cell viability and GFP fluorescencea
| Cell treatment
|
GFP total cell concn (cells ml−1)b | G-mean for GFP total cellsc | PI-stained cell concn (cells ml−1)d | G-mean for PI+GFP cellse | % PI+GFP cellsh (PI-stained cells) | G-mean for GFP-PI cellsf | GFP increase for supernatantg | |
|---|---|---|---|---|---|---|---|---|
| Temp (0°C) | Time | |||||||
| 50 | 0 min | 3.95 × 109 | 3.01 | 1.53 × 107 | 2.36 | 31.76 | 3.02 | 0 |
| 5 min | 2.76 × 109 | 2.09 | 1.56 × 108 | 2.01 | 29.25 | 2.07 | 743 | |
| 10 min | 1.91 × 109 | 1.99 | 7.65 × 108 | 1.86 | 16.01 | 2.04 | 1,808 | |
| 15 min | 1.16 × 109 | 1.86 | 1.96 × 109 | 1.67 | 10.37 | 1.95 | 3,268 | |
| 20 min | 7.09 × 108 | 1.75 | 2.19 × 109 | 1.60 | 7.03 | 1.90 | 3,863 | |
| 30 min | 5.01 × 108 | 1.65 | 2.52 × 109 | 1.57 | 6.33 | 1.84 | 4,140 | |
| 39 | 0 h | 1.65 × 109 | 3.38 | 8.75 × 106 | 2.48 | 47.51 | 3.41 | 0 |
| 1 h | 1.76 × 109 | 2.47 | 3.90 × 107 | 2.03 | 26.56 | 2.48 | 539 | |
| 2 h | 1.76 × 109 | 2.46 | 4.70 × 107 | 2.00 | 22.75 | 2.50 | 667 | |
| 3 h | 1.57 × 109 | 2.45 | 4.75 × 107 | 2.07 | 27.54 | 2.50 | 717 | |
| 4 h | 1.81 × 109 | 2.45 | 5.14 × 107 | 2.07 | 24.59 | 2.50 | 839 | |
| 5 h | 2.00 × 109 | 2.48 | 5.85 × 107 | 2.09 | 25.36 | 2.50 | 1,256 | |
The data are averages based on duplicate samples.
Concentration of GFP fluorescent cells as determined by flow cytometry.
Geometric mean fluorescence intensity of the cell population gated by flow cytometry.
Concentration of PI-stained cells (dead cells) as determined by flow cytometry (dead cells).
PI+GFP cells, GFP fluorescent cells gated by flow cytometry that were also stained with PI (dead fluorescent cells).
GFP−PI cells, GFP-stained cells gated by flow cytometry that were not stained with PI (live fluorescent cells).
Relative increase in GFP fluorescence intensity compared to an untreated control culture, expressed as a percentage.
See Table 1.
When cells were gently heat treated at 39°C, fewer cells were stained with PI compared to cells that were heat treated at 50°C (Table 1). Also, the total number of GFP fluorescent cells did not decrease as markedly at 39°C as at 50°C. Still, there was a high correlation between the number of PI-stained cells and the release of GFP from the cells (r = 0.95). By contrast, there was significantly less negative correlation between the number of PI-stained cells and the number of GFP fluorescent cells than at 50°C, indicating that more PI-stained cells retained GFP fluorescence (r = 0.55). This was confirmed by calculating the proportion of dead cells that were also GFP fluorescent, which was higher after heat treatment at 39°C than after heat treatment at 50°C (Table 1). It is important to note that even if a portion of the dead cells retained GFP fluorescence after heat treatment at either temperature, the majority of the cells lost GFP fluorescence.
Comparison of the effects of UV light and 50°C heat treatments on cell parameters.
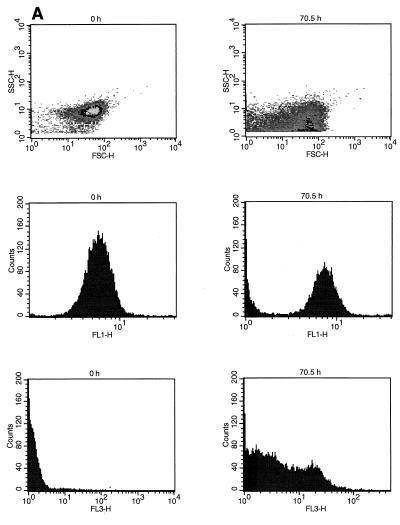
UV light treatment resulted in changes in the size and shape of the cells (Fig. 5A). At 70.5 h after UV light treatment the GFP fluorescence intensity properties (FL1) had changed, and the proportion of cells with low fluorescence intensity was larger (Fig. 5A). The PI-stained cells also exhibited great variation in fluorescence intensity (FL3) after UV light treatment (Fig. 5A). This variation might have been due to dying cells that excluded the PI stain to a greater extent than completely dead cells excluded it.
FIG. 5.
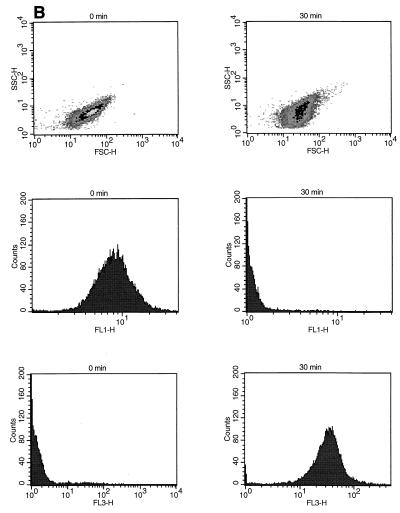
Effects of UV treatment (A) and heat treatment (B) on P. fluorescens A506::gfp2 cell population size (FSC-H), shape and granularity (SSC-H), GFP fluorescence intensity (FL1-H), and PI fluorescence intensity (FL3-H).
After heat treatment at 50°C, there was a more drastic decrease in the GFP fluorescence intensity (FL1) of the cells compared to the UV light treatment, and there was a corresponding increase in the number of strongly PI-stained cells (Fig. 5B). However, the cell size and shape did not change as dramatically as they changed following UV treatment, as demonstrated by analysis of the FSC and SSC properties of the cells (Fig. 5B).
DISCUSSION
In this study, a combination of GFP tagging and different viability assays provided detailed information about the status of P. fluorescens A506 cell populations under various conditions. One problem with the use of viability stains alone is that they are not equally effective under all conditions but depend on the growth phase of the culture, the staining time, dye uptake, etc. (12, 20). Also, a given stain depends on functioning or lack of functioning of certain cellular parameters that are considered necessary for cell viability, such as membrane integrity or membrane potential (12). As a result, depending on the stain used to determine viability, different results may be obtained. Therefore, a combination of viability stains with a stable marker (GFP) is an option for more accurate determination of cell viability.
As we have previously observed, the total counts of gfp-tagged P. fluorescens cells remain at relatively high levels during long-term starvation (23, 25). In the present study, when P. fluorescens A506::gfp2 cells were incubated at 37.5°C under starvation conditions, about 104 cells/ml remained viable in the population, although the number of culturable cells dropped below the level of detection (<0.1 CFU/ml). In contrast, increases in cell number were observed initially for microcosms incubated under starvation conditions at 5 and 30°C, before each population reached a plateau. Such growth may have been due to utilization of the residual nutrients present in the microcosms or to the reductive division normally seen in a starved population (18). It is important to note that the gfp-tagged strain and the nontagged strain behaved similarly under the different incubation conditions, indicating that the presence of this gene and/or its protein product do not alter how P. fluorescens A506 responds to these particular stresses. Similar results have been observed in other studies (13).
In order for gfp to be used as a marker gene, GFP fluorescence must remain at a level that allows tagged cells to be detected. Throughout this study, the tagged cells were easily detected by flow cytometry, spectrofluorometry, and fluorescence microscopy. Cells starved at 5 and 30°C maintained their GFP fluorescence, as measured by flow cytometry, for at least 11 months, while VBNC cells remained fluorescent for at least 6 months (data not shown). Recently, a GFP-based direct viable count method was used to detect VBNC Salmonella typhi cells in groundwater (4), which demonstrated that GFP detection-based methods are applicable to other VBNC bacteria as well.
In a VBNC culture the majority of the cells do not appear to be viable as determined by direct viability assays. When GFP is used as a marker, it is not apparent whether these cells are dead or not due to the stability of the GFP (25). After continued fluorescence of stressed cells was observed for an extended period of time, it was necessary to determine whether there was a difference between the stressed cells and dead cells with regard to their fluorescence properties (i.e., whether it was possible that fluorescent dead cells were skewing the results).
Heat treatment and UV exposure were used to kill cells without destroying the protein. The GFP is stable at temperatures up to 70°C (2), and we found that fluorescence was not bleached by the UV exposure treatments used in our experiments. Therefore, GFP released into the solution remained fluorescent after the treatments and could be quantified by spectrofluorometry.
One problem with viability studies of cell populations is that there is no clear definition of a dead cell. However, one possibility is to consider cells that have lost membrane integrity to be dead (12). This is the basis for using PI staining for quantitation of dead cells. After heat treatment most cells completely lost GFP fluorescence (Table 1). Since these cells also were stained with PI, they were considered to be dead. However, a portion of the PI-stained cells retained their GFP fluorescence, especially cells treated at 39°C. The PI-stained and GFP fluorescent cells probably represented a dying population whose cells had slightly damaged membranes, which resulted in the cells becoming stained with PI without losing all GFP fluorescence. Therefore, combining GFP tagging of cells and PI staining could provide a way not only for distinguishing between live and dead bacterial populations but also for studying damaged and dying populations.
The UV light and 50°C heat treatments affected the cell populations in different ways (Fig. 5). Heat treatment at 50°C for 30 min killed most of the cells, which resulted in a sharp distinction between live and dead cells both with respect to GFP fluorescence and PI staining (Fig. 5). UV treatment, on the other hand, apparently killed or damaged the cells without immediately destroying the cell membrane, since no obvious changes in the SSC, FSC, and fluorescence properties of the cell populations occurred immediately after the treatment (results not shown). However, when the cells were examined 70.5 h after UV treatment, the size and fluorescence properties of the population had changed, presumably due to degradation of the cell membranes. The gradient in the PI fluorescence intensity of the cell population 70.5 h after treatment (Fig. 5) is similar to the pattern which we observed previously for starving cells (Unge and Jansson, unpublished data). We predict that in starving populations (as in nature) cell death does not occur uniformly throughout an entire population but the cell membrane is damaged to different extents and in different fractions of the cell population over time. A population consisting of weakly PI-stained cells after UV treatment, therefore, most likely consists of cells that are dying and have slightly damaged membranes. Jepras et al. observed a similar buildup of Escherichia coli cells with intermediate PI fluorescence intensity after heat treatment and speculated that this was due to cells which had not been affected fully by the heat treatment and into which PI had not fully entered (12).
Interestingly, there was always a clear separation between the peak for GFP fluorescing cells and the increasing peak for nonfluorescent background (dead) cells as observed by flow cytometry. This could be explained if completely dead cells lose all GFP, whereas dying cells gradually leak GFP from increasingly permeable membranes but are still clearly fluorescent. At this point we cannot explain why we do not see an entire range of fluorescence intensity in a population, as would be the case if the cells continuously lost GFP fluorescence until they became nonfluorescent. One explanation could be that there is a threshold between dying cells and dead cells in terms of membrane integrity and once that threshold is passed, the cells rapidly leak all remaining GFP.
In conclusion, we considered PI-stained cells that had lost GFP fluorescence dead cells, PI-stained, GFP fluorescent cells damaged (probably dying) cells, and GFP fluorescent cells that were not stained with PI viable cells. We found that there was a clear relationship between cellular GFP fluorescence and viability. Therefore, GFP can reliably be used as a marker for detection of viable GMMs in environmental samples in which cells are often starved or in a VBNC state.
REFERENCES
- 1.Anderson J B, Sternberg C, Poulsen L K, Bjørn S P, Givskov M, Mølin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokman S H, Ward W W. Renaturation of Aequorea green-fluorescent protein. Biochem Biophys Res Commun. 1981;101:1372–1380. doi: 10.1016/0006-291x(81)91599-0. [DOI] [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Eushirchen G, Ward W, Prasher D. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Cho J-C, Kim S-J. Viable, but non-culturable, state of a green fluorescence protein-tagged environmental isolate of Salmonella typhi in groundwater and pond water. FEMS Microbiol Lett. 1999;170:257–264. doi: 10.1111/j.1574-6968.1999.tb13382.x. [DOI] [PubMed] [Google Scholar]
- 5.De Weger L A, Dunbar P, Mahafee W F, Lugtenberg B J J, Sayler G S. Use of bioluminescence markers to detect Pseudomonas spp. in the rhizosphere. Appl Environ Microbiol. 1991;57:3641–3644. doi: 10.1128/aem.57.12.3641-3644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flemming C A, Leung K T, Lee H, Trevors J T, Greer C W. Survival of lux-lac-marked biosurfactant-producing Pseudomonas aeruginosa UG2L in soil monitored by nonselective plating and PCR. Appl Environ Microbiol. 1994;60:1606–1616. doi: 10.1128/aem.60.5.1606-1613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gustafsson K, Jansson J K. Ecological risk assessment of the deliberate release of genetically modified microorganisms. Ambio. 1993;22:236–242. [Google Scholar]
- 8.Heim R, Prasher D C, Tsien R Y. Wavelength mutations and postranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansson J K. Tracking genetically engineered microorganisms in nature. Curr Opin Biotechnol. 1995;6:275–283. doi: 10.1016/0958-1669(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 10.Jansson J K, Prosser J. Quantification of the presence and activity of specific microorganisms in nature. Mol Biotechnol. 1997;7:103–120. doi: 10.1007/BF02761746. [DOI] [PubMed] [Google Scholar]
- 11.Jansson J K, de Bruijn F J. Biomarkers and bioreporters. In: Demain A, Davies J, editors. Manual of industrial microbiology and biotechnology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 651–665. [Google Scholar]
- 12.Jepras R I, Carter J, Pearson S C, Paul F E, Wilkinson M J. Development of a robust flow cytometric assay for determining numbers of viable bacteria. Appl Environ Microbiol. 1995;61:2696–2701. doi: 10.1128/aem.61.7.2696-2701.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leff L G, Leff A A. Use of green fluorescent protein to monitor survival of genetically engineered bacteria in aquatic environments. Appl Environ Microbiol. 1996;62:3486–3488. doi: 10.1128/aem.62.9.3486-3488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindemann J, Suslow T V. Competition between ice-nucleation active wild-type and ice nucleation-deficient deletion mutant strains of Pseudomonas syringae and P. fluorescens biovar I and biological control of frost injury on strawberry blossoms. Phytopathology. 1987;77:882–886. [Google Scholar]
- 15.Lindow S E. The use of reporter genes in the study of microbial ecology. Mol Ecol. 1995;4:555–566. [Google Scholar]
- 16.Matin A. Molecular analysis of the starvation stress in Escherichia coli. FEMS Microbiol Ecol. 1990;74:185–196. [Google Scholar]
- 17.Nebe-Von Caron G, Badley R A. Viability assessment of bacteria in mixed populations using flow cytometry. J Microsc. 1995;179:55–65. [Google Scholar]
- 18.Oliver J D. Formation of viable but nonculturable cells. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 239–272. [Google Scholar]
- 19.Oliver J D, McDougald D, Barrett T, Glover L A, Prosser J I. Effect of temperature and plasmid carriage on nonculturability in organisms targeted for release. FEMS Microbiol Ecol. 1995;17:229–238. [Google Scholar]
- 20.Porter J, Edwards C, Pickup R W. Rapid assessment of physiological status in Escherichia coli using fluorescent probes. J Appl Bacteriol. 1995;79:399–408. doi: 10.1111/j.1365-2672.1995.tb03154.x. [DOI] [PubMed] [Google Scholar]
- 21.Prasher D C, Eckenrode V E, Ward W W, Prendergast F G, Cormier M J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez G C, Phipps C, Ishiguro K, Ridgway H F. Use of fluorescent redox probe for direct visualization of actively respiring bacteria. Appl Environ Microbiol. 1992;58:1801–1808. doi: 10.1128/aem.58.6.1801-1808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tombolini R, Unge A, Davey M E, Bruijn F J, Jansson J K. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol Ecol. 1997;22:17–28. [Google Scholar]
- 24.Unge A, Tombolini R, Möller A, Jansson J K. Optimization of GFP as a marker for detection of bacteria in environmental samples. In: Hastings J W, Kricka L J, Stanley P E, editors. Bioluminescence and chemiluminescence: molecular reporting with photons. Sussex, United Kingdom: John Wiley & Sons; 1997. pp. 391–394. [Google Scholar]
- 25.Unge A, Tombolini R, Mølbak L, Jansson J K. Simultaneous monitoring of bacterial number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl Environ Microbiol. 1999;65:813–821. doi: 10.1128/aem.65.2.813-821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf P W, Oliver J D. Temperature effects on the viable but nonculturable state of Vibrio vulnificus. FEMS Microbiol Ecol. 1992;101:33–39. [Google Scholar]