Summary
Recent studies showed that SARS-CoV-2 can infect adult human pancreas and trigger pancreatic damage. Here, using human fetal pancreas samples and 3D differentiation of human pluripotent cells into pancreatic endocrine cells, we determined that SARS-CoV-2 receptors ACE2, TMPRSS2, and NRP1 are expressed in precursors of insulin-producing pancreatic β-cells, rendering them permissive to SARS-CoV-2 infection. We also show that SARS-CoV-2 enters and undergoes efficient replication in human multipotent pancreatic and endocrine progenitors in vitro. Moreover, we investigated mechanisms by which SARS-CoV-2 enters pancreatic cells, and found that ACE2 mediates the entry, while NRP1 and TMPRSS2 do not. Surprisingly, we found that in pancreatic progenitors, SARS-CoV-2 enters cells via cathepsin-dependent endocytosis, which is a different route than in respiratory tract. Therefore, pancreatic spheroids might serve as a model to study candidate drugs for endocytosis-mediated viral entry inhibition and to investigate whether SARS-CoV-2 infection may affect pancreas development, possibly causing lifelong health consequences.
Subject areas: Model organism, Virology
Graphical abstract
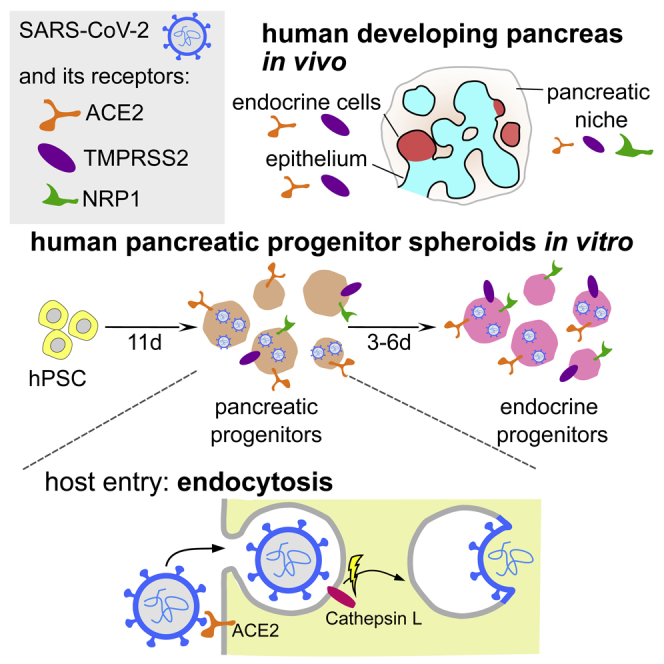
Highlights
-
•
SARS-CoV-2 receptors are present in human developing pancreas in vivo and in vitro
-
•
SARS-CoV-2 infects multipotent and endocrine pancreatic progenitors in vitro
-
•
SARS-CoV-2 enters the progenitors via alternate cathepsin-mediated endocytosis
Model organism; Virology
Introduction
COVID-19 pandemic is caused by SARS-CoV-2, a recently identified betacoronavirus closely related to the SARS-CoV that emerged in 2002 (Zhang and Holmes, 2020; Wu et al., 2020). The two human SARS-like coronaviruses share several characteristics, including receptor usage and cell tropism (Li et al., 2003; Milewska et al., 2020a; Wrapp et al., 2020), and the respiratory tract being the main gate of viral entry (Ye et al., 2020). Although life-threatening pneumonia is the most obvious complication of SARS-CoV-2 infection (Zhu et al., 2020), COVID-19 is a systemic disease leading to multi-organ damage and still has unknown health consequences for recovered patients (Synowiec et al., 2021). The systems most affected by SARS-CoV-2 include cardiovascular (Puelles et al., 2020; Tavazzi et al., 2020; Varga et al., 2020), gastrointestinal (Wang et al., 2020; Xiao et al., 2020; Zhang et al., 2020), renal (Puelles et al., 2020; Su et al., 2020), neurological (Di Carlo et al., 2020; Helms et al., 2020), and blood clotting (Connors and Levy, 2020).
Diabetes is a key risk factor for developing severe COVID-19 and disease-related mortality (Barron et al., 2020; CDC COVID-19 Response Team, 2020; Cummings et al., 2020; Onder et al., 2020; Lim et al., 2021). Interestingly, multiple studies pointed to the reverse association, namely, COVID-19 may impair the pancreas function (Croft et al., 2020; Kumaran et al., 2020; Szatmary et al., 2020; Wang et al., 2020; Li et al., 2020a; Ebekozien et al., 2020; Akarsu et al., 2022) and trigger diabetes (Chee et al., 2020; Kamrath et al., 2020; Caruso et al., 2020; Rubino et al., 2020; Hollstein et al., 2020, in children - Unsworth et al., 2020). Many recovered patients suffer from post-COVID-19 syndrome (Nalbandian et al., 2021), which also includes a several fold increase in diabetes prevalence compared to the general population (Ayoubkhani et al., 2021). Interestingly, the closest relative to SARS-CoV-2, SARS-CoV was also found in pancreatic tissue and associated with a higher risk of diabetes development (Lang et al., 2003; Yang et al., 2010).
Diabetes develops when glucose homeostasis is disrupted, either due to loss or dysfunction of pancreatic β-cells that produce insulin, or due to diminished body sensitivity to insulin. There are multiple scenarios by which SARS-CoV-2 infection might induce diabetes (Accili, 2021), and the most straightforward is direct infection and, consequently, loss of β-cells. Indeed, multiple studies showed that SARS-CoV-2 infects multiple exocrine and endocrine pancreatic cells, including β-cells, in postmortem pancreas of patients with COVID-19 (Coate et al., 2020; Fignani et al., 2020; Kusmartseva et al., 2020; Samanta et al., 2020; Müller et al., 2021; Shaharuddin et al., 2021; Steenblock et al., 2021), human β-cells derived from pluripotent stem cells (hPSCs) (Yang et al., 2020), and primary ex vivo islets, leading to a decreased β-cell function in vitro (Müller et al., 2021). However, these studies point to high variability regarding infection rate and cell type affected between patients, as well as to variable expression of SARS-CoV-2 entry receptors without straightforward correlation with infection susceptibility. Moreover, how the virus presence affects pancreatic cells is not clear. Cell death was not commonly reported in affected β-cells. In one study, β-cells infection was rare and apoptosis was not observed; however, the authors found that SARS-CoV-2 nucleocapsids are present more frequently in NKX6-1+ but INS- cells (Müller et al., 2021). Since the only NKX6-1+ cells in the adult pancreas are β-cells, this would suggest dedifferentiation of β-cells to immature, fetal-like, and nonfunctional states upon SARS-CoV-2 infection. Another work has also found that some infected β-cells in patients lose their identity and transdifferentiate via eIF2 pathway (Tang et al., 2021). Importantly, β-cells dedifferentiation can lead to diabetes (Talchai et al., 2012). Therefore, biological short- and long-term consequences of the SARS-CoV-2 presence in human islets cells remain still not well understood.
In utero exposure to various environmental factors, including viral infection, might predispose to obesity, type 2 or type 1 diabetes in later life (Craig et al., 2019; Shulman et al., 2014). These environmental factors probably interfere with proper pancreatic development. Recent studies revealed SARS-CoV-2 receptors expression in pre-implantation embryos (Weatherbee et al., 2020), as well as in placental and fetal tissues later in development (Li et al., 2020b; Cao et al., 2020), rendering the possibility of fetal infection. Indeed, several reports have indicated in utero mother-to-fetus transmission of the SARS-CoV-2 (Fenizia et al., 2020; Hosier et al., 2020; Vivanti et al., 2020; Shende et al., 2021). High loads of the virus in placental tissue could have an immense impact on development of the fetal pancreas, its maturation, and future physiological capacity to produce insulin and glucagon. Therefore, it is critical to establish when during development cells are vulnerable to SARS-CoV-2 infection and whether fetal or developing pancreatic cells are susceptible to SARS-CoV-2 is yet unknown.
Coronaviruses can use two routes to enter target cells: they either fuse their lipid envelope with the cell membrane on the cell surface or are internalized by endocytosis (Hoffmann et al., 2020a; Milewska et al., 2018; Owczarek et al., 2018). Both routes require angiotensin converting enzyme 2 (ACE2) in the initial steps, while the next steps depend on the availability of proteases, which are responsible for the activation of the S protein. While in the respiratory tract the SARS-CoV-2 seems to exclusively utilize the cell surface membrane fusion route, the question remains whether in other tissues the mechanism is similar. This is important as ACE2 plays a critical role in regulating several vital parameters, including blood pressure, and its targeting could come with serious risks. On the other hand, targeting host proteases particularly responsible for the entry of SARS-CoV-2 into a given tissue could have fewer side effects.
Here, we investigate the susceptibility of developing human pancreatic cells to SARS-CoV-2 infection. Furthermore, we identify mechanisms through which SARS-CoV-2 enters pancreatic progenitors. Our data provide insights into SARS-CoV-2 cell infectivity of pancreatic endocrine cell progenitors and define a potential target for pancreatic cell-type-specific antiviral intervention.
Results
SARS-CoV-2 receptors are expressed in the developing human pancreas and its in vitro counterparts
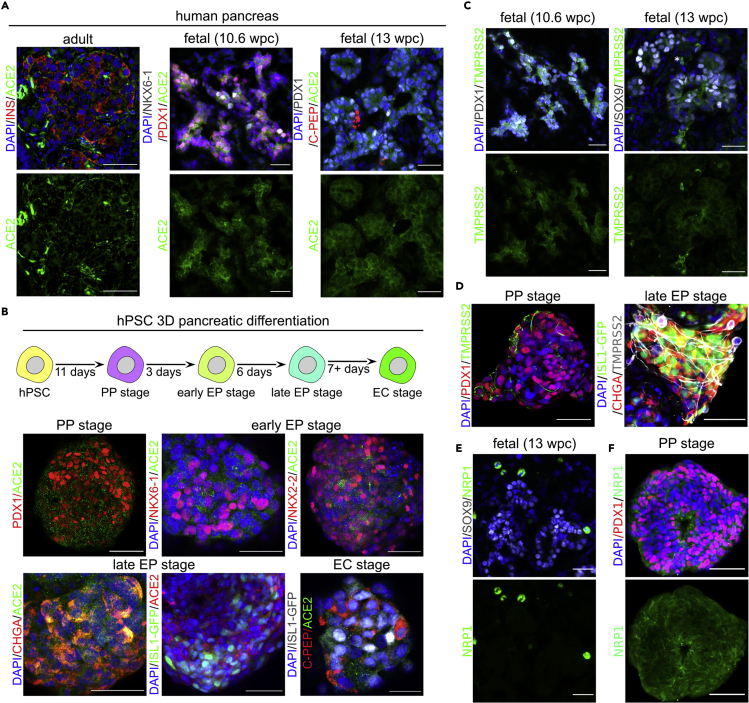
ACE2 serves as the SARS-CoV-2 receptor and is necessary to render cells permissive to infection (Hoffmann et al., 2020a). To test whether SARS-CoV-2 can infect the developing human pancreas, we first analyzed ACE2 expression in the developing organ compared to the adult pancreas. Using previously validated anti-ACE2 antibodies (Fignani et al., 2020; Kusmartseva et al., 2020; Coate et al., 2020; Müller et al., 2021), we observed ACE2 expression in the cytoplasm of adult pancreatic islet cells, which is consistent with previous reports (Fignani et al., 2020; Müller et al., 2021). Immunostaining of the human pancreas at 10.6 and 13 weeks post conception (wpc) revealed broad expression of the ACE2 protein in PDX1+ and NKX6.1+ pancreatic progenitors (Figure 1A). Analysis of single-cell RNA-sequencing (scRNA-seq) datasets of the human fetal pancreas at week 16 and 17 (Cao et al., 2020) showed ACE2 mRNA presence in a subset of pancreatic cells, including endocrine cells (Figure S1). Together, these data show that ACE2 is present in developing human pancreas.
Figure 1.
SARS-CoV-2 receptors are expressed in the human fetal pancreas and in pancreatic progenitors derived from hPSCs
(A) Left: A representative confocal image of the human pancreas of healthy adult donors stained with antibodies against ACE2 (green) and insulin (red). Middle: ACE2 (green) expression in the human fetal pancreas at 10.6 wpc. PDX1+ (red) and NKX6-1+ (gray) mark pancreatic progenitors. Right: ACE2 (green) expression in the human fetal pancreas at 13 wpc. PDX1+ (gray) marks pancreatic progenitors and endocrine cells, and C-PEP (red) marks β-cells. Scale bars = 50 μm (adult tissue), 25 μm (fetal tissue). DAPI (blue) was used as a counterstain.
(B) Top: Scheme of 3D differentiation of hPSCs toward pancreatic endocrine cells. Bottom: ACE2 (green or red) is expressed in hPSC-derived 3D spheroids at multiple differentiation stages, i.e. pancreatic progenitors (PP; marked by PDX1 in red), early endocrine progenitors (early EP; marked by NKX6-1 nad NKX2-2 in red), late EPs (marked by CHGA in red or ISL1-GFP in green), and endocrine cells (EC, marked by C-PEP in red and ISL1-GFP in gray). Scale bars = 50 μm or 25 μm (EC). DAPI (blue) marks the nuclei. N = 3 independent differentiation experiments.
(C) TMPRSS2 (green) is expressed in the human fetal pancreas at 10.6 wpc (left) and 13 wpc (right). PDX1+ (gray) and SOX9 (gray) mark pancreatic progenitors. Scale bars = 25 μm. DAPI (blue) marks the nuclei.
(D) TMPRSS2 (green or gray) is expressed in hPSC-derived PP (left) and late EP (right) stage spheroids. PP and EP cells are marked by PDX1 (red) or CHGA (red) and ISL-GFP (green), respectively. Scale bar = 50 μm. DAPI (blue) marks the nuclei. N = 3 independent differentiation experiments.
(E) Lack of NRP1 (green) expression in the human fetal pancreatic epithelium at 13 wpc, marked by SOX9 (gray). NRP1 is strongly expressed in a subset of pancreatic niche cells. Scale bars = 25 μm. DAPI (blue) marks the nuclei.
(F) NRP1 expression (green) in hPSC-derived PP stage spheroids, PDX1 (red) marks PPs. NRP1 is strongly expressed in a subset of pancreatic niche cells. Scale bars = 50 μm. DAPI (blue) marks the nuclei. N = 3 independent differentiation experiments. See also Figures S1 and S2.
To test if SARS infects the developing pancreas, we utilized the in vitro model of human development, the pancreatic differentiation of hPSCs. We asked if hPSC-derived pancreatic cells also express ACE2, making them vulnerable to SARS-CoV-2 infection. Several protocols to generate human β-cells from hPSCs have been developed (Nair et al., 2019; Nostro et al., 2015; Pagliuca et al., 2014; Rezania et al., 2014), in which hPSCs are progressively coaxed toward pancreatic endocrine cell fate through definitive endoderm, gut tube endoderm, pancreatic progenitor (PP), and endocrine progenitor (EP) stages to become β-cells (EC). We applied a 3D sphere differentiation protocol to drive hPSCs through consecutive stages of endocrine pancreas development (Figure 1B, scheme) (see STAR Methods). Immunofluorescent staining for stage-specific markers was used to confirm the presence of pancreatic progenitors and endocrine cells (Figure S2A). Endocrine cells at the final stage of differentiation have been shown to express ACE2 and can be infected by SARS-CoV-2 (Yang et al., 2020), which we confirmed herein for ISL+ (expressed in all endocrine cells) and C-PEP+ (a by-product of insulin processing) cells (Figure 1B, bottom right). However, ACE2 expression has not been evaluated at any progenitor stages. Analysis of the scRNA-seq datasets of hPSC differentiated toward β-cells using a different 3D protocol (Sharon et al., 2019; Weng et al., 2020) suggests low ACE2 mRNA expression, detectable in a small subset of progenitors (Figures S2B and S2C). To further address this gap, we performed immunostaining and observed the ACE2 protein in the cytoplasm and at the cell membrane of PDX1+ PP cells (Figure 1B, top left), early NKX6-1+ and NKX2-2+ EP cells (Figure 1B, top right), and late CHGA+ and ISL1-GFP+ EP cells (Figure 1B, bottom left). Therefore, the SARS-CoV-2 receptor ACE2 is expressed in different human pancreatic progenitors during in vitro differentiation.
SARS-CoV-2 also uses proteins other than ACE2 to enter cells. Transmembrane protease serine 2 (TMPRSS2) activates the viral attachment spike protein (S) bound by ACE2 that allows viral entry via membrane fusion (Hoffmann et al., 2020a). Recently, another cell surface molecule, neuropilin-1 (NRP1) has been reported to enhance the virus entry into some host cells after the S protein is proteolytically cleaved by furin (Hoffmann et al., 2020b), revealing a short amino acid sequence of the S protein that can be recognized by NRP1 (Cantuti-Castelvetri et al., 2020; Daly et al., 2020; Synowiec et al., 2021). The scRNA-seq of 16–17 wpc human fetal pancreas shows enrichment of TMPRSS2 in the exocrine pancreas, while NRP1 in ductal, endothelial, and stromal cells (Figure S1). Using immunofluorescence, we assayed the expression of TMPRSS2 and NRP1 in the human fetal pancreas and hPSC-derived pancreatic progenitors. We detected TMPRSS2 protein in 10.6 and 13 wpc human fetal pancreas (Figure 1C), specifically in SOX9+ epithelial pancreatic progenitors. In contrast, NRP1 protein was very rarely detectable in the fetal pancreas and did not colocalize with SOX9 expression (Figure 1E). The scRNA-seq data for in vitro differentiating hPSCs (Figure S2B and S2C) suggest TMPRSS2 and NRP1 are expressed in different pancreatic progenitors at higher levels than ACE2. Indeed, we found strong immunofluorescent signals when detecting these proteins in PP and EP stage hPSC-derived spheroids (Figures 1D and 1F). In conclusion, SARS-CoV-2 receptors are expressed in the developing human pancreas, which might render it permissive to the viral infection. Furthermore, these receptors are also expressed in a 3D in vitro model of human pancreas development, suggesting that it is feasible for research on SARS-CoV-2 infection.
SARS-CoV-2 infects developing human pancreatic cells in vitro
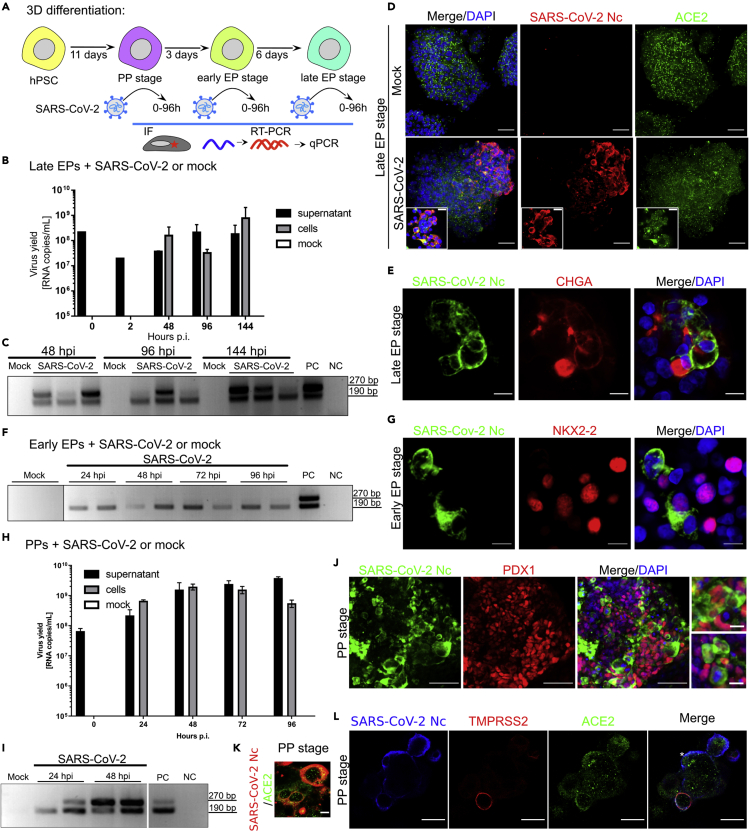
As we found widespread expression of SARS-CoV-2 receptors in hPSC-derived pancreatic progenitors, we next tested for SARS-CoV-2 infection along the developmental trajectory (Figure 2A). First, we infected pancreatic spheroids at the end of EP with SARS-CoV-2 and cultured them for 144 h. At 0, 2, 48, 96, and 144 h post inoculation, we evaluated the viral yield by RT-qPCR amplification of the N gene. We detected an increase in viral RNA copy number at all time points tested in the culture supernatant and after 48, 96, and 144 h in infected pancreatic spheroids (Figure 2B). Furthermore, using RT-PCR, we verified the presence of SARS-CoV-2 subgenomic mRNA (sg mRNA) in infected pancreatic spheroids at these time points (Figure 2C). Next, we used an antibody specific to the viral Nc protein (Figure S3A) and detected SARS-CoV-2 colocalization with ACE2 in pancreatic spheroids 48 h after infection (Figure 2D). At 72 h post inoculation, immunofluorescent analyses confirmed the SARS-CoV-2 presence in CHGA+ endocrine cells (Figure 2E). We often found the infected cells adjacent to CHGA+ and ISL1+ cells (Figure S3B), suggesting these cells might be less advanced, developing EPs. Thus, we infected pancreatic spheroids at the early EP stage and found SARS-CoV-2 sg mRNAs by RT-PCR (Figure 2F) as well as nucleocapsids in NKX2-2+ cells by immunofluorescence (Figure 2G), confirming that EPs can be infected by SARS-CoV-2.
Figure 2.
SARS-CoV-2 infects developing hPSC-derived pancreatic cells
(A) Experimental outline to test developing human pancreatic progenitors permissiveness for SARS-CoV-2 infection.
(B) RT-qPCR analysis of SARS-CoV-2 replication in late endocrine progenitor (EP) spheroids at 0–144 h post inoculation. Data are presented as the mean of viral RNA copies per milliliter ± SEM. N = 3 independent experiments.
(C) SARS-CoV-2 sg mRNAs in late EP spheroids derived from hPSCs. The presence of the N sg mRNAs at different time points post inoculation in the pancreatic endocrine cells infected with SARS-CoV-2 or mock controls was evaluated using RT-PCR. NC, negative control; PC, positive control. Two bands correspond to the products of the 1st and 2nd round of the nested PCR.
(D) SARS-CoV-2 nucleocapsid (Nc, in red) and ACE2 (green) in hPSC-derived late EP spheroids infected with SARS-CoV-2 or mock controls. DAPI, in blue, marks the nuclei. Scale bar = 50 μm (insets 25 μm). N = 3 independent experiments.
(E) SARS-CoV-2 (green) infects CHGA+ (red) human endocrine progenitors. DAPI (blue) marks the nuclei. Scale bar = 10 μm, Nc = nucleocapsid. N = 3 independent experiments.
(F) RT-PCR analysis of subgenomic mRNAs in hPSC-derived endocrine progenitors infected with SARS-CoV-2 at 24, 48, 72, and 96 h post inoculation or mock controls. NC, negative control; PC, positive control.
(G) NKX2.2+ (red) early endocrine progenitors are permissive to SARS-CoV-2 infection (green). DAPI (blue) marks the nuclei. Scale bar = 10 μm, Nc = nucleocapsid. N = 3 independent experiments.
(H) RT-qPCR of SARS-CoV-2 replication in human pancreatic progenitors at 0, 24, and 48 h post inoculation. Data are presented as the mean of viral RNA copies per milliliter ± SEM. N = 3 independent experiments.
(I) RT-PCR analysis of subgenomic mRNAs in pancreatic progenitors infected with SARS-CoV-2 or mock controls. NC, negative control; PC, positive control.
(J) SARS-CoV-2 (green) effectively infects hPSC-derived PDX1+ (red) pancreatic progenitors. DAPI, in blue, marks the nuclei. Scale bar = 50 μm, insets 10 μm. N = 6 independent experiments.
(K) ACE2 (green) colocalizes with SARS-CoV-2 Nc (red) in infected pancreatic progenitor spheroids. Scale bar = 10 μm. N = 3 independent experiments.
(L) SARS-CoV-2 Nc (blue) can infect pancreatic progenitors without TMPRSS2 expression. Anti-TMPRSS2 antibody is shown in red, while anti-ACE2 antibody is shown in green. Scale bar = 25 μm. N = 3 independent experiments. See also Figure S3.
Next, we stepped back in the progression of hPSC pancreatic differentiation and inoculated spheroids at the PP stage to investigate the SARS-CoV-2 presence in these progenitor cells by RT-PCR, RT-qPCR, and immunofluorescence staining (Figures 2H–2J). RT-qPCR showed high virus replication in PPs at 24 to 96 h post inoculation (Figure 2H). Concomitantly, RT-PCR revealed the presence of sg mRNA in SARS-CoV-2-infected PPs, but not in mock-infected controls (Figure 2I). Immunofluorescent staining with antibodies against viral Nc protein detected widespread infection with SARS-CoV-2 in PDX1+ PPs (Figure 2J). Cells infected at the PP stage expressed ACE2 (Figure 2K) and frequently co-expressed TMPRSS2 (Figure 2I). Yet, we observed ACE2+ infected cells without detectable TMPRSS2 expression (Figure 2I, asterisk). This would suggest that in pancreatic progenitors SARS-CoV-2 might use a cell entry pathway other than TMPRSS2-mediated membrane fusion.
Together, we determined that the developing pancreatic and endocrine progenitors are infected in vitro by SARS-CoV-2.
Impact of SARS-CoV-2 infection on pancreatic progenitors
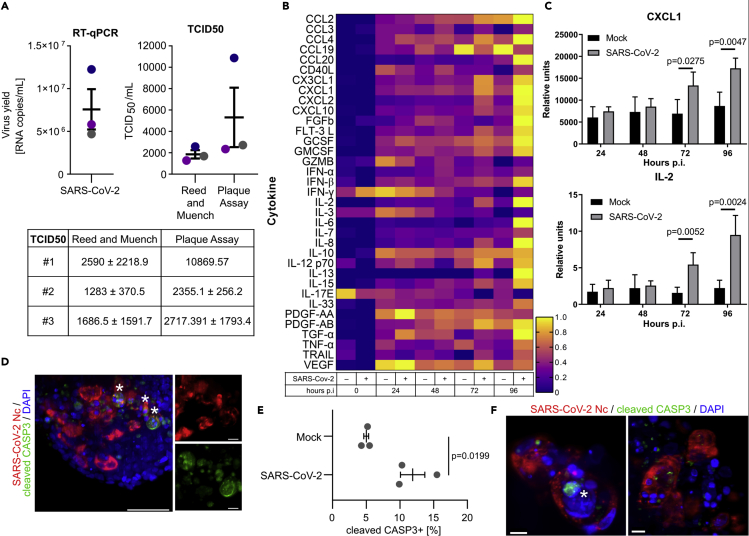
We then investigated consequences of SARS-CoV-2 infection in pancreatic progenitors. First, we confirmed that PDX1+ pancreatic progenitors are able to produce progeny virions by re-infecting Vero cells with titrated supernatant collected from spheroids 72 h after infection (Figure 3A). To assess innate antiviral response in PP spheroids, we profiled a cohort of cytokines and inflammatory factors that are known to be important for innate or adaptive immune responses in the culture medium 0, 24, 48, 72, and 96 h after SARS-CoV-2 infection. Correlating with the abundant SARS-CoV-2 presence in pancreatic spheroids, the levels of several cytokines, including IL2, CXCL1, or IFN-α were upregulated at 96 h after SARS-CoV-2 infection compared to mock controls (Figures 3B and 3C). Next, we assessed whether SARS-CoV-2 infection results in cell death of pancreatic progenitors. We have assessed apoptosis after 7 days post-infection by immunostaining for cleaved caspase 3 and we found a 2.4-fold increase in apoptosis in infected versus non-infected cells (Figures 3D and 3E). We have also assessed necroptosis, which was shown to be present in islets of patients with COVID-19 (Steenblock et al., 2021). For this, we stained spheroids 7 days post infection against phospho-MLKL. Yet, we very rarely observed positive staining of SARS-CoV-2+ cells. In our experiments, we also observed rare multinucleated clusters with strong continuous SARS-CoV-2 nucleocapsid signal, suggesting syncytia formation (Figure 3F). These results together suggest that only some infected pancreatic progenitors undergo cell death, via apoptosis.
Figure 3.
Antiviral response in hPSC-derived pancreatic progenitors after SARS-CoV-2 infection
(A) SARS-CoV-2 virions are produced in pancreatic progenitors. (Left) Replication of virus was evaluated at 72 h post inoculation using RT-qPCR. Data are presented as the mean of viral RNA copies per milliliter ± SEM. (Right) Calculation of TCID50/mL in collected samples by Reed and Muench and Plaque Assay methods. Colors of the dots indicate independent experiments (N = 3) corresponding to the RT-qPCR graph. Titer is presented as mean TCID50/mL ± SEM. (Bottom) Table with TCID50 ± SD values from each replicate.
(B) Heatmap of normalized cytokine and chemokine levels in SARS-CoV-2-infected hPSC-derived pancreatic spheroids compared to the mock control. Protein levels were measured 24–96 h post infection using Luminex assay. See also Table S1.
(C) CXCL1 and IL-2 protein levels in the cell culture medium of SARS-CoV-2-infected or mock control human pancreatic spheroids at 24–96 h post infection, measured by Luminex. Data are presented as mean ± SEM. The p values were calculated by unpaired two-tailed Student’s t test. N = 4 independent experiments.
(D) Apoptotic cells within SARS-CoV-2-infected (red) pancreatic progenitors. Apoptosis was assessed by cleaved Caspase 3 staining (green) 7 days post infection. DAPI marks nuclei. Maximum intensity projection of image captured by the Zeiss Lightsheet 7 microscope is shown. SARS-CoV-2 Nc+, cleaved CASP3+ double-positive cells are marked by asterisks, and magnified on the right. Scale bar = 50 μM (left) and 10 μM (right panels). N = 3 independent experiments.
(E) Quantification of cleaved Caspase 3+ cells within SARS-CoV-2 Nc+ and non-infected pancreatic progenitors, performed on spheroid images captured by Zeiss Lightsheet 7. Data are shown as mean % cleaved Caspase 3+ cells ±SEM, and unpaired two-tailed t-test was used to count the p-value. N = 3 independent experiments.
(F) Example of multinucleated clusters with strong continuous SARS-CoV-2 nucleocapsid (red) signal, suggesting syncytia formation. Apoptotic cells were observed within these clusters (cleaved CASP3, green, marked by the asterisk). DAPI stains nuclei. Scale bar = 10 μM.
SARS-CoV-2 utilizes ACE2 as a receptor and requires endocytic internalization to enter hPSC-derived pancreatic progenitors
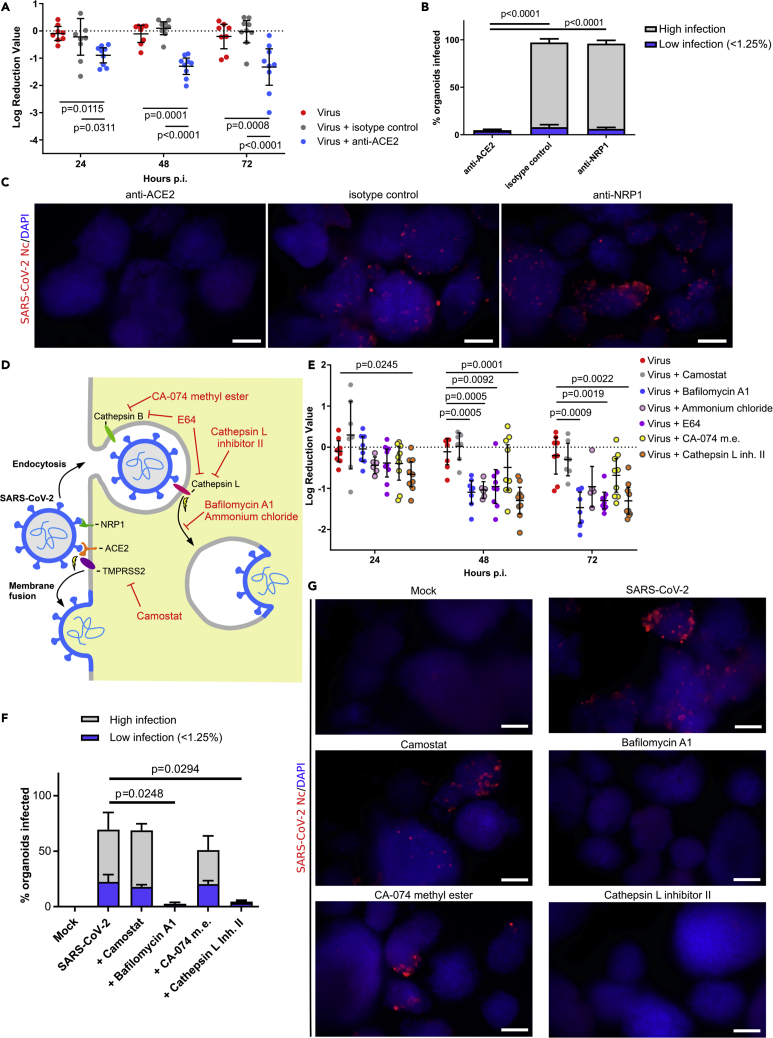
Given the efficient infection in PP spheroids, we investigated the route of SARS-CoV-2 entry into these cells. We first used antibodies against ACE2 and NRP1 to hinder the interaction between the virus and its potential receptor. Pancreatic spheroids were infected with the SARS-CoV-2 virus in the presence or absence of anti-ACE2, anti-NRP1, or IgG isotype antibodies. The virus replication was assessed using RT-qPCR and immunodetection of the viral Nc protein in infected cells. In SARS-CoV-2-infected spheroids, we observed a 10-fold reduction in virus replication in the presence of anti-ACE2 antibodies, as measured by RT-qPCR (Figure 4A), and almost complete depletion of infected cells, as measured by immunofluorescence staining (Figures 4B and 4C). Despite NRP1 expression in pancreatic cells, treatment of pancreatic spheroids with anti-NRP1 antibodies did not significantly affect virus entry (Figures 4B and 4C). We therefore concluded that NRP1 is not essential for SARS-CoV-2 entry to hPSC-derived pancreatic spheroids.
Figure 4.
SARS-CoV-2 infects human pluripotent stem cell-derived pancreatic progenitors by endocytosis
(A) RT-qPCR analysis of SARS-CoV-2 infection blocking experiments with anti-receptor antibodies treatment in pancreatic progenitors at 24–72 h post infection. Treatment with anti-ACE2 antibody prior to SARS-CoV-2 exposure reduces infection by a log10 factor compared to virus-only or IgG isotype control. Whiskers denote 95% CI and mean value, p values were counted using Tukey multiple comparison test. N = 9 independent experiments.
(B) Blocking ACE2, but not NRP1, by respective antibodies decreases SARS-CoV-2 infection in pancreatic progenitors, as assessed by immunofluorescence. SARS-CoV-2 Nc positive spheroids with “low” infection rate (<1.25% of cells infected at the organoid surface) and “high” infection rate (≥ 1.25% infected cells, mean 7.05%, 95% CI: 3.83–10.26 95% CI) were quantified, as described in STAR Methods, and shown as mean percentages of total spheroids ± SEM. p values were counted using the Tukey multiple comparison test. N = 4 independent experiments.
(C) Representative images for the quantifications of Figure 4B, with SARS-CoV-2 Nc stained in red and DAPI-stained the nuclei in blue. Scale bars = 200 μm.
(D) Overview of SARS-CoV-2 entry strategies. SARS-CoV-2 can enter the cells using ACE2 or NRP1 cell surface receptors binding with the S spike protein. ACE2-bound virus can enter the cell via TMPRSS2-mediated cleavage, which induces cell membrane fusion, or alternatively by endocytosis, where release of the virus into cytoplasm is mediated by cathepsins. The small molecule inhibitors used in the study are denoted in red.
(E) RT-qPCR analysis of SARS-CoV-2 entry blocking experiments with inhibitor treatment in pancreatic progenitors 24–72 h post infection. Treatment with bafilomycin A1, E64, and cathepsin L reduces infection by a log10 factor compared to the virus-only control. Whiskers denote 95% CI and mean value, p values were counted using the Dunnett multiple comparison test. N = 9 independent experiments.
(F) Inhibition of endocytosis by the respective inhibitors (bafilomycin A1 and cathepsin L) decreases the infection rate of SARS-CoV-2 in pancreatic progenitors as assessed by immunofluorescence. SARS-CoV-2 Nc positive spheroids with “low” infection rate and “high” infection rate (as in Figure 4B) were quantified and shown as mean percentages of total spheroids ± SEM. p-values were counted using the Dunnett multiple comparison test and significant values are shown. N = 4 independent experiments.
(G) Representative images for quantifications in Figure 4F, with SARS-CoV-2 Nc stained in red and DAPI-stained nuclei in blue. Scale bars = 200 μm.
The second factor predetermining the susceptibility of the cell to infection is the presence of proteolytic enzymes, which cleave and activate the S protein and enable virus entry to the target cells (Figure 4D). To map the next steps of SARS-CoV-2 entry route into the pancreatic cells, we first determined whether the infection depends on TMPRSS2 or cathepsins. TMPRSS2 is a serine protease present on the cell surface and the main protease responsible for the activation of the S protein in the lungs, while cathepsins are proteases active only in the acidified endocytic compartment.
Both types of proteases were targeted with well-described inhibitors of TMPRSS2 (camostat mesylate) or cathepsins (E64, cathepsin L Inhibitor II, and CA-074 methyl ester) and viral replication was estimated with RT-qPCR. The results obtained clearly show that three endocytosis inhibitors affected SARS-CoV-2 replication in pancreatic spheroids by 10-fold for the broad cysteine protease inhibitor E64 and the cathepsin L inhibitor, and by 5-fold for the cathepsin B inhibitor (Figure 4E). In contrast, the TMPRSS2 inhibitor, camostat, did not affect viral infection. These data suggest that SARS-CoV-2 enters pancreatic cells using a slower endocytic gateway. To further ensure that the entry of SARS-CoV-2 requires endocytosis in the pancreatic tissue, two inhibitors of endosomal acidification were used. Treatment of cells with bafilomycin A (an inhibitor of the proton pump in endosomes) and ammonium chloride (buffering acidification of endosomes) inhibited virus replication by 10-fold (Figure 4E). Furthermore, we stained spheroids treated with protease inhibitors and SARS-CoV-2 infected at 72 h post infection, for SARS-CoV-2 Nc protein (Figures 4F and 4G). Coherently with RT-qPCR results, bafilomycin A1, and cathepsin L inhibitor almost completely abolished the number of SARS-CoV-2 Nc+ spheroids, while camostat had no effect. Thus, we further confirmed the endosomal pathway as the main route of infection in pancreatic progenitor spheroids.
Discussion
The longitudinal health consequences of systemic SARS-CoV-2 infection in adults and children remain unclear. For example, SARS-CoV-2 infection can cause hyperglycemia in people without preexisting diabetes, and hyperglycemia was found to persist for 3 years after recovery from SARS-CoV-1 during the 2002 epidemics, suggesting long-term damage to pancreatic β-cells (Yang et al., 2010). Environmental stress and viral infection during the development and maturation of pancreas can predispose to diabetes (Allen et al., 2018; Perng et al., 2019). In this study, we demonstrate that SARS-CoV-2 receptors are expressed in human fetal tissues and that the virus effectively infects and replicates in hPSC-derived pancreatic progenitors. These progenitors are progeny for endocrine cells that were shown to be susceptible to SARS-CoV-2 infection in previous in vitro (Yang et al., 2020) or ex vivo studies (Müller et al., 2021). We did not observe a broad cell death in infected pancreatic progenitors, which might suggest that these cells are somehow protected from cell death due to their plasticity. Indeed, other studies suggest that upon SARS-CoV-2 infection β-cells might dedifferentiate into a pancreatic progenitor-like stage without induction of cell death (Steenblock et al., 2021; Tang et al., 2021) but likely affecting function of the organ.
To fully understand the infection and its potential consequences, it is crucial to delineate the entry pathways of SARS-CoV-2. To investigate a route of entry into pancreatic cells, ACE2 and NRP1 antibodies were used to hinder the interaction between the virus and its potential receptor. The ACE2 receptor was consistently shown to be present in pancreatic pericytes and ducts (Fignani et al., 2020; Coate et al., 2020; Kusmartseva et al., 2020), while its presence in mature β-cells seems to be more variable and controversial (Fignani et al., 2020; Coate et al., 2020; Kusmartseva et al., 2020; Müller et al., 2021), which we have also observed. Yet, we found that ACE2 is expressed in PPs and EPs and that it colocalizes with the SARS-CoV-2 nucleocapsid in infected cells. NRP1, which serves as an alternative SARS-CoV-2 receptor (Cantuti-Castelvetri et al., 2020), was also expressed in pancreatic islets and outside islets in our study, consistently with a previous report (Hasan et al., 2010) and scRNA-seq data. However, we did not observe NRP1 expression in the epithelial compartment of the fetal pancreas, while it was present in surrounding cells. Consistent with the low expression of NRP1 in PPs, we showed that the SARS-CoV-2 virus utilizes the ACE2 receptor to enter pancreatic progenitors, while the importance of NRP1 is marginal during infection.
The SARS-CoV-2 entry route was further delineated using selective inhibitors of the virus entry pathway. Human coronaviruses enter cells by fusion on the cell surface or after internalization by endocytosis (Shirato et al., 2018; Tang et al., 2020). The entry pathway depends on the availability of proteases, which are responsible for proteolytic activation of the S protein. While cell surface serine proteases, like TMPRSS2, serve as the activators in the respiratory tract, in other models, endosomal cathepsins are required for SARS-CoV-2 entry (Kawase et al., 2012). Camostat, a serine protease inhibitor, which blocks serine protease activity (Hoffmann et al., 2021; Kawase et al., 2012; Milewska et al., 2018), was used to evaluate the TMPRSS2 importance during the SARS-CoV-2 entry to the pancreatic spheroids. We found broad expression of TMPRSS2 in the epithelium of the fetal pancreas and in in vitroderived pancreatic progenitors. However, our results showed that the virus is not using TMPRSS2 in this tissue, and bypasses this route by engaging endosomal entry, similarly to the human coronavirus OC43 (Owczarek et al., 2018). By employing inhibitors of endosomal acidification or cysteine proteases, we further verified that SARS-CoV-2 enters pancreatic cells by the endosomal route, while fusion on the cell surface is not used.
These observations are important, as the use of the endocytic route allows us to test some interventions that were previously abandoned. Inhibition of endocytic virus entry was initially the concept for drugs such as chloroquine. Although they are ineffective in general treatment, the question remains whether they may play a role in systemic infection.
Limitations of study
The results were obtained using PSC-derived spheroids, which may differ from the actual in vivo conditions. Careful data validation using primary cells or animal models should be carried out before the final conclusions are drawn.
Future studies should determine the molecular or developmental changes induced by SARS-CoV-2 in developing pancreatic cells, for which hPSC-derived 3D spheroids can be utilized. As PDX1+ pancreatic progenitors give rise to all types of pancreatic cells, the consequences of SARS-CoV-2 infection could be broader than just correct islet formation and functional maturation. Furthermore, our immunohistological data and the available scRNA-seq of the human fetal pancreas show that ACE2, TMPRSS2, and NRP1 are expressed in non-epithelial cells in the developing pancreas, such as the endothelium, which can further influence the development. As existing protocols for in vitro generation of endocrine pancreatic cells, including herein utilized 3D spheroids, generally lack cellular components of the pancreatic niche, such research in vitro would require models that more faithfully recapitulate fetal development. Longitudinal research on the clinical and epidemiologic implications of SARS-CoV-2 presence in the developing pancreas will be critical to understanding the effects of in utero infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anit-ACE2 | Abcam | Cat# ab15348; RRID: AB_301861 |
| Goat anti-ACE2 | R&D | Cat# AF933; RRID: AB_355722 |
| Mouse anti-CHGA | Santa Cruz | Cat# sc-393941; RRID: AB_2801371 |
| Rabbit anti-INS | Cell Signaling | Cat# 3014; RRID: AB_2126503 |
| Rat anti-C-peptide | DSHB | Cat# GN-ID4; RRID: AB_2255626 |
| Mouse anti-NKX2-2 | DSHB | Cat# 74.5A5; RRID: AB_531794 |
| Mouse anti-NKX6-1 | DSHB | Cat# F55A10; RRID: AB_532378 |
| Mouse anti-NRP1 | Santa Cruz | Cat# sc-5307; RRID: AB_2282634 |
| Goat anti-PDX1 | R&D | Cat# AF2419; RRID: AB_355257 |
| Rabbit anti-SARS-CoV-2 nucleocapsid (Nc) | Homemade: Pyrc Lab | N/A |
| Mouse anti-SARS-CoV-2 Nc | Thermo Fisher | Cat# MA5-29981; RRID: AB_2785780 |
| Rabbit anti-SOX9 | Millipore | Cat# AB5535; RRID: AB_2239761 |
| Mouse anti-TMPRSS2 | DSHB | Cat# P5H9-A3; RRID: AB_2205599 |
| Rabbit anti-cleaved Caspase 3 (Asp175) | Cell Signaling | Cat# 9664; RRID: AB_2070042 |
| Mouse anti-phospho- MLKL (Ser538) | St. John’s Laboratory | Cat# STJ97776 |
| Goat isotype control | GeneTex | Cat# GTX35039; RRID: AB_10623176 |
| Sheep isotype control | R&D | Cat# 5-001-A; RRID: AB_10141430 |
| Alexa Fluor 350 Donkey Anti-Rabbit | Thermo Fisher | Cat# A10039; RRID: AB_2534015 |
| Alexa Fluor 488 Donkey Anti-Goat | Jackson ImmunoResearch | Cat# 705-545-147; RRID: AB_2336933 |
| Alexa Fluor 488 Donkey Anti-Mouse | Jackson ImmunoResearch | Cat# 715-545-150; RRID: AB_2340846 |
| Alexa Fluor 488 Donkey Anti-Rabbit | Jackson ImmunoResearch | Cat# 711-545-152; RRID: AB_2313584 |
| TRITC Donkey Anti-Rabbit | Jackson ImmunoResearch | Cat# 711-025-152; RRID: AB_2340588 |
| TRITC Donkey Anti-Goat | Jackson ImmunoResearch | Cat# 705-025-147; RRID: AB_2340389 |
| TRITC Donkey Anti-Mouse | Jackson ImmunoResearch | Cat# 715-025-150; RRID: AB_2340766 |
| TRITC Donkey Anti-Rat | Jackson Immuno Research | Cat# 712-025-153; RRID: AB_2340636 |
| Alexa Fluor 647 Donkey Anti-Mouse | Jackson ImmunoResearch | Cat# 715-605-151; RRID: AB_2340863 |
| Alexa Fluor 647 Donkey Anti-Goat | Jackson Immuno Research | Cat# 705-605-147; RRID: AB_2340437 |
| Bacterial and virus strains | ||
| SARS-CoV-2 strain: BavPat1/2020 | EVAg | Ref-SKU: 026V-03883 |
| Chemicals, peptides, and recombinant proteins | ||
| StemFlex Medium | Thermo Fisher | Cat# A3349401 |
| rhVTN | Thermo Fisher | Cat# A31804 |
| RPMI 1640 | Corning | Cat# 15-041-CVR |
| DMEM | Corning | Cat# 15-013-CV |
| CMRL 1066 | Corning | Cat# 15-110-CV |
| GlutaMAX Supplement | Thermo Fisher | Cat# 35050038 |
| HyClone Characterized FBS | GE Healthcare | Cat# SH30071.02 |
| Insulin-Transferrin-Selenium (ITS -G) | Thermo Fisher | Cat# 41400045 |
| MEM Non-Essential Amino Acids Solution (NEAA) | Thermo Fisher | Cat # 11140035 |
| B-27 Supplement | Thermo Fisher | Cat# 17504044 |
| Recombinant Human/Murine/Rat Activin A | Peprotech | Cat# 120-14P |
| Recombinant Human KGF | Peprotech | Cat# AF-100-19 |
| Recombinant Human EGF | Peprotech | Cat# AF-100-15 |
| CHIR 99021 | Peprotech | Cat# 2520691 |
| TTNPB | Tocris | Cat# 0761 |
| Alk5i II | Adooq | Cat# A11762 |
| LDN193189 | Peprotech | Cat# 1066208 |
| LY-411575 | Adooq | Cat# A11176 |
| T3 (3,3′,5-Triiodo-L-thyronine sodium salt) | Sigma Aldrich | Cat# T6397 |
| vitamin C (L-ascorbic acid) | Sigma Aldrich | Cat# A4544 |
| N-acetyl-L-cysteine | Sigma Aldrich | Cat# A7250 |
| zinc sulfate | Merck Millipore | Cat# 1.08883 |
| Heparin sodium salt | Sigma Aldrich | Cat# H3149 |
| Camostat mesylate | Sigma-Aldrich | Cat# SML0057 |
| E64 | Sigma-Aldrich | Cat# E3132 |
| Bafilomycin A1 | Sigma-Aldrich | Cat# B1793-2UG |
| Cathepsin B inhibitor CA-074 methyl ester | Sigma-Aldrich | Cat# C5857 |
| Cathepsin L inhibitor II | Sigma-Aldrich | Cat# 21942 |
| NH4Cl | Bioshop | Cat# AMC303 |
| Critical commercial assays | ||
| Viral DNA/RNA kit | A&A Biotechnology | Cat# 034-100 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher | Cat# 4368814 |
| GoTaq Probe 1-Step RT-qPCR System | Promega | Cat# A6121 |
| Human XL Cytokine Luminex Performance Assay Fixed Panel | R&D | Cat# LKTM014 |
| DreamTaq Green PCR Master Mix (2X) | Thermo Fisher | Cat# K1081 |
| Experimental models: Cell lines | ||
| Vero E6 | ATCC | CRL-1586 |
| ISL1-GFP hPSCs | A gift from Drs. Lei Bu, Kenneth Chien (Lui et al., 2018) | N/A |
| Oligonucleotides | ||
| Forward primer for RT-qPCR CAC ATT GGC ACC CGC AAT C |
This paper | N/A |
| Reverse primer for RT-qPCR GAG GAA CGA GAA GAG GCT TG |
This paper | N/A |
| Probe for RT-qPCR ACT TCC TCA AGG AAC AAC ATT GCC A (FAM/BHQ1) |
This paper | N/A |
| Forward primer for sg mRNA TAT ACC TTC CCA GGT AAC AAA CCA |
This paper | N/A |
| Reverse primer for sg mRNA, first PCR reaction GTA GCT CTT CGG TAG TAG CCA AT |
This paper | N/A |
| Reverse primer for sg mRNA, second PCR reaction TCT TCC TTG CCA TGT TGA GTG A |
This paper | N/A |
| Software and algorithms | ||
| Graph Pad Prism 8 software | Graph Pad Prism | https://www.graphpad.com/scientific-software/prism/ |
| Nikon NIS Elements AR 5.21.01 64 bi | Nikon | https://www.microscope.healthcare.nikon.com/en_EU/products/software/nis-elements/nis-elements-advanced-research |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Malgorzata Borowiak (malbor3@amu.edu.pl).
Materials availability
This study did not generate new unique reagents.
Experimental models and subject details
Human tissue samples
The formalin-fixed paraffin-embedded (FFPE) pancreatic tissues specimens were retrieved from the archival collections of the Department of Clinical Pathology, Collegium Medicum in Bydgoszcz, Nicolaus Copernicus University in Torun (Poland) under protocols approved by the Bioethical Commission. Informed consent was obtained from all subjects. Clinical characteristics of patients were extracted from the medical records and all the data were analyzed anonymously. Only material with confirmed normal histology was included in the present study. We obtained human fetal pancreas 10.6 and 13 weeks after conception, in accordance with Institutional Review Board (IRB3065 to MB) guidelines.
Cell lines
Vero E6 (Cercopithecus aethiops; kidney epithelial; ATCC: CRL-1586) cells were maintained in DMEM (Thermo Fisher Scientific) supplemented with 3% FBS (heat-inactivated fetal bovine serum; Thermo Fisher Scientific) and streptomycin (100 μg/mL), penicillin (100 U/mL) and ciprofloxacin (5 μg/mL).
ISL1-GFP hPSCs, 46XY (a gift from Drs Kenneth Chien and Lei Bu) (Lui et al., 2018) were cultured on vitronectin (rhVTN; Thermo Fisher Scientific)-coated plates in StemFlex (Thermo Fisher) medium. Cells were routinely passaged every 3–4 days using PBS-EDTA.
Cells were cultured at 37°C in an atmosphere containing 5% CO2. All cell lines were tested monthly for mycoplasma with a PCR assay.
Virus
SARS-CoV-2 (isolate 026V-03883) was kindly granted by Dr. Christian Drosten, Charité – Universitätsmedizin Berlin, Germany and provided by the European Virus Archive - Global (EVAg); https://www.european-virus-archive.com/).
Method details
Human tissue processing and immunofluorescence staining
FFPE adult pancreas tissue samples were fixed in 10% neutral buffered formalin for 24 h (Alpinus Chemia), and further processed using the automated histoprocessor (Excelsior™ AS; Thermo Fisher Scientific), according to routine protocol used in pathology laboratories. The 4-μm FFPE sections adhered to Superfrost Plus coated glass slides were deparaffinized and subjected to the standard citrate buffer antigen retrieval protocol before proceeding to immunofluorescence staining (IF). Human fetal pancreatic tissue was fixed in 4% paraformaldehyde/PBS for 4 h, washed with PBS, soaked with 30% sucrose and embedded in TissueTek. The 12-μm frozen sections were adhered to Superfrost Plus coated glass slides and stored at - 80°C.
Pancreatic differentiation
For 3D differentiation, we used a protocol based on Nair et al. (2019). Briefly, ready-to-passage cells were detached using PBS-EDTA, washed and resuspended at the 1:2.5 split ratio in 5 mL of StemFlex per 6-well plate well. Differentiation was carried out in non-treated 6-well plates (Eppendorf) in a 5% CO2, 37°C incubator with constant mixing. After 24 h, when 100–200 μM spheres formed, they were washed with PBS and the medium was exchanged to day 1 medium to initiate differentiation. The media were as follows – day 1: RPMI 1640 (Corning) + 1x Glutamax (Thermo Fisher Scientific) + 1x pen/strep (Thermo Fisher)+ 0.2% FBS (HyClone characterized, GE Healthcare) + 1:5,000 ITS-G (Thermo Fisher) + 3 μM CHIR99021 (Peprotech) + 100 ng/mL Activin A (Peprotech); day 2: RPMI + 1x Glutamax + pen/strep +0. 2% FBS +1: 2,000 ITS-G + 100 ng/mL Activin A; day 3: RPMI + 1x Glutamax + 1x pen/strep +0.2% FBS +1:1,000 ITS-G + 100 ng/mL Activin A; day 4–5: RPMI + 1x Glutamax + 1x pen/strep +0.4% FBS +1:1,000 ITS-G + 25 ng/mL KGF (Peprotech); day 6–7: DMEM (Corning) + 1x Glutamax + 1x pen/strep +1% B27 (Thermo Fisher) + 3 nM TTNPB (Tocris); day 8: DMEM + Glutamax + pen/strep +1% B27 + 3 nM TTNPB +50 ng/mL EGF (Peprotech); day 9–11: DMEM + Glutamax + pen/strep +1% B27 + 50 ng/mL EGF +50 ng/mL KGF; day 12–20: DMEM + Glutamax + pen/strep +1% B27 + 1x NEAA (Thermo Fisher Scientific) + 10 μM Alk5i II (Adooq) + 500 nM LDN193189 (Peprotech) + 1 μM LY-411575 (Adooq) + 1 μM T3 (Sigma Aldrich)+ 0.5 mM vitamin C (Sigma Aldrich) + 1 mM N-acetyl cysteine (Sigma Aldrich) + 10 μM zinc sulfate heptahydrate (Merck) + 10 μg/mL heparin (Sigma Aldrich); day 20 +: CMRL 1066 (Corning) + Glutamax + pen/strep +1% B27 + 1x NEAA (Thermo Fisher) + 10 μM Alk5i II + 1 μM T3 + 0.5 mM vitamin C + 1 mM N-acetyl cysteine +10 μM zinc sulfate heptahydrate +10 μg/mL heparin. The medium was changed daily on days 1–19 and every other day from day 20+.
Virus
SARS-CoV-2virus stock was prepared by infecting fully confluent Vero E6 cells at a TCID50 of 400 per mL. Three days after inoculation, the cultures’ supernatant was aliquoted and stored at −80 °C. The control Vero E6 cell supernatant from mock-infected cells was prepared in the same manner. The virus yield was evaluated by titration on fully confluent Vero E6 cells in 96-well plates, according to the method of Reed and Muench. Plates were incubated at 37 °C for 2 days and the cytopathic effect (CPE) was scored by observation under an inverted microscope.
Virus infection
Spheroids were inoculated at a specific differentiation stage with SARS-CoV-2 at a TCID50 of 5000 per mL (as determined in Vero E6 cells). After 2 h of incubation at 37 °C, the unbound virions were removed by washing with 1 × PBS, and the cultures were supplemented with fresh medium, depleted of differentiation inducers. To analyze the kinetics of virus replication, 100 μL of medium or 400 μL of cell lysate were collected at given time points. All samples were stored at −80°C and analyzed using RT-qPCR.
Isolation of nucleic acids, reverse transcription, and quantitative PCR
A viral DNA/RNA kit (A&A Biotechnology, Poland) was used for nucleic acid isolation from cell culture supernatants. RNA was isolated according to the manufacturer’s instructions. Viral RNA was quantified using quantitative PCR coupled with reverse transcription (RT-qPCR) (GoTaq Probe 1-Step RT-qPCR System, Promega, Poland) using CFX96 Touch real time PCR detection system (Bio-Rad, Poland). The reaction was carried out in the presence of the probe (ACT TCC TCA AGG AAC AAC ATT GCC A (FAM/BHQ1) and primers (forward CAC ATT GGC ACC CGC AAT C and reverse GAG GAA CGA GAA GAG GCT TG). The heating scheme was as follows: 15 min at 45°C and 2 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 58°C or 60°C. To assess the copy number for the N gene, standards were prepared and serially diluted.
Detection of SARS-CoV-2 N sg mRNA
Total nucleic acids were isolated from virus or mock-infected cells with Viral DNA/RNA Kit (A&A Biotechnology), according to the manufacturer’s instructions. Reverse transcription was performed using a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Viral cDNA was amplified in a 20 μL reaction mixture containing 1 × DreamTaq Green PCR master mix (Thermo Fisher Scientific), and primers (500 nM each). The following primers were used to amplify the subgenomic mRNA of SARS-CoV-2 (sg mRNA): common sense primer (leader sequence), 5 – TATACCTTCCCAGGTAACAAACCA-3′; nucleocapsid antisense, 5’-GTAGCTCTTCGGTAGTAGCCAAT-3′ (first PCR reaction), and 5’-TCTTCCTTGCCATGTTGAGTGA-3′ (second PCR reaction). The conditions were as follows: 3 min at 95 °C, 35 cycles (30 cycles for second PCR) of 30 s at 95 °C, 30 s at 55°C, and 20 s at 72°C, followed by 5 min at 72°C and 10 min at 4°C. PCR products were run on 1% agarose gels (1x Tris-acetate EDTA [TAE] buffer) and analyzed using molecular imaging software (Thermo Fisher Scientific).
Immunofluorescent analysis
Tissue samples were blocked and permeabilized with antibody dilution solution (ADS), i.e. 3% normal donkey serum (Jackson Immunoresearch), 0.1% Triton X-100 (BioShop) in PBS for 30 min at room temperature. The samples were then incubated with primary antibodies diluted in ADS overnight at 4°C. Primary antibodies used in the study are listed in Table 1. After two 10-min washes with PBS-Tween20 (0.1% v/v) (BioShop), cells were incubated with secondary antibodies conjugated with Alexa Fluor 350 (Thermo Fisher), Alexa Fluor 488, TRITC or Alexa Fluor 647 (all Jackson Immunoresearch) diluted 1:800 in ADS for 2 h at room temperature. Secondary antibodies were then washed out twice for 5 min with PBS-Tween and incubated with DAPI (Sigma Aldrich) as a counterstain. Samples were mounted with Pro-Long Diamond Antifade (Thermo Fisher Scientific) prior to imaging.
Table 1.
List of primary antibodies
| No | Target | Vendor | Catalog | Dilution | Host | RRID |
|---|---|---|---|---|---|---|
| 1 | ACE2 | Abcam | ab15348 | 1:200 | Rabbit | AB_301861 |
| 2 | ACE2 | R&D | AF933 | 1:200 | Goat | AB_355722 |
| 3 | CHGA | Santa Cruz | sc-393941 | 1:200 | Mouse | AB_2801371 |
| 4 | C-peptide | DSHB | GN-ID4 | 1:40 | Rat | AB_2255626 |
| 5 | INS | Cell Signaling | 3014 | 1:200 | Rabbit | AB_2126503 |
| 6 | NKX2-2 | DSHB | 74.5A5 | 1:40 | Mouse | AB_531794 |
| 7 | NKX6-1 | DSHB | F55A10 | 1:40 | Mouse | AB_532378 |
| 8 | NRP1 | Santa Cruz | sc-5307 | 1:50 | Mouse | AB_2282634 |
| 9 | PDX1 | R&D | AF2419 | 1:200 | Goat | AB_2845433 |
| 10 | SARS-CoV-2 nucleocapsid (Nc) | Homemade Pyrc Laboratorya |
1:1,000 | Rabbit | ||
| 11 | SARS-CoV-2 Nc b | Thermo Fisher | MA5-29981 | 1:100 | Mouse | AB_2785780 |
| 12 | SOX9 | Millipore | AB5535 | 1:250 | Rabbit | AB_2239761 |
| 13 | TMPRSS2 | DSHB | P5H9-A3 | 1:10 | Mouse | AB_2205599 |
| 14 | cleaved Caspase 3 (Asp175) | Cell Signaling | 9664 | 1:400 | Rabbit | AB_2070042 |
| 15 | phospho- MLKL (Ser538) | St. John’s Laboratory | STJ97776 | 1:100 | Mouse |
Hu = human, Mus = mouse, Porc = porcine.
See Figure S3A for validation data.
Used only in experiments with anti-cleaved Caspase 3 antibody.
The 3D spheroids were removed from the medium, washed with ice-cold 1% bovine serum albumin (BioShop), fixed in 4% PFA for 45 min at 4°C, and washed with ice-cold PBS-Tween. Spheroids were permeabilized and blocked in ADS with addition of 0.2% SDS for 45 min at 4°C. Spheroids were incubated with primary antibodies diluted in ADS/0.2% SDS and subsequently with secondary antibodies, overnight at 4°C. The spheroids were washed after antibodies incubation in ADS/SDS for three times, 1 h each. DAPI counterstain was added to the secondary antibody mix. The confocal imaging was performed in a high content imaging 96 Well plate (Corning) in water. For lightsheet imaging spheroids were embedded in 1% agarose dissolved in water in capillaries with 1 mm diameter.
Imaging
Epifluorescence microscopy was performed using Leica DM IL-Led (Leica, Germany), with lenses: N Plan Fluor 4x/0.12, N Plan Fluor 10x/0.30, N Plan Fluor 20x/0.40, N Plan Fluor 40x/0.60 and JENOPTIK Progres Gryphax camera (JENOPTIK, Germany). Confocal microscopy was performed with a Nikon A1Rsi (Nikon, Germany), with lenses: Plan Fluor 4x/0.13, Plan Apo 10x/0.45 DIC N1, Plan Apo VC 20x/0.75 DIC N2, Apo 40x/1.25 WI λS DIC N2, Plan Apo VC 60x/1.4 Oil DIC N2; Software: Nikon NIS Elements AR 5.21.01 64 bit (Nikon, Germany). For quantification of apoptotic cells spheroids were imaged using Zeiss Lightsheet 7 microscope with 20x lens and 0.83x zoom in water, and Zeiss Zen Black software. Quantification was performed using Zen Blue software.
Virus titration
Pancreas organoids prepared using two different differentiation protocols were infected with SARS-CoV-2 virus at 5000 50% tissue culture infectious dose (TCID50)/mL. Infection was performed in two technical replicates. After 2 h of incubation at 37°C, cells were rinsed twice with PBS, and a fresh medium was added. Culture supernatants were collected 72 h post-infection for subsequent analyses. Virus yields were assessed by two methods: Reed and Muench titration and plaque assay. First, collected samples were titrated with serial dilutions of samples on fully confluent Vero cells in 96-well plates. Infected cells were incubated at 37°C, and after 72 h the cytopathic effect (CPE) was scored using an inverted microscope. The titer was calculated as TCID50.
For a plaque assay, Vero E6 cells were seeded in 24 well plates 24 h prior to the inoculation. At the day of infection, 80–90% confluent cells were overlaid with serial dilutions of samples. Following 1 h incubation at 37°C, cells were overlaid with 1 mL of DMEM media supplemented with 10% heat-inactivated FBS, penicillin/streptomycin and 1% methylcellulose (Sigma-Aldrich, Poland). 72 h p.i. cells were fixed overnight with 4% PFA. Next, the buffer was discarded, and cells were stained using 0.1% crystal violet solution in 50:50 water/methanol solution for 10 min at room temperature. Finally, cells were washed with water and plaques were counted.
The titer of samples from a plaque assay was calculated by following formulas:
Plaque assay was performed in duplicate for each sample.
Cytokine analysis
The experiments were carried out in a 24-well plate and cells were infected with the virus and mock control at TCID50 of 5,000 per milliliter. After 2 h, the unbound virions were washed off by rinsing the cells thrice with PBS and fresh medium was added. After 5 min, the supernatants were collected from each well and fresh medium was added. The experiment was carried out for 96 h and every 24 h the supernatants were collected. The collected samples were heat inactivated for 10 min at 60°C. Samples were stored at −80°C for further analysis. Evaluation of the levels of selected proteins in cell culture supernatants was performed using xMAP technology, Luminex assay (Human XL Cytokine Luminex Performance Assay; Bio-Techne, Minneapolis, MN, USA), and the MAGPIX fluorescent-based detection system (Luminex, Austin, TX, USA) according to the manufacturer’s protocol. A cohort of chemokines and cytokines including CCL2, CCL3, CCL4, CCL19, CCL20, CD40L, CX3CL1, CXCL1, CXCL10, CXCL2, FGFb, FLT3L, GCSF, GMCSF, GZMB, IFN-β, IFN-y, IL-10, IL-12p70, IL-13, IL-15, IL-17E, IL-2, IL-3, IL-33 IL-6, IL-7, IL-8, IFN-α, PDGF-AA, PDGF-AB, TGF-α, TNF-α, TRAIL, and VEGF were tested 24, 48, 72 and 96 h after SARS-CoV-2 infection. All results were interpreted from the calibration curves in pg/mL or ng/mL. The heatmap was prepared with GraphPad Prism 8 with mean cytokine levels for each time point and normalized by subtracting the minimum value and dividing by the maximum value.
Inhibition of virus entry
To delineate the entry routes of SARS-CoV-2 in the pancreas, spheroids were cultured in round bottom 96-well or 48-well plates and were incubated for 2 h at 37°C in the presence (20 μg/mL) of anti-ACE2 (goat anti-ACE2, R&D; cat. no.: AF933), anti-neuropilin-1 (sheep anti-neuropilin-1, R&D; cat. no.: AF3870), or isotype antibodies (goat isotype control, GeneTex, cat. no.: GTX35039; sheep isotype control, R&D; cat. no.: 5-001-A). Alternatively, the spheroids were incubated with one of the following compounds: 50 μM camostat mesylate (Sigma-Aldrich; cat. no.: SML0057), 50 μM E64 (Sigma-Aldrich; cat. no.: E3132), 50 nM bafilomycin A1 (Sigma-Aldrich; cat. no.: B1793-2UG), 30 μM cathepsin B inhibitor CA-074 methyl ester (Sigma-Aldrich; cat. no.: C5857), 25 μM cathepsin L inhibitor II (Sigma-Aldrich; cat. no.: 21942), or 10 mM NH4Cl (Bioshop; cat. no.: AMC303). All compounds were diluted in the appropriate culture medium. Following incubation, cells were overlaid with viral stocks at 5000 TCID50/mL and incubated for 2 h at 37°C. The cells were then washed thrice with PBS and the spheroids were further cultured in the presence of compounds. The infection was carried out for 3 days and every 24 h the supernatants were collected, and subsequently fresh medium was added to each well. The effects of antibodies and compounds were analyzed using RT-qPCR and counting of IF-labeled organoids using epifluorescence. For IF quantification, for each spheroid we quantified the number of infected cells on the visible surface (N = 4 independent experiments with at least 30 organoids per experiment, mean = 48.8) and the organoid diameter, from which we estimated % of infected cells per hemisphere surface. We arbitrarily created a threshold of 1.25% infected cells per spheroid to separate “low” infection rate (<1.25% cells, at least one cell per organoid) from “high” infection rate (mean 7.05% infected cells; 3.83–10.26 95% CI) spheroids.
Quantification and statistical analysis
Each IF image is representative for at least 3 independent differentiation and virus infection experiments or biological specimens (tissue samples). Statistical analyses were performed using GraphPad Prism 8. Statistical analysis of Luminex data was performed with unpaired two-tailed Student’s t test. Statistical analysis of the SARS-CoV-2 receptor and entry inhibition by RT-qPCR and IF was performed using Tukey (receptor blocking) or Dunnett (entry inhibition) multiple comparison test. The details of replicate number, statistical test used, dispersion and precision measures for each experiment are given in the corresponding Figure legend.
Acknowledgments
We would like to thank Dr. Christian Drosten (Charité – Universitätsmedizin Berlin, Germany) for sharing SARS-CoV-2, and Drs. Kenneth Chien and Lei Bu for sharing ISL1-GFP cell line. The authors are grateful to all members of the Borowiak and Pyrc laboratories for helpful discussions. We would like to thank Dr. Artur Jankowski, Dr. Anna Klimaszewska-Wisniewska, Paulina Antosik, Justyna Durslewicz, and Joanna Gerc for excellent technical support. This work was supported by the funds provided by the Ministry of Science and Higher Education for the research on SARS-CoV-2, the Polish National Science Centre UMO-2017/27/B/NZ6/02488 to KP, the EU-Horizon 2020 ITN OrganoVir grant 812673 to KP and the Foundation for Polish Science, POIR.04.04.00-00-20C5/16-00, Polish National Science Centre UMO-2020/39/B/NZ3/01408, and Adam Mickiewicz University Poznan COVID grant 5/2020 to MB.
Author contributions
W. J. Sz, A. D., A. M. - experimental design, carrying out experiments, data analysis, figure preparation. N. Z., K.B., E. B.-D., K.O. - carrying out experiments, technical support. M. Sa., M. Su – Luminex assay. D. G. - shared tissue samples. J. D. - study and experimental design, manuscript editing. K. P. and M. B. - study and experimental design, data analysis, obtaining funding and manuscript writing.
Declaration of interests
We declare no conflict of interest.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104594.
Contributor Information
Krzysztof Pyrc, Email: k.a.pyrc@uj.edu.pl.
Malgorzata Borowiak, Email: malbor3@amu.edu.pl.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Accili D. Can COVID-19 cause diabetes? Nat. Metab. 2021;3:123–125. doi: 10.1038/s42255-020-00339-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akarsu C., Karabulut M., Aydin H., Sahbaz N.A., Dural A.C., Yegul D., Peker K.D., Ferahman S., Bulut S., Dönmez T., et al. Association between acute pancreatitis and COVID-19: could pancreatitis be the missing piece of the puzzle about increased mortality rates? J. Invest. Surg. 2022;35:119–125. doi: 10.1080/08941939.2020.1833263. [DOI] [PubMed] [Google Scholar]
- Allen D.W., Kim K.W., Rawlinson W.D., Craig M.E. Maternal virus infections in pregnancy and type 1 diabetes in their offspring: systematic review and meta-analysis of observational studies. Rev. Med. Virol. 2018;28:e1974. doi: 10.1002/rmv.1974. [DOI] [PubMed] [Google Scholar]
- Ayoubkhani D., Khunti K., Nafilyan V., Maddox T., Humberstone B., Diamond I., Banerjee A. Post-covid syndrome in individuals admitted to hospital with covid-19: retrospective cohort study. BMJ. 2021;372:n693. doi: 10.1136/bmj.n693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron E., Bakhai C., Kar P., Weaver A., Bradley D., Ismail H., Knighton P., Holman N., Khunti K., Sattar N., et al. Associations of type 1 and type 2 diabetes with COVID-19-related mortality in England: a whole-population study. Lancet Diabetes Endocrinol. 2020;8:813–822. doi: 10.1016/s2213-8587(20)30272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., Van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., O’Day D.R., Pliner H.A., Kingsley P.D., Deng M., Daza R.M., Zager M.A., Aldinger K.A., Blecher-Gonen R., Zhang F., et al. A human cell atlas of fetal gene expression. Science. 2020;370:eaba7721. doi: 10.1126/science.aba7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso P., Longo M., Esposito K., Maiorino M.I. Type 1 diabetes triggered by covid-19 pandemic: a potential outbreak? Diabetes Res. Clin. Pract. 2020;164:108219. doi: 10.1016/j.diabres.2020.108219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC COVID-19 Response Team Preliminary estimates of the prevalence of selected underlying health conditions among patients with coronavirus disease 2019 — United States, February 12–March 28, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:382–386. doi: 10.15585/mmwr.mm6913e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee Y.J., Ng S.J.H., Yeoh E. Diabetic ketoacidosis precipitated by Covid-19 in a patient with newly diagnosed diabetes mellitus. Diabetes Res. Clin. Pract. 2020;164:108166. doi: 10.1016/j.diabres.2020.108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate K.C., Cha J., Shrestha S., Wang W., Gonçalves L.M., Almaça J., Kapp M.E., Fasolino M., Morgan A., Dai C., et al. SARS-CoV-2 cell entry factors ACE2 and TMPRSS2 are expressed in the microvasculature and ducts of human pancreas but are not enriched in β cells. Cell Metabol. 2020;32:1028–1040.e4. doi: 10.1016/j.cmet.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors J.M., Levy J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020;135:2033–2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig M.E., Kim K.W., Isaacs S.R., Penno M.A., Hamilton-Williams E.E., Couper J.J., Rawlinson W.D. Early-life factors contributing to type 1 diabetes. Diabetologia. 2019;62:1823–1834. doi: 10.1007/s00125-019-4942-x. [DOI] [PubMed] [Google Scholar]
- Croft A., Bucca A., Jansen J.H., Motzkus C., Herbert A., Wang A., Hunter B.R. First-time diabetic ketoacidosis in type 2 diabetics with covid-19 infection: a novel case series. J. Emerg. Med. 2020;59:193–197. doi: 10.1016/j.jemermed.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings M.J., Baldwin M.R., Abrams D., Jacobson S.D., Meyer B.J., Balough E.M., Aaron J.G., Claassen J., Rabbani L.E., Hastie J., et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020;395:1763–1770. doi: 10.1016/s0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo D.T., Montemurro N., Petrella G., Siciliano G., Ceravolo R., Perrini P. Exploring the clinical association between neurological symptoms and COVID-19 pandemic outbreak: a systematic review of current literature. J. Neurol. 2020;268:1561–1569. doi: 10.1007/s00415-020-09978-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebekozien O.A., Noor N., Gallagher M.P., Alonso G.T. Type 1 diabetes and COVID-19: preliminary findings from a multicenter surveillance study in the U.S. Diabetes Care. 2020;43:e83–e85. doi: 10.2337/dc20-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenizia C., Biasin M., Cetin I., Vergani P., Mileto D., Spinillo A., Gismondo M.R., Perotti F., Callegari C., Mancon A., et al. Analysis of SARS-CoV-2 vertical transmission during pregnancy. Nat. Commun. 2020;11:5128. doi: 10.1038/s41467-020-18933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fignani D., Licata G., Brusco N., Nigi L., Grieco G.E., Marselli L., Overbergh L., Gysemans C., Colli M.L., Marchetti P., et al. SARS-CoV-2 receptor angiotensin I-converting enzyme type 2 (ACE2) is expressed in human pancreatic β-cells and in the human pancreas microvasculature. Front. Endocrinol. 2020;11:596898. doi: 10.3389/fendo.2020.596898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan N.M., Kendrick M.A., Druckenbrod N.R., Huelsmeyer M.K., Warner T.F., MacDonald M.J. Genetic association of the neuropilin-1 gene with type 1 diabetes in children: neuropilin-1 expression in pancreatic islets. Diabetes Res. Clin. Pract. 2010;87:29–32. doi: 10.1016/j.diabres.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., Collange O., Boulay C., Fafi-Kremer S., Ohana M., et al. Neurologic features in severe SARS-CoV-2 infection. N. Engl. J. Med. 2020;382:2268–2270. doi: 10.1056/nejmc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Smith J.C., Krüger N., Arora P., Sørensen L.K., Søgaard O.S., Hasselstrøm J.B., Hempel T., Raich L., et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;65:103255. doi: 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein T., Schulte D.M., Schulz J., Glück A., Ziegler A.G., Bonifacio E., Wendorff M., Franke A., Schreiber S., Bornstein S.R., Laudes M. Autoantibody-negative insulin-dependent diabetes mellitus after SARS-CoV-2 infection: a case report. Nat. Metab. 2020;2:1021–1024. doi: 10.1038/s42255-020-00281-8. [DOI] [PubMed] [Google Scholar]
- Hosier H., Farhadian S.F., Morotti R.A., Deshmukh U., Lu-Culligan A., Campbell K.H., Yasumoto Y., Vogels C.B., Casanovas-Massana A., Vijayakumar P., et al. SARS-CoV-2 infection of the placenta. J. Clin. Investig. 2020;130:4947–4953. doi: 10.1172/jci139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamrath C., Mönkemöller K., Biester T., Rohrer T.R., Warncke K., Hammersen J., Holl R.W. Ketoacidosis in children and adolescents with newly diagnosed type 1 diabetes during the COVID-19 pandemic in Germany. JAMA. 2020;324:801. doi: 10.1001/jama.2020.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/jvi.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran N.K., Karmakar B.K., Taylor O.M. Coronavirus disease-19 (COVID-19) associated with acute necrotising pancreatitis (ANP) BMJ Case Rep. 2020;13:e237903. doi: 10.1136/bcr-2020-237903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmartseva I., Wu W., Syed F., Van Der Heide V., Jorgensen M., Joseph P., Tang X., Candelario-Jalil E., Yang C., Nick H., et al. Expression of SARS-CoV-2 entry factors in the pancreas of normal organ donors and individuals with COVID-19. Cell Metabol. 2020;32:1041–1051.e6. doi: 10.1016/j.cmet.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z., Zhang L., Zhang S., Meng X., Li J., Song C., Sun L., Zhou Y. Pathological study on severe acute respiratory syndrome. Chinese Med J. 2003;116:976–980. [PubMed] [Google Scholar]
- Li J., Wang X., Chen J., Zuo X., Zhang H., Deng A. COVID-19 infection may cause ketosis and ketoacidosis. Diabetes Obes. Metabol. 2020;22:1935–1941. doi: 10.1111/dom.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Chen L., Zhang J., Xiong C., Li X. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PLoS One. 2020;15 doi: 10.1371/journal.pone.0230295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Bae J.H., Kwon H.-S., Nauck M.A. COVID-19 and diabetes mellitus: from pathophysiology to clinical management. Nat. Rev. Endocrinol. 2021;17:11–30. doi: 10.1038/s41574-020-00435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui K.O., Zangi L., Silva E.A., Bu L., Sahara M., Li R.A., Mooney D.J., Chien K.R. Driving vascular endothelial cell fate of human multipotent Isl1+ heart progenitors with VEGF modified mRNA. Cell Res. 2018;23:1172–1186. doi: 10.1038/cr.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A., Nowak P., Owczarek K., Szczepanski A., Zarebski M., Hoang A., Berniak K., Wojarski J., Zeglen S., Baster Z., et al. Entry of human coronavirus NL63 into the cell. J. Virol. 2018;92:e01933-17. doi: 10.1128/jvi.01933-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A., Kula-Pacurar A., Wadas J., Suder A., Szczepanski A., Dabrowska A., Owczarek K., Marcello A., Ochman M., Stacel T., et al. Replication of severe acute respiratory syndrome coronavirus 2 in human respiratory epithelium. J. Virol. 2020;94 doi: 10.1128/jvi.00957-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J.A., Groß R., Conzelmann C., Krüger J., Merle U., Steinhart J., Weil T., Koepke L., Bozzo C.P., Read C., et al. SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. Nat. Metab. 2021;3:149–165. doi: 10.1038/s42255-021-00347-1. [DOI] [PubMed] [Google Scholar]
- Nair G.G., Liu J.S., Russ H.A., Tran S., Saxton M.S., Chen R., Juang C., Li M.l., Nguyen V.Q., Giacometti S., et al. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nat. Cell Biol. 2019;21:263–274. doi: 10.1038/s41556-018-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S., Cook J.R., Nordvig A.S., Shalev D., Sehrawat T.S., et al. Post-acute COVID-19 syndrome. Nat. Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostro M.C., Sarangi F., Yang C., Holland A., Elefanty A.G., Stanley E.G., Greiner D.L., Keller G. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Rep. 2015;4:591–604. doi: 10.1016/j.stemcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Owczarek K., Szczepanski A., Milewska A., Baster Z., Rajfur Z., Sarna M., Pyrc K. Early events during human coronavirus OC43 entry to the cell. Sci. Rep. 2018;8:7124. doi: 10.1038/s41598-018-25640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca F.W., Millman J.R., Gürtler M., Gürtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng W., Oken E., Dabelea D. Developmental overnutrition and obesity and type 2 diabetes in offspring. Diabetologia. 2019;62:1779–1788. doi: 10.1007/s00125-019-4914-1. [DOI] [PubMed] [Google Scholar]
- Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/nejmc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A., O'Dwyer S., Quiskamp N., Mojibian M., Albrecht T., et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- Rubino F., Amiel S.A., Zimmet P., Alberti G., Bornstein S., Eckel R.H., Mingrone G., Boehm B., Cooper M.E., Chai Z., et al. New-onset diabetes in covid-19. N. Engl. J. Med. 2020;383:789–790. doi: 10.1056/nejmc2018688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J., Gupta R., Singh M.P., Patnaik I., Kumar A., Kochhar R. Coronavirus disease 2019 and the pancreas. Pancreatology. 2020;20:1567–1575. doi: 10.1016/j.pan.2020.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaharuddin S.H., Wang V., Santos R.S., Gross A., Wang Y., Jawanda H., Zhang Y., Hasan W., Garcia G., Arumugaswami V., Sareen D. Deleterious effects of SARS-CoV-2 infection on human pancreatic cells. Front. Cell. Infect. Microbiol. 2021;11:678482. doi: 10.3389/fcimb.2021.678482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N., Vanderhooft J., Straubhaar J., Mueller J., Chawla R., Zhou Q., Engquist E.N., Trapnell C., Gifford D.K., Melton D.A. Wnt signaling separates the progenitor and endocrine compartments during pancreas development. Cell Rep. 2019;27:2281–2291.e5. doi: 10.1016/j.celrep.2019.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shende P., Gaikwad P., Gandhewar M., Ukey P., Bhide A., Patel V., Bhagat S., Bhor V., Mahale S., Gajbhiye R., Modi D. Persistence of SARS-CoV-2 in the first trimester placenta leading to transplacental transmission and fetal demise from an asymptomatic mother. Hum. Reprod. 2021;36:899–906. doi: 10.1093/humrep/deaa367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman L.M., Hampe C.S., Ben-Haroush A., Perepliotchikov Y., Vaziri-Sani F., Israel S., Miller K., Bin H., Kaplan B., Laron Z. Antibodies to islet cell autoantigens, rotaviruses and/or enteroviruses in cord blood and healthy mothers in relation to the 2010–2011 winter viral seasons in Israel: a pilot study. Diabet. Med. 2014;31:681–685. doi: 10.1111/dme.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Yang M., Wan C., Yi L.X., Tang F., Zhu H.Y., Yi F., Yang H.C., Fogo A.B., Nie X., Zhang C. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synowiec A., Szczepański A., Barreto-Duran E., Lie L.K., Pyrc K. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a systemic infection. Clin. Microbiol. Rev. 2021;34 doi: 10.1128/cmr.00133-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenblock C., Richter S., Berger I., Barovic M., Schmid J., Schubert U., Jarzebska N., von Mässenhausen A., Linkermann A., Schürmann A., et al. Viral infiltration of pancreatic islets in patients with COVID-19. Nat. Commun. 2021;12:3534. doi: 10.1038/s41467-021-23886-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmary P., Arora A., Thomas Raraty M.G., Joseph Dunne D.F., Baron R.D., Halloran C.M. Emerging phenotype of severe acute respiratory syndrome-coronavirus 2-associated pancreatitis. Gastroenterology. 2020;159:1551–1554. doi: 10.1053/j.gastro.2020.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Uhl S., Zhang T., Xue D., Li B., Vandana J.J., Acklin J.A., Bonnycastle L.L., Narisu N., Erdos M.R., et al. SARS-CoV-2 infection induces beta cell transdifferentiation. Cell Metabol. 2021;33:1577–1591.e7. doi: 10.1016/j.cmet.2021.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazzi G., Pellegrini C., Maurelli M., Belliato M., Sciutti F., Bottazzi A., Sepe P.A., Resasco T., Camporotondo R., Bruno R., et al. Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur. J. Heart Fail. 2020;22:911–915. doi: 10.1002/ejhf.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth R., Wallace S., Oliver N.S., Yeung S., Kshirsagar A., Naidu H., Kwong R.M.W., Kumar P., Logan K.M. New-onset type 1 diabetes in children during COVID-19: multicenter regional findings in the U.K. Diabetes Care. 2020;43:170–171. doi: 10.2337/dc20-1551. [DOI] [PubMed] [Google Scholar]
- Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/s0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanti A.J., Vauloup-Fellous C., Prevot S., Zupan V., Suffee C., Do Cao J., Benachi A., De Luca D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020;11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang H., Fan J., Zhang Y., Wang H., Zhao Q. Pancreatic injury patterns in patients with coronavirus disease 19 pneumonia. Gastroenterology. 2020;159:367–370. doi: 10.1053/j.gastro.2020.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee B.A.T., Glover D.M., Zernicka-Goetz M. Expression of SARS-CoV-2 receptor ACE2 and the protease TMPRSS2 suggests susceptibility of the human embryo in the first trimester. Open Biology. 2020;10:200162. doi: 10.1098/rsob.200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng C., Xi J., Li H., Cui J., Gu A., Lai S., Leskov K., Ke L., Jin F., Li Y. Single-cell lineage analysis reveals extensive multimodal transcriptional control during directed beta-cell differentiation. Nat. Metab. 2020;2:1443–1458. doi: 10.1038/s42255-020-00314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.K., Lin S.S., Ji X.J., Guo L.M. Binding of SARS coronavirus to its receptor damages islets and causes acute diabetes. Acta Diabetol. 2010;47:193–199. doi: 10.1007/s00592-009-0109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffré F., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.W., Yuan S., Yuen K.S., Fung S.Y., Chan C.P., Jin D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Shi L., Wang F.S. Liver injury in COVID-19: management and challenges. The Lancet Gastroenterol. Hepatol. 2020;5:428–430. doi: 10.1016/s2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.