Summary
The Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) exhibits reduced susceptibility to vaccine-induced neutralizing antibodies, requiring a boost to generate protective immunity. We assess the magnitude and short-term durability of neutralizing antibodies after homologous and heterologous boosting with mRNA and Ad26.COV2.S vaccines. All prime-boost combinations substantially increase the neutralization titers to Omicron, although the boosted titers decline rapidly within 2 months from the peak response compared with boosted titers against the prototypic D614G variant. Boosted Omicron neutralization titers are substantially higher for homologous mRNA vaccine boosting, and for heterologous mRNA and Ad26.COV2.S vaccine boosting, compared with homologous Ad26.COV2.S boosting. Homologous mRNA vaccine boosting generates nearly equivalent neutralizing activity against Omicron sublineages BA.1, BA.2, and BA.3 but modestly reduced neutralizing activity against BA.2.12.1 and BA.4/BA.5 compared with BA.1. These results have implications for boosting requirements to protect against Omicron and future variants of SARS-CoV-2. This trial was conducted under ClincalTrials.gov: NCT04889209.
Key Words: COVID-19, SARS-CoV-2, booster, Omicron variant, neutralizing antibody, mRNA vaccine, recombinant adenovirus vaccine, sublineage, BA.2.12.1, BA.4/BA.5
Graphical abstract
Highlights
-
•
Vaccine boost substantially increases Omicron neutralizing antibody titers
-
•
Boosted neutralization titers to Omicron but not prototypic D614G decline rapidly
-
•
Ad26.COV2.S is better as prime or boost with mRNA vaccines than as homologous boost
-
•
Omicron sublineages exhibit 5–12 times reduced neutralization by mRNA-1273 boost sera
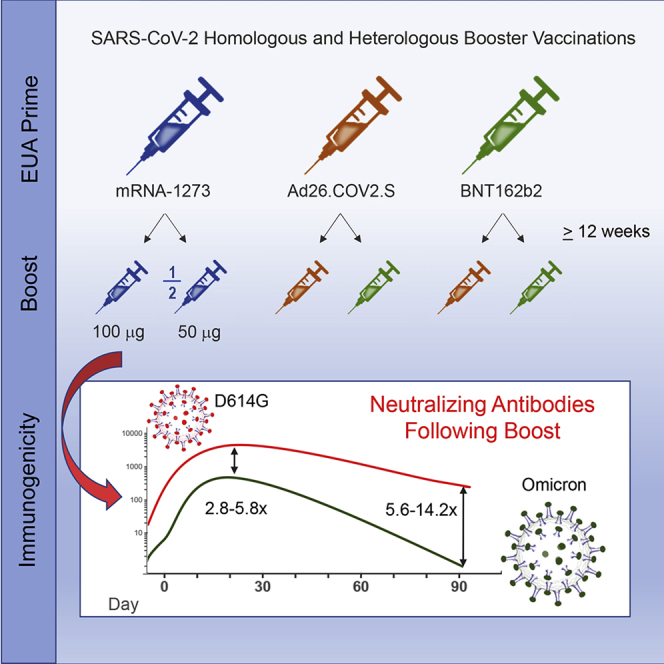
Following COVID-19 vaccine prime and boost, Lyke et al. find higher Omicron neutralization titers for homologous mRNA boost and heterologous mRNA and Ad26.COV2.S boost compared with homologous Ad26.COV2.S boost. Omicron titers rapidly decline by day 91 compared with prototypic D614G. Moderate differences in neutralization (<3-fold) were noted among Omicron sublineages.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant (B.1.1.529) is currently the dominant circulating variant in the COVID-19 pandemic. An unusually large number of mutations, including ∼30 in the spike protein, resulted in increased transmissibility, partial resistance to natural- and vaccine-induced immunity, and increased vaccine breakthrough infections.1, 2, 3 The Omicron wave was initially dominated by the BA.1 sublineage and, more recently, was dominated by sublineages BA.2 and the closely related BA.2.12.1. A fourth sublineage, BA.3, remains relatively rare, whereas the more recent BA.4 and BA.5 sublineages, which share identical spike sequences, are rapidly becoming the dominant variants in the pandemic. These Omicron sublineages share a subset of spike mutations but individually also contain unique spike mutations.4
A single homologous vaccine boost after a primary series of either the BNT162b2 (Pfizer-BioNTech) or mRNA-1273 (Moderna) vaccines generates high titers of neutralizing antibodies to Omicron;5, 6, 7 however, little is known about the durability of the response and the impact of heterologous vaccine boosting. As part of an ongoing study evaluating homologous and heterologous booster vaccines,8 we assessed the magnitude and short-term durability of neutralizing activity against Omicron.
Results
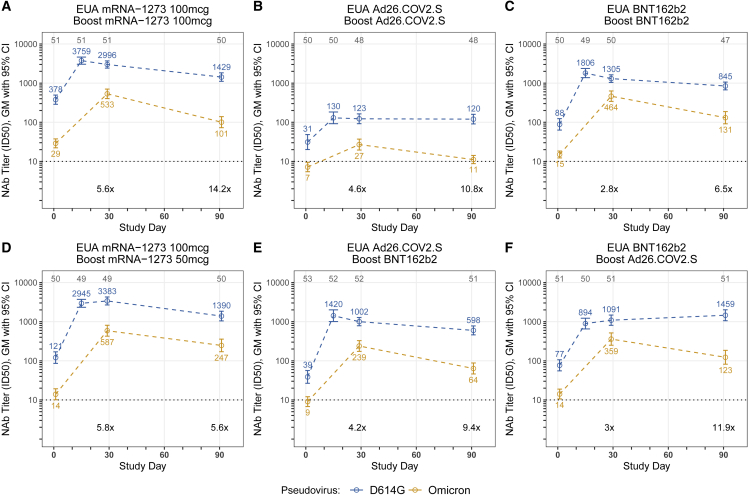
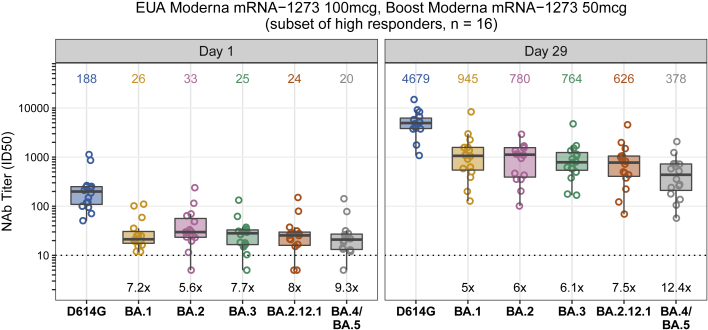
Samples from six groups with ∼50 participants per vaccine group who received either homologous primary/booster combinations (mRNA1273, 50 and 100 μg; BNT162b2; Ad26.COV2.S) (Figures 1A–1D) or two heterologous primary-booster combinations (Ad26.COV2.S prime-BNT162b2 boost; BNT162b2 prime-Ad26.COV2.S boost) were assessed for neutralizing activity (Table S1). The groups were inoculated in successive stages, and mean boost intervals ranged from 16.4 to 28.5 weeks. Baseline (day 1 pre-boost) pseudovirus neutralization antibody (PsVNA) titers to the prototypic D614G variant (Wuhan-1 containing a single D614G spike mutation) were detected in 78% to 100% of participants, with geometric mean titers (GMTs) of 31 to 378 across the different vaccine groups. PsVNA titers to Omicron were detected in 4.3% to 92% of participants across the vaccine groups, with GMTs of only 7 to 29 (Figures 1A–1F; Tables S2 and S3). The boost increased the PsVNA GMTs to above 1,000 for the D614G variant and above 200 for Omicron at day 29 for all groups except the homologous prime-boost Ad26.COV2.S group (D614G GMT 123 and Omicron GMT 27). Day 29 GMTs were 2.8- to 5.8-fold lower for Omicron compared with the D614G variant across the vaccine groups. We also evaluated GMTs to D614G at day 15. Titers peaked at either day 15 (Figure 1A–1C and 1E; Tables S2 and S3) or 29 (Figures 1D and 1F) and differed by less than 1.4-fold between the two time points. In an additional subset evaluation, PsVNA titers to Omicron sublineages BA.2 and BA.3 were similar to BA.1, whereas sublineages BA.2.12.1 and BA.4/BA.5 were 1.5 and 2.5 times less susceptible than BA.1, respectively, when assayed with serum samples from day 29 after homologous 50 μg mRNA-1273 boosting (Figure 2; Table S4).
Figure 1.
Pseudovirus neutralization expressed as 50% inhibitory dilution (ID50) to the D614G variant and Omicron at day 1 (pre-booster) and days 29 and 91 post-booster
(A) mRNA-1273 vaccine boosted with mRNA-1273 100 μg.
(B) Ad26.COV2.S vaccine boosted with Ad26.COV2.S.
(C) BNT162b2 vaccine boosted with BNT162b2.
(D) mRNA-1273 vaccine boosted with mRNA-1273 50 μg.
(E) Ad26.COV2.S vaccine boosted with BNT162b2.
(F) BNT162b2 vaccine boosted with Ad26.COV2.S.
Each group consisted of ∼50 participants (n = ∼25 age 18–55 years old; n = ∼25 age ≥ 56 years old); the actual number of samples assayed for each study are shown at the top of each panel as gray text. The PsVNA (neutralizing antibody [nAb]) results for the D614G variant were previously reported for all groups except the homologous 50 μg mRNA-1273-boosted group. Values and colored text represent the geometric means; error bars represent 95% confidence intervals of ID50 titers. The geometric mean fold reduction in ID50 for Omicron relative to the D614G variant is depicted in black text for days 29 and 91 post-booster. Technical duplicates were performed.
Figure 2.
Neutralization of Omicron sublineages BA.1, BA.2, BA.2.12.1, BA.3, and BA.4/BA.5
Omicron sublineage neutralization titers (ID50) in serum samples obtained before (day 1) and 29 days after homologous mRNA-1273 boosting (50 μg) in 16 study participants who received two inoculations of mRNA-1273 (100 μg) under emergency-use authorization. Box plots represent median (horizontal line within the box) and 25th and 75th percentiles (lower and upper borders of the box), with the whiskers drawn to the value nearest to, but within, 1.5× interquartile range above and below the borders of the box and individual results depicted in open circles. GMTs are shown above each box plot. The fold change neutralization titers relative to D614G are depicted in black text at the bottom of the panels. Technical duplicates were performed.
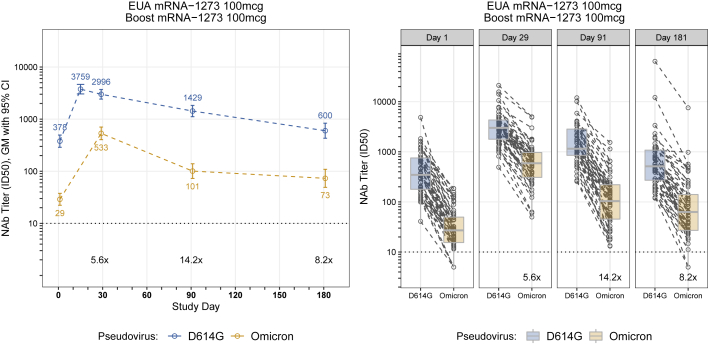
Between days 29 and 91, neutralizing GMTs among the groups decreased 2.4- to 5.3-fold for Omicron and no more than 2.4-fold for the D614G variant. Most of the decrease in Omicron neutralization in the homologous 100 μg mRNA-1273-boosted group occurred rapidly by day 91 (5.3-fold) compared with day 181 (7.3-fold) (Figure 3; Table S3). Statistically significant evidence of a steeper decline of PsVNA titers against Omicron relative to D614G, during the same interval, was found for 5 of 6 groups (p < 0.001 and adjusted for multiple comparisons). An exception was the 50 μg mRNA-1273-boosted group, which had similar declines in PsVNA titers between days 29 and 91 (2.4-fold reduction in neutralizing GMTs) for both variants (p = 0.93) (Figures 1D; Tables S2 and S3). In the two Ad26.COV2.S-boosted groups, neutralizing GMTs to the D614G variant but not Omicron remained stable, or slightly increased, between days 29 and 91 (Figures 1B and 1F). Results were similar between age groups. Two participants reported a positive SARS-CoV-2 test (days 4 and 59). Three participants (including one with a positive SARS-CoV-2 test) were noted to have an unusual increase in PsVNA titers from days 29 to 91 and one from days 91 to 181. No important differences were noted in any of the estimates or testing results reported here following sensitivity analyses that excluded these potentially biased results (Data S1).
Figure 3.
Six month durability of SARS-CoV-2 pseudovirion neutralizing antibody titers after three inoculations of mRNA-1273 (100 μg dose)
Results for participants with available PsVNA data up to day 181 visit (n = 49), from participants who received the two standard inoculations of mRNA-1273 100 μg and were boosted with mRNA-1273 (100 μg).
Left: Values and colored text represent the geometric means; error bars represent 95% confidence intervals of ID50 titers. The geometric mean fold reduction in ID50 for Omicron relative to D614G is depicted in black text for days 29,91, and 181 post-booster.
Right: Spaghetti plot by study day showing differences in PsVNA levels for D614G and Omicron. Box plots represent median (horizontal line within the box) and 25th and 75th percentiles (lower and upper borders of the box), with the whiskers drawn to the value nearest to, but within, 1.5× interquartile range above and below the borders of the box. The geometric mean fold reduction in ID50 for Omicron relative to D614G is depicted in black text.
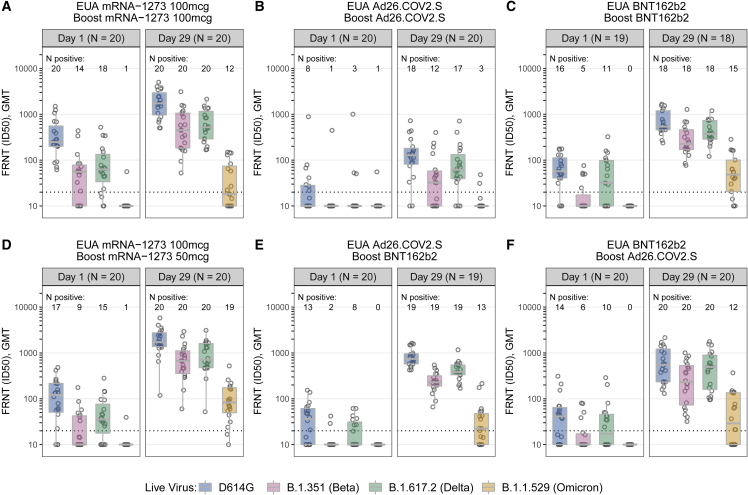
Focus reduction neutralization testing (FRNT) was performed in a subset of 20 participants (10 aged 18–55 years and 10 aged ≥56 years) per study group. Overall, the titers and percentage of FRNT responders, in the subset analysis, were lower compared with PsVNA, although trends were similar between boosted groups (Figure 4; Tables S5 and S6). At day 29 post-boost, detectable FRNT titers to the D614G variant and Omicron were measured in most participants, with the exception of homologous prime-boost Ad26.COV2.S (D614G GMT 110 and Omicron GMT 11.7). Titers to Beta and Delta variants were intermediate to those of the D614G variant and Omicron, with titers to Beta being lower than those against Delta (Tables S7). There was a general concordance between the FRNT and PsVNA assays in how they rank ordered the 50% inhibitory dilution (ID50) neutralizing antibody (nAb) titers against the D614G and Omicron variants but with a marked reduction in Omicron titers in the FRNT assay (Figure S1).
Figure 4.
Live virus focus-reduction neutralization expressed ID50 in Vero-TMPRSS2 cells to D614G, Beta, Delta, and Omicron variants at days 1 (pre-booster) and 29 post-booster
(A) mRNA-1273 vaccine boosted with mRNA-1273 100 μg.
(B) Ad26.COV2.S vaccine boosted with Ad26.COV2.S.
(C) BNT162b2 vaccine boosted with BNT162b2.
(D) mRNA-1273 vaccine boosted with mRNA-1273 50 μg.
(E) Ad26.COV2.S vaccine boosted with BNT162b2.
(F) BNT162b2 vaccine boosted with Ad26.COV2.S.
Each point represents the GMT from two duplicates per specimen (within the same assay run). A value equivalent to half the lower limit of detection (LLOD = 20) was assigned to observations with no detectable response. A specimen was considered as having a positive response if at least one of the duplicates was above the LLOD. Box plots represent median (horizontal line within the box) and 25th and 75th percentiles (lower and upper borders of the box), with the whiskers drawn to the value nearest to, but within, 1.5× interquartile range above and below the borders of the box.
Discussion
Although homologous mRNA boosting and heterologous boosting with mRNA or Ad26.COV2.S vaccines elicited high PsVNA titers to Omicron BA.1, the response waned substantially within 3 months from the boost. Nonetheless, neutralization titers were approximately 2 times higher 6 months after an mRNA-1273 boost (day 181) than they were at day 1, a time point approximately 3.5 months after the second inoculation (Figure 3A), suggesting modest improvement in durability of the response. The PsVNA response to Omicron BA.1 after homologous Ad26.COV2.S boosting was low and short lived, which contrasts with high PsVNA titers when a single inoculation of Ad26.COV2.S was used as either a prime or a boost with an mRNA vaccine. Additionally, the PsVNA titers to Omicron sublineages BA.1, BA.2, and BA.3 measured in sera after a homologous 50 μg mRNA-1273 boost were similar to one another, being 5 to 6.1 times lower than against D614G, while the BA.2.12.1 and BA.4/BA.5 neutralization titers were 7.5 and 12.4 times lower than against D614G. This suggests that Omicron is gaining increased neutralization resistance over time.
In most cases, the boosted PsVNA titers against the D614G variant declined more slowly than those against Omicron BA.1, perhaps reflecting greater maturation of humoral immunity between closely matched vaccine and variant spike proteins.9 This was particularly evident when Ad26.COV2.S was used as a homologous boost (Figure 1B) or as a heterologous boost after BNT162b2 vaccination (Figure 1F). The Ad26.COV2.S vaccine has been associated with greater durability of nAb responses compared with mRNA vaccines,10 and while this was also true for post-boost neutralization titers to the D614G variant, the post-boost response to Omicron BA.1 exhibited a rapid decline. It remains to be determined whether the rate of decline of nAbs against Omicron can be improved with additional homologous or heterologous vaccine boosting or with a longer rest interval between inoculations.
Peak GMTs against D614G and Omicron were similar between the 50 and 100 μg homologous mRNA-1273-boosted groups (Figures 1A, 1D, 4A, and 4D). In a previous study of participants primed with two inoculations of 100 μg mRNA-1273 in the Moderna phase 3 trial and subsequently boosted with either 50 or 100 μg mRNA-1273, GMTs against D614G and Omicron were 2.5 to 2.8 times higher in the 100 μg boosted group than in the 50 μg boosted group.7 It is also noteworthy that Omicron neutralization titers declined more slowly by day 91 in the 50 μg than in the 100 μg homologous mRNA-1273-boosted group (Figures 1A and 1D). These differences in the magnitude and rate of decline of nAbs may be partially explained by the boosting interval, which was approximately 3 months longer in the half-dose mRNA-1273 group (3.5 versus 6.5 months) owing to the staged recruitment study design.
These data support that homologous mRNA and heterologous boost combinations will increase humoral immunity to Omicron as a strategy to mitigate risk from this and possibly future variants. The kinetics of the nAb response post-boost suggest a rapid decay in GMT to Omicron by day 91 that is consistent with waning vaccine effectiveness11,12 and may impact decisions regarding the need for additional boosters.
Limitations of the study
Limitations to this study include the lack of randomization to study groups, the individual variability in boosting intervals, and regional variability in SARS-CoV-2 transmission rates across the study sites. Although this study focused solely on nAbs and utilized a PsVNA assay correlated with vaccine efficacy,13 clinical effectiveness against variants and an understanding of the breadth of immune responses, including cell-mediated immunity, are urgently needed.
Consortia
The members of the DMID 21-0012 Study Group are Jennifer S. Husson, Angie Price, Jennifer A. Whitaker, Wendy A. Keitel, Ann R. Falsey, Ian Shannon, Daniel Graciaa, Nadine Rouphael, Evan J. Anderson, Satoshi Kamidani, Gysella B. Muniz, Sonika Bhatnagar, Anna Wald, Megan Berman, Laura Porterfield, Amber Stanford, Jennifer Lee Dong, Steven E. Carsons, Diana Badillo, Susan Parker, Michelle Dickey, Sasha E. Larsen, John Hural, Brian Ingersoll, Marina Lee, Lilin Lai, Katharine Floyd, Madison Ellis, Kathryn M. Moore, Kelly Manning, Stephanie L. Foster, and Mit Patel.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexaflour-647 | Novus Biologicals | ab269823; RRID:AB_2892763 |
| CR3022 | Sigma-Aldrich | ZHU1077; RRID:AB_2848080 |
| Bacterial and virus strains | ||
| EHC-83E | Mid-turbinate nasal swab | SARS-CoV-2/human/USA/GA-EHC-083E/2020 |
| B.1.617.2 | Mid-turbinate nasal swab | hCoV-19/USA/PHC658/2021 |
| B.1.351 | Dr. Andy Pekosz (John Hopkins University, Baltimore, MD) | hCoV-19/South Africa/KRISP-K005325/2020 |
| B.1.1.529 | Mid-turbinate nasal swab | hCoV19/EHC_C19_2811C |
| Chemicals, peptides, and recombinant proteins | ||
| Methylcellulose | Sigma-Aldrich | Cat. #: M0512-250G |
| Critical commercial assays | ||
| Bright-Glo Luciferase Assay System | Promega | Cat# E2610 |
| Opti-MEM | Life Technologies | Cat# 31,985,062 |
| Experimental models: Cell lines | ||
| HEK 293T/17 cells | American Type Culture Collection | Cat# CRL-11268 |
| 293T/ACE2 cell | Drs. Michael Farzan and Huihui Mu | N/A |
| VeroE6 C1008 cells | ATCC | Cat# CRL-1586, RRID:CVCL_0574 |
| Recombinant DNA | ||
| VRC5601: pHR' CMV Luc | Vaccine Research Center, NIH | N/A |
| VRC5602: pCMV ΔR8.2 | Vaccine Research Center, NIH | N/A |
| VRC9260: TMPRSS2 | Vaccine Research Center, NIH | N/A |
| VRC7480 | Vaccine Research Center, NIH | N/A |
| VRC7480.D614G | This paper | N/A |
| VRC7480.Omicron.BA.1 | This paper | N/A |
| VRC7480.Omicron.BA.2 | Genscript | N/A |
| VRC7480.Omicron.BA.3 | Genscript | N/A |
| VRC7480.Omicron.BA.2.12.1 | This paper | N/A |
| VRC7480.Omicron.BA.4/BA.5 | Genscript | N/A |
| Software and algorithms | ||
| GraphPad Prism 9.2.0 | San Diego, CA | N/A |
| Viridot Program | Katzelnick et al.14 | https://github.com/leahkatzelnick/Viridot |
| R Version 4.1.3 | R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/ | N/A |
| SAS Version 9.4 | SAS Institute Inc., Cary NC, USA | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contacts Kirsten E. Lyke (klyke@som.umaryland.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability statement
-
•
The safety and immunogenicity data reported in this study cannot be deposited in a public repository because they are being collected and analyzed as part of the DMID 21-0012 study, which is ongoing and for which the dataset has not yet been locked. To request access, contact the IDCRC Secondary Research Project Manager (IDCRC.sec.research@fredhutch.org, section 18 of IDCRC Manual of Procedures, https://idcrc.org/about/procedures/index.html). Summary statistics describing previously published data is available at the NEJM: https://doi.org/10.1056/NEJMoa2116414.
-
•
This paper does not report original code.
-
•
Any additional information required to analyze the data reported in this paper is available from the Lead Contact upon request.
Experimental model and subject details
This phase 1/2 open-label, non-randomized, adaptive design, clinical trial was performed in sequential stages at ten U.S. clinical sites.8 Healthy adults (aged ≥18 years) who had received a full Covid-19 vaccine regimen available under EUA (Ad26.COV2.S [Janssen], mRNA-1273 [Moderna, Inc], or BNT162b2 [Pfizer/BioNTech]) at least 12 weeks earlier and reported no prior history of SARS-CoV-2 infection or monoclonal antibody infusion, were eligible for inclusion. Participants were enrolled and assigned a booster regimen until recruitment goals were met (approximately 50 participants per EUA prime group and ∼150 per stage) in five sequential stages. A sample size of 50 per group, 25 per age stratum, was targeted, consistent with Phase 1/2 studies. No tests of hypothesis for comparison between groups were planned nor conducted. Six hundred and ninety-six adults were enrolled over five stages. Four homologous primary/booster combinations (mRNA1273, 50-μg and 100-μg; BNT162b2; Ad26.COV2.S) (Figures 1A–1D) and two heterologous primary-booster combinations (Ad26.COV2.S prime-BNT162b2 boost; BNT162b2 prime-Ad26.COV2.S boost) (Figures 1E and 1F) representing 305 participants were selected to be reported here. Prime-boost intervals ranged from 3 (Stage 1) to 6.6 (Stage 5) months. Samples from available participants per group (∼25 participants 18–55 years old and ∼25 participants ≥56 years) were assayed. We determined 50% serum neutralizing antibody (NAb) titers using lentivirus-based PsVNA12 and live virus FRNT.3 PsVNA assays were performed in all participants (∼50 per group) with the D614G variant (Wuhan-1 containing a single D614G spike mutation) and Omicron (BA.1 sublineage) for Day 1 (pre-boost) and Days 15, 29 and 91 post-boost for all groups, in addition to Day 181 post-boost for the homologous Moderna 100-μg group. Trial participants were predominantly non-Hispanic and white with equal representation of male and females. A full description of the demographic information of study participants has previously been reported.8 A subset (n = 16) of serum samples from this later group, chosen from those with robust responses detected at Day 29, was also assayed against the BA.2, BA.2.12.1, BA.3, and BA.4/BA.5 sublineage of Omicron. FRNT assays were performed in a subset of 20 participants per group (10 participants aged 18–55 years and 10 participants aged >56 years), for the D614G, Beta (B.1.351), Delta (B.1.617.2) and Omicron variants at Days 1 and 29.
Methods details
Cells
HEK 293T/17 cells (American Type Culture Collection, cat. no. CRL-11268) were used for transfection to produce spike-pseudotyped viruses. The cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum. A 293T cell line stably overexpressing the human ACE2 cell surface receptor protein (293T/ACE2 cell line) was used as targets for infection in the PsVNA. 293T/ACE2 cells were obtained from Drs. Michael Farzan and Huihui Mu (Scripps) and were maintained DMEM supplemented with 10% FBS and 3 μg/mL puromycin. VeroE6-TMPRSS2 cells used in the FRNT assay were generated and cultured as previously described.15
Plasmids and viruses
SARS-CoV-2 spike-pseudotyped viruses were prepared by using a sequence-modified version of a plasmid (VRC7480) encoding codon-optimized full-length spike of the ancestral Wuhan-1 strain. The D614G spike mutation was introduced by site-directed mutagenesis. The omicron sublineage spike plasmids were either produced by site-directed mutagenesis or were synthesized by GenScript using VRC7480 as template. All mutations were confirmed by full-length spike gene sequencing. Spike-pseudotyped viruses were produced in HEK293T/17 cells by transfection using Fugene 6 (Promega Cat#E2692) and a combination of spike plasmid, lentiviral backbone plasmid (pCMV ΔR8.2) and firefly Luc reporter gene plasmid (pHR' CMV Luc) in a 1:17:17 ratio in Opti-MEM (Life Technologies) as described.13 The variant spike-pseudotyped viruses contained the mutations listed:
| Variant | Spike mutations |
|---|---|
| D614G | D614G |
| Omicron (BA.1) | A67V, Δ69-70, T95I, G142D, Δ143-145, Δ211, L212I, +214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
| Omicron (BA.2) | T19I, Δ21-23, A24S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron (BA.3) | A67V, Δ69-70, G142D, Δ143-145, Δ211, L212I, G339D, S373P, S375F, D405N, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron (BA.2.12.1) | T19I, L24S, ΔP25, ΔP26, ΔA27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452Q, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, S704L, N764K, D796Y, Q954H, N969K |
| Omicron (BA.4/BA.5) | T19I, L24S, ΔP25, ΔP26, ΔA27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
For the FRNT assay, the EHC-083E (SARS-CoV-2/human/USA/GA-EHC-083E/2020) variant was isolated and propagated as previously described.16 B.1.351 variant isolate (hCoV-19/USA/MD-HP01542/2021), kindly provided by Dr. Andy Pekosz (John Hopkins University, Baltimore, MD), was propagated once in VeroE6-TMPRSS2 cells to generate a working stock. hCoV-19/USA/PHC658/2021 (herein referred to as the B.1.617.2 variant) was derived from a nasal swab collected in May 2021.15 hCoV19/EHC_C19_2811C (referred to as the B.1.1.529 variant) was derived from a mid-turbinate nasal swab collected in December 2021. This SARS-CoV-2 genome is available under GISAID accession number EPI_ISL_7171744. All viruses used in the FRNT assay were deep sequenced and confirmed as previously described.15
| Live virus variant | Spike mutations |
|---|---|
| EHC-083E | D614G |
| B.1.351 | L18F, D80A, D215G, L241-, L242-, A243-, K417N, E484K, N501Y, D614G, A701V |
| B.1.617.2 | T19R, K77T, G142D, Del (E156-R158); Ins G, L452R, T478K, D614G, P681R, D950N |
| Omicron (BA.1) | A67V, Δ69-70, T95I, G142D, Δ143-145, Δ212, N211I, +214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
Serum samples
As part of a larger study,8 participants were enrolled with approximately equal numbers in two age strata (18–55 years and >56 years) and the three EUA-dosed vaccine groups (N = 50/group) across multiple groups. Blood was collected for serum at Day 1 pre-vaccination and days 15, 29, 91 and 181 following booster dosing, with follow-up planned to 12 months. Samples were processed within 8 h and aliquoted for cryopreservation prior to shipment to Fisher Bioservices (Germantown, MD). Serum samples were selected from eligible persons (N = 50/group stratified into two age strata across six groups) who received the designated study booster vaccine, as previously described. Only participants with sufficient serum collected to enable testing were considered for selection. Serum samples were sent to designated immunologic reference laboratories (Duke University for PsVNA and Emory University for FRNT) for analysis. A subset of 16 participants with the highest response at Day 29 to D614G were selected from Group 13E (EUA mRNA-1273 100mcg, boost mRNA-1273 50mcg) to be assayed for Omicron sublineages (BA.1, BA.2, BA.2.12.1, BA.3, and BA.4/BA.5). All serum samples were heat-inactivated for 30 min at 56°C prior to assay.
Pseudovirus neutralization assay
Neutralization of Spike-pseudotyped viruses was assessed in 293T/ACE2 cells as a function of reductions in firefly luciferase reporter activity.13 All serum samples were assayed against Spike-pseudotyped SARS-CoV-2 D614G and Omicron (BA.1 sublineage). A subset of samples was also assayed with Omicron sublineages BA.2, BA.2.12.1, BA.3 and BA.4/BA.5. Results are reported as Inhibitory Dilution 50 (ID50); these values represent the serum dilution that reduces relative luminescence units (RLU) by 50% relative to the RLU in the virus control wells after subtraction of background RLU. These assays were conducted under the oversight of the Quality Assurance Unit for Duke Vaccine Immunogenicity Programs (QADVIP).
Live virus focus reduction neutralization (FRNT) assay
FRNT assays were performed as previously described.15, 17, 18 Briefly, samples were diluted at 3-fold in 8 serial dilutions using DMEM in duplicates with an initial dilution of 1:10 in a total volume of 60 μL. Serially diluted samples were incubated with an equal volume of D614G, B.1.351, B.1.617.2 or B.1.1.529 (100–200 foci per well based on the target cell) at 37°C for 45 min in a round-bottomed 96-well culture plate. The antibody-virus mixture was then added to VeroE6-TMPRSS2 cells and incubated at 37°C for 1 h. Post-incubation, the antibody-virus mixture was removed and 100 μL of pre-warmed 0.85% methylcellulose (Sigma-Aldrich, #M0512-250G) overlay was added to each well. Plates were incubated at 37°C for either 18 h (D614G, B.1.351, B.1.617.2) or 40 h (B.1.1.529) and the methylcellulose overlay was removed and washed six times with PBS. Cells were fixed with 2% paraformaldehyde in PBS for 30 min. Following fixation, plates were washed twice with PBS and permeabilization buffer (0.1% BSA and 0.1% Saponin in PBS) was added to permeabilized cells for at least 20 min. Cells were incubated with an anti-SARS-CoV spike primary antibody directly conjugated to Alexaflour-647 (CR3022-AF647) for 4 h at room temperature or overnight at 4°C. Cells were washed three times in PBS and foci were visualized on an ELISPOT reader. Antibody neutralization was quantified by counting the number of foci for each sample using the Viridot program.14 The neutralization titers were calculated as follows: 1 - (ratio of the mean number of foci in the presence of sera and foci at the highest dilution of respective sera sample). Each specimen was tested in duplicate. The FRNT-50 titers were interpolated using a 4-parameter nonlinear regression in GraphPad Prism 9.2.0. Samples that do not neutralize at the limit of detection at 50% are plotted at 10 and was used for geometric mean and fold-change calculations.
Quantification and statistical analysis
Statical support was provided by the Statistical Center for HIV/AIDS Research and Prevention (SCHARP) at the Fred Hutchinson Cancer Research Center, University of Washington. Descriptive statistics, such as quantiles (min, max, median and 25th and 75th percentiles) and Geometric Mean (GM) are reported for observations collected at each visit, along with percent of participants with positive response. For post-boost visits, the Geometric Mean Fold Difference (GMFD), relative to baseline (Day 1, pre-boost) levels are also provided, along with proportion of participants with a 2-fold (or higher) and a 4-fold (or higher) increase relative to baseline. Neutralization titers reported as below the lower limit of detection (LLOD) were assigned a value equivalent to half the LLOD. For FRNT, the unit of analysis was obtained as the Geometric Mean of the duplicate titers. Positive response (at the sample level) was defined as titers above the LLOQ or at least one of the duplicate titers above the LLOQ. Ninety five percent confidence intervals (CI) for GM and GMFD are obtained based on a t-distribution. The fold change in the Omicron Antibody levels, relative to D614G, was obtained for participants with positive response to D614G. The GM of this decrease fold is reported only for Day 29 and Day 91. A statistical analysis was performed to compare the change in PsVNA titers against Omicron (ID50) from Day 29 to Day 91 to the change in PsVNA titers (ID50) against D614G. A regression model on the change in titers was fit using Generalized Estimating Equations, to account for correlation of within-participant observations, and including the type of Pseudovirus and vaccine-boost combination as predictors of interest, along with their interaction. Adjustment for the interval between collection times, which could differ between participants, as also included. Estimates of the GMFD for both Omicron and D614G are provided for each group. Statistical testing of decline for Omicron relative to D614F was performed, under a family-wise significance level of 1%, with adjustment for multiple comparisons using the Hochberg approach.
Participants for whom a positive SARS-CoV-2 test has been reported, or for whom unexplained increase in titers, defined as change of a highly outlier magnitude (2.5 times IQR above the 75th percentile) within the intervals reported here, were identified. A sensitivity analysis was conducted by excluding these participants.
Details on statistical comparisons and sensitivity analyses
Potentially influential observations and sensitivity analyses
No participants reported receiving a booster vaccination outside of the study within the period reported. The following set of participants were identified to have reported a positive SARS-CoV-2 test within the period reported or to show a highly outlier increase in PsVNA titers after Day 29 (defined as 2.5 times the IQR above the 75th percentile of the change between visits, within their vaccination group):
-
•
One participant in Group 2E (mRNA-1273 100mcg prime-mRNA-1273 100mcg boost) showed a sharp increase in PsVNA levels both against D614G and Omicron between Day 91 and Day 181, along with a sharp increase in Binding IgG Antibody levels to the N-protein (N-protein data and Binding IgG data not included).
-
•
One participant in Group 6E (BNT162b2 prime-Ad26.COV2.S prime) had a molecular positive test recorded 55 days post-boost, with a subsequent molecular negative test recorded 59 days post-boost. This participant also showed a sharp increase in PsVNA levels against D614G and Omicron between Day 29 and Day 91.
-
•
One participant in Group 9E (BNT162b2 prime-BNT162b2 boost) had a molecular positive test reported 4 days post-boost, although not showing any noticeable increase PsVNA levels relative to the rest of participants in the same group.
-
•
One other participant in Group 9E (BNT162b2 prime-BNT162b2 boost) showed an outlier increase in PsVNA against D614G from Day 29 to Day 91. However, no increase in PsVNA against Omicron, nor in their Binding IgG Antibody levels to the N-protein, was observed.
-
•
One participant in Group 13E (mRNA-1273 100mcg prime-mRNA-1273 50mcg boost) showed a sharp increase in PsVNA against Omicron from Day 29 to Day 181, with moderate increase observed against D614G. Levels for Binding Ab show a sharp increase, but this is observed between Day 1 and Day 15.
A sensitivity analysis was performed by excluding the five participants identified above. The results from this sensitivity analysis, relative to the analysis of the full set of participants, are as follows:
-
•
Estimates of the Geometric Mean PSV-Ab levels fall between 90.7% and 108.7% of the estimates from the full set analysis.
-
•
For non-baseline visits, estimates of the Geometric Mean Fold Change in PSVNA levels (relative to Baseline) fall between 89.1% and 104.0% of the estimates from the full set analysis.
-
•
For Omicron, estimates of the Geometric Mean Fold Decrease in PSVNA levels (relative to D614G) fall between 99.1% and 104.7% of the estimates from the full set analysis.
In summary, no important differences in the various estimates were observed in a sensitivity analysis that excluded participants reporting positive SARS-CoV-2 tests or showing outlier high magnitude of change in PsVNA titers between study visits.
Additional resources
The clinical trial was sponsored by the Infectious Diseases Clinical Research Consortium through the National Institute for Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) as protocol DMID 21-0012 and registered as ClinicalTrials.gov # NCT04889209.
Acknowledgments
The trial was sponsored and primarily funded by the Infectious Diseases Clinical Research Consortium through the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers UM1AI48372, UM1AI148373, UM1AI148450, UM1AI148452, UM1AI148573, UM1AI148574, UM1AI148575, UM1AI148576, UM1AI148684, and UM1 AI148689 and with support from the NIAID Collaborative Influenza Vaccine Innovation Centers (CIVICs) contract 75N93019C00050 and NIH Vaccine Research Center. Funding for the Suthar laboratory included the following: NIH P51 OD011132, 3U19AI057266-17S1, 1U54CA260563, and HHSN272201400004C, NIH/NIAID CEIRR under contract 75N93021C00017 to Emory University from NIAID and Emory School of Medicine, and Woodruff Health Sciences Center 2020 COVID-19 CURE Award. We would like to acknowledge Moderna, Inc., Johnson & Johnson/Janssen, and Pfizer/BioNTech Pharmaceuticals for their collaboration, scientific input, and sharing of documents needed to implement this trial. All products were acquired through the government procurement process.
Author contributions
Conceptualization, K.E.L., R.L.A., P.C.R., J.H.B., and D.C.M.; methodology, C.D.I., D.S., P.C.R., M.S.S., and D.C.M.; data curation, J.A.Z.; investigation, K.E.L., R.L.A., R.C.P., M.E.D., A.E., L.A.J., A.R.B., H.M.E.S., C.A.R., J.M.M., C.J., R.E.R., M.J.M., R.C.B., R.W.F., M.B., A.C.K., T.M.B., K.R., S.E., D.D., R.N.C., M.S.S., and D.C.M.; writing – original draft, K.E.L., R.L.A., S.D.I., M.S.S., P.C.R., J.H.B., and D.C.M.; formal analysis and visualization, C.D.I., D.S., and E.R.B.; funding acquisition, S.U.N., P.C.R., K.M.N., D.S.S., and J.H.B.; resources, C.M.P., M.S.S., P.C.R., and D.C.M.; project administration, J.I.A. and S.C.; supervision, K.M.N., D.S.S., P.C.R., A.E., J.H.B., M.S.S., and D.C.M.
Declaration of interests
R.L.A., C.D.I., C.M.P., D.S., R.P.C., M.E.D., A.E., H.M.E.S., R.E.R., M.B., A.C.K., T.M.B., D.D., R.N.C., J.I.A., S.C., J.A.Z., S.U.N., E.R.B., and D.J.P. report no competing interests. K.E.L. receives grant awards from Pfizer, Inc., COVID-19 vaccine research. L.A.J.’s institution receives grant funding from NIH and CDC for vaccine-related assessments, including those of COVID-19 vaccines. A.R.B. has grant funding from Pfizer, Janssen, Merck, and Cyanvac for non-COVID-19-related work and serves as a consultant for GSK and Janssen. C.A.R.'s institution has received funds to conduct clinical research from the National Institutes of Health, CDC, BioFire, Inc., Genentech, GSK, Janssen, MedImmune, Merck, Micron, Moderna, Novavax, PaxVax, Pfizer, Regeneron, and Sanofi-Pasteur. She is co-inventor of patented RSV vaccine technology, which has been licensed to Meissa Vaccines, Inc. J.M.M. has served as a consultant for Merck, Sharp, and Dohme for non-Covid-related work. C.J. receives funding from the Bill and Melinda Gates Foundation, NIH, and CDC, consults for Gilead and Abbvie, serves on a DSMB for MedPace, and receives royalties from UpToDate. M.J.M. has laboratory research and clinical trials contracts for vaccines or MAB versus SARS-CoV-2 with Lilly, Pfizer (exclusive of the current work), and Sanofi and personal fees for Scientific Advisory Board service from Merck, Meissa Vaccines, Inc., and Pfizer. R.C.B. receives funding for vaccine trials from Path Nipah and Pfizer. R.W.F. receives funding to perform clinical trials from Pfizer, Moderna, Astra Zeneca, and Emergent Health, and he serves on advisory boards for Johnson & Johnson, Merck, Sanofi Pasteur, and Seqirus. S.E. receives funding to her institution from Sanofi Pasteur for a non-COVID-19 vaccine study. K.M.N. holds a grant from Pfizer, without salary support, for a COVID-19 vaccine study and salary support from the National Institutes of Health (NIH) for work on multiple COVID-19 vaccine trials. D.S.S. is supported by grant awards from NIH/NIAID. P.C.R. and J.H.B. report a pending US patent application no. 63/025918 entitled “Coronavirus RNA vaccines and methods of use.” D.C.M. receives funding from NIH and Moderna for laboratory studies of COVID-19 vaccine antibody responses. M.S.S. receives funding from Moderna, Inc., and Ocugen. D.C.M. receives funding from Moderna, Inc.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: June 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100679.
Contributor Information
Kirsten E. Lyke, Email: klyke@som.umaryland.edu.
Robert L. Atmar, Email: ratmar@bcm.edu.
David C. Montefiori, Email: monte@duke.edu.
the DMID 21-0012 Study Group:
Jennifer S. Husson, Angie Price, Jennifer A. Whitaker, Wendy A. Keitel, Ann R. Falsey, Ian Shannon, Daniel Graciaa, Nadine Rouphael, Evan J. Anderson, Satoshi Kamidani, Gysella B. Muniz, Sonika Bhatnagar, Anna Wald, Megan Berman, Laura Porterfield, Amber Stanford, Jennifer Lee Dong, Steven E. Carsons, Diana Badillo, Susan Parker, Michelle Dickey, Sasha E. Larsen, John Hural, Brian Ingersoll, Marina Lee, Lilin Lai, Katharine Floyd, Madison Ellis, Kathryn M. Moore, Kelly Manning, Stephanie L. Foster, and Mit Patel
Supplemental information
References
- 1.CDC . US Department of Health and Human Services, CDC; 2022. Science Brief: Omicron (B.1.1.529) Variant.https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html [Google Scholar]
- 2.CDC . US Department of Health and Human Services; 2022. Potential Rapid Increase of Omicron Variant Infection in the United States.https://www.cdc.gov/coronavirus/2019-ncov/science/forecasting/mathematical-modeling-outbreak.html [Google Scholar]
- 3.Edara V.V., Manning K.E., Ellis M., Lai L., Moore K.M., Foster S.L., Floyd K., Davis-Gardner M.E., Mantus G., Nyhoff L.E., et al. mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Rep. Med. 2022;3:100529. doi: 10.1016/j.xcrm.2022.100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houriiyah T., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Moyo S., Amoako D.G., Baxter C., et al. Continued Emergence and Evolution of Omicron in South Africa: New BA.4 and BA.5 Lineages. medRxiv. 2022 doi: 10.1101/2022.05.01.22274406. Preprint at. [DOI] [Google Scholar]
- 5.Ariën K.K., Heyndrickx L., Michiels J., Vereecken K., Van Lent K., Coppens S., Willems B., Pannus P., Martens G.A., Van Esbroeck M., et al. Three doses of BNT162b2 vaccine confer neutralising antibody capacity against the SARS-CoV-2 Omicron variant. NPJ Vaccines. 2022;7:35. doi: 10.1038/s41541-022-00459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz S.R., Hoffmann M., Roth E., Pracht K., Burnett D.L., Mazigi O., Schuh W., Manger B., Mielenz D., Goodnow C.C., et al. Augmented neutralization of SARS-CoV-2 Omicron variant by boost vaccination and monoclonal antibodies. Eur. J. Immunol. 2022 doi: 10.1002/eji.202249841. https://www.ncbi.nlm.nih.gov/pubmed/35253229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pajon R., Doria-Rose N.A., Shen X., Schmidt S.D., O’Dell S., McDanal C., Feng W., Tong J., Eaton A., Maglinao M., et al. SARS-CoV-2 Omicron variant neutralization after mRNA-1273 booster vaccination. N. Engl. J. Med. 2022:1088–1091. doi: 10.1056/NEJMc2119912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atmar R.L., Lyke K.E., Deming M.E., Jackson L.A., Branche A.R., El Sahly H.M., Rostad C.A., Martin J.M., Johnston C., Rupp R.E., et al. DMID 21-0012 Study Group Homologous and heterologous covid-19 booster vaccinations. N. Engl. J. Med. 2022:1046–1057. doi: 10.1056/NEJMoa2116414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokal A., Broketa M., Barba-Spaeth G., Meola A., Fernández I., Fourati S., Azzaoui I., de La Selle A., Vandenberghe A., Roeser A., et al. Analysis of mRNA vaccination-elicited RBD-specific memory B cells reveals strong but incomplete immune escape of the SARS-CoV-2 Omicron variant. Immunity. 2022:1096–1104.e4. doi: 10.1016/j.immuni.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collier A.R.Y., Yu J., McMahan K., Liu J., Chandrashekar A., Maron J.S., Atyeo C., Martinez D.R., Ansel J.L., Aguayo R., et al. Differential kinetics of immune responses elicited by Covid-19 vaccines. N. Engl. J. Med. 2021;385:2010–2012. doi: 10.1056/NEJMc2115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferdinands J.M., Rao S., Dixon B.E., Mitchell P.K., DeSilva M.B., Irving S.A., Lewis N., Natarajan K., Stenehjem E., Grannis S.J., Han J. Waning 2-dose and 3-dose effectiveness of mRNA vaccines against COVID-19–associated emergency department and urgent care encounters and hospitalizations among adults during periods of Delta and omicron variant predominance — VISION network, 10 states, August 2021–January 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:255–263. doi: 10.15585/mmwr.mm7107e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O’Connell A.-M., et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. 2022:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katzelnick L.C., Coello Escoto A., McElvany B.D., Chávez C., Salje H., Luo W., Rodriguez-Barraquer I., Jarman R., Durbin A.P., Diehl S.A., et al. Viridot: an automated virus plaque (immunofocus) counter for the measurement of serological neutralizing responses with application to dengue virus. PLoS Negl Trop Dis. Oct. 2018;12:e0006862. doi: 10.1371/journal.pntd.0006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edara V.V., Pinsky B.A., Suthar M.S., Lai L., Davis-Gardner M.E., Floyd K., Flowers M.W., Wrammert J., Hussaini L., Ciric C.R., et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N. Engl. J. Med. 2021;385:664–666. doi: 10.1056/NEJMc2107799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edara V.V., Hudson W.H., Xie X., Ahmed R., Suthar M.S. Neutralizing Antibodies Against SARS-CoV-2 Variants After Infection and Vaccination. JAMA. 2021 doi: 10.1001/jama.2021.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanderheiden A., Edara V.V., Floyd K., Kauffman R.C., Mantus G., Anderson E., Rouphael N., Edupuganti S., Shi P.Y., Menachery V.D., et al. Development of a rapid focus reduction neutralization test assay for measuring SARS-CoV-2 neutralizing antibodies. Curr Protoc Immunol. 2020;131:e116. doi: 10.1002/cpim.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edara V.V., Norwood C., Floyd K., Lai L., Davis-Gardner M.E., Hudson W.H., Mantus G., Nyhoff L.E., Adelman M.W., Fineman R., et al. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe. 2021;29:516–521.e3. doi: 10.1016/j.chom.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The safety and immunogenicity data reported in this study cannot be deposited in a public repository because they are being collected and analyzed as part of the DMID 21-0012 study, which is ongoing and for which the dataset has not yet been locked. To request access, contact the IDCRC Secondary Research Project Manager (IDCRC.sec.research@fredhutch.org, section 18 of IDCRC Manual of Procedures, https://idcrc.org/about/procedures/index.html). Summary statistics describing previously published data is available at the NEJM: https://doi.org/10.1056/NEJMoa2116414.
-
•
This paper does not report original code.
-
•
Any additional information required to analyze the data reported in this paper is available from the Lead Contact upon request.