Summary
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) monoclonal antibodies (mAbs) are among the treatments recommended for high-risk ambulatory persons with coronavirus 2019 (COVID-19). Here, we study viral culture dynamics post-treatment in a subset of participants receiving the mAb bamlanivimab in the ACTIV-2 trial (ClinicalTrials.gov: NCT04518410). Viral load by qPCR and viral culture are performed from anterior nasal swabs collected on study days 0 (day of treatment), 1, 2, 3, and 7. Treatment with mAbs results in rapid clearance of culturable virus. One day after treatment, 0 of 28 (0%) participants receiving mAbs and 16 of 39 (41%) receiving placebo still have culturable virus (p < 0.0001). Recrudescence of culturable virus is detected in three participants with emerging mAb resistance and viral RNA rebound. While further studies are necessary to fully define the relationship between shed culturable virus and transmission, these results raise the possibility that mAbs may offer immediate (household) and public-health benefits by reducing onward transmission.
Keywords: COVID, COVID-19, monoclonal antibodies, COVID therapies, SARS-CoV-2, viral culture, resistance, mAbs
Graphical abstract
Highlights
-
•
Longitudinal sampling of participants treated with monoclonal antibody bamlinivimab
-
•
Treatment with bamlanivimab results in rapid clearance of culturable SARS-CoV-2
-
•
Culturable virus detected upon viral rebound is linked to emergent mutations
Using longitudinal samples from the ACTIV-2 clinical trial of the monoclonal antibody bamlinivimab, Boucau et al. investigate the duration of shedding culturable virus. Treatment with monoclonal antibody results in rapid clearance of culturable virus. The emergence of mutations in a subset of participants coincides with viral rebound and resurgent culturable virus.
Introduction
As the coronavirus 2019 (COVID-19) pandemic progress, interventions have been developed to prevent transmission and progression to severe disease in infected persons. Monoclonal antibodies (mAbs) were among the first therapies to receive emergency-use authorization (EUA) for the treatment of COVID-19 and remain among the first-line therapy options for the outpatient management of high-risk individuals with mild to moderate COVID-19 (https://www.covid19treatmentguidelines.nih.gov). While the initial circulating strains of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were susceptible to all mAbs deployed clinically, recently emerging strains, in particular the Omicron variant, are substantially less susceptible to some mAbs.1,2 Each variant has a unique mAb-susceptibility profile, and guidelines for clinical management have serially changed to account for the resistance pattern of the dominant circulating variant at any given time (https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html). Currently, Bebtelovimab is the mAb of choice given its demonstrated activity against BA.1 and BA.2.3,4 Interestingly, data suggest that emerging variants are not inevitably more broadly resistant to mAbs but may regain sensitivity to mAbs not active against earlier variants5 (https://www.phe.gov/emergency/events/COVID19/investigation-MCM/Bamlanivimab-etesevimab/Pages/resumption-in-distribution-bamlanivimabetesevimab.aspx). All mAbs used to date for COVID-19 target the interaction between the SARS-CoV-2 spike protein and the ACE2 receptor on host cells, effectively blocking viral uptake. mAbs differ primarily in their binding site on the spike protein and potentially the affinity with which they bind; all mAbs currently in clinical use are of the immunoglobulin G1 (IgG1) subclass. Given that all mAbs have the same target and mechanism of action and are of the same subclass, clinical phenotypes observed upon treatment with one effective mAb are highly likely to be common to all effective mAbs.
mAbs have been shown to accelerate the decay of SARS-CoV-2 levels in the upper respiratory tract,6,7 but their effects on duration of shedding culturable virus is unknown. While viral RNA is commonly used to assess viral burden, shedding of culturable virus could be a more sensitive indicator of antiviral activity. Further, in the absence of a proven correlate of infectiousness, culturable virus has been considered the best available proxy for the ability to transmit infection.8 We hypothesized that reduction in shedding of culturable virus might occur more rapidly than reduction in anterior nasal SARS-CoV-2 RNA levels following mAb treatment. A full understanding of the potential benefits and limitations of mAbs and other treatments would help determine their optimal use for preventing and treating SARS-CoV-2 infection.
Bamlanivimab is a neutralizing mAb that received EUA as a treatment for individuals 12 years of age and older with mild to moderate COVID-19 in November 2020.9 We performed viral culture analysis of participants enrolled in the ACTIV-2 randomized placebo-controlled trial of bamlanivimab monotherapy for non-hospitalized adults with mild to moderate COVID-1910 (ClinicalTrials.gov: NCT04518410). In that study, bamlanivimab treatment reduced respiratory tract (nasopharyngeal) viral RNA levels by 3 days post-treatment. In this work, we sought to understand how mAb treatment impacts the dynamics of shedding culturable SARS-CoV-2.
Results
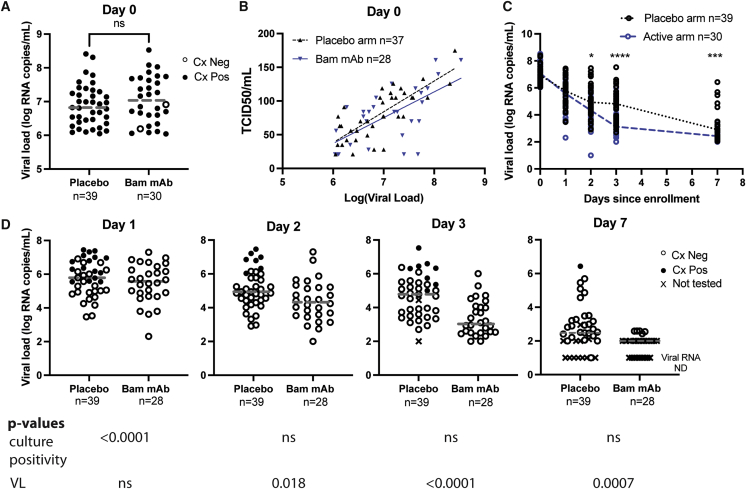
To compare shedding of culturable virus and change in anterior nasal (AN) sample SARS-CoV-2 RNA over time after treatment with mAbs, we cultured virus from AN swabs collected from participants enrolled in the ACTIV-2 study10 who had a baseline (pre-treatment, day 0) viral load of ≥6 log10 SARS-CoV-2 RNA copies/mL and available swab samples from study days 0, 1, 2, 3, and 7. Participants with evidence of bamlanivimab resistance mutations at baseline or during follow up based on our previous viral sequencing work11 were excluded for the primary analysis. Of the 317 participants in the ACTIV-2 study, 69 met inclusion criteria for the primary analysis in this study: 310 had available day 0 AN swabs, 94 had baseline viral load ≥6 log10 SARS-CoV-2 RNA copies/mL, and 73 had swabs available at days 0, 1, 2, 3, and 7. Four participants were excluded from the primary analysis due to emergent resistance identified in our previous work.11 Of the 69 participants meeting inclusion criteria, 39 were in the placebo arm and 30 were in the bamlanivimab arm (20 received the 7,000 mg dose and 10 received the 700 mg dose). Baseline participant characteristics, including age, race, comorbidities, days of symptoms before enrollment, and serostatus, were similar between groups (Table S1). Spike sequencing demonstrated that the majority of strains fell into SARS-CoV-2 clade 20A, with the remainder in clades 20B, 20C, 20E, and 20G (Figures S1A and S1B). No strains were variants of concern. Baseline AN RNA level was similar between groups (Figure 1A). Baseline viral culturability, as determined by cytopathic effect (CPE), was also similar between groups, with 39/39 (100%) participants in the placebo arm and 28/30 (93%) participants in the mAb arm with culture-positive baseline sample (Figure 1A). The two participants with baseline negative cultures were excluded from further analysis. For samples showing CPE (39 placebo and 28 mAb samples), we calculated the semiquantitative viral culture titer (tissue culture infectious dose 50 [TCID50]); the relationship between SARS-CoV-2 RNA and semiquantitative viral TCID50 was similar between groups at the time of enrollment (Figure 1B).
Figure 1.
Bamlanivimab treatment results in rapid SARS-CoV-2 culture conversion
(A) Pre-treatment culture positivity and viral load. Horizontal line indicates median.
(B) Pre-treatment TCID50 values versus viral load for individuals with CPE. Spearman correlations: placebo r = 0.7382, p < 0.0001; Bam mAb r = 0.6012, p value = 0.0007.
(C) Decay in qPCR-determined viral load over time post-treatment. Horizontal lines connect the medians for each experimental group for each timepoint.
(D) Culture positivity and viral load over time post-treatment. . Horizontal line indicates median. Cx, culture; Bam mAb, bamlanivimab monoclonal antibody; ND, not detected. X indicates samples not tested either because of lack of sample availability (one placebo sample day 3) or because VL was at or below the limit of detection.
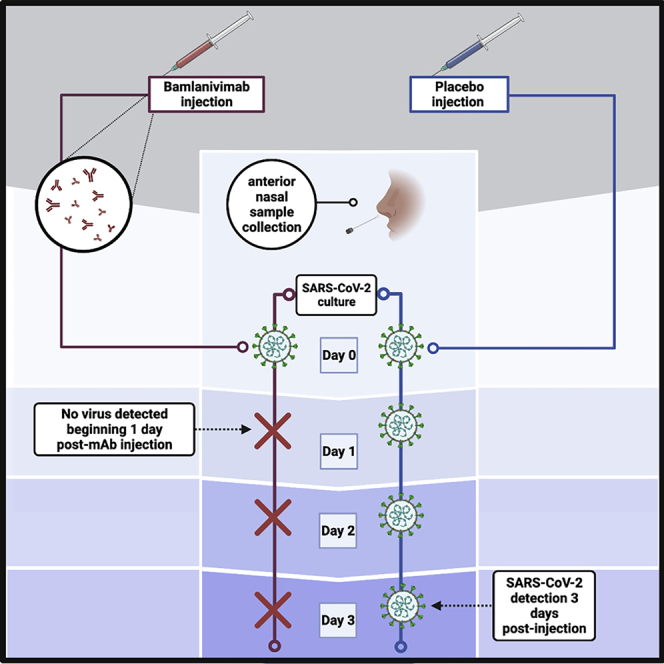
Participants received either placebo or bamlanivimab on day 0. AN sample SARS-CoV-2 RNA was assessed prior to treatment (day 0) and at study days 1, 2, 3, and 7 post-treatment. Shedding of culturable virus was assessed prior to treatment (day 0) and at study days 1–7 post-treatment for all samples with viral loads >2 log10. While study day 1 AN swab viral RNA levels were similar between arms (Figures 1C and S2A), a significant difference in culture positivity was observed on day 1. In the placebo arm, the culture-positivity rate was 16/39 (41%) versus 0/28 (0%) in the bamlanivimab arm (p < 0.0001; Figures 1D and S2B). In the placebo arm on day 1, the lowest viral load associated with a positive culture was 5.5 log10 RNA copies/mL, and 16/25 (64%) of placebo-arm samples with a viral load ≥5.5 log10 RNA copies/mL were also found to be culture positive. In contrast, all 18 samples from the bamlanivimab arm with viral loads ≥5.5 log10 RNA copies/mL were culture negative. By day 2 post-treatment, 7 of 39 (18%) participants in the placebo arm were still culture positive; all participants in the bamlanivimab arm remained culture negative. On day 3, eight of 37 tested placebo participants remained culture positive, and on study day 7, one placebo participant remained culture positive. All bamlanivimab-treated participants remained culture negative from day 1 onward.
To assess whether treatment with other mAbs would drive similarly rapid culture conversion, we queried a database developed as part of the POSITIVES study.12, 13, 14, 15 The POSITIVES study is a longitudinal cohort of patients newly diagnosed with COVID-19; following enrollment, nasal swabs are collected thrice weekly for 2 weeks. Each sample is simultaneously analyzed by whole-genome sequencing, spike sequencing, viral load determination, and viral culture. As of April 20, 2022, 109 immunocompetent patients had been enrolled in the cohort; analysis of the dataset revealed that six individuals had received mAb therapy after a positive baseline culture (4 Delta variant treated with casirivimab/imdevimab, 1 Delta variant treated with bamlanivimab/etesevimab, and 1 Omicron variant infection treated with sotrovimab). Analyzing viral load (VL) and TCID50 for those patients revealed that all six had durable viral culture conversion at the sample taken subsequent to mAb treatment (Figure S3). Sampling was performed less frequently than for ACTIV-2 participants, but these results support the idea that mAb treatment drives viral culture conversion.
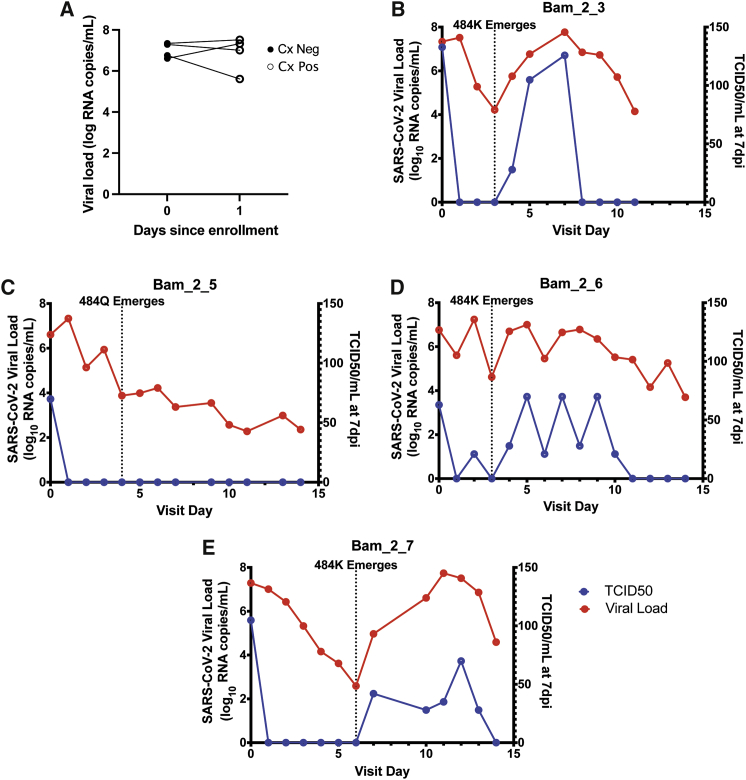
Viral resistance to bamlanivimab monotherapy has been described both in vitro and clinically, with resistance attributed to defined mutations in the SARS-CoV-2 spike protein.2,11,16 We previously identified participants from ACTIV-2 with emergent bamlanivimab resistance mutations,11 and these individuals were excluded from our primary analysis. We hypothesized that early viral culture clearance observed following bamlanivimab treatment was due to mAb binding and neutralization of virions and that mAb-resistance emergence would lead to renewed shedding of culturable virus. To test this hypothesis, we evaluated four participants with treatment-emergent bamlanivimab resistance mutations identified in our previous work;11 virus from three participants had emergent E484K mutation and one had emergent E484Q mutation. All four participants had positive viral cultures at day 0; similar to other participants in the bamlanivimab treatment arm of this substudy, all four individuals converted their cultures to negative on day 1 (Figure 2A). However, the emergence of the E484K mutation following bamlanivimab treatment in the three participants was associated with rebound in VLs and return of positive viral cultures with rising TCID50 levels (Figures 2B–2D). The participant with emergent E484Q mutation in their infecting virus had only modest increases in VL and no re-emergence of positive viral cultures (Figure 2E).
Figure 2.
Emergence of bamlanivimab resistance mutations correlates with recrudescent shedding of culturable virus
(A) Viral load and culture positivity at baseline and day 1 post-treatment for four study participants with recrudescent shedding of culturable virus. Cx, culture.
(B–E) Viral load and TCID50 for four study participants whose infecting virus developed E484 mutations of the spike protein following bamlanivimab monotherapy.
Discussion
Our results demonstrate that mAb treatment drives a rapid reduction in AN shedding of culturable virus that precedes a detectable reduction in viral RNA level. To date, most experimental estimates of SARS-CoV-2 transmission have been based on population-level studies. Very few studies have prospectively evaluated transmission, and to our knowledge, no correlates of infectiousness have been definitively identified. Shedding viable virus is highly likely to be a necessary condition for infectiousness, and previous studies have suggested a duration of shedding culturable virus that correlates with an epidemiologically informed understanding of the period of infectiousness.8 In the absence of proven correlates of infectiousness, shedding culturable virus is a reasonable proxy for the potential to transmit infection. While targeted prospective studies will be needed to precisely define the relationship between SARS-CoV-2 culture positivity in any given assay and disease transmission, our results suggest that treatment with mAbs may markedly reduce the period that an individual with COVID-19 infection remains infectious.
While bamlanivimab is not currently in clinical use, the identical molecular targets and mechanisms of action of all SARS-CoV-2-targeting mAbs that have been used clinically make it likely that our finding of rapid culture clearance will translate similarly to other mAb treatments. Indeed, our limited data for other variants and mAbs support this proposition (Figure S3). Whether this reduction in shedding of culturable virus is unique to mAbs or would be similarly observed with other treatments, such as antiviral drugs with different mechanisms of action, is incompletely known. A recent study demonstrating reduction in culturable SARS-CoV-2 by study day 3 in participants treated with molnupiravir relative to placebo suggests that this benefit may not be specific to mAb treatment but may hold across a range of COVID-19 therapies.17 Targeted studies of the impact of various treatments on culture conversion will be needed to definitively determine those most likely to impact shedding of culturable virus.
Our results additionally have implications for the potential of treated individuals to evolve and transmit SARS-CoV-2 strains with acquired resistance mutations. Re-emergence of culturable virus in a subset of ACTIV-2 participants with treatment-emergent mAb resistance suggests that the development of resistance mutations may lead to a return of infectiousness and the risk of transmission of mAb-resistant strains. The recrudescent shedding of culturable virus in a subset of mAb-treated individuals also suggests that time to initial culture conversion and sustained culture conversion provide complementary information. While rapid culture conversion may reflect a reduced initial period of infectiousness, sustained culture conversion would reflect durable loss of infectiousness. Lessons can potentially be taken from optimized management of HIV, in which combination therapy has been shown to be the most effective means of preventing viral evolution and the consequent emergence of resistant virus. Focused studies explicitly testing combinations of agents would potentially be helpful in defining optimal treatment regimens for COVID-19. While bamlanivimab is not currently used clinically, our resistance-related findings remain relevant given the recent reliance on the single mAb regimen sotrovimab and the report of emerging resistance against this regimen.18 Bebtelovimab is the most recent mAb to be granted EUA status (https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-new-monoclonal-antibody-treatment-covid-19-retains); whether resistance will emerge as it is more widely used to treat infections with newly emerging variants remains to be determined.
Moving COVID-19 from a pandemic disease to an endemic disease will require making optimal use of the full arsenal of available interventions for prevention and treatment. While COVID-19 therapies have, to date, been evaluated based on benefit to the treated individual, our results suggest that mAbs may have an additional role to play in reducing the period of infectiousness and consequently reducing the risk of secondary transmission. CDC recommendations for the duration of isolation following COVID-19 infection have recently changed (https://www.cdc.gov/media/releases/2021/s1227-isolation-quarantine-guidance.html); the optimal duration of isolation remains unclear. If mAbs reduce the period that an individual remains infectious, such interventions could ultimately be used to reduce the recommended time of isolation, conferring significant individual and societal benefits. Understanding the virologic consequences of therapeutic interventions for SARS-CoV-2 infection is critical for informing the optimal deployment of COVID-19 therapies as we seek to shift toward an endemic phase of the disease and for informing public-health recommendations after treatment.
Limitations of the study
Our cohort is comprised of individuals with symptomatic infection who were unvaccinated based on study timing. In this substudy, we focused on the subset of individuals most likely to have positive baseline cultures, namely those with baseline viral RNA level of ≥6 log10 SARS-CoV-2 RNA copies/mL. Based on the association we have previously observed between SARS-CoV-2 AN RNA level and likelihood of shedding culturable virus,15,19,20 individuals with lower baseline viral RNA would be more likely to be culture negative at presentation. The precise relationship between baseline AN-swab-culture positivity, RNA level, and viral transmissibility remains to be precisely defined; whether with further optimization, virus could be reliably cultured from samples with lower viral RNA levels remains to be determined. However, the potential benefits of rapid culture conversion and reduced transmission might be greatest for individuals with high viral RNA levels and those presenting early in the course of illness, who are most likely to be culture positive.8
Because of available specimens, each individual was only sampled once for each time point. While this is typical for clinical cohort sampling, our measurements are likely noisier than they would be if duplicate sampling were possible. The nature of our samples makes positive controls internal to each specimen impossible. However, the randomized, placebo-controlled study design offers a solid comparison, as any sampling variability would be expected to be evenly distributed between groups. While we anticipate that results with bamlanivimab will translate to other mAbs, bamlanivimab, like all mAbs previously or currently in use as COVID-19 therapies, is an IgG1 subclass antibody. Not all antibody subclasses have the same distribution within the body, and it is possible that any future mAbs of different subclasses may not have similar effects.
Consortia
The members of the POSITIVES study team who are not otherwise listed as authors on the manuscript are Grace C. Chamberlin, Geoffrey Chen, Rebecca F. Gilbert, Surabhi L. Iyer, Jacob E. Lemieux, May Y. Liew, Taryn Lipiner, Caitlin Marino, Josh Mathews, Zahra Reynolds, Mark J. Siedner, Arshdeep Singh, Ashley M. Stuckwisch, Rockib Uddin, Tammy D. Vyas, and Jatin M. Vyas.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Isolate USA-WA1/2020, GenBank: MN985325 | BEI Resources | Cat# NR-52281 |
| Biological samples | ||
| Remnant nasal swab samples in viral transport media – ACTIV2 trial | Chew et al., 2021 (ref.10) | N/A |
| Nasal swab samples in viral transport media – POSITIVES study | Boucau et al., 2022 (ref.12) or This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s Modified Eagle Medium | Fisher Scientific | Cat# MT10013CV |
| HEPES | Fisher Scientific | Cat# MT25060CI |
| Penicillin/Streptomycin | Fisher Scientific | Cat# MT30001CI |
| Glutamine | Thermo Scientific | Cat# 35050061 |
| Fetal Bovine serum | Sigma | Cat# F4135 |
| Trypsin-EDTA | Fisher Scientific | Cat# 25-200-072 |
| Polybrene | Santa Cruz Biotechnology | Cat# sc-134220 |
| Deposited data | ||
| Gene Expression Omnibus | Accession Number | PRJNA816433 |
| Experimental models: Cell lines | ||
| Vero-E6 | ATCC | Cat# CRL-1586 |
| Software and algorithms | ||
| GraphPad Prism v9.1.0 | GraphPad Software, https://www.graphpad.com/ | N.A. |
Resource availability
Lead contact
Further information and requested for reagents should be directed to and will be fulfilled by the lead contact, Amy K. Barczak (abarczak@mgh.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Study participants - ACTIV-2
ACTIV-2 is a multi-center randomized, blinded placebo-controlled phase 2/3 platform trial in non-hospitalized adults10 (ClinicalTrials.gov Identifier: NCT04518410). ACTIV-2 evaluated the safety and efficacy of mAb bamlanivimab infusion compared to saline placebo in non-hospitalized participants with positive SARS-CoV-2 antigen or nucleic acid test within 7 days and less than 10 days of COVID-19 symptoms. Participants were enrolled at 39 sites in the U.S. between August 19 and November 17 2020. There were two cohorts with different dosages of bamlanivimab (700 and 7000mg, each compared to placebo) in ACTIV-2, however due to low numbers, the two intervention groups were analyzed together. The study protocol was approved by the Mass General Brigham IRB as a secondary use protocol. The parent ACTIV-2 protocol was approved by a central IRB, Advarra (Pro00045266), with additional local IRB review and approval as required by participating sites. All participants provided written informed consent for ACTIV-2.
ACTIV-2 is an AIDS Clinical Trials Group (ACTG) study (ACTG A5401). As with all ACTG studies, at the time samples are collected, additional samples are collected, banked, and made accessible to the scientific community to enable others to ask distinct scientific questions with those samples. To ask the specific questions posed in this work, we applied to ACTG leadership and obtained permission to access these samples for secondary use.
Participant selection- ACTIV-2 substudy
Inclusion criteria for this substudy included baseline (pre-treatment, day 0) AN viral load of ≥6 log10 SARS-CoV-2 RNA copies/mL, available AN swab samples from study days 0, 1, 2, 3 and 7. Participants otherwise meeting criteria for inclusion but with evidence of bamlanivimab resistance mutations at baseline or during follow-up based on our previous viral sequencing work11 were excluded from the primary analysis and were analyzed as a separate emergent resistance subset.
Study participants - POSITIVES
The POSITIVES study is a longitudinal cohort study of individuals newly diagnosed with COVID-19 within the MassGeneral Brigham system.12, 13, 14, 15 Individuals are enrolled following a positive test, and AN samples are collected thrice weekly for two weeks.
Method details
Sample collection - ACTIV-2
Serial AN samples were self-collected by participants daily between enrollment (day 0) and day 14 using standardized swabs and collection procedures. Following self-collection, AN swabs were stored at cool temperatures (refrigerated or in a study-provided cooler with a combination of refrigerated and frozen gel packs). Swabs were returned to a study site and frozen at −80°C (−65°C to −95°C) within 7 days of collection. Samples were shipped on dry ice to a central laboratory where it was eluted in 3 mL of viral transport media. Each sample was cultured once using 0.3 mL of elution fluid. The collection, storage, processing, and assay methods have previously been validated.15,21
Study participants and sample collection- POSITIVES
AN swabs are self-collected by participants and placed into 4 mL volume of viral culture media. Collected samples are picked up by a study coordinator within 3 hr of collection and delivered directly to the lab. Samples are then divided into 8 aliquots and frozen at −80C until used. Each sample is analyzed by whole genome sequencing, spike sequencing, viral RNA level determination, and viral culture. As of 4/20/2022, 6 immunocompetent individuals in the POSITIVES cohort had positive baseline cultures and had been treated with mAbs; VL and culture results are reported in Figure S3.
Cell lines
Vero-E6 cells (ATCC) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with HEPES, Penicillin/Streptomycin, Glutamine, and 10% Fetal Bovine serum (FBS), detached using Trypsin-EDTA. Cells were kept in culture for no longer than one month or ten passages, whichever came first.
Viral culture
Viral culture experiments were performed as previously reported in the BSL3 laboratory of the Ragon Institute of MGH, MIT, and Harvard.15,19,20 Vero-E6 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with HEPES, Penicillin/Streptomycin, Glutamine, and 10% Fetal Bovine serum (FBS), detached using Trypsin-EDTA and seeded at 75,000 cells per well in 24-well plates or 20,000 cells per well in 96-well plates 16-20 hours before infection. AN specimens were thawed on ice, filtered through a Spin-X 0.45 or 0.65um centrifugal filters at 10,000 × g for 5min and diluted them 1:10 in DMEM supplemented with HEPES, Penicillin/Streptomycin and Glutamine. 100uL of the diluted solution was used to inoculate triplicate wells in a 24-well plate for large scale culture experiments. After 1 hour of incubation at 37°C and 5% CO2, the viral inoculum was removed and 1mL of DMEM supplemented with HEPES, Penicillin/Streptomycin and Glutamine and 2% FBS (D2+) was added to each well. For the TCID50 experiments, 25ul of the undiluted filtrate was added to four wells of a 96-well plate and serially diluted (1:5) in D2+ media containing 5ug/mL of polybrene. The 96-well plates were then spinfected for 1 hour at 2000 × g at 37°C. The SARS-CoV-2 isolate USA-WA1/2020 strain and DMEM supplemented with HEPES, Penicillin/Streptomycin and Glutamine were used as positive and negative controls, respectively. We observed viral culture plates at 3- and 7-days post-infection with a light microscope and documented wells showing cytopathic effect (CPE). TCID50 titers were calculated using the Spearman-Karber method. In previous work, we had found that our assay was highly likely to grow virus for samples with RNA levels ≥6 log10 RNA copies/mL.15,19,20 All samples with viral RNA levels at or above the limit of detection (2 log10) were subjected to viral culture, with the exception of one Day 3 sample from the placebo group (VL 4.4 log10) that was not available for viral culture testing.
Culture positivity
Specimens were defined as culture positive if at least 1 out of 3 wells showed CPE in the 24-well culture experiments or 1 out of 24 wells showed CPE in the TCID50 experiments. Specimens with no observable CPE in either 24 well or TCID50 experiments were defined as culture negative. The USA WA-1/2020 strain was used as a positive control in all experiments.
Quantification and statistical analysis
Statistical analyses were performed using PRISM software v9.2.0. Comparison of viral RNA levels between the placebo and bamlanivimab treatment arms at the different time points was performed using the non-parametric two-tailed Mann-Whitney test. Comparison of proportion of culture conversion between the placebo and bamlanivimab treatment arms at the different time points was performed using two-sided Fisher’s exact test on a 2 × 2 contingency table.
Additional resources
Parent study ACTIV-2: ClinicalTrials.gov Identifier NCT04518410.
Website for ACTIV-2 https://actgnetwork.org/activ-2/.
Acknowledgments
The authors would like to thank the participants, site staff, site investigators, and the entire ACTIV-2/A5401 study team. The authors would additionally like to thank Amanda K. Marsh for her help as well as the Ragon BSL3 core staff. Biorender.com was used to generate the graphical abstract. This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers 3UM1AI068636-14S2, UM1 AI068634, UM1 AI068636, and UM1 AI106701. The viral culture work was performed in the Ragon Institute BSL3 core, which is supported in part by the NIH-funded Harvard University Center for AIDS Research (P30 AI060354). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Bamlanivimab was donated for the ACTIV-2 study by Eli Lilly and Company.
Author contributions
Conceptualization, J.Z.L. and A.K.B.; methodology, investigation, data curation, and formal analysis, J.B., J. Regan, M.C.C., R.D., J.P.F., C.R.C., J.Z.L., and A.K.B.; resources, M.D.H., J. Ritz, C.M., J.A.D., A.C.J., A.N., P.K., A.L.G., R.W.C., W.A.F., E.S.D., D.A.W., J.J.E., J.S.C., D.M.S., and the POSITIVES study team; supervision and project management, J.Z.L. and A.K.B.; writing – original draft, J.B., J.Z.L., and A.K.B.; writing – reviewing & editing, J.B., J. Regan, M.C.C., R.D., J.P.F., M.D.H., J. Ritz, C.M., C.R.C., J.A.D., A.C.J., A.N., P.K., A.L.G., R.W.C., W.A.F., E.S.D., D.A.W., J.J.E., J.S.C., D.M.S., the POSITIVES study team, J.Z.L., and A.K.B.
Declaration of interests
J.B., M.C.C., R.D., J. Regan, J.P.F., C.R.C., M.D.H., J. Ritz, C.M., J.A.D., A.C.J., R.W.C., J.S.C., and A.K.B. report no competing interests. K.W.C. reports research grant support to the institution from Merck Sharp & Dohme. A.N. and P.K. are employees and shareholders of Eli Lilly. A.L.G. declares central testing contracts with Abbott and research support from Gilead and Merck outside of the submitted work. W.A.F. reports research funding from Ridgeback Biopharmaceuticals and consultancy fees from Roche and Merck and serves on adjudication committees for Janssen and Syneos. E.S.D. reports consulting fees from Gilead Sciences and Merck and research support to the institution from Gilead Sciences and ViiV. J.J.E. reports serving as an ad hoc consultant to GSK/VIR and as data monitoring committee (DMC) chair for Adagio Phase III studies. D.M.S. reports consulting fees from the following companies: Fluxergy, Kiadis, Linear Therapies, Matrix BioMed, Arena Pharmaceuticals, VxBiosciences, Model Medicines, Bayer Pharmaceuticals, Signant Health, and Brio Clinical. J.Z.L. reports consulting for Abbvie and Recovery Therapeutics.
Published: June 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100678.
Contributor Information
Jonathan Z. Li, Email: jli@bwh.harvard.edu.
Amy K. Barczak, Email: abarczak@mgh.harvard.edu.
Supplemental information
Data and code availability
-
•
Sequencing data has been deposited in Gene Expression Omnibus: PRJNA816433.
-
•
The clinical data reported in this study cannot be deposited in a public repository because of ethical restrictions. To request access, contact sdac.data@sdac.harvard.edu with the written agreement of the AIDS Clinical Trials Group and the manufacturer of the investigational product. Sequence data for strains is available upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
References
- 1.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 2.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 3.Dougan M., Azizad M., Chen P., Feldman B., Frieman M., Igbinadolor A., Kumar P., Morris J., Potts J., Baracco L., et al. Bebtelovimab, alone or together with bamlanivimab and etesevimab, as a broadly neutralizing monoclonal antibody treatment for mild to moderate, ambulatory COVID-19. medRxiv. 2022 doi: 10.1101/2022.03.10.22272100. Preprint at. [DOI] [Google Scholar]
- 4.Westendorf K., Zentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39:110812. doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., Iwatsuki-Horimoto K., Halfmann P., Watanabe S., Maeda K., et al. Efficacy of antiviral agents against the SARS-CoV-2 omicron subvariant BA.2. N. Engl. J. Med. 2022;386:1475–1477. doi: 10.1056/NEJMc2201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dougan M., Nirula A., Azizad M., Mocherla B., Gottlieb R.L., Chen P., Hebert C., Perry R., Boscia J., Heller B., et al. Bamlanivimab plus etesevimab in mild or moderate covid-19. N. Engl. J. Med. 2021;385:1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 9.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew K.W., Moser C., Daar E.S., Wohl D.A., Li J.Z., Coombs R., Ritz J., Giganti M., Javan A.C., Li Y., et al. Bamlanivimab reduces nasopharyngeal SARS-CoV-2 RNA levels but not symptom duration in non-hospitalized adults with COVID-19. medRxiv. 2021 doi: 10.1101/2021.12.17.21268009. Preprint at. [DOI] [Google Scholar]
- 11.Choudhary M.C., Chew K.W., Deo R., Flynn J.P., Regan J., Crain C.R., Moser C., Hughes M., Ritz J., Ribeiro R.M., et al. Emergence of SARS-CoV-2 resistance with monoclonal antibody therapy. medRxiv. 2021 doi: 10.1101/2021.09.03.21263105. Preprint at. [DOI] [Google Scholar]
- 12.Boucau J., Marino C., Regan J., Uddin R., Choudhary M.C., Flynn J.P., Chen G., Stuckwisch A.M., Mathews J., Liew M.Y., et al. Duration of viable virus shedding in SARS-CoV-2 omicron variant infection. medRxiv. 2022 doi: 10.1101/2022.03.01.22271582. Preprint at. [DOI] [Google Scholar]
- 13.Regan J., Flynn J.P., Choudhary M.C., Uddin R., Lemieux J., Boucau J., Bhattacharyya R.P., Barczak A.K., Li J.Z., Siedner M.J. Detection of the omicron variant virus with the Abbott BinaxNow SARS-CoV-2 rapid antigen assay. Open Forum Infect. Dis. 2022;9:ofac022. doi: 10.1093/ofid/ofac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seaman M.S., Siedner M.J., Boucau J., Lavine C.L., Ghantous F., Liew M.Y., Mathews J., Singh A., Marino C., Regan J., et al. Vaccine breakthrough infection with the SARS-CoV-2 delta or omicron (BA.1) variant leads to distinct profiles of neutralizing antibody responses. medRxiv. 2022 doi: 10.1101/2022.03.02.22271731. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siedner M.J., Boucau J., Gilbert R.F., Uddin R., Luu J., Haneuse S., Vyas T., Reynolds Z., Iyer S., Chamberlin G.C. Duration of viral shedding and culture positivity with post-vaccination SARS-CoV-2 delta variant infections. JCI Insight. 2022;7:e155483. doi: 10.1172/jci.insight.155483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 17.Fischer W., Eron J.J., Holman W., Cohen M.S., Fang L., Szewczyk L.J., Sheahan T.P., Baric R., Mollan K.R., Wolfe C.R., et al. Molnupiravir, an oral antiviral treatment for COVID-19. medRxiv. 2021 doi: 10.1101/2021.06.17.21258639. Preprint at. [DOI] [Google Scholar]
- 18.Rockett R.J., Basile K., Maddocks S., Fong W., Agius J.E., Mackinnon J.J., Arnott A., Chandra S., Gall M., Draper J., et al. Resistance conferring mutations in sars-cov-2 delta following sotrovimab infusion. medRxiv. 2021 doi: 10.1101/2021.12.18.21267628. Preprint at. [DOI] [Google Scholar]
- 19.North C.M., Barczak A., Goldstein R.H., Healy B.C., Finkelstein D.M., Ding D.D., Kim A., Boucau J., Shaw B., Gilbert R.F., et al. Determining the incidence of asymptomatic SARS-CoV-2 among early recipients of COVID-19 vaccines (DISCOVER-COVID-19): a prospective cohort study of healthcare workers before, during and after vaccination. Clin. Infect. Dis. 2022;74:1275–1278. doi: 10.1093/cid/ciab643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yonker L.M., Boucau J., Regan J., Choudhary M.C., Burns M.D., Young N., Farkas E.J., Davis J.P., Moschovis P.P., Bernard Kinane T., et al. Virologic features of severe acute respiratory syndrome coronavirus 2 infection in children. J. Infect. Dis. 2021;224:1821–1829. doi: 10.1093/infdis/jiab509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg M.G., Zhen W., Lucic D., Degli-Angeli E.J., Anderson M., Forberg K., Olivo A., Sheikh F., Toolsie D., Greninger A.L., et al. Development of the RealTime SARS-CoV-2 quantitative Laboratory Developed Test and correlation with viral culture as a measure of infectivity. J. Clin. Virol. 2021;143:104945. doi: 10.1016/j.jcv.2021.104945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Sequencing data has been deposited in Gene Expression Omnibus: PRJNA816433.
-
•
The clinical data reported in this study cannot be deposited in a public repository because of ethical restrictions. To request access, contact sdac.data@sdac.harvard.edu with the written agreement of the AIDS Clinical Trials Group and the manufacturer of the investigational product. Sequence data for strains is available upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.