Summary
Efficient quantitative assays for measurement of viral replication and infectivity are indispensable for future endeavors to develop prophylactic or therapeutic antiviral drugs or vaccines against SARS-CoV-2. We developed a SARS-CoV-2 cell-cell transmission assay that provides a rapid and quantitative readout to assess SARS-CoV-2 spike hACE2 interaction in the absence of pseudotyped particles or live virus. We established two well-behaved stable cell lines, which demonstrated a remarkable correlation with standard cell-free viral pseudotyping for inhibition by convalescent sera, small-molecule drugs, and murine anti-spike monoclonal antibodies. The assay is rapid, reliable, and highly reproducible, without a requirement for any specialized research reagents or laboratory equipment and should be easy to adapt for use in most investigative and clinical settings. It can be effectively used or modified for high-throughput screening for compounds and biologics that interfere with virus-cell binding and entry to complement other neutralization assays currently in use.
Keywords: SARS-CoV-2, cell-cell transmission, pseudotyping, quantitative assay, neutralization, stable cell line, infectivity assay, convalescent sera, post-vaccination sera
Graphical abstract
Highlights
-
•
An efficient cell-cell transmission assay for SARS-CoV-2 spike-hACE2 interaction
-
•
Uses two stable cell lines and a single, helper-dependent adenovirus vector
-
•
Reliably tests effects of small-molecule drugs, monoclonal antibodies, and vaccines
-
•
Rapid, reproducible assay, easily modifiable to encode spike variants of concern
Motivation
Reproducible and facile methods of measuring viral replication are indispensable in the development of vaccines and therapeutic medications against SARS-CoV-2. We sought to develop a rapid, time-saving, cell-cell transmission assay that can be used to measure and quantify SARS-CoV-2 virus-cell binding and entry to circumvent the challenges faced by the assays currently used as gold standards for SARS-CoV-2 neutralization studies.
Ssenyange et al. describe an efficient, reproducible cell-cell transmission assay for rapid quantification of SARS-CoV-2 spike interaction with hACE2 that can be used to test the effects of convalescent/post-vaccination serum, monoclonal antibodies, and small-molecule drugs. It can be adapted for clinical settings and to test spike variants of concern.
Introduction
Coronavirus disease 2019 (COVID-19), due to the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become an ongoing, persistent global public health challenge since its emergence in December 2019 (Vogel, 2020), with over 260 million confirmed cases worldwide and close to 5.2 million COVID-19-related deaths as of November 30, 2021 (https://coronavirus.jhu.edu/), despite the wide availability of multiple fully approved safe and efficacious vaccines.
SARS-CoV-2 is a betacoronavirus, belonging to the genus Coronavirus, in the family Coronaviridae. It is an enveloped, non-segmented, positive-sense single-stranded RNA virus (Li et al., 2020a; Chan et al., 2020). It has a genome nearly 30 kb in length, including many open reading frames (ORFs), which express at least 27 proteins (Chan et al., 2020; Wu et al., 2020a). The surface spike glycoprotein (S) plays a key role in viral entry into target host cells (Wrapp et al., 2020; Bosch et al., 2003). The receptor-binding subunit S1 attaches to the host cell via the cellular receptor human angiotensin-converting enzyme 2 (hACE2), triggering proteolytic activation of S and subsequent conformational changes of the S2 subunit, which facilitates the fusion of viral and cellular membranes (Huang et al., 2020; Li et al., 2003, 2020b).
Currently, the development of numerous prophylactic and therapeutic strategies is in progress to mitigate this global public health crisis (Klasse and Moore, 2020; Lythgoe and Middleton, 2020; Sarma et al., 2020; Wang et al., 2020a), including the use of small-molecule drugs (Riva et al., 2020); biologics, including interferon (Shalhoub, 2020); convalescent sera (Bloch et al., 2020; Casadevall and Pirofski, 2020); monoclonal antibodies (Zost et al., 2020; Zhou and Zhao, 2020; Wu et al., 2020b; Ho et al., 2020; Pinto et al., 2020; Liu et al., 2020; Ju et al., 2020; Jiang et al., 2020; Cao et al., 2020b); oligonucleotides (Rehman and Tabish, 2020); peptides (VanPatten et al., 2020); and vaccines (Yu et al., 2020; Graham, 2020; Wang and Zhang, 2020). Remdesivir remains the only US Food and Drug Administration (FDA)-approved antiviral drug for use in adults and pediatric patients for treatment of COVID-19 requiring hospitalization (https://www.fda.gov/drugs/news-events-human-drugs/fdas-approval-veklury-remdesivir-treatment-covid-19-science-safety-and-effectiveness). Three vaccines have emergency use authorization (EUA) from the FDA or are fully approved (https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines), and four vaccines have been formally approved by the European Medicines Agency (EMA) (https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines/covid-19-vaccines). A few SARS-CoV-2-targeting monoclonal antibodies (mAbs) have also been approved through an EUA for use in COVID-19 patients meeting eligibility criteria (https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs). No other specific antiviral drug against SARS-CoV-2, however, has been formally approved by the FDA or EMA.
Quantitative assays for the measurement of viral replication and infectivity are indispensable for future endeavors to develop prophylactic or therapeutic antiviral drugs or vaccines against SARS-CoV-2. The current gold standards for SARS-CoV-2 neutralization include pseudotyping using S and a suitable virus core encoding a reporter (Jiang et al., 2020; Schmidt et al., 2020; Ou et al., 2020; Zhao et al., 2021; Crawford et al., 2020) or inhibition of live virus replication in vitro or in animal models (Zhao et al., 2021; Johansen et al., 2020; Bao et al., 2020). Although it has been previously reported that inhibition of replication-competent virus or pseudotyped particles correlates well with protection from SARS-CoV-2 virus challenge in pre-clinical models of infection (Yu et al., 2020; Chandrashekar et al., 2020; Mercado et al., 2020), these assays require high-level biocontainment and continued production, testing, and cryostorage of virus vector supernatants, which may be variable in quality and differ remarkably from lab to lab. These assays are typically very time consuming, requiring several days to produce and concentrate the virus to achieve a suitably high titer, with assay readout on susceptible cells after a few days (Quinonez and Sutton, 2002); live coronavirus requires a BSL3 laboratory and readout by plaque or similar quantitative assay several days after cell or animal infection, performed in vitro or in vivo, respectively.
To circumvent some of these issues, we developed a cell-cell transmission-based assay that utilizes producer cells expressing S, an HIV packaging vector, and an HIV-based transfer vector, which, when co-cultured together with hACE2-expressing target cells, provide a rapid and quantitative readout to assess S-hACE2 interaction in the absence of producing or using pseudotyped particles or live virus. The assay is very rapid, reliable, and highly reproducible; it is easy to implement in most laboratories without need for special reagents or equipment, compared with other assays that rely upon virus production and testing. It can be effectively used or modified for high-throughput screening for compounds and biologics that interfere with virus-cell binding and entry to complement other neutralization assays that are currently in use.
Results
Development of a quantitative cell-cell transmission assay for SARS-CoV-2 in transiently transfected cells
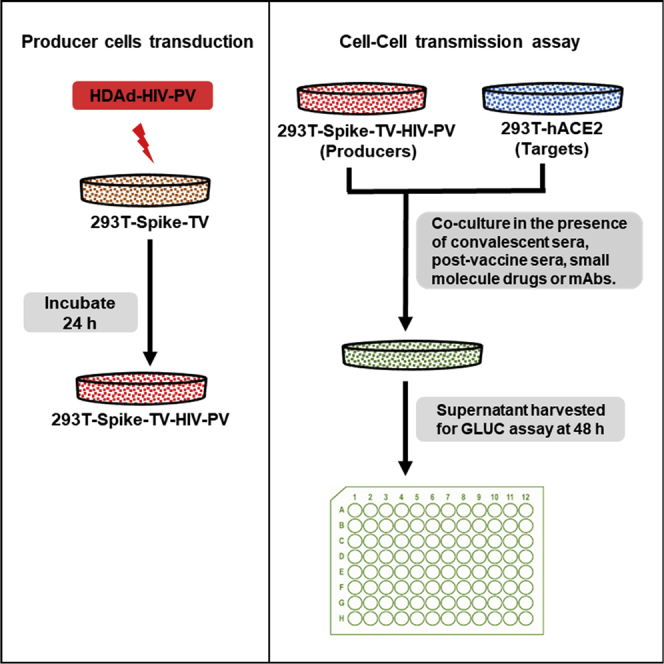
Viruses can spread via either a cell-free or a cell-associated route, the latter involving direct cell-cell contact (Mothes et al., 2010). Cell-cell transmission can occur via a virologic synapse and, at least in the case of HIV, is typically much more efficient than cell-free infection (Sattentau, 2008; Agosto et al., 2015; Chen et al., 2007; Jolly et al., 2004; Jolly and Sattentau, 2004; Martin et al., 2010; Zhong et al., 2013a, 2013b; Abela et al., 2012). Like other coronaviruses, the S protein mediates cell-surface receptor recognition, cell attachment, and fusion during SARS-CoV-2 virus infection of susceptible cells expressing hACE2, its cognate receptor (Li et al., 2003; Ou et al., 2020; Hoffmann et al., 2020; Belouzard et al., 2012; Shang et al., 2020). To measure cell-cell transmission of SARS CoV-2, we employed a system developed by the late David Derse (Mazurov et al., 2010b). We generated target cells expressing hACE2, the cognate receptor for SARS-CoV-2, and producer cells that were transiently transfected with plasmid DNAs to introduce S, along with all requisite trans-acting factors for HIV core production (HIV packaging vector or HIV-PV) and an HIV-based transfer vector (HIV-TV), as described under STAR Methods and illustrated in Figure 1A.
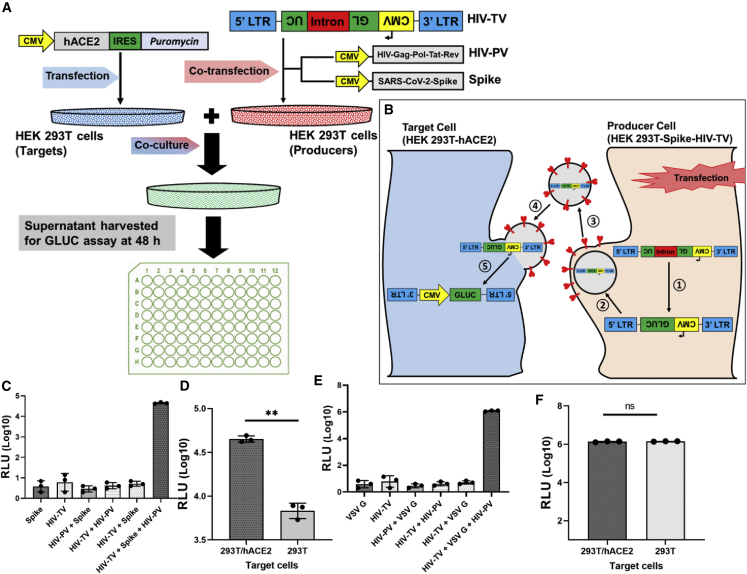
Figure 1.
Cell-cell transmission assay is dependent upon the expression of hACE2 in target cells and the HIV packaging vector (HIV-PV), HIV transfer vector (HIV-TV), and viral glycoprotein in producer cells
(A) Schematic illustration of cell-cell transmission assay experimental design.
(B) Schematic illustration of cell-cell transmission at the cellular level, showing how HIV-TV (with a backward or reverse luciferase gene) is activated in the target cells. The Gaussia luciferase gene has an intron that is spliced out in the producer cell following transfection (1) and packaged into pseudoparticles (2), which are then released into the culture medium (3). S-pseudotyped particles infect adjacent 293T-hACE2 target cells (4). In the target cell, the vector RNA (now spliced) is reverse transcribed and integrated into the target cell genome, activating the CMV-driven Gaussia luciferase gene (5). Note that in the cytosol of the producer cell the vector is RNA, which is also true for released pseudoparticles; only in the target cell is the vector RNA converted to dsDNA and then integrated into the target cell genome.
(C–F) pHIV-TV was transfected into 293T cells in the presence or absence of HIV-PV and either empty plasmid, plasmid encoding spike, or VSV G. After 48 h these cells were mixed with 293T or 293T-hACE2 targets, and 48 h later the culture supernatant was assessed for Gaussia luciferase activity. The paired t tests for two-group comparison were analyzed using GraphPad Prism software. ns, not significant; ∗∗p < 0.01.
To validate the cell-cell transmission assay, we initially tested whether the assay is dependent on the expression of S and HIV-PV by producer cells and the expression of hACE2 by target cells. HIV-TV was transfected into 293T cells in the presence or absence of HIV-PV and either empty plasmid, plasmid encoding Wuhan S, or a vesicular stomatitis virus (VSV) G expression plasmid. After 48 h these cells were mixed with 293T or 293T-hACE2 cells, and 48 h later the culture supernatant was assessed for relative light unit (RLU) activity. As shown in Figure 1C, RLU values were close to background (<10) without any glycoprotein added; in the presence of S, RLUs increased ∼4,000-fold. Compared with target cells without hACE2, RLUs increased ∼10-fold in the presence of hACE2 (Figure 1D). In the absence of HIV-PV, RLU activity was <10, essentially a background value (Figure 1C). As anticipated, cell-cell transmission in the presence of VSV G was not dependent on hACE2, since its cognate receptor is ubiquitous in mammalian eukaryotic cells (Figure 1F).
Convalescent sera from COVID-19+ patients and post-vaccination sera inhibit cell-cell transmission in transiently transfected cells
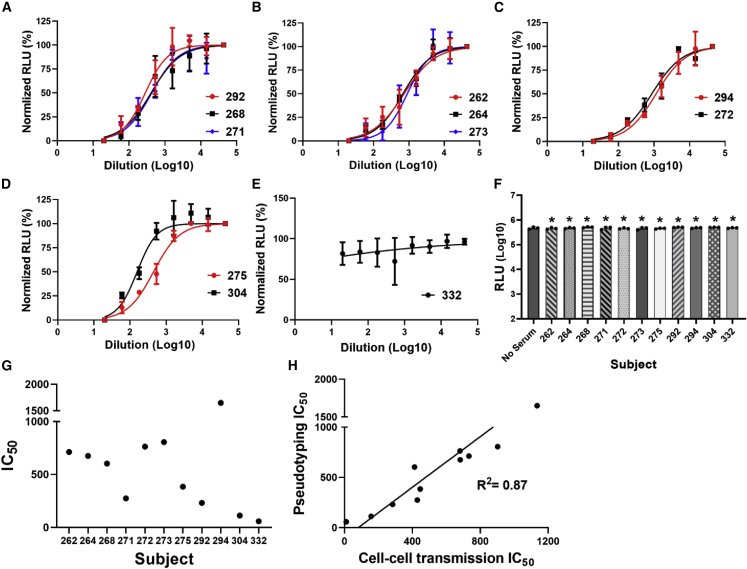
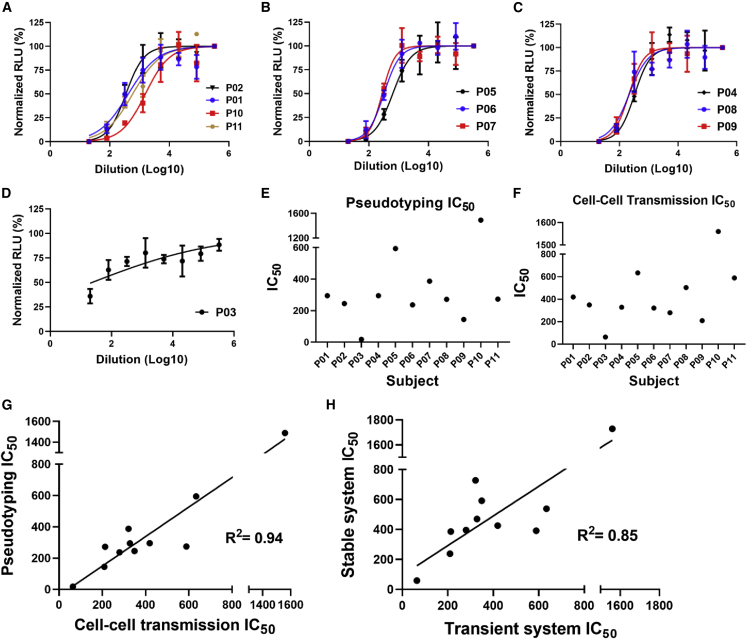
We next investigated the effects of convalescent sera from COVID-19+ patients and post-vaccination sera on cell-cell transmission, initially using transiently transfected cells. Convalescent sera from 11 COVID-19+ patients and sera from 11 subjects at 1 month post vaccination were selected at random for testing. Demographic and relevant clinical information of the subjects is shown in Tables S1 and S2 for convalescent and post-vaccination sera, respectively. Four-fold serial dilutions of convalescent and post-vaccination sera were pre-incubated with S-expressing producer cells for 1 h, and then hACE2-expressing target cells were added, performed in triplicate. RLU was measured after 48 h, and IC50 values were calculated. Apart from two sera (one convalescent, Figure 2E, and one post vaccination, Figure 3E), we observed that convalescent (Figures 2A–2D) and post-vaccination (Figures 3A–3D) sera inhibited cell-cell transmission of S in the transient system, with a strong positive correlation with the results we observed with S pseudotyping using the same sera (Figures 2H and 3H for convalescent and post-vaccination sera, respectively). No inhibition by convalescent or post-vaccination sera was observed with cell-cell transmission using VSV G, as anticipated (Figures 2F and 3F).
Figure 2.
COVID-19+ convalescent sera inhibit spike-mediated cell-cell transmission in the transient system
(A–D) Four-fold serial dilutions of convalescent sera were pre-incubated with spike-expressing producer cells for 1 h, and then hACE2-expressing target 293T cells were added, performed in triplicate. RLU was measured after 48 h, and IC50 values were calculated.
(E) One serum sample showed no inhibitory effect.
(F) No inhibition of VSV G pseudotyped virus was observed at the lowest serum dilution tested (1:20). ∗Not significant compared with no serum control.
(G) IC50 values calculated for S pseudotyping using same sera, performed as shown in Figure S1.
(H) Correlation between IC50 values for cell-cell transmission and pseudotyping assay for convalescent sera. See also Figures S1 and S4. The paired t tests for two-group comparison were analyzed using GraphPad Prism software.
Figure 3.
Post-vaccination sera inhibit spike-mediated cell-cell transmission in the transient system
(A–D) Four-fold serial dilutions of post-vaccination sera were pre-incubated with spike-expressing producer cells for 1 h, and then hACE2-expressing target cells were added, performed in triplicate. RLU was measured after 48 h, and IC50 values were calculated.
(E) One serum sample showed no inhibitory effect.
(F) No inhibition of VSV G pseudotyped virus was observed at the lowest serum dilution tested (1:20). ∗Not significant compared with no serum control.
(G) IC50 values calculated for S pseudotyping using same sera, performed as in Figure S1.
(H) Correlation between IC50 values for the cell-cell transmission and pseudotyping assay for post-vaccination sera. See also Figures S1 and S4. The paired t tests for two-group comparison were analyzed using GraphPad Prism software.
Studies with LCB1 peptide and small-molecule drugs in the transient system
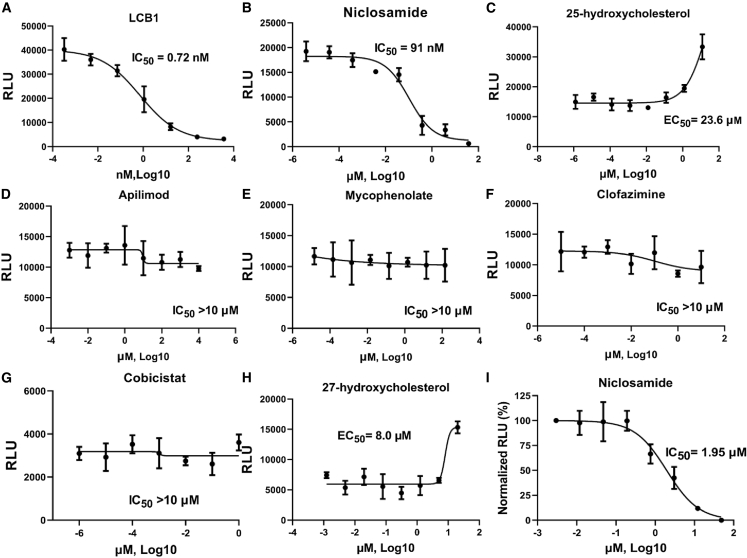
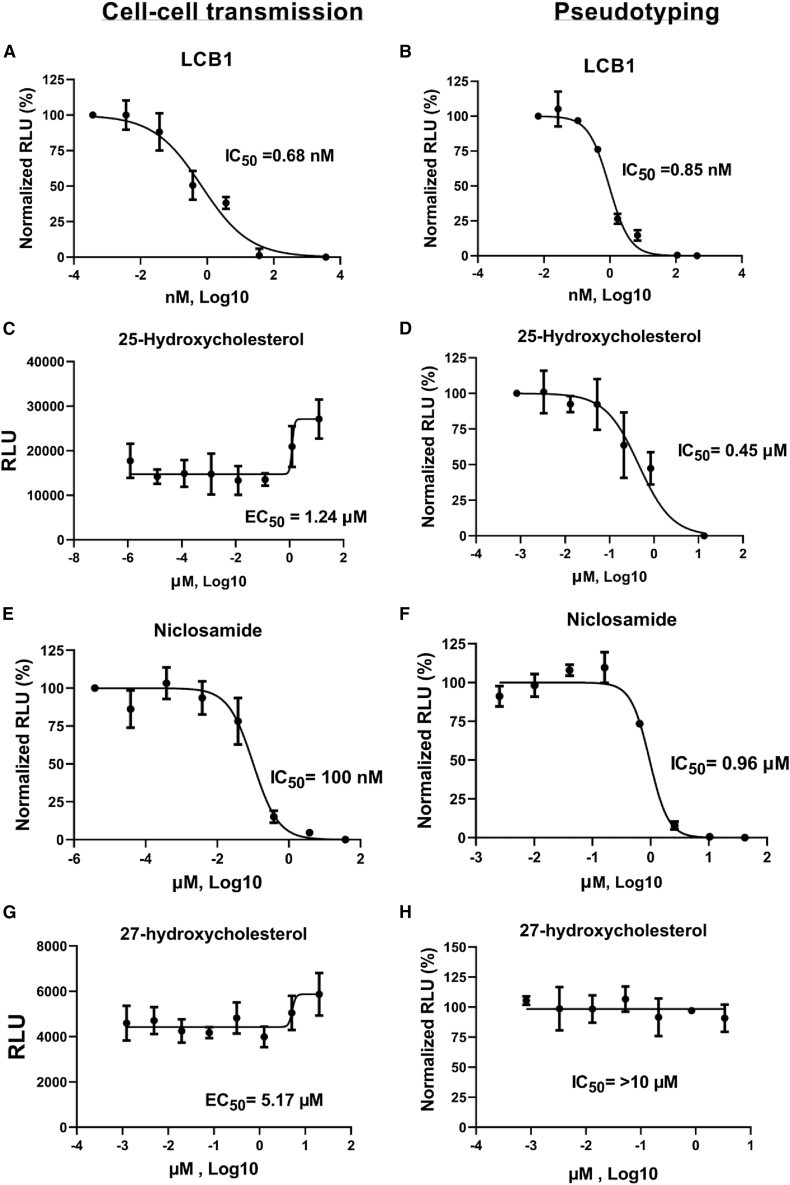
We then investigated the effect of LCB1 (56-mer peptide) and small-molecule drugs, including apilimod, cobicistat, clofazimine, niclosamide, 25-hydroxycholesterol, 27-hydroxycholesterol, and mycophenolate (MPA), all previously reported to be inhibitory to SARS-CoV-2 infection (Prabhakara et al., 2021; Xie et al., 2020; Yuan et al., 2021; Wang et al., 2020b), on cell-cell transmission of S in the transient system as described under STAR Methods. LCB1 has been reported to efficiently neutralize pseudotyped SARS-CoV-2 virus entry (Cao et al., 2020a). When we tested its ability to inhibit cell-cell transmission of S, the IC50 was 0.72 nM (Figure 4A), which is essentially the same as the IC50 value for pseudotyping (0.85 nM, Figure 6B). Among the small-molecule drugs, niclosamide was inhibitory of cell-cell transmission of S (IC50 of 91 nM; Figure 4B), whereas 25-hydroxycholesterol and 27-hydroxycholesterol were stimulatory, enhancing cell-cell transmission of S with EC50 of 23.6 and 8 μΜ (Figures 4C–4H), respectively. We observed no obvious inhibitory or stimulatory effect of either 25-hydroxycholesterol or 27-hydroxycholesterol on cell-cell transmission when VSV G instead of S was employed as the viral glycoprotein (Figures S3E and S3F, respectively). We observed no obvious inhibitory effect on S cell-cell transmission when using MPA, cobicistat, clofazimine, or apilimod, with each of them having IC50 > 10 μM (Figures 4D–4G).
Figure 4.
Effect of peptide and small-molecule drugs in the transient system
(A–I) Ten-fold serial dilutions of LCB1 and other small-molecule drugs were pre-incubated with producer cells in parallel for 1 h, and then hACE2-expressing target cells were added, performed in triplicate. LCB1 (A), niclosamide (B), 25-hydroxycholesterol (C), apilimod (D), mycophenolate (E), clofazimine (F), cobicistat (G), and 27-hydroxycholesterol (H) are shown. Niclosamide inhibited cell fusion. Serial dilutions of niclosamide were incubated with TZMbl-Spike producer cells for 1 h. Target cells (HOS-3734) were then added, and RLU was measured the following day (I). See also Figure S4.
Figure 6.
Characterization and validation of the stable cell line system
(A–H) Ten-fold serial dilutions of LCB1 (A), 25-hydroxycholesterol (C), niclosamide (E), and 27-hydroxycholesterol (G) were pre-incubated with spike-expressing 293T producer cells for 1 h, and then hACE2-expressing target cells were added, performed in triplicate. RLU was measured at 48 h, and IC50 values were calculated for each serum. Results were compared with S pseudotyping assay for each compound: LCB1 (B), 25-hydroxycholesterol (D), niclosamide (F), and 27-hydroxycholesterol (H). See also Figures S5 and S6.
Establishment of stable cell lines to quantify SARS-CoV-2 cell-cell transmission
The results described above demonstrate quantitation of S-mediated cell-cell transmission in transiently transfected 293T cells. Transient transfection of cells is associated with many drawbacks—it is relatively complicated, unreliable, occasionally irreproducible, and not amenable to high-throughput use. We thus sought to develop and establish stable cell lines that can be used to quantify S cell-cell transmission rapidly and reliably with highly reproducible results. We generated a target cell line stably expressing hACE2 (293T-hACE2) and a producer cell line stably expressing Wuhan S and the HIV-TV, inGLUC (cell line termed 293T-Spike-TV), as described under STAR Methods. Expression of S and hACE2 in these stable cell lines was confirmed by immunoblotting using an anti-FLAG antibody and anti-hACE2 antibody, as shown in Figures S5C and S5D, respectively. Stable expression of HIV-TV was confirmed by nested PCR to show the presence of 3′ long terminal repeat (LTR) sequences (a part of the transfer vector expression cassette, with an expected PCR product of 1.5 kb) using genomic DNA extracted from the stable producer cell line, 293T-Spike-TV (Figure S5E). Despite repeated efforts, we were unable to make a producer cell line stably expressing functional HIV-PV. To circumvent this problem, we utilized a previously described helper-dependent adenovirus (HDAd) vector (Hu et al., 2015) to transiently introduce HIV-PV, as described under STAR Methods. HDAd vectors have shown remarkable consistency and efficiency in the transduction of 293T cells compared with plasmid transfection, with higher levels of HIV structural and enzymatic protein expression (Hu et al., 2015). Expression of HIV Gag by the HDAd-transduced producer cells was confirmed by immunoblot (Figure S5A). This HDAd also encodes the LacZ gene, which serves as a reporter. We transduced 293T-Spike-TV cells with increasing amounts of HDAd-HIV-PV and fixed and stained cells for LacZ expression at 48 h. As the MOI increased, the percentage of blue cells also rose, such that essentially all the cells were transduced at high MOI (Figures S6A and S6B). We titrated the HDAd-HIV-PV to use optimal amounts that resulted in high transduction efficiency with minimal to no cytopathic effect on both the producer and the target stable cell lines. When increasing MOI of the HDAd-HIV-PV was employed, the RLU values following co-culture using the stable producer and target cell lines were ∼5-fold less compared with the transient system, but still ∼10,000-fold above background, with values directly correlated to the amount of HDAd-HIV-PV used for transduction (Figure S6C).
Additional characterization and validation of the stable cell line system
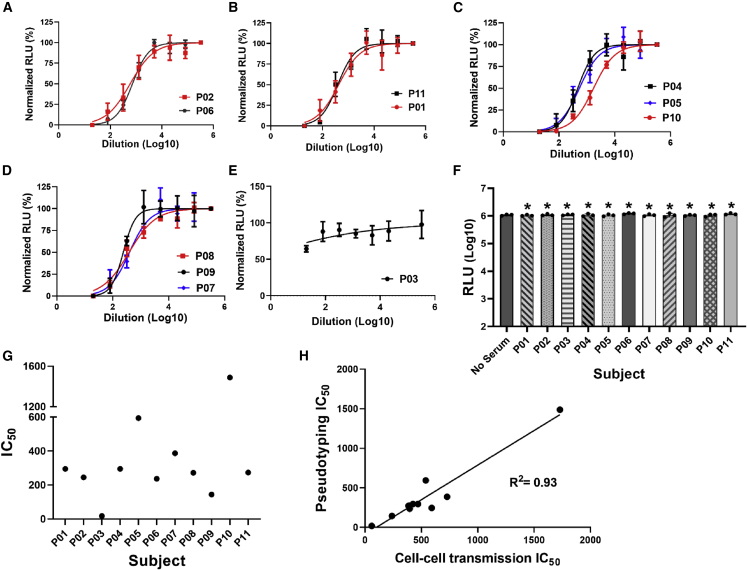
To further characterize and validate the stable cell lines, we tested the effects of LCB1, post-vaccination sera, and small-molecule drugs, including cobicistat, niclosamide, 25-hydroxycholesterol, and 27-hydroxycholesterol, on cell-cell transmission, in comparison with pseudotyping. We tested 11 samples of post-vaccination sera from the same subjects used in the transient system. Demographic and relevant clinical information of the subjects is shown in Table S2. With a single exception, all tested sera inhibited cell-cell transmission at varying titers (Figures 5A–5C). Apart from outlier P03 (Figure 5D), IC50 titers for the cell-cell transmission assay varied between 63.8 and 1,560 (Figure 5F). Sera from the same subjects were also tested in the pseudotyping assay (Figure 5E). Although the cell-cell transmission IC50 titers were generally higher compared with those of pseudotyping, there was a strong positive correlation between cell-cell transmission and pseudotyping results (Figure 5G), with an R2 value of 0.94. This is added proof that the cell-cell transmission assay is indeed measuring S-hACE2 interaction and virus infectivity. Unsurprisingly, the post-vaccination IC50 titers for the transient and stable cell-cell transmission systems were highly correlated, with an R2 value of 0.85 (Figure 5H).
Figure 5.
Post-vaccination sera inhibit cell-cell transmission of spike in the stable system
(A–C) Four-fold serial dilutions of post-vaccination sera were pre-incubated with spike-expressing producer cells for 1 h, and then hACE2-expressing target cells were added, performed in triplicate. RLU was measured after 48 h, and IC50 values calculated.
(D) One serum sample showed no inhibitory effect.
(E and F) IC50 values were calculated for pseudotyping (E) and cell-cell transmission (F), using the same sera.
(G) Correlation between IC50 values for cell-cell transmission and pseudotyping assay for post-vaccination sera in the stable system.
(H) Correlation of IC50 values for post-vaccination sera in the cell-cell transmission assay between the transient and the stable system. See also Figures S5 and S6.
Niclosamide was inhibitory in the stable cell-cell transmission assay, with an IC50 value of 0.10 μM, ∼10-fold lower than that of the pseudotyping (0.96 μM; Figure 6) and ∼20-fold lower than that of the cell fusion assay (1.95 μM; Figure 4I). 25-Hydroxycholesterol and 27-hydroxycholesterol were both stimulatory. 25-Hydroxycholesterol enhanced cell-cell transmission ∼2-fold, with an EC50 value of 1.24 μM, whereas 27-hydroxycholesteol had an EC50 value of 5.17 μM, enhancing cell-cell transmission ∼1.5-fold. This was in contrast to what we observed with S pseudotyping, in which 25-hydroxycholesterol was inhibitory, with an IC50 value of 0.45 μM, and 27-hydroxycholesterol had no obvious inhibitory or stimulatory effect on S pseudotyping (Figure 6).
LCB1 peptide was inhibitory, with an IC50 of 0.68 nM, which is ∼1.3-fold less than that of pseudotyping at 0.85 nM. Moreover, a time-of-addition experiment demonstrated that LCB1 efficiently inhibits cell-cell transmission at −1 and 0 h (with IC50 values of 0.68 and 1.37 nM, respectively) before target-producer cell co-culture. The inhibitory effect was reduced when LCB1 was added 1 h after co-culture (IC50 of 2.25 nM), and no effect was observed with addition after 2 h, as shown in Figure S2F. A similar trend was observed when we compared LCB1 time-of-addition results with pseudotyping (Figure S2E).
Studies with tetherin and murine anti-spike monoclonal antibodies
Tetherin is an interferon-induced membrane-associated host protein whose expression is known to block the release of HIV-1 and other enveloped viral particles (Perez-Caballero et al., 2009). We tested the effect of increasing amounts of tetherin on cell-cell transmission of SARS-CoV-2 S and how it compared with cell-free S pseudotyping. Producer cells were transiently transfected with increasing amounts of a cytomegalovirus (CMV)-driven plasmid encoding tetherin in a 12-well format for cell-cell transmission and 10-cm plates for pseudotyping, with results normalized to the amount of DNA transfected per surface area (ng/cm2). We observed an inhibitory effect of increasing amounts of tetherin on both S cell-cell transmission and pseudotyping. However, a more profound effect was seen on S pseudotyping, with a 10-fold decrement in RLU values at the highest amount of tetherin plasmid transfected (48 ng/cm2; Figure S3B), whereas cell-cell transmission RLU values were reduced only ∼2-fold at an even higher tetherin plasmid amount (normalized for plate surface area at 72 ng/cm2; Figure S3A). A similar trend of effect with tetherin was observed when VSV G instead of S was employed as the viral glycoprotein (Figures S3C and S3D).
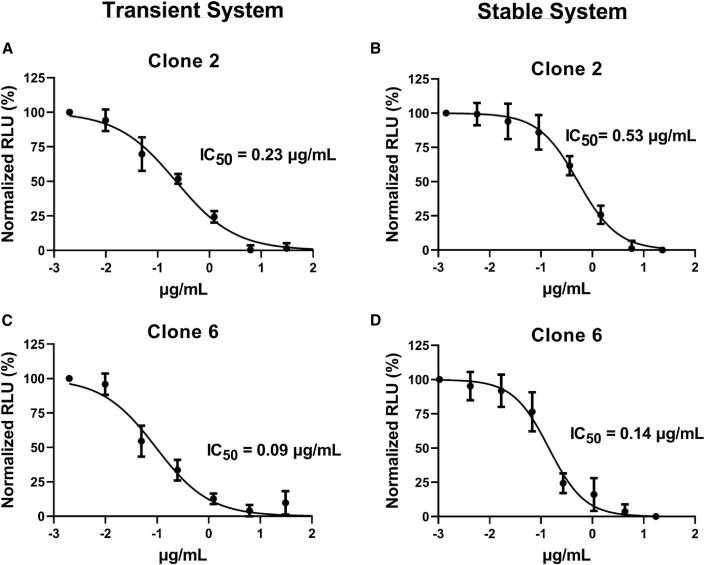
We also tested two anti-S murine monoclonal antibodies (clones 2 and 6, both of which bound the receptor-binding domain [RBD]), produced by co-author S.C. Four-fold serial dilutions of each monoclonal antibody were pre-incubated with S-expressing producer cells for 1 h, and then hACE2-expressing target cells were added, performed in triplicate. RLU was measured after 48 h, and IC50 values were calculated. Both clone 2 and 6 antibodies had good activity, inhibiting cell-cell transmission with IC50 values of 0.53 and 0.14 μg/mL, respectively. Clone 6 was consistently more inhibitory by several fold, compared with clone 2. Results in the transient cell-cell transmission system showed a similar relationship, with IC50 values of 0.23 and 0.09 μg/mL for clones 2 and 6, respectively (Figure 7).
Figure 7.
Anti-spike monoclonal antibodies inhibit cell-cell transmission
(A–D) Four-fold serial dilutions of monoclonal antibody were pre-incubated with spike-expressing producer cells for 1 h, and then hACE2-expressing target cells were added, performed in triplicate. RLU was measured after 48 h, and IC50 values were calculated. Clones 2 and 6 were assessed in the transient (A and C) and stable (B and D) cell-cell transmission assay.
Further studies of cell-cell transmission
It is possible that some of the resultant RLUs observed for the cell-cell transmission assay are due to cell-free virus being released from the producers and then infecting targets. To quantify this, prior to co-culturing the stable producers and targets, we removed increasing amounts of culture supernatant from the producers to infect the targets separately. We then co-cultured the producers and the targets. As can be seen in Figure S6D, there are some RLUs produced in the cell-free aspect of the experiment, but significantly less compared with the amount in the co-culture aspect of the experiment. These results are consistent with most of the RLUs in the cell-cell transmission assay coming from cell-cell and not cell-free transmission.
We also decided to construct producer cell lines expressing other S variants. We obtained codon-optimized versions of the Delta and Omicron variant S genes, both driven by the CMV IE enhancer/promoter. These cassettes were separately inserted into the base HIV-TV and stably introduced by transfection into 293T cells. A cell-cell transmission assay was performed in parallel with 293T-Spike-TV cells, which express Wuhan S. All three producer cell lines gave high RLUs, suggesting that this cell-cell transmission assay will work with S variants of concern as illustrated in Figure S6E.
Discussion
Quantitative assays for the measurement of viral replication and infectivity are indispensable to future endeavors to develop prophylactic or therapeutic antiviral drugs or vaccines for SARS-CoV-2. We describe herein a cell-cell transmission assay that provides a quantitative readout to assess SARS-CoV-2 S-hACE2 interaction, in the absence of producing or using pseudotyped particles or live virus. At most this assay requires only BSL2 biocontainment and is very reliable, relatively rapid, and highly reproducible. The current gold standards for SARS-CoV-2 neutralization include pseudotyping using S and a suitable virus core encoding a reporter (Schmidt et al., 2020; Ou et al., 2020; Jiang et al., 2020; Zhao et al., 2021; Crawford et al., 2020) or inhibition of live virus replication in vitro or in animal models (Johansen et al., 2020; Bao et al., 2020; Zhao et al., 2021). Although it has been previously reported that inhibition of replication-competent virus or pseudotyped particles correlates well with protection from SARS-CoV-2 virus challenge in pre-clinical models of infection (Yu et al., 2020; Chandrashekar et al., 2020; Mercado et al., 2020), these assays necessitate higher level biocontainment and continued production, testing, and cryostorage of virus vector supernatants, which can be variable in quality and differ based on the laboratory. These assays may be time consuming, requiring several days to produce and concentrate pseudotyped particles to achieve high titer, with assay readout on susceptible cells after a few days (Quinonez and Sutton, 2002). This entire process typically may take a minimum of 7 days from the transfection of producer cells to the readout following transduction of the target cells with concentrated pseudoparticle vector supernatants, compared with this cell-cell transmission assay that takes a maximum of 4 days to complete. Hence, we thought it useful to develop this rapid, time-saving, cell-cell transmission assay that can be used to measure and quantify SARS-CoV-2 virus cell binding and entry to circumvent the challenges faced by the assays that are currently used as gold standards for SARS-CoV-2 neutralization.
Viruses can spread in a cell-free or a cell-associated manner, the latter involving direct cell-cell contact, also known as cell-cell transmission (Mothes et al., 2010). Currently, there is no direct evidence of cell-cell transmission in humans. On the other hand, it is not precisely clear how virus spread occurs, either in vitro or in vivo, whether it is cell-free, cell-associated, or due to cell fusion, although cell syncytia have been observed in the lung tissues of COVID-19+ patients (Bradley et al., 2020; Tian et al., 2020; Polak et al., 2020). Cell-cell transmission can occur via a virologic synapse and, at least in the case of HIV, is typically much more efficient than cell-free infection (Agosto et al., 2015; Zhong et al., 2013a, 2013b; Jolly et al., 2010; Jolly and Sattentau, 2004; Martin et al., 2010; Abela et al., 2012). The ability of viruses to utilize and manipulate cell-cell contact likely contributes to the success of viral infections; cell-cell infection and spread not only facilitates rapid viral dissemination but may also promote immune evasion and influence disease progression (Zhong et al., 2013b; Aubert et al., 2009; Rudnicka et al., 2009). Whether any of the circulating S variants have enhanced cell-cell transmission is an open area of investigation. We developed a cell-cell transmission assay based on this concept, which is dependent on target cells expressing hACE2 and producer cells expressing S, along with all the requisite trans-acting factors for HIV core production (HIV-PV) and an HIV-based transfer vector, as illustrated in Figure 1C. Initial testing performed by transient transfection demonstrated that the assay is dependent on producer 293T cells expressing S, the transfer vector, and HIV-PV. The assay is also dependent on target cells expressing hACE2, with RLU numbers increasing ∼10-fold when we used 293T-hACE2 versus unmodified 293T cells as targets. This is consistent with 293Ts expressing very low levels of hACE2 or a substitute receptor, as observed previously (Sun et al., 2021; Temmam et al., 2022).
Since transient transfection is relatively complex, occasionally unreliable, and not entirely amenable to high-throughput use, we sought to develop and successfully established stable cell lines that could be used to quantify cell-cell transmission rapidly and reliably, with highly reproducible and quantitative results. We thus generated a target cell line stably expressing hACE2 (293T-hACE2) and a producer cell line stably expressing S and the transfer vector, inGLUC (293T-Spike-TV), with HIV-PV introduced by transduction using HDAd-HIV-PV 24 h prior to target-producer cell co-culture. We titrated and optimized the HDAd-HIV-PV to use amounts that showed highest transduction efficiency with minimal to no cytopathic effect on the 293T cells. Of note, this HDAd can be propagated and amplified using a specialized helper adenovirus, such that the titers are more than 1012 vp/mL, and the HDAd is quite stable when cryostored for years at −80°C.
To further validate and characterize the cell-cell transmission assay, we tested the effects of convalescent sera and post-vaccination sera on cell-cell transmission of S in the transient and stable systems, comparing the results with pseudotyping. A robust positive correlation between cell-cell transmission and pseudotyping IC50 titers for both convalescent and post-vaccination sera strongly suggests that the cell-cell transmission assay is indeed measuring S-hACE2 interaction and infectivity. Unsurprisingly, there was also a high correlation between the post-vaccination cell-cell transmission IC50 titers for the transient versus the stable system. The IC50 titers were generally higher with cell-cell transmission than those seen with pseudotyping, suggesting that S cell-cell transmission is more easily inhibited than cell-free transmission.
In addition to sera, we tested the effects of peptide LCB1 and small-molecule drugs, including cobicistat, niclosamide, apilimod, MPA, clofazimine, 25-hydroxycholesterol, and 27-hydroxycholesterol, all previously reported to be inhibitory of SARS-CoV-2 infection (Prabhakara et al., 2021; Xie et al., 2020; Yuan et al., 2021; Wang et al., 2020b). LCB1 has been reported to efficiently neutralize pseudotyped SARS-CoV-2 virus entry (Cao et al., 2020a), and we observed a similar effect on both pseudotyping and cell-cell transmission of Wuhan S. Comparison testing of peptide against the different S variants of concern (notably B.1.1.7, B.1.617.2, B.1.351, and B.1.1.529 https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/) showed inhibition with variants B.1.1.7 and B.1.617.2, and to our surprise, no effect was seen with variants B.1.351 and B.1.1.529 at peptide concentrations similar to those shown in Figures S4A–S4D. Comparison testing of sera against the S variants of concern showed no substantial difference in the IC50 titers compared with Wuhan S (Figures S4E–S4L). Among the small molecules, only niclosamide, 25-hydroxycholesterol, and 27-hydroxycholesterol had significant effects on cell-cell transmission. Niclosamide was inhibitory, with an IC50 value of 0.10 μM, whereas both 25-hydroxycholesterol and 27-hydroxycholesterol were stimulatory in the stable cell line system, enhancing S-mediated cell-cell transmission with EC50 of 1.23 and 5.17 μM, respectively. These two cholesterol derivatives are both known to inhibit cholesterol biosynthesis and reduce cholesterol content in membranes; precisely how they would differentially affect cell-free versus cell-cell SARS-CoV-2 cell infection is uncertain at this juncture but worthy of further study.
We also tested murine anti-S monoclonal antibodies and tetherin. Tetherin is an interferon-induced membrane-associated protein whose expression is known to block the full release of HIV-1 and other enveloped viral particles from cells (Perez-Caballero et al., 2009). Tetherin showed a profound inhibitory effect against pseudotyped particles with an ∼10-fold decrease in RLU when very low amounts were introduced into the producer cells. It had a less impressive effect on cell-cell transmission, with just an ∼2-fold decrease in RLU at the highest normalized concentration of tetherin we tested (72 ng/cm2), as illustrated in Figure S3. In general, it is felt that tetherin has a much greater impact on cell-free virus production, since the viruses produced remain attached to the producer cell and not released, whereas in the case of cell-cell transmission the large amount of extracellular virus present may counteract the inhibitory effect of the particle tethering, especially if the target is in contact with the producer cell. The impressive potency of the murine anti-S monoclonal antibodies in both cell-free and cell-cell transmission assays suggests that humanization of these antibodies for further evaluation, including clinical testing, may be of use.
This cell-cell transmission assay is also highly reproducible: repeated testing of an inhibitory peptide yielded very consistent results from three different experiments performed weeks apart, with minimal variation in IC50 values (Figure S2). Compared with pseudotyping, in which many laboratories use assorted plasmids to produce pseudotyped particles to titer on a range of targets using a variety of formats and readouts, here we employed just two stable cell lines and a single HDAd, the latter of which can be amplified to very high titer of >1012 vp/mL. Thus, our cell-cell transmission assay, which is based on three relatively simple, readily available, and easily renewable reagents, should allow for facile comparisons when used by qualified investigators anywhere on the planet. The producer cell line was also easily modified to encode two S variants of concern that have emerged in the last 12 months.
In conclusion, we have developed and established a rapid, reliable, and reproducible SARS-CoV-2 S-hACE2 cell-cell transmission assay. The assay requires two stable cell lines that are well behaved and HDAd-HIV-PV, which has remarkable efficiency in transducing 293T cells. This assay does not require any specialized research reagents or laboratory equipment and should be easy to adapt for use in most investigative and clinical settings. It can be effectively used or modified for high-throughput screening for compounds and biologics that interfere with virus-cell binding and entry to complement other neutralization assays currently in use.
Limitations of the study
We attempted to make a stable cell line expressing HIV-PV; despite repeated efforts, we were unable to do so. Thus, at present we utilize an HDAd-HIV-PV that needs to be separately prepared. In the future we hope to modify the stable cell line to encode and express HIV-PV. In addition, as the pandemic continues, more S variants of concern are likely to arise; in each case we would need to construct a new stable producer cell line. Here, it took us roughly 6 weeks to make the Delta and Omicron S-encoding HIV transfer vectors and stably transfect and hygromycin B select the 293T cells and perform initial validation studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-hACE2 | R&D systems | Cat# AF933-SP |
| Mouse anti-FLAG | Sigma-Aldrich | Cat# F1804-50UG |
| Rabbit anti-goat-HRP | Sigma-Aldrich | Cat# AP106P |
| Rabbit anti-mouse HRP | Cell Signaling | Cat# 58802 |
| Goat anti-rabbit HRP | Sigma-Aldrich | Cat# 12-348 |
| Rabbit anti-GAPDH | Sigma-Aldrich | Cat# SAB5600208 |
| Anti-human HRP | Sigma-Aldrich | Cat# AP112P |
| Anti-HIV human sera | NIH AIDS Reagent Repository | Cat# 3957 |
| Biological samples | ||
| Murine anti-spike monoclonal antibodies | Sidi Chen | https://doi.org/10.1038/s41467-022-29288-3 |
| Post-vaccine sera | West Haven VA Medical Center | N/A |
| Convalescent Sera | YNHH hospital (IMPACT research team) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Coelenterazine | Gold Bio | CAS #55779-48-1 |
| Sodium Iodide | Sigma-Aldrich | CAS #7681-82-5 |
| Deposited data | ||
| Raw and Analyzed Data | Mendeley Data | https://doi.org/10.17632/m7twyzc4yj.3 |
| Experimental models: Cell lines | ||
| Human Embryonic Kidney 293T cells | ATCC | Cat# CRL-3216 |
| 293T-Spike-HIV-TV | This paper | N/A |
| 293T-hACE2 | This paper | N/A |
| Oligonucleotides | ||
| LCB1 peptide (56-mer) DKEWILQKIYEIMRLLDELGHAEASMR VSDLIYEFMKKGDERLLEEAERLLEEVER |
ABI Scientific Inc | Sequence ID: 206841 |
| Nested PCR; Inside-Reverse primer: 5′-GCAGTGAGCGCAACGCAATTAATGTGA-3′ | This paper | N/A |
| Nested PCR; Outside-Reverse primer: 5′-GCGTTGGCCGATTCATTAATGCAGCT-3′ | This paper | N/A |
| Nested PCR; Outside-Forward primer: 5′-TTCATGCCTTCTTCTCTTTCCTACAGGGG-3′ | This paper | N/A |
| Nested PCR; Inside-Forward primer: 5′-GGCCACGATGTTGAAGTCTTCGTTGTTC-3′ | This paper | N/A |
| Recombinant DNA | ||
| pHIV-CMV-hACE2-IRES-Puro | This paper | N/A |
| pT-PB-SARS-CoV-2-Spike-IRES-Blasti | This paper | N/A |
| pUCHR-inGLuc-Beta | Walther Mothes | https://doi.org/10.1371/journal.pone.0053138 |
| pUCHR-inGLuc-pgk-hygro | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 9.3.1 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| HDAd-HIV-PV | This paper | N/A |
| Clofazimine | Sigma-Aldrich | Cas# 2030-63-9 |
| Cobicistat | Sigma-Aldrich | Cas# 1004316-88-4 |
| Apilimod | Sigma-Aldrich | Cas# 541550-19-0 |
| Mycophenolate | Sigma-Aldrich | Cas# 128794-94-5 |
| Niclosamide | Sigma-Aldrich | Cas# 50-65-7 |
| 27-hydroxycholesterol | Sigma-Aldrich | Cas# 20380-11-4 |
| 25-hydroxycholesterol | Sigma-Aldrich | Cas# 2140-46-7 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Richard Sutton (richard.sutton@yale.edu).
Materials availability
Plasmids and the stable cell lines generated during this study are available from the lead contact upon request.
Experimental model and subject details
Ethics statement
Deidentified COVID-19+ convalescent sera samples were obtained from YNHH hospital (IMPACT research team). IMPACT study was approved by the Yale University institutional review board (IRB). Deidentified Post-vaccine serum samples were obtained from West Haven VA Medical Center (approved by both the Yale IRB and WHVA IRB). Written, informed consent was obtained and documented for all subjects. Convalescent sera from 11 COVID-19+ patients and sera from 11 subjects at one month post vaccination were selected at random for testing. Demographic and relevant clinical information of the subjects is shown in Tables S1 and S2 for convalescent and post-vaccine sera, respectively.
Cell lines
Human embryonic kidney cell line 293T (#CRL-3216), was originally purchased from ATCC. Cell lines were maintained in Dulbecco’s modified Eagle’s high-glucose medium supplemented with 10% fetal calf serum, penicillin, streptomycin, and 10 μg/mL of ciprofloxacin (complete DMEM), with other antibiotics or supplements as indicated in the text. Cells were grown in 5% CO2, 37°C water-jacketed incubators and passaged every 3 to 5 days.
Method details
Vectors and plasmids
CMV-driven expression plasmid for S with an FLAG tag at the COOH terminus (pcDNA-SARS-CoV-2-S) was as previously described, as was plasmid encoding human ACE2 (hACE2) (Zhao et al., 2021). The hACE2 2.6 kbp ORF was blunt-cloned into a third generation HIV vector 3′ of CMV promoter and 5′ of an IRES-puror cassette to generate pHIV-CMV-hACE2-IRES-Puro. pSV-Tat, pCMV-Tat, and pLTR-LUC were as previously described (Zhao et al., 2021). Full length codon optimized Spike (S) was used in the development of this assay, S from pcDNA-SARS-CoV-2-S was inserted into a piggybac transposon (originally obtained from Matt Wilson of Baylor, along with the transposase plasmid pCMV-piggybac) that had been modified to encode a CMV-IRES-bsdr cassette; resultant plasmid was named pT-PB-SARS-CoV-2-Spike-IRES-Blasti. HIV-TV plasmid, encoding the intron-regulated HIV-based Gaussia luciferase pUCHR-inGLuc-Beta (HIV-inGLuc), was obtained from Dr. Walther Mothes, originally generated in the Derse/Heidecker lab as previously described (Mazurov et al., 2010a). Transcription of GLuc is antisense relative to transcription of the viral genomic RNA and is interrupted by a γ-globin intron inserted in sense orientation relative to the genomic RNA (Zhong et al., 2013a); this plasmid was modified to encode a hygromycin Br expression cassette (pUCHR-inGLuc-pgk-hygro). Plasmid expressing human tetherin, pcDNA3.1-tetherin, was also from Dr. Walther Mothes. HIV-PV was as described (Hu et al., 2015). HDAd-HIV-PV was produced as previously described (Suzuki et al., 2010; Coskun et al., 2006; Palmer and Ng, 2003). Variant B.1.1.7-expressing plasmid was obtained from Dr. Walther Mothes; it is codon-optimized and contains eight S protein amino acid mutations (Δ69-70, Y144Del, N501Y, A570D, P681H, T716I, S982A and D1118H) in addition to D614G (Tada et al., 2021a). B.1.351 variant was initially isolated in South Africa and has nine S protein amino acid mutations (L18F, D80A, D215G, L242-244del, R246I, K417N, E484K, N501Y and A701V) three of which (K417N, E484K and N501Y) are in the receptor binding domain (RBD) (Tegally et al., 2020). To construct this variant the latter three mutations were introduced into pcDNA-SARS-CoV-2-S using gene synthesis. Variant B.1.617.2 was initially isolated in India, its spike protein has L452R and T478K mutations in the RBD in addition to D614G and a P681R mutation near the proteolytic processing site (Tada et al., 2021b). To construct this VOC, L452R and T478K were introduced into pcDNA-SARS-CoV-2-S using gene synthesis. Codon-optimized Delta S variant plasmid was from the Mothes’ lab; codon-optimized Omicron S variant plasmid was originally purchased from Sino-Biological.
Cell-cell transmission assay
To measure cell-cell transmission of SARS CoV-2, we employed a system developed by the late David Derse (Mazurov et al., 2010b). In one cell (the target cell), hACE2 was stably introduced by VSV G-mediated HIV-based transduction using pHIV-CMV-hACE2-IRES-Puro to produce target HEK 293T-hACE2 cells, maintained in selection using 10 μg/mL puromycin (Sigma-Aldrich). From Walther Mothes we obtained a second-generation cell-cell transmission HIV-TV, in which an improved intron in the forward orientation was used to disrupt a backwards Gaussia luciferase that is secreted from infected cells. For the transient cell-cell transmission assay this HIV-TV, inGLUC, was transiently co-transfected into 293T cells along with S and HIV-PV. After 48 h these cells were mixed 1:1 with 293T-hACE2 cells, and 48 h later cell culture supernatant was removed to measure Gaussia luciferase activity.
For the stable system, producer 293T cells were made by first transfecting linearized pUCHR-inGLuc-pgk-hygro using calcium phosphate co-precipitation method and selecting stably transfected cells using 200–400 μg/mL hygromycin B (Calbiochem). Next, linearized pT-PB-SARS-CoV-2-Spike-IRES-Blasti was co-transfected along with piggybac transposase using calcium phosphate co-precipitation method and S-modified cells were selected in both blasticidin (10 μg/mL; GoldBio) and hygromycin B (200 μg/mL; Calbiochem). To express HIV-PV, 2 × 105 stable producer 293T-Spike-TV cells were seeded in a 12-well format and transduced with 40 nL of concentrated HDAd-HIV-PV (Hu et al., 2015) at an MOI of ∼4 IU/cell. At 24 h post-transduction cells were refed with fresh media, then harvested after another 24 h and co-cultured at ratio of 1:1 with target 293T-hACE2 cells. For both the stable and transient systems, cells were co-cultured in triplicate in 0.2 mL of complete DMEM per well in a 96-well format. After 48 h cell culture supernatant was harvested and assayed for Gaussia luciferase activity.
The Delta and Omicron S DNA cassettes, both CMV IE/promoter-driven, were separately blunt-end ligated into pUCHR-inGLuc-pgk-hygro into a unique Sfi1 restriction site downstream of the 3′ LTR; cloning details are available upon request. These two plasmids were linearized and individually transfected into 293T cells using the calcium-phosphate method, and stably transfected cells were selected using hygromycin B as described above.
X-gal staining for LacZ activity
2 × 105 293T- Spike-TV cells were seeded in a 12-well format then transduced with increasing amounts of HDAd-HIV-PV as indicated. At 24 h cells were refed with fresh complete DMEM media, incubated further for 24 h and then fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 10 min. X-gal staining solution (1 mg/mL X-gal from Gold Bio, 6 mM potassium ferrocyanide, and 6 mM potassium ferricyanide in PBS) was incubated with fixed cells for 24 h before plates were washed with PBS and photographs taken.
Gaussia luciferase assay
Gaussia luciferase (GLUC) assay buffer was prepared using 10 μM coelenterazine (Gold Bio), 50 mM Sodium Iodide (Sigma-Aldrich) in phosphate-buffered saline (PBS). 100 μL of culture supernatant was harvested from the co-cultured cells as described above and transferred into a white 96-well microplate to which 100 μL of the GLUC assay buffer was present. RLU was quantified on a Biotek plate reader. Data was analyzed with non-linear regression using GraphPad Prism to generate neutralization curves and IC-50 values.
Pseudotyped virus neutralization assay
Pseudotyped HIV-FFLUC was produced as previously described (Zhang et al., 2019) using pcDNA-SARS-CoV-2-S (Zhao et al., 2021). Pseudotyped particles were concentrated by ultracentrifugation. Convalescent and post-vaccine sera, monoclonal antibodies, and LCB1 were serially diluted as indicated and pre-incubated with S-pseudotyped particles for 1 h at 37°C then added to 293T-hACE2 target cells in 96-well format. After an overnight incubation fresh medium was added. After another 48 h cells were lysed and RLU measured as described previously (Zhao et al., 2021). IC-50 values were calculated using GraphPad Prism software. Non-linear regression with normalized response model was applied.
Sera, LCB1, murine anti-spike monoclonal antibodies, small molecule drugs, and tetherin
COVID-19 + convalescent sera were obtained from YNHH hospital (IMPACT research team). Post-vaccine sera were obtained from West Haven VA Medical Center. 56-mer peptide LCB1 was as previously described (Zhao et al., 2021) and stored at −80°C prior to use. Clofazimine, cobicistat, apilimod, niclosamide, 25-hydroxycholesterol, 27-hydroxycholesterol, and mycophenolate were purchased from Sigma-Aldrich. Murine anti-spike monoclonal antibodies, clones 2 and 6 were a gift of Sidi Chen and were as previously described (Peng et al., 2022). Producer and target 293T cells were generated as described above. Producer cells were harvested 48 h post-transfection or transduction for the transient or stable system, respectively. 4 × 104 producer cells were resuspended in 50 μL complete DMEM per well in 96-well plates. Serially diluted amounts of anti-spike mAbs, sera, LCB1, or small molecule drugs were then added to producer cells and incubated at 37°C for 1 h, then an equal amount of target cells in 50μL were added per well. The cells were co-cultured at 37°C, after 48 h supernatants were harvested and Gaussia luciferase activity measured as described above. Studies with tetherin were done by transiently transfecting the producer 293T cells with increasing amounts of tetherin expression plasmid DNA in 12-well format for cell-cell transmission. After 48 h cells were co-cultured with target 293T-hACE2 cells and supernatants harvested after 48 h of GLUC activity measured as described above.
Time course experiment for LCB1
96-well plates were seeded with 4 × 104 producer 293T cells in 100 μL per well. 10-fold serially diluted LCB1 was added at −1, 0, +1 and +2 h relative to the addition of 293T-hACE2 targets. After 48 h GLUC activity was measured as described above.
Cell fusion assay
HOS cells stably expressing HIV Tat and hACE2 were mixed 1:1 with TZMbl cells stably expressing S as described (Zhao et al., 2021). At 48 h transfected cells were lifted, mixed 1:1, and after another 16–24 h cells were lysed and RLU measured by plate reader in 96-well format as described (Zhao et al., 2021).
Western blotting and nested PCR
Expression of hACE2, S, and HIV Gag proteins in the stable 293T cells was confirmed by immunoblotting. Cells were lysed using RIPA buffer. Samples were boiled for 10 min in the presence of SDS and DTT, size-separated on pre-made SDS-PAGE gradient gels (Bio-Rad) and transferred onto PVDF filter membranes as previously described (Zeng et al., 2020). hACE2 and S were detected by goat anti-hACE2 (R&D systems) and mouse anti-FLAG (Sigma-Aldrich) primary antibodies and rabbit anti-goat-HRP (Sigma-Aldrich) and rabbit anti-mouse HRP (Cell Signaling) secondary antibodies, respectively. Gag-pol protein expression was detected by using anti-HIV human sera as primary antibody (#3957 from NIH AIDS Reagent Repository) and anti-human HRP secondary antibody (Sigma-Aldrich). In parallel cellular GAPDH was detected by immunoblotting using rabbit anti-GAPDH primary antibody (Sigma-Aldrich) and goat anti-rabbit HRP secondary antibody (Sigma-Aldrich).
To confirm presence of the HIV transfer vector in the stable cell line, nested PCR was performed as previously described (Lorenz, 2012), using extracted genomic DNA as template and size-separating DNA products by horizontal agarose gel electrophoresis.
Quantification and statistical analysis
All statistical analyses were conducted using GraphPad Prism 9.3.1. Luciferase assays were performed, and luciferase expression quantified, in triplicate as indicated in the figure legends with the mean being the average value of the three readings. In all the experiments, data are represented as the mean ± SD. The significance of the differences between the groups was analyzed with paired t-test. p values < 0.05 were considered statistically significant. The specific p value is depicted in the respective figure panel.
Acknowledgments
We thank Dr. Walther Mothes (Yale) and Matthew Wilson (Baylor College of Medicine) for generous reagent gifts. The following reagent was obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: Polyclonal Anti-Human Immunodeficiency Virus Immune Globulin, Pooled Inactivated Human Sera, ARP-3957, contributed by NABI and the National Heart Lung and Blood Institute (Dr. Luiz Barbosa). We thank the Yale IMPACT Research Team Membership for post-COVID-19 sera (Abeer Obaid, Akiko Iwasaki, Allison Nelson, Arnau Casanovas-Massana, Angela Nunez, Anjelica Martin, Bertie Geng, Codruta Todeasa, Denise Shepard, Elizabeth B. White, Erin Silva, Giuseppe De-Iuliis, Harold Rahming, Hong-Jai Park, Irene Matos, Jessica Nouws, Kadi-Ann Rose, Kelly Anastasio, Kristina Brower, Laura Glick, Lokesh Sharma, Maksym Minasyan, Maria Batsu, Maxine Kuang, Melissa Linehan, Michael H. Askenase, Mikhail Smolgovsky, Nicole Sonnert, Pavithra Vijayakumar, Santos Bermejo, Sofia Velazquez, Tyler Rice, William Khoury-Hanold, Xiaohua Peng, Yiyun Cao, John Fournier, M. Catherine Muenker, Adam J. Moore, Molly L. Bucklin, David McDonald, Camila Odio, and Yvette Strong). We also thank Ching Ying “Vanessa” Li for her work performing some GLUC assays. This work was supported in part by NIH grant R01 AI150334.
Author contributions
Conceptualization, G.S. and R.E.S.; methodology, G.S. and R.E.S.; validation, G.S. and M.K.; formal analysis, G.S. and R.E.S.; investigation, G.S., M.K., M.Z., and R.E.S.; resources, S.G., S.F.F., C.S.D.C., S.C., L.P., and P.R.; writing – original draft, G.S. and R.E.S.; writing – review & editing, G.S., S.G., and R.E.S.; visualization: R.E.S.; supervision, R.E.S.; project administration, R.E.S.; funding acquisition, R.E.S.
Declaration of interests
The authors declare no competing interests.
Published: June 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100252.
Contributor Information
Shaili Gupta, Email: shaili.gupta@yale.edu.
Richard E. Sutton, Email: richard.sutton@yale.edu.
Supplemental information
Data and code availability
All the Raw datasets generated during this study have been deposited at Mendeley Data and are publicly available as of the date of publication. DOIs are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abela I.A., Berlinger L., Schanz M., Reynell L., Günthard H.F., Rusert P., Trkola A. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog. 2012;8:e1002634. doi: 10.1371/journal.ppat.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto L.M., Uchil P.D., Mothes W. HIV cell-to-cell transmission: effects on pathogenesis and antiretroviral therapy. Trends Microbiol. 2015;23:289–295. doi: 10.1016/j.tim.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert M., Yoon M., Sloan D.D., Spear P.G., Jerome K.R. The virological synapse facilitates herpes simplex virus entry into T cells. J. Virol. 2009;83:6171–6183. doi: 10.1128/jvi.02163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L.N., Deng W., Huang B.Y., Gao H., Liu J.N., Ren L.L., Wei Q., Yu P., Xu Y.F., Qi F.F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses-Basel. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B., Winters J.L., Van Buskirk C., Grossman B.J., Joyner M., Henderson J.P., et al. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Invest. 2020;130:2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Van Der Zee R., De Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.X., Goreshnik I., Coventry B., Case J.B., Miller L., Kozodoy L., Chen R.E., Carter L., Walls A.C., Park Y.J., et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130:1545–1548. doi: 10.1172/jci138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., Mcmahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Hübner W., Spinelli M.A., Chen B.K. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 2007;81:12582–12595. doi: 10.1128/jvi.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun A.K., Van Maanen M., Nguyen V., Sutton R.E. Human chromosome 2 carries a gene required for production of infectious human immunodeficiency virus type 1. J. Virol. 2006;80:3406–3415. doi: 10.1128/jvi.80.7.3406-3415.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- Ho H.L., Wang F.Y., Lee H.R., Huang Y.L., Lai C.L., Jen W.C., Hsieh S.L., Chou T.Y. Seroprevalence of COVID-19 in Taiwan revealed by testing anti-SARS-CoV-2 serological antibodies on 14, 765 hospital patients. Lancet Reg. Health West Pac. 2020;3:100041. doi: 10.1016/j.lanwpc.2020.100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., O'boyle K., Palmer D., Ng P., Sutton R.E. High-level production of replication-defective human immunodeficiency type 1 virus vector particles using helper-dependent adenovirus vectors. Mol. Ther. Methods Clin. Dev. 2015;2:15004. doi: 10.1038/mtm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.X., Chen Y.L., Li S.P., Shen J.P., Zuo K., Zhou S.C., Chang C. Development and validation of a simple-to-use nomogram for predicting the upgrade of atypical ductal hyperplasia on core needle biopsy in ultrasound-detected breast lesions. Front. Oncol. 2020;10:609841. doi: 10.3389/fonc.2020.609841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:545. doi: 10.1016/j.it.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen M.D., Irving A., Montagutelli X., Tate M.D., Rudloff I., Nold M.F., Hansbro N.G., Kim R.Y., Donovan C., Liu G., et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020;13:877–891. doi: 10.1038/s41385-020-00340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Booth N.J., Neil S.J.D. Cell-cell spread of human immunodeficiency virus type 1 overcomes tetherin/BST-2-mediated restriction in T cells. J. Virol. 2010;84:12185–12199. doi: 10.1128/jvi.01447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Kashefi K., Hollinshead M., Sattentau Q.J. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 2004;199:283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Sattentau Q.J. Retroviral spread by induction of virological synapses. Traffic. 2004;5:643–650. doi: 10.1111/j.1600-0854.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Klasse P.J., Moore J.P. Antibodies to SARS-CoV-2 and their potential for therapeutic passive immunization. Elife. 2020;9:e57877. doi: 10.7554/elife.57877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., et al. Coronavirus infections and immune responses. J. Med. Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.H., Moore M.J., Vasilieva N., Sui J.H., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou W., Yang L., You R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol. Res. 2020;157:104833. doi: 10.1016/j.phrs.2020.104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lorenz T.C. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. J. Vis. Exp. 2012:e3998. doi: 10.3791/3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. 2020;41:363–382. doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N., Welsch S., Jolly C., Briggs J.A.G., Vaux D., Sattentau Q.J. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J. Virol. 2010;84:3516–3527. doi: 10.1128/jvi.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurov D., Ilinskaya A., Heidecker G., Lloyd P., Derse D. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 2010;6:e1000788. doi: 10.1371/journal.ppat.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurov D., Ilinskaya A., Heidecker G., Lloyd P., Derse D. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 2010;6:e1000788. doi: 10.1371/journal.ppat.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., Mcmahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W., Sherer N.M., Jin J., Zhong P. Virus cell-to-cell transmission. J. Virol. 2010;84:8360–8368. doi: 10.1128/jvi.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D., Ng P. Improved system for helper-dependent adenoviral vector production. Mol. Ther. 2003;8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Peng L., Hu Y., Mankowski M.C., Ren P., Chen R.E., Wei J., Zhao M., Li T., Tripler T., Ye L., et al. Monospecific and bispecific monoclonal SARS-CoV-2 neutralizing antibodies that maintain potency against B.1.617. Nat. Commun. 2022;13:1638. doi: 10.1038/s41467-022-29288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D., Zang T., Ebrahimi A., Mcnatt M.W., Gregory D.A., Johnson M.C., Bieniasz P.D. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Polak S.B., Van Gool I.C., Cohen D., Von Der Thusen J.H., Van Paassen J. A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression. Mod. Pathol. 2020;33:2128–2138. doi: 10.1038/s41379-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakara C., Godbole R., Sil P., Jahnavi S., Gulzar S.e.J., Van Zanten T.S., Sheth D., Subhash N., Chandra A., Shivaraj A., et al. Strategies to target SARS-CoV-2 entry and infection using dual mechanisms of inhibition by acidification inhibitors. PLoS Pathog. 2021;17:e1009706. doi: 10.1371/journal.ppat.1009706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinonez R., Sutton R.E. Lentiviral vectors for gene delivery into cells. DNA Cell Biol. 2002;21:937–951. doi: 10.1089/104454902762053873. [DOI] [PubMed] [Google Scholar]
- Rehman S.U., Tabish M. Alternative splicing of ACE2 possibly generates variants that may limit the entry of SARS-CoV-2: a potential therapeutic approach using SSOs. Clin. Sci. 2020;134:1143–1150. doi: 10.1042/cs20200419. [DOI] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicka D., Feldmann J.R.M., Porrot F.O., Wietgrefe S., Guadagnini S.P., Prévost M.-C., Estaquier J.R.M., Haase A.T., Sol-Foulon N., Schwartz O. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. J. Virol. 2009;83:6234–6246. doi: 10.1128/jvi.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Prajapat M., Avti P., Kaur H., Kumar S., Medhi B. Therapeutic options for the treatment of 2019-novel coronavirus: an evidence-based approach. Indian J. Pharmacol. 2020;52:1. doi: 10.4103/ijp.ijp_119_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.-H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalhoub S. Interferon beta-1b for COVID-19. Lancet. 2020;395:1670–1671. doi: 10.1016/s0140-6736(20)31101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y.S., Luo C.M., Ye G., Geng Q.B., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Cui Q., Garcia G., Wang C., Zhang M., Arumugaswami V., Riggs A.D., Shi Y. Comparative transcriptomic analysis of SARS-CoV-2 infected cell model systems reveals differential innate immune responses. Scientific Rep. 2021;11:17146. doi: 10.1038/s41598-021-96462-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Cela R., Clarke C., Bertin T.K., Mouriño S., Lee B. Large-scale production of high-quality helper-dependent adenoviral vectors using adherent cells in cell factories. Hum. Gene Ther. 2010;21:120–126. doi: 10.1089/hum.2009.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Dcosta B.M., Samanovic M.I., Herati R.S., Cornelius A., Zhou H., Vaill A., Kazmierski W., Mulligan M.J., Landau N.R., Goff S.P. Convalescent-phase sera and vaccine-elicited antibodies largely maintain neutralizing titer against global SARS-CoV-2 variant spikes. mBio. 2021;12:e0069621. doi: 10.1128/mbio.00696-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Mulligan M.J., Landau N.R. Partial resistance of SARS-CoV-2 Delta variants to vaccine-elicited antibodies and convalescent sera. iScience. 2021;24:103341. doi: 10.1016/j.isci.2021.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. Preprint at medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Temmam S., Vongphayloth K., Baquero E., Munier S., Bonomi M., Regnault B., Douangboubpha B., Karami Y., Chrétien D., Sanamxay D., et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022;604:330–336. doi: 10.1038/s41586-022-04532-4. [DOI] [PubMed] [Google Scholar]
- Tian S.F., Xiong Y., Liu H., Niu L., Guo J.C., Liao M.Y., Xiao S.Y. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod. Pathol. 2020;33:1007–1014. doi: 10.1038/s41379-020-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanPatten S., He M., Altiti A., F Cheng K., Ghanem M.H., Al-Abed Y. Evidence supporting the use of peptides and peptidomimetics as potential SARS-CoV-2 (COVID-19) therapeutics. Future Med. Chem. 2020;12:1647–1656. doi: 10.4155/fmc-2020-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel L. What's next now that the WHO has declared a COVID-19 pandemic? CMAJ. 2020;192:E349–E350. doi: 10.1503/cmaj.1095855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Huang Y., Hu Q., Li C., Liu H., Wang X., Li W., Ma W., Pu Y., Du Y., et al. A simulated dosimetric study of contribution to radiotherapy accuracy by fractional image guidance protocol of Halcyon system. Front. Oncol. 2020;10:543147. doi: 10.3389/fonc.2020.543147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang L. Broadly neutralizing antibodies and vaccine design against HIV-1 infection. Front. Med. 2020;14:30–42. doi: 10.1007/s11684-019-0721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li W., Hui H., Tiwari S.K., Zhang Q., Croker B.A., Rawlings S., Smith D., Carlin A.F., Rana T.M. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39:e106057. doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N.S., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., Mclellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., Gu C., Zhang R., Tu C., Xie Y., et al. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898.e5. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., Mcmahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Yin X., Meng X., Chan J.F.-W., Ye Z.-W., Riva L., Pache L., Chan C.C.-Y., Lai P.-M., Chan C.C.-S., et al. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature. 2021;593:418–423. doi: 10.1038/s41586-021-03431-4. [DOI] [PubMed] [Google Scholar]
- Zeng C., Evans J.P., Pearson R., Qu P., Zheng Y.M., Robinson R.T., Hall-Stoodley L., Yount J., Pannu S., Mallampalli R.K., et al. Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI Insight. 2020;5:143213. doi: 10.1172/jci.insight.143213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chapman J.H., Ulcay A., Sutton R.E. Neutralization synergy between HIV-1 attachment inhibitor fostemsavir and anti-CD4 binding site broadly neutralizing antibodies against HIV. J. Virol. 2019;93:e01446-18. doi: 10.1128/jvi.01446-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Su P.-Y., Castro D.A., Tripler T.N., Hu Y., Cook M., Ko A.I., Farhadian S.F., Israelow B., Dela Cruz C.S., et al. Rapid, reliable, and reproducible cell fusion assay to quantify SARS-Cov-2 spike interaction with hACE2. PLoS Pathog. 2021;17:e1009683. doi: 10.1371/journal.ppat.1009683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong P., Agosto L.M., Ilinskaya A., Dorjbal B., Truong R., Derse D., Uchil P.D., Heidecker G., Mothes W. Cell-to-cell transmission can overcome multiple donor and target cell barriers imposed on cell-free HIV. PLoS One. 2013;8:e53138. doi: 10.1371/journal.pone.0053138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong P., Agosto L.M., Munro J.B., Mothes W. Cell-to-cell transmission of viruses. Curr. Opin. Virol. 2013;3:44–50. doi: 10.1016/j.coviro.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Zhao Q. Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. Int. J. Biol. Sci. 2020;16:1718–1723. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the Raw datasets generated during this study have been deposited at Mendeley Data and are publicly available as of the date of publication. DOIs are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.