Abstract
Respiratory diseases are among the leading causes of morbidity and mortality worldwide, coupled with the ongoing coronavirus disease 2019 (COVID-19) pandemic. mRNA lipid nanoparticle (LNP) vaccines have been developed, but their intramuscular delivery limits pulmonary bioavailability. Inhalation of nanoparticle therapeutics offers localized drug delivery that minimizes off targeted adverse effects and has greater patient compliance. However, LNP platforms require extensive reformulation for inhaled delivery. Lung-derived extracellular vesicles (Lung-Exo) offer a biological nanoparticle alternative that is naturally optimized for mRNA translation and delivery to pulmonary cells. We compared the biodistribution of Lung-Exo against commercially standard biological extracellular vesicles (HEK-Exo) and LNPs (Lipo), where Lung-Exo exhibited superior mRNA and protein cargo distribution to and retention in the bronchioles and parenchyma following nebulization administration. This suggests that inhaled Lung-Exo can deliver mRNA and protein drugs with enhanced pulmonary bioavailability and therapeutic efficacy.
Keywords: Lung, Extracellular vesicles, Exosomes, Nebulizers
1. Introduction
Respiratory diseases are becoming increasingly prevalent,1 with diseases such as lower respiratory infections and chronic obstructive pulmonary disease substantially driving patient burden across all ages.2 Inhaled therapeutics provide an attractive drug delivery method that offers localized and noninvasive treatments. By directly inhaling drugs, pulmonary bioavailability can be optimized, while minimizing subsequent adverse effects.3 , 4 However, drug formulation and aerosol deposition are critical obstacles that impede therapeutic efficacy. Nanomaterials offer a solution by altering drug size, solubility, and surface chemistry to become compatible with the pulmonary microenvironment.3 , 5 , 6 Through encapsulation using lipid nanoparticles (LNPs), drug cargo is protected from rapid degradation.7 The distribution of these drug-loaded LNPs needs to be well characterized to ensure drug targeting to affected lung tissue. Magnetic aerosols have been developed and applied to in vitro airway models 8 and computer simulations 9 to track droplet delivery. However, aerosol distribution of therapeutic nanoparticles is not universal across formulations. Developing a standardized loadable nanoparticle for a range of pulmonary therapeutic applications and characterizing its distribution upon inhaled delivery would elucidate cellular targeting and optimize drug dosage. mRNA loaded LNPs have demonstrated therapeutic efficacy against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection as an intramuscular injection vaccine,[10], [11], [12], [13] opening the application of mRNA vaccines to treat other lung diseases across different delivery methods such as nebulization.14 , 15 However, extensive LNP formulation is required to optimize mRNA translation and pulmonary bioavailability for inhaled delivery. Therefore, biological nanoparticles such as extracellular vesicles (EVs) offer an alternative to synthetic LNPs that are naturally optimized for mRNA encapsulation and cellular delivery. EVs, including exosomes, are nanosized vesicles secreted by various cells types into almost all biological fluids.16 Exosomes have known therapeutic properties across various disease applications.15 , 17 Exosomes contain a variety of molecular components including RNAs and proteins that differ depending on their parent-cell origination.18 , 19 Exosomes secreted from pulmonary cells may contain molecular components and membrane features that are recognized and favored in the lung microenvironment.18 Utilizing lung-derived exosomes (Lung-Exo) as an inhaled drug delivery vesicle may increase drug retention and efficacy by more efficiently evading immune clearance and targeting pulmonary cells. Along with drug delivery, Lung-Exo themselves have demonstrated therapeutic benefits. In a rodent model of idiopathic pulmonary fibrosis, Lung-Exo better restores lung function and reduces fibrotic severity than its mesenchymal stem cell exosome counterpart.20 However, the biodistribution of Lung-Exo after inhaled delivery has yet to be determined. In this study, we developed fluorescently labeled Lung-Exo by loading green fluorescent protein (GFP)-encoding mRNA and red fluorescent protein (RFP) through an established electroporation method 21 , 22 and compared its distribution against commercial standard biological nanoparticles, HEK293T-derived exosomes (HEK-Exo), and synthetic LNPs (Lipo). We hypothesize that lung-derived exosomes are naturally optimized for the distribution and retention of mRNA and protein cargo components in the lung following inhaled delivery.
2. Results
2.1. mRNA and protein loading into exosomes and liposomes
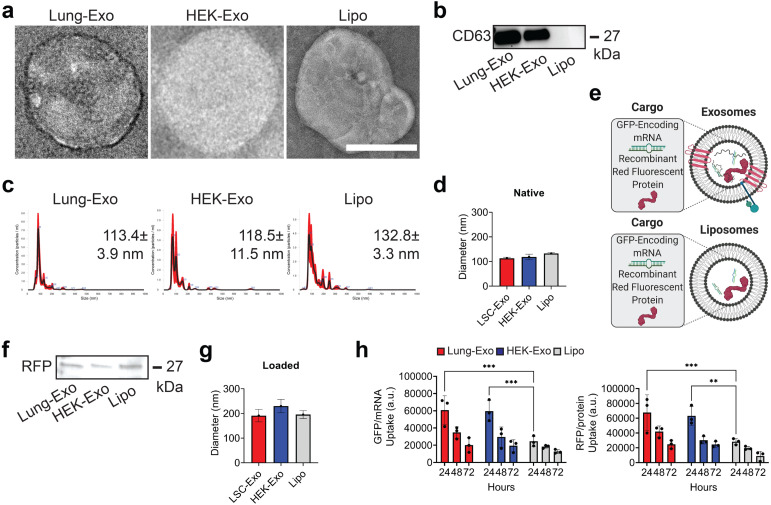
Lung-Exo, HEK-Exo, and Lipo were used to assess how NP derivation affects biodistribution in the lung. NP morphology was assessed by transmission electron microscopy (TEM), confirming that the isolation of exosomes and liposomes did not disrupt vesicular membrane integrity (Fig. 1A). An immunoblot confirmed that Lung-Exo and HEK-Exo have an exosomal phenotype (CD63+), while Lipo lacks an exosomal phenotype (Fig. 1B). Nanoparticle tracking analysis (NTA) determined that the mode diameter of Lung-Exo, HEK-Exo, and Lipo were 113.4 ± 3.9 nm, 118.5 ± 11.5 nm, and 132.8 ± 3.3 nm (Fig. 1C, Fig. 1D). To label the NPs, green fluorescent protein (GFP)-encoding mRNA and red fluorescent protein (RFP) were loaded into exosomes and liposomes (Fig. 1E). Protein loading was confirmed by an immunoblot (Fig. 1F). NTA of loaded NPs showed an increase in mode diameter of Lung-Exo, HEK-Exo, and Lipo (Fig. 1G). When co-cultured with lung parenchymal cells, Lung-Exo and HEK-Exo had significantly greater mRNA (2.5-fold and 2.4-fold) and protein cargo (2.4-fold and 2.2-fold) uptake than their liposome counterpart after 24 h, respectively (Fig. 1H, Figure S1). This suggests that cellular targeting is greater enhanced through biologically derived nanoparticles than synthetic nanoparticles.
Fig. 1.
Characterization of mRNA and protein-loaded nanoparticles. (A) TEM images of native Lung-Exo, HEK-Exo, and Lipo. (B) Immunoblot of CD63 in exosome and liposome lysate. (C) NTA size distribution analysis and mode nanoparticle diameters. (D) Quantification of NTA size distribution analysis of the average mean standard error of five replicates; n 1 per group. (E) Schematic of mRNA and protein loading into exosomes and liposomes. Created with BioRender.com. (F) Immunoblot of RFP in exosome and liposome lysate. (G) Quantification of NTA size distribution analysis of the average mean standard error of five replicates; n 1 per group. (H) Quantification of Lung-Exo, HEK-Exo, and Lipo pixel intensity normalized to nuclei in lung parenchymal cells; n 3 per group.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2. Nebulized lung-derived exosomes have superior distribution and retention in the lung
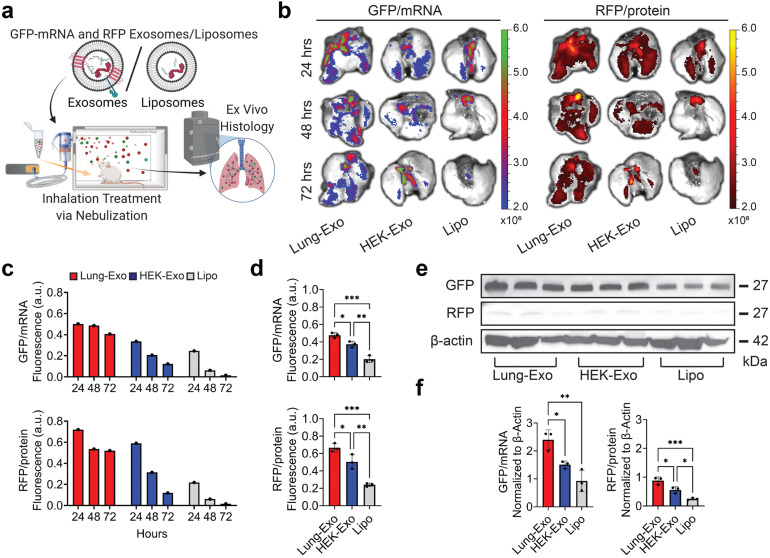
To test if mRNA and protein drugs can be delivered by inhalation, a single dose of labeled NPs were delivered to healthy mice by jet nebulization (Fig. 2A). Ex vivo imaging revealed the distribution of nanoparticles at 24, 48, and 72 h (Fig. 2B), with notable clearance or degradation of liposomes over time (Fig. 2C). At 24 h, Lung-Exo had 1.3-fold and 2.4-fold greater mRNA distribution, and 1.3-fold and 2.8-fold greater protein distribution than its HEK-Exo and Lipo counterparts (Fig. 2D), respectively, baselined to their native nanoparticle controls (Figure S2). mRNA and protein cargo delivery were verified by immunoblots (Fig. 2E), where mice who received Lung-Exo had the highest mRNA translation and protein expression in lung tissue (Fig. 2F). Together, these data suggest the superior distribution and retention of inhaled exosomes in the lung than inhaled liposomes. Notably, exosomes of lung origin outperformed the HEK exosome control by having greater nanoparticle distribution, retention, mRNA translation, and protein expression in the lung. These suggest the enhanced bioavailability of Lung-Exo as an inhaled therapeutic and drug delivery vesicle for respiratory diseases.
Fig. 2.
Biodistribution of nebulized labeled nanoparticles in the lung. (A) Schematic of mRNA and protein loading, nebulization administration, and ex vivo histology. Created with BioRender.com. (B) Representative ex vivo images of mouse lungs after mRNA and protein loaded Lung-Exo, HEK-Exo, and Lipo nebulization. (C) Quantification of the integrated density of GFP and RFP fluorescence in ex vivo mouse lungs; n 1 per group. (D) Quantification of the integrated density of GFP and RFP fluorescence in ex vivo mouse lungs 24 h after nebulization; n 3 per group. (E) Immunoblots of GFP and RFP in mouse lung lysate. (F) Quantification of immunoblots normalized to -actin; n 3 per group.
2.3. Lung-derived exosomes have superior delivery of mRNA and protein to the bronchioles and parenchyma
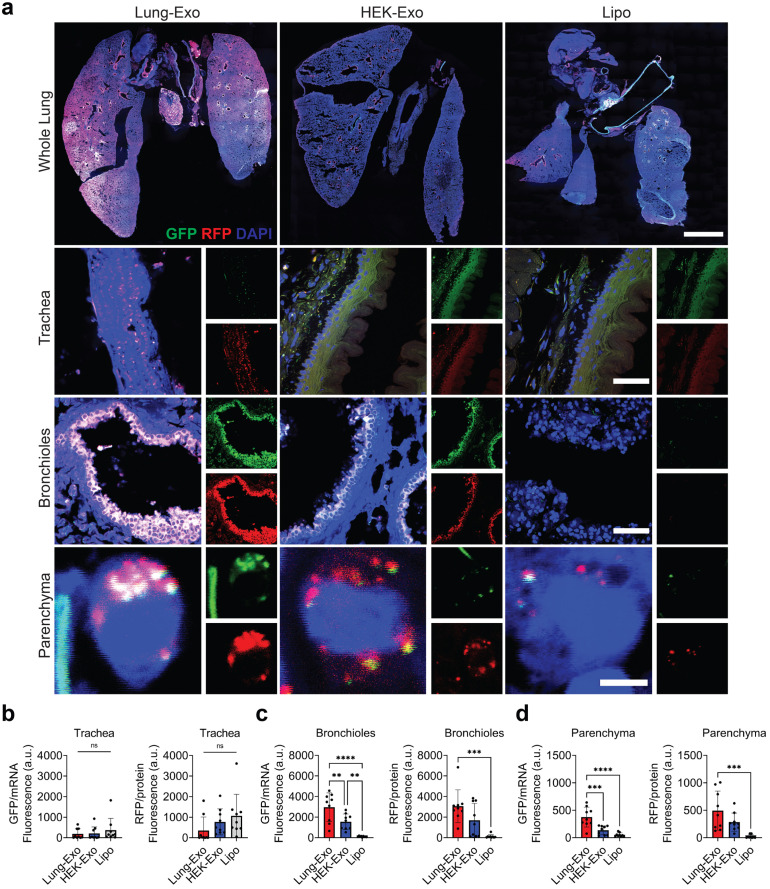
Nanoparticles are an attractive inhaled therapeutic in that their innate size distributions (<) are immediately respirable and allow for alveolar deposition.23 To track inhaled nanoparticles into the deep lung, we segmented the whole lung into its three main areas: the trachea, bronchioles, and parenchyma (Fig. 3A). Liposomes showed trends of tracheal deposition (Fig. 3B), while exosomes showed significantly greater deposition into the bronchioles (Fig. 3C) and parenchyma (Fig. 3D). Lung-Exo had the greatest protein expression in the bronchioles (24.1-fold) and parenchyma (22.9-fold) compared to Lipo. Notably, Lung-Exo had the greatest mRNA translation in the bronchioles (1.9-fold and 27.5-fold) and parenchyma (2.8-fold and 7.2-fold) than both HEK-Exo and Lipo, respectively. These data suggest that mRNA and protein delivery and translation may be significantly impacted by its nanoparticle phenotype. The native lung signature of Lung-Exo may provide superior delivery and retention of cargo components to the lung than exosomes derived from different cell types or synthetic nanoparticles.
Fig. 3.
mRNA and protein delivery to the trachea, bronchioles, and parenchyma. (A) Representative immunostaining images of whole lung, tracheal, bronchiole, and parenchymal sections for GFP (green), RFP (red) and DAPI (blue); scale bar in whole lung images; scale bar in tracheal and bronchiole sections; scale bar in parenchymal sections. (B) Quantification of Lung-Exo, HEK-Exo, and Lipo pixel intensity normalized to nuclei in tracheal sections; n 9 per group. (C) Quantification of Lung-Exo, HEK-Exo, and Lipo pixel intensity normalized to nuclei in bronchiole sections; n 9 per group. (D) Quantification of Lung-Exo, HEK-Exo, and Lipo pixel intensity normalized to nuclei in parenchymal sections; n 9 per group.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.4. Off-targeted uptake of lung-derived exosomes
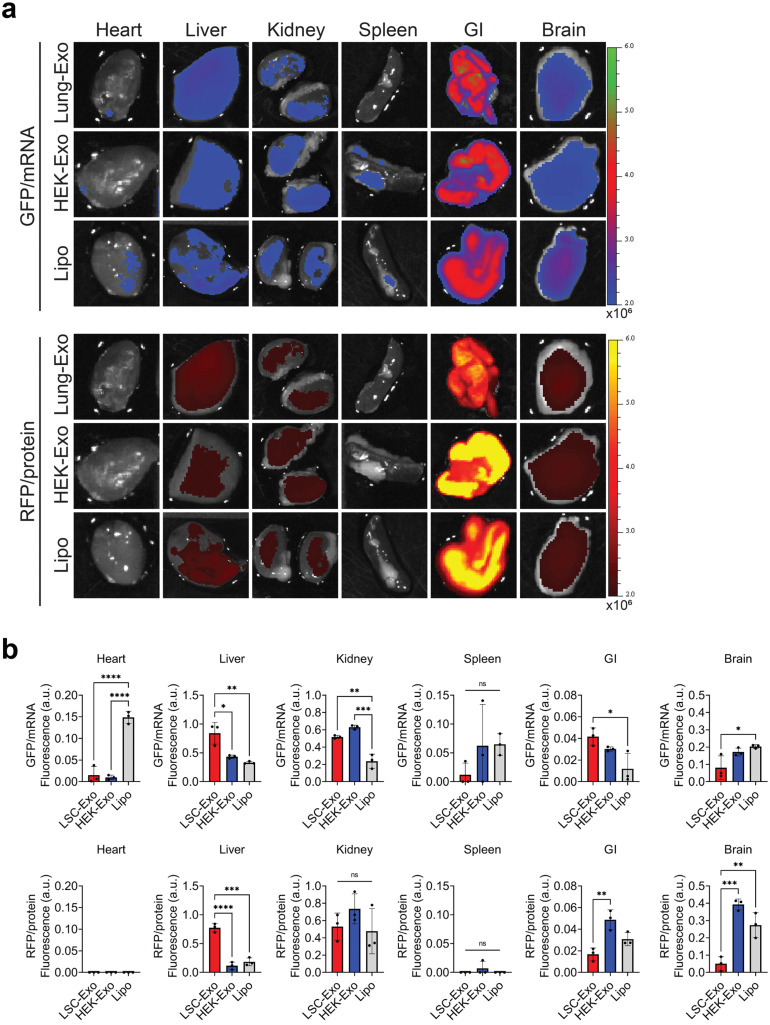
Off-targeted uptake of the nanoparticles was tracked in the other major organs (Fig. 4A), where mice who received inhaled Lipo showed rapid clearance and mRNA uptake through the pulmonary circulation into the heart (Fig. 4B, Figure S2). The gastrointestinal (GI) tract showed absorption of both exosomes and liposomes, with consideration of tissue autofluorescence (Fig. 4B, Figure S2). Interestingly, Lung-Exo showed significant metabolism to the liver (Fig. 4B, Figure S2). Our previous studies have demonstrated significant improvement in liver function through decreased alanine transaminase (ALT) enzymes and decreased serum monocyte chemoattractant protein-1 (MCP-1) in a rat model of pulmonary fibrosis that received nebulized Lung-Exo.20 Federally-approved small molecule drugs nintedanib 24 and pirfenidone 25 slow the progression of pulmonary fibrosis, but have been associated with adverse events involving an elevation of liver enzymes that may result in a termination of drug use.24 , 26 Mesenchymal stem cell nanoparticles promote hepatic regeneration and reduce liver enzymes in a liver fibrosis model,27 suggesting that applying Lung-Exo as an inhaled drug delivery vehicle may too mitigate liver enzyme elevation through its enhanced bioavailability.
Fig. 4.
((A) Ex vivo images of mouse heart, liver, kidney, spleen, GI, and brain 24 h after loaded Lung-Exo, HEK-Exo, and Lipo nebulization. (B) Quantification of the integrated density of GFP and RFP fluorescence in ex vivo mouse heart, liver, kidney, spleen, GI, and brain normalized to native nanoparticle controls (see Supplementary Figure 2).
3. Materials and methods
Cell Culture: Human LSCs were generated from healthy whole lung samples from the Cystic Fibrosis and Pulmonary Diseases Research and Treatment Center at the University of North Carolina at Chapel Hill and expanded as previously described.[28], [29], [30], [31] LSCs were plated on a fibronectin-coated (Corning Incorporated, Corning, NY, USA) flask and maintained in Iscove’s Modified Dulbecco’s Media (IMDM; ThermoFisher Scientific, Waltham, MA, USA) containing 20% fetal bovine serum (FBS; Corning Incorporated, Corning, NY, USA), 1% L-glutamine (ThermoFisher Scientific, Waltham, MA, USA), 0.5% Gentamicin (ThermoFisher Scientific, Waltham, MA, USA), and 0.18% 2-mercaptoethanol (ThermoFisher Scientific, Waltham, MA, USA). Human embryonic kidney (HEK) 293T cells were purchased from American Type Culture Collection (ATCC; American Type Culture Collection, Manassas, VA, USA). HEK cells were plated on a flask and maintained in Minimum Essential Media (MEM; ThermoFisher Scientific, Waltham, MA, USA) containing 10% FBS, 1% L-glutamine, 0.5% Gentamicin, and 0.18% 2-mercaptoethanol. Media changes on all cultures were performed every other day. LSCs and HEK cells were allowed to reach 70%–80% confluence before generating serum-free secretome (Lung-Secretome, HEK-Secretome) as previously described.20 Lung- and HEK-Secretome were collected and filtered through a 0. filter to remove cellular debris. All procedures performed in this study involving human samples were in accordance with the ethical standard of the institutional research committee and with the guidelines set by the Declaration of Helsinki.
Exosome isolation and characterization: Lung-Exo and HEK-Exo were collected and isolated from Lung-Secretome and HEK-Secretome using an ultrafiltration method.32 Filtered secretome was pipetted into a 100kDa Amicon centrifugal filter unit (MilliporeSigma, Burlington, MA, USA) and centrifuged at 400 RCF and 10 °C. After all media passed through the centrifugal filter unit, remaining exosomes were detached from the filter and resuspended using 1X Dulbecco’s phosphate-buffered saline (DPBS; ThermoFisher Scientific, Waltham, MA, USA) with 25 mM trehalose (MilliporeSigma, Burlington, MA, USA) for further analysis.33 Pegylated Remote Loadable Liposomes (Lipo) were purchased from Avanti Polar Lipids (Avanti Polar Lipids, Inc, Alabaster, AL, USA). LSC-Exo, HEK-Exo, and Lipo were fixed with 4% paraformaldehyde (PFA; Electron Microscopy Sciences, Hatfield, PA, USA) and 1% glutaraldehyde (Sigma-Aldrich, St. Louis, MO, USA) onto 100 mesh copper grids (Electron Microscopy Sciences, Hartfield, PA, USA) for transmission electron microscopy imaging (JEOL JEM-2000FX, Peabody, MA, USA). Samples were stained with Vanadium Negative Stain (ab172780; Abcam, Cambridge, United Kingdom). Sample concentrations and mean diameters were quantified by nanoparticle tracking analysis before and after fluorescent label loading (NanoSight NS3000, Malvern Panalytical, Malvern, UK).
Nanoparticle fluorescent label loading: GFP-encoding mRNA and RFP protein labeled nanoparticles were generated by loading mRNA and protein into exosome and liposome particles via electroporation, yielding labeled Lung-Exo, HEK-Exo, and Lipo.21,22 1 billion nanoparticles from each sample were diluted in Gene Pulser® Electroporation Buffer (Bio-Rad, Hercules, CA, USA) at a 1:9 ratio of nanoparticles to buffer. Particle numbers were calculated from the mean concentration calculations from three replicates by NTA. 10 ug of GFP-encoding mRNA DasherGFP® (Aldevron, Fargo, ND, USA) and RFP (ab268535; Abcam, Cambridge, United Kingdom) were added to the nanoparticle-buffer solution and transferred to an ice-cold 0.4 cm Gene Pulser/MicroPulser Electroporation Cuvette (Bio-Rad, Hercules, CA, USA). The electroporation cuvette was inserted into the Gene Pulser Xcell™ Total System (Bio-Rad, Hercules, CA, USA) and electroporated under the following conditions: pulse type: square waveforms; voltage: 200 V; pulse length: 10 msec; number of pulses: 5; pulse interval: 1 s Electroporation buffer was filtered out of the fluorescently labeled nanoparticles by the ultrafiltration method described above. 10 ug of GFP-encoding mRNA DasherGFP (Aldevron, Fargo, ND, USA) and 10 ug of RFP were loaded into LSC-Exo, HEK-Exo, and Lipo nanoparticles via the electroporation method described above.
SDS-PAGE and western blot: Native and fluorescently labeled LSC-Exo, HEK-Exo, and Lipo were further characterized by immunoblotting. Samples were lysed, denatured, and reduced by Laemmli sample buffer (Bio-Rad, Hercules, CA, USA) and -mercaptoethanol (Bio-Rad, Hercules, CA, USA) at 90 °C for 5 min. Protein samples and molecular ladder (Precision Plus Protein Unstained Standards; Bio-Rad, Hercules, CA, USA) were loaded into a 10% acrylamide precast Tris-Glycine gel (Bio-Rad, Hercules, CA, USA) for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) separation. Gels were run at a stacking voltage of 100 V until samples ran out of the wells, followed by a constant voltage of 200 V. Gels were visualized and imaged in a Bio-Rad Imager (Bio-Rad, Hercules, CA, USA). Gels were transferred onto polyvinylidene fluoride membranes (PVDF; Bio-Rad, Hercules, CA, USA) using the Bio-Rad wet electroblotting transfer system (Bio-Rad, Hercules, CA, USA). Following transfer, membranes were washed three times in 1X phosphate-buffered saline with 0.1% Tween detergent (PBS-T; MilliporeSigma, Burlington, MA, USA) for 5 min each and blocked using 5% milk in PBS-T for one hour at room temperature. Membranes were blotted against anti--Actin (ab6276; Abcam, Cambridge, United Kingdom), anti-CD63 (PA5-100713; ThermoFisher Scientific, Waltham, MA, USA), anti-GFP (ab290; Abcam, Cambridge, United Kingdom), and anti-RFP (ab62341; Abcam, Cambridge, United Kingdom) primary antibodies in 5% milk in PBS-T and incubated at 4 °C for one week. After incubation, membranes were incubated with the corresponding goat anti-rabbit (ab6721; Abcam, Cambridge, United Kingdom) and goat anti-mouse (ab6789; Abcam, Cambridge, United Kingdom) HRP-conjugated secondary antibodies for 1 h at room temperature. Membranes were then visualized using Clarity Western ECL Substrate (Bio-Rad, Hercules, CA, USA) and imaged in a Bio-Rad Imager (Bio-Rad, Hercules, CA, USA). Band intensities were analyzed using ImageJ analysis software (NIH; https://imagej.nih.gov/ij/).
Animal procedures: Seven-week-old male CD1 mice (022) were obtained from Charles River Laboratory (Wilmington, MA, USA). Lung-Exo, HEK-Exo, and Lipo were administered via jet nebulization (Pari Trek S Portable 459 Compressor Nebulizer Aerosol System, PARI, Starnberg, Germany). Fluorescently labeled nanoparticles were given in a single dose of 109 particles per kg of body weight. Immediately after sacrifice, the lungs, heart, liver, kidneys, spleen, cecum, and brain were excised and imaged using a Xenogen Live Imager (PerkinElmer, Waltham, MA, USA). Blood was collected in Vacuette ethylenediaminetetraacetic acid (EDTA) tubes (Greiner Bio-One, Kremsmünster, Austria) and centrifuged at maximum speed for 5 min to separate out serum. All animal studies complied with the requirements of the Institutional Animal Care and Use Committee (IACUC) of North Carolina State University.
Histology: Immunostaining was performed on tissue slides and chamber slides fixed in 4% PFA (Electron Microscopy Sciences, Hatfield, PA, USA) in DPBS for 30 min, followed by permeabilization and blocking with Dako Protein blocking solution (Aglient Technologies, Santa Clara, CA, USA) with 0.1% saponin (Sigma-Aldrich, St. Louis, MO, USA) at room temperature for 1 h. Slides were mounted with ProLong Gold Antifade Mountant (Invitrogen, Waltham, MA, USA) and ProLong Gold Antifade Mountant with DAPI (Invitrogen, Waltham, MA, USA). Membrane and slides were imaged on the Olympus FLUOVIEW CLSM (Olympus; FV3000, Shinjuku, Tokyo, Japan) with an Olympus UPlanSAPO 10x objective (Olympus; 1-U2B824, Shinjuku, Tokyo, Japan) and Olympus UPlanSAPO 60x objective (Olympus; 1-U2B832, Shinjuku, Tokyo, Japan). Images were analyzed using ImageJ analysis software.
Statistical analysis: Statistical analysis was performed using GraphPad Prism analysis software (GraphPad Software Inc., San Diego, CA, USA). Results are shown as the mean standard deviation. Comparisons among two groups were performed using an unpaired t-test, followed by Welch’s correction test. Comparisons among more than two groups were performed using a parametric one-way ANOVA test, followed by Bonferroni’s multiple comparisons test. p0.05 was considered statistically significant. The legend is as follows: * p-values 0.05; ** p-values 0.01; *** p-values 0.001; **** p-values 0.0001.
4. Discussion
Our present study reveals that lung-derived exosomes display superior pulmonary distribution and retention following inhaled delivery. Biological and synthetic nanoparticles can be loaded with exogenous molecular components, while maintaining their membrane integrities and phenotypes. Lung parenchymal cells cultured with exosomes had significantly greater mRNA and protein cargo uptake, suggesting that biological nanoparticles better facilitate cellular targeting and molecular drug function than synthetic LNPs. We showed that exosomes and liposomes deliver functional mRNA and protein cargo following jet nebulization administration to mice. Lung-Exo displayed enhanced pulmonary bioavailability through greater lung distribution and retention than its HEK-Exo and Lipo counterparts. Mice who received nebulized Lung-Exo had the highest mRNA translation and protein expression in lung tissue, as well as the highest NP deposition to the bronchioles and parenchyma, compared to HEK-Exo and Lipo. These data suggest that lung-derived exosomes are optimized drug delivery vesicles for inhaled therapeutics.
5. Conclusion
Interstitial lung disorders, lower respiratory infections, and small airway diseases require drug delivery to the deep lung to target affected lung tissue. Lung-Exo are naturally optimized for the pulmonary microenvironment and can better distribute and retain molecular drugs in the deep lung than synthetic nanoparticles. Lung-derived exosomes offer a unique nanoparticle drug delivery system, with enhanced bioavailability, that can serve as an mRNA and protein drug delivery vesicle tailored for lung diseases.
CRediT authorship contribution statement
Kristen D. Popowski: Methodology, Formal analysis, Investigation, Writing. Blanca López de Juan Abad: Methodology, Investigation, Writing. Arianna George: Investigation, Writing. Dylan Silkstone: Methodology, Investigation. Elizabeth Belcher: Methodology, Investigation. Jaewook Chung: Methodology, Investigation. Asma Ghodsi: Investigation, Writing. Halle Lutz: Investigation. Jada Davenport: Investigation. Mallory Flanagan: Investigation. Jorge Piedrahita: Resources. Phuong-Uyen C. Dinh: Conceptualization, Resources, Supervision. Ke Cheng: Conceptualization, Resources, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: North Carolina State University has filed a patent on the technologies related to this study. K.C. is an equity holder and consultant of Xsome Biotech Inc. Xsome has entered an exclusive license agreement with North Carolina State University.
Acknowledgments
We thank the Analytical Instrumentation Facility at North Carolina State University for TEM imaging. This work was supported by grants from the National Institutes of Health (HL123920, HL137093, HL144002, HL146153, HL147357, and HL149940 to K.C.) and the American Heart Association (18TPA34230092 and 19EIA34660286 to K.C.).
Footnotes
Given his role as Editor in Chief, Professor Ke Cheng had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to Professor Zhen Gu.
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.vesic.2022.100002.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
.
References
- 1.Labaki W.W., Han M.K. Chronic respiratory diseases: a global view. Lancet Resp Med. 2020;8(6):531–533. doi: 10.1016/S2213-2600(20)30157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the global burden of disease study 2019. Lancet (London, England) 2020;396(10258):1204–1222. doi: 10.1016/S0140-6736(20)30925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson C.F., Grimmett M.E., Domalewski C.J., Cui H. Inhalable nanotherapeutics to improve treatment efficacy for common lung diseases. Wiley Interdiscip Rev. 2020;12(1) doi: 10.1002/wnan.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resnier P., Mottais A., Sibiril Y., Le Gall T., Montier T. Challenges and successes using nanomedicines for Aerosol delivery to the airways. Curr Gene Therapy. 2016;16(1):34–46. doi: 10.2174/1566523216666160104142013. [DOI] [PubMed] [Google Scholar]
- 5.Beck-Broichsitter M., Merkel O.M., Kissel T. Controlled pulmonary drug and gene delivery using polymeric nano-carriers. J Control Release Off J Control Release Soc. 2012;161(2):214–224. doi: 10.1016/j.jconrel.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Henning A., Schneider M., Nafee N., Muijs L., et al. Influence of particle size and material properties on mucociliary clearance from the airways. J Aeros Med Pulm Drug Deliv. 2010;23(4):233–241. doi: 10.1089/jamp.2009.0806. [DOI] [PubMed] [Google Scholar]
- 7.d’Angelo I., Conte C., Miro A., Quaglia F., Ungaro F. Pulmonary drug delivery: a role for polymeric nanoparticles? Curr Top Med Chem. 2015;15(4):386–400. doi: 10.2174/1568026615666150108123256. [DOI] [PubMed] [Google Scholar]
- 8.Ally J., Martin B., Behrad Khamesee M., Roa W., Amirfazli A. Magnetic targeting of aerosol particles for cancer therapy. J Magn Magn Mater. 2005;293(1):442–449. doi: 10.1016/j.jmmm.2005.02.038. [DOI] [Google Scholar]
- 9.Dames P., Gleich B., Flemmer A., Hajek K., Seidl N., et al. Targeted delivery of magnetic aerosol droplets to the lung. Nat Nanotechnol. 2007;2(8):495–499. doi: 10.1038/nnano.2007.217. [DOI] [PubMed] [Google Scholar]
- 10.Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., et al. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182(5):1271–1283. doi: 10.1016/j.cell.2020.07.024. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polack F.P., Thomas S.J., Kitchin N., Absalon J., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh E.E., Frenck R.W.J., Falsey A.R., Kitchin N., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. 2020;383(25):2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang R.Y.K., Chan H.-K. Lipid nanoparticles for the inhalation of mRNA. Nat Biomed Eng. 2021;5(9):949–950. doi: 10.1038/s41551-021-00794-x. [DOI] [PubMed] [Google Scholar]
- 15.Li Z., Wang Z., Dinh P.-U.C., Zhu D., et al. Cell-mimicking nanodecoys neutralize SARS-CoV-2 and mitigate lung injury in a non-human primate model of COVID-19. Nature Nanotechnol. 2021;16(8):942–951. doi: 10.1038/s41565-021-00923-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Théry C., Zitvogel L., Amigorena S. Exosomes: composition biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 17.Hu S., Li Z., Shen D., Zhu D., et al. Exosome-eluting stents for vascular healing after ischaemic injury. Nat Biomed Eng. 2021;5(10):1174–1188. doi: 10.1038/s41551-021-00705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popowski K., Lutz H., Hu S., George A., Dinh P.-U., Cheng K. Exosome therapeutics for lung regenerative medicine. J Extracell Vesicles. 2020;9(1) doi: 10.1080/20013078.2020.1785161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao C.-Y., Papoutsakis E.T. Extracellular vesicles: exosomes, microparticles, their parts, and their targets to enable their biomanufacturing and clinical applications. Curr Opin Biotechnol. 2019;60:89–98. doi: 10.1016/J.COPBIO.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Dinh P.-U.C., Paudel D., Brochu H., Popowski K.D., et al. Inhalation of lung spheroid cell secretome and exosomes promotes lung repair in pulmonary fibrosis. Nature Commun. 2020;11(1):1064. doi: 10.1038/s41467-020-14344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakase I., Noguchi K., Aoki A., Takatani-Nakase T., Fujii I., Futaki S. Arginine-rich cell-penetrating peptide-modified extracellular vesicles for active macropinocytosis induction and efficient intracellular delivery. Sci Rep. 2017;7(1):1991. doi: 10.1038/s41598-017-02014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naseri Z., Oskuee R.K., Jaafari M.R., Forouzandeh Moghadam M. Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. Int J Nanomedicine. 2018;13:7727–7747. doi: 10.2147/IJN.S182384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fennelly K.P. Particle sizes of infectious aerosols: implications for infection control. Lancet Resp Med. 2020;8(9):914–924. doi: 10.1016/S2213-2600(20)30323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flaherty K.R., Wells A.U., Cottin V., Devaraj A., et al. Nintedanib in progressive fibrosing interstitial Lung diseases. N Engl J Med. 2019;381(18):1718–1727. doi: 10.1056/NEJMoa1908681. [DOI] [PubMed] [Google Scholar]
- 25.King T.E.J., Bradford W.Z., Castro-Bernardini S., Fagan E.A., et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 26.Lancaster L.H., de Andrade J.A., Zibrak J.D., Padilla M.L., et al. Pirfenidone safety and adverse event management in idiopathic pulmonary fibrosis. Eur Resp Rev Off J Eur Resp Soc. 2017;26(146) doi: 10.1183/16000617.0057-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang H., Huang K., Su T., Li Z., et al. Mesenchymal stem cell/red blood cell-inspired nanoparticle therapy in mice with carbon tetrachloride-induced acute liver failure. ACS Nano. 2018;12(7):6536–6544. doi: 10.1021/acsnano.8b00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry E., Cores J., Hensley M.T., Anthony S., et al. Adult lung spheroid cells contain progenitor cells and mediate regeneration in rodents with bleomycin-induced pulmonary fibrosis. Stem Cells Transl Med. 2015;4(11):1265–1274. doi: 10.5966/sctm.2015-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cores J., Hensley M.T., Kinlaw K., Rikard S.M., et al. Safety and efficacy of allogeneic lung spheroid cells in a mismatched rat model of pulmonary fibrosis. Stem Cells Transl Med. 2017;6(10):1905–1916. doi: 10.1002/sctm.16-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cores J., Dinh P.C., Hensley T., Adler K.B., Lobo L.J., Cheng K. A pre- <scp>investigational new drug</scp> study of lung spheroid cell therapy for treating pulmonary fibrosis. STEM CELLS Transl Med. 2020;9(7):786–798. doi: 10.1002/sctm.19-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinh P.-U.C., Cores J., Hensley M.T., Vandergriff A.C., et al. Derivation of therapeutic lung spheroid cells from minimally invasive transbronchial pulmonary biopsies. Resp Res. 2017;18(1):132. doi: 10.1186/s12931-017-0611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greening D.W., Xu R., Ji H., Tauro B.J., Simpson R.J. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Molecular Biol (Clifton, N.J.) 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 33.Bosch S., de Beaurepaire L., Allard M., Mosser M., et al. Trehalose prevents aggregation of exosomes and cryodamage. Sci Rep. 2016;6(1):36162. doi: 10.1038/srep36162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.