Despite their varied purposes, many indispensable molecules in medicine, materials, and asymmetric catalysis share a biaryl core.1,2 The necessity of joining arene building blocks to access these valuable compounds has inspired multiple approaches for biaryl bond formation and challenged chemists to develop increasingly concise and robust methods for this task.3 Oxidative coupling of two C–H bonds offers an efficient strategy for the formation of a biaryl C–C bond, however, fundamental challenges remain in controlling the reactivity and selectivity for uniting a given pair of substrates.4,5 Biocatalytic oxidative cross-coupling reactions have the potential to overcome limitations inherent to numerous small molecule-mediated methods by providing a paradigm with catalyst-controlled selectivity.6 In this article, we disclose a strategy for biocatalytic cross-coupling through oxidative C–C bond formation using cytochrome P450 enzymes. We demonstrate the ability to catalyze cross-coupling reactions on a panel of phenolic substrates using natural P450 catalysts. Moreover, we engineer a P450 to possess the desired reactivity, site-selectivity, and atroposelectivity by transforming a low-yielding, unselective reaction into a highly efficient and selective process. This streamlined method for constructing sterically hindered biaryl bonds provides a programmable platform for assembling molecules with catalyst-controlled reactivity and selectivity.
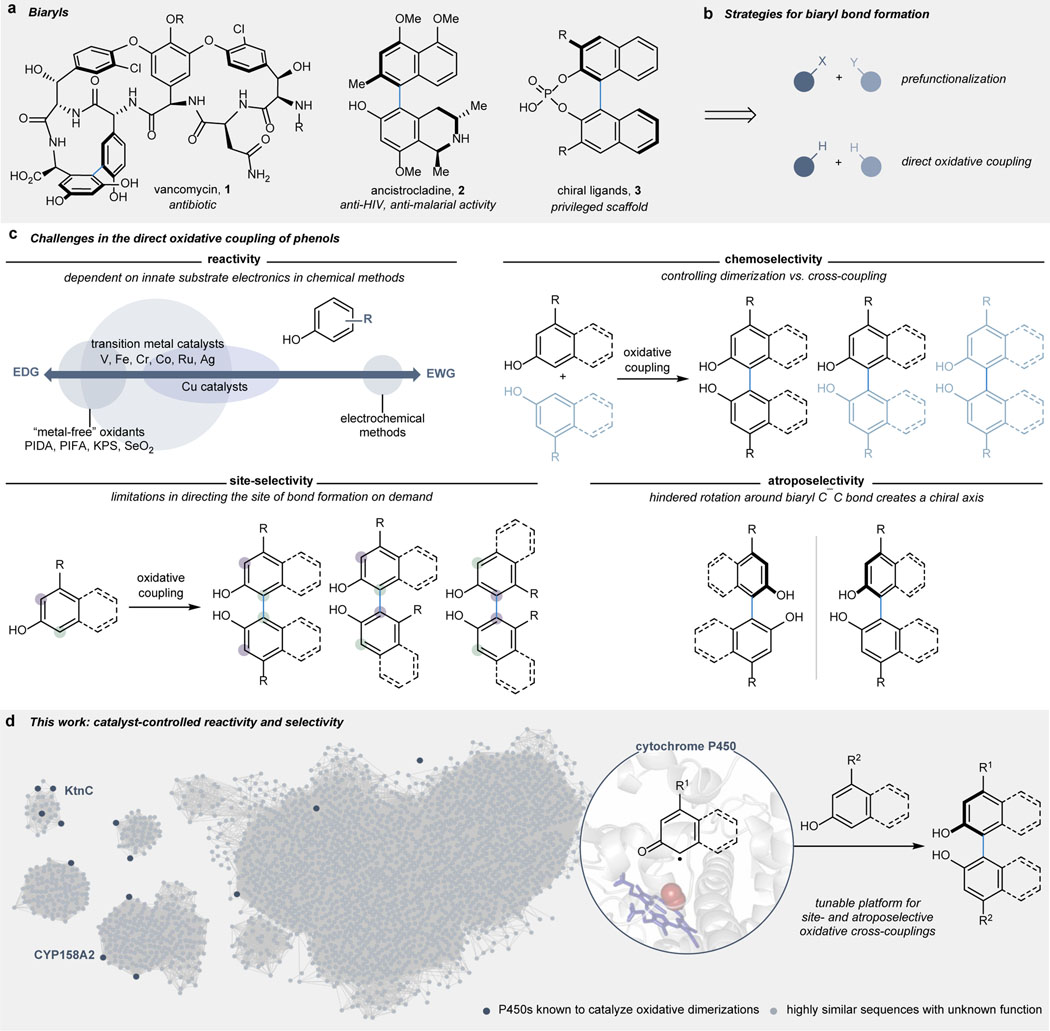
Biaryl bonds are ubiquitous in drugs, materials, and ligands for asymmetric catalysis (Figure 1a).1,2 The most commonly employed strategy to construct these molecules relies on metal-catalyzed cross-coupling of prefunctionalized aryl fragments, thereby offering preprogrammed site-selectivity, albeit at the expense of adding extra steps in a synthetic campaign (Figure 1b).7,8 These reactions can require extensive screening to identify suitable conditions for each new pair of coupling partners and are still restricted in the connectivities which are readily accessible,9–13 with the formation of tetra-ortho-substituted biaryl bonds remaining an outstanding challenge.14,15 Oxidative coupling provides an alternative strategy for biaryl bond formation through the net transformation of two C–H bonds into a C–C bond (Figure 1b).4,5 However, these advantages come at the expense of the efficiency and selectivity of this transformation, which is typically dictated by the intrinsic steric and electronic properties of each substrate, thereby limiting the versatility of this transformation (Figure 1c).4,16–18 Numerous metal catalysts and metal-free oxidants have been reported for the dimerization and cross-coupling of phenolic substrates; however, the application of these methods is commonly restricted to electron-rich phenols while oxidative coupling with electron deficient phenols remains more broadly challenging.16–27 When general reactivity can be achieved, controlling the chemo-, site-, and atroposelectivity of the oxidative coupling presents a considerable hurdle, particularly in cases where the intrinsic steric and electronic properties of the reacting phenols prohibit access to the desired product isomer.17,18,28–32
Fig. 1 |. Biaryl bond formation through direct oxidative cross-coupling.
a, The biaryl scaffold is a privileged substructure in drug discovery and asymmetric catalysis. b, Biaryl bonds can be access through metal-catalyzed cross-coupling of prefunctionalized starting materials or direct oxidative coupling. c, Oxidative biaryl bond formation faces fundamental challenges in reactivity and selectivity. d, In this work, we report the first biocatalytic platform for the selective cross-coupling of phenolic substrates through oxidative C–C bond formation.
Biocatalytic oxidative cross-coupling reactions have the potential to overcome the limitations inherent to small molecule-mediated methods for direct oxidative coupling by providing a paradigm with catalyst-controlled selectivity. Nature has evolved oxidative enzymes, including laccases and cytochromes P450 (P450s), that mediate the dimerization of phenolic compounds in the biosynthesis of biaryl natural products (Supplemental Table S2).6,33 The application of laccases in selective catalysis has been limited by the scarcity of examples in which these enzymes exert control over the selectivity of the bond formation following the initial oxidation.34–36 In contrast, an ever-expanding repertoire of P450s mediate highly selective oxidation reactions,37 among which exist a small subset of enzymes that catalyze site- and atroposelective dimerizations.6 This subset of wild-type P450s that perform oxidative coupling chemistry are yet to be explored as biocatalysts for unnatural cross-coupling reactions. Moreover, these catalysts exist within a large pool of related enzyme sequences with untapped potential to catalyze a diverse array of oxidative coupling reactions (see sequence similarity network, SSN, in Figure 1d). We hypothesized that this class of enzymes could be developed into tunable biocatalysts for convergent oxidative cross-coupling reactions with catalyst-controlled reactivity, site-selectivity, and atroposelectivity.
Among the P450s previously identified as catalysts for oxidative dimerization en route to biaryl natural products (Supplemental Table S2), we were initially drawn to a small cluster of Aspergillus enzymes that each impart unique site-selectivity in the dimerization of coumarin 4 (Supplemental Figure S27).38,39 To assess the activity and scalability of this enzymatic reaction, analytical scale biotransformations were conducted in Saccharomyces cerevisiae heterologously expressing the gene encoding KtnC.39 As previously demonstrated by Müller and coworkers, the formation of the expected dimeric product 7 was observed; however, constraints on substrate loading in S. cerevisiae were not amenable to preparative scale reactions (Supplemental Figure S28). To develop a more scalable whole-cell biocatalytic platform in yeast, the gene encoding KtnC was incorporated into the genome of Pichia pastoris. This new whole-cell system allowed for more than a three-fold increase in the total formation of biaryl product, enabling preparative scale reactions (see Supplemental Section VI).40
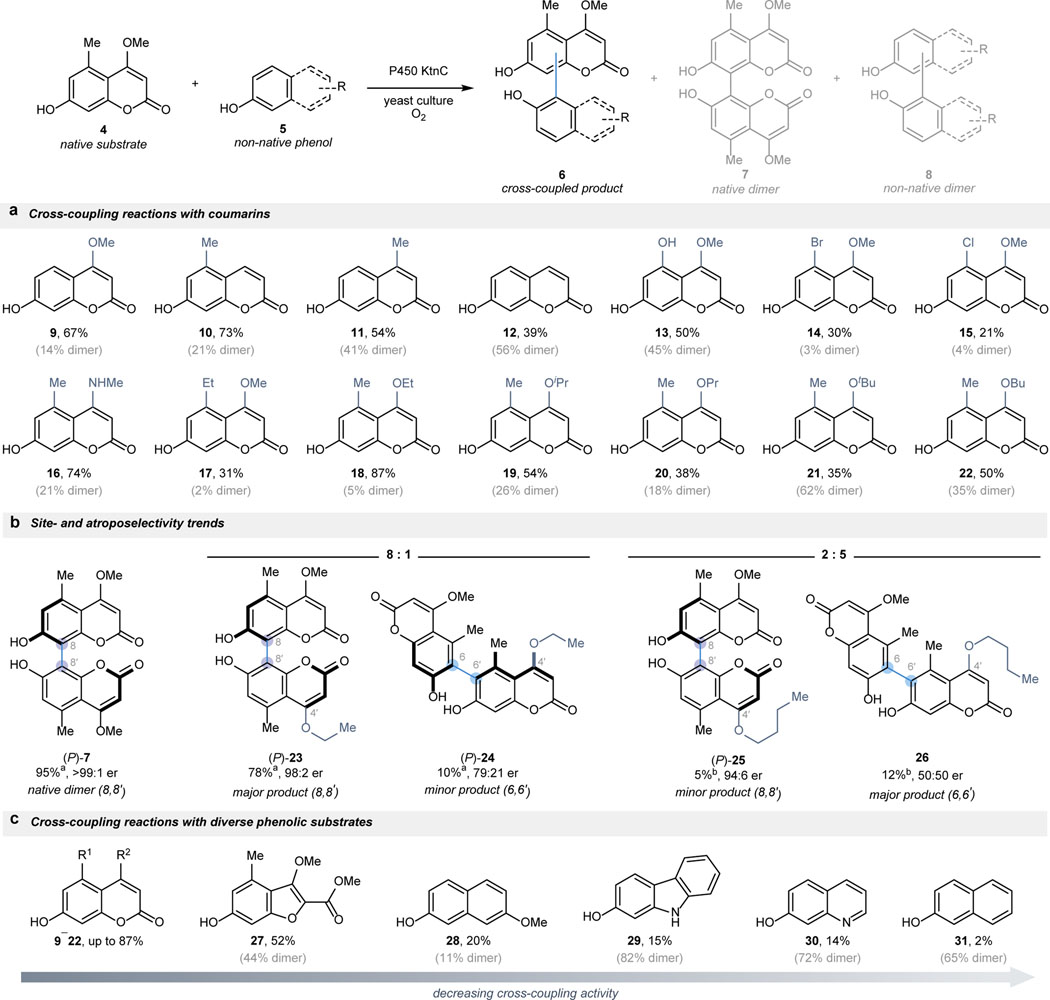
With the optimized biocatalytic platform in hand, we sought to move beyond natural dimerization reactions to unnatural cross-coupling chemistry. We hypothesized that the binding of two non-equivalent phenolic substrates in the enzyme active site would overcome traditional steric and electronic limitations in small molecule-mediated oxidative cross-coupling reactions. In a proof-of-concept experiment, coumarins 4 and 16 were both added to a culture producing KtnC, leading to the formation of unnatural cross-coupled products. Further experimentation demonstrated that tuning the stoichiometry of the cross-coupling partners could maximize cross-coupled product formation over dimerization (Supplemental Figure S30). Using this strategy for KtnC-catalyzed cross-coupling reactions, substrate promiscuity was observed with a panel of coumarin substrates (Figure 2a). Initial cross-coupling reactions between 4 and 9–12 demonstrated that maintaining similar substitution patterns to 4 at the C4 and C5 positions is beneficial but not required for activity. Additionally, KtnC tolerated a range of coumarins bearing electron-rich and electron-deficient substituents (13–16), diverging from the steric and electronic restrictions that typically govern small molecule-catalyzed oxidative cross-coupling reactions.17,18 Added steric bulk at the C4 and C5 positions of coumarins 17–22 was also readily accepted. In contrast to the high levels of reactivity achieved in coumarin cross-coupling reactions with KtnC, modest to no cross-coupling reactivity was observed for these substrate pairs when subjected to established methods for the oxidative coupling of phenolic substrates (Supplemental Table S1).5,22,41–43
Fig. 2 |. Scope of oxidative cross-coupling reactions catalyzed by wild-type P450 KtnC.
a, KtnC displayed promiscuity in the cross-coupling of native coumarin 4 with a range of non-equivalent coumarins with varying steric and electronic properties. b, Most cross-coupling reactions maintained the site- and atroposelectivity displayed by the native product 7, with the exception of the formation of a rare 6,6ʹ product (24 and 26). c, KtnC catalyzed the cross-coupling with a diverse set of phenolic substrates, albeit with decreased reactivity and selectivity as the substrate structures deviated further from the native coumarin scaffold. Reaction conditions: P. pastoris cells expressing KtnC in the presence of 4 and 5 in up to a 1:10 molar ratio with 25–100 μM 4 at 30 ˚C with shaking at 235 rpm for 48–72 h; cross-coupling reactions reported as percent conversions, apercent yields, or brelative percent yields; competing dimerization of substrate 4 reported as percent conversions in parentheses.
The majority of the cross-coupling reactions examined maintained the site- and atroposelectivity exhibited in the natural dimerization reaction. For example, the major product resulting from the cross-coupling between 4 and 18 harbored the 8,8ʹ-connectivity and was formed with excellent atroposelectivity (see 23). However, in addition to the expected 8,8ʹ-product, a minor cross-coupled product was observed in reactions with 4 and coumarins bearing increased steric bulk at the C4 position (18–22; Figure 2b). Further characterization of this product revealed a rare 6,6ʹ-connectivity, with cross-coupling to the butyl ester (22) completely shifting the preference from the formation of the 8,8ʹ-product (25) to the 6,6ʹ-product (26) in a 2:5 ratio. Notably, both prefunctionalization and direct oxidative coupling strategies were unsuccessful in generating the 6,6’-isomer in these cross-coupling reactions, highlighting the steric and electronic biases that impede access to specific connectivities using small molecule-based strategies (Supplemental Figure S8 and S10).17,18
In addition to forming a range of unsymmetrical bicoumarin products, a panel of more diverse phenolic substrates were tested as cross-coupling partners for coumarin 4 (Figure 2c). In general, superior reactivity was observed with phenolic substrates that resembled native substrate 4; however, as the substrate structures diverged further from the native coumarin scaffold, significant decreases in the cross-coupling activity and site-selectivity occurred. We hypothesized that the limitations in substrate scope and chemo-, site-, and atroposelectivity could be overcome with protein engineering.
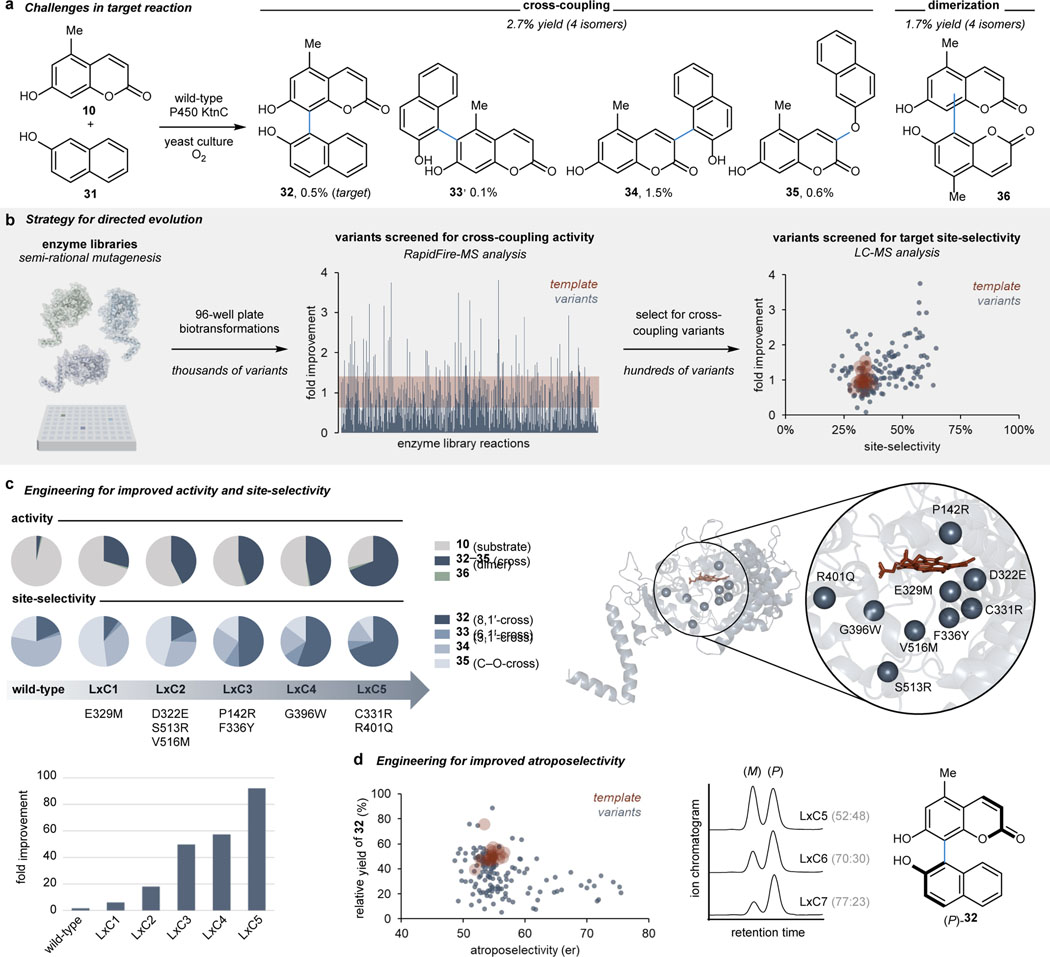
We anticipated that the successful engineering of a cross-coupling catalyst (LxC) could transform a low-yielding, unselective biocatalytic reaction into a practical method for synthesis. The cross-coupling between coumarin 10 and 2-naphthol (31) was selected as the target reaction due to the low levels of reactivity and site-selectivity (<3% combined yield of four cross-coupled products) displayed by wild-type KtnC (Figure 3a and Supplemental Figure S66). Protein libraries were generated through semi-rational mutagenesis, resulting in thousands of variants with one or multiple substitutions within 12 Å of the active site. Variants were screened in high-throughput yeast biotransformations and analyzed for total cross-coupled product formation by RapidFire mass spectrometry (MS). Reaction mixtures containing higher levels of the cross-coupled product were then analyzed for cross-coupling site-selectivity in a second-tier screen using LC-MS (Figure 3b). Ultimately, this two-tier MS screen enabled rapid identification of variants with improved activity and site-selectivity for the targeted cross-coupling reaction.
Fig. 3 |. Engineering P450 biocatalysts for improved activity and selectivity.
a, Target reaction selected for directed evolution campaign, with wild-type KtnC catalyzing the cross-coupling at a 2.7% yield to four different cross-coupled isomers, in addition to dimeric products. b, Strategy for the directed evolution of P450s for improved activity and site-selectivity employing semi-rational mutagenesis and a two-tier mass spectrometry screen, as represented by data from round three of evolution above. c, Over five rounds of evolution, a total of nine active site mutations were incorporated to provide LxC5 with 92-fold improvement in activity and site-selectivity toward the target cross-coupled product 32. d, In addition to site-selectivity, the atroposelectivity of the reaction was demonstrated to be tunable with active site mutations.
Through iterative rounds of directed evolution, the total activity and site-selectivity in the desired cross-coupling reaction was progressively improved (Figure 3c). Initially, improvements in total activity were targeted, resulting in a 19-fold increase in total cross-coupled product formation over the first two rounds of engineering, while keeping competing levels of coumarin dimerization low. However, the site-selectivity of the reaction was undesirable, with the C–O cross-coupled product (35) representing nearly half of all the cross-coupled products formed by LxC2. Therefore, variants with improved site-selectivity were targeted in subsequent engineering rounds, leading to greater site-selectivity for the desired 8,1ʹ-product (32). Ultimately, a biocatalyst with a 92-fold improvement in activity was engineered through five rounds of evolution to form cross-coupled product 32. Preparative scale biotransformations with LxC5 resulted in the isolation of cross-coupled product 32 in a 50% yield (Supplemental Section VI). The efficiency of this reaction was further exemplified in comparison to tradition synthetic approaches toward 32, with a direct oxidative cross-coupling using a copper catalyst giving a 19% yield (Supplemental Table S1, Entry 2a)41 and a Suzuki cross-coupling strategy providing a 26% yield over four steps (Supplemental Scheme S2).
As variants were engineered selecting for increased cross-coupling yield and site-selectivity, a decrease in the atroposelectivity of the reaction was observed from an 80:20 er with the wild-type enzyme to a 52:48 er with LxC5 (Supplemental Figure S72). To regain catalyst control over atroposelectivity, two additional rounds of engineering were performed using a combination of RapidFire and chiral LC-MS (Figure 3d). A variant (LxC7) that catalyzed the cross-coupling to 32 in a 77:23 er was identified, albeit with a reduction in yield. These results demonstrate that this biocatalytic platform is highly engineerable for the desired activity, site-selectivity, and atroposelectivity, thereby providing a programmable platform for catalyst-controlled oxidative cross-coupling reactions.
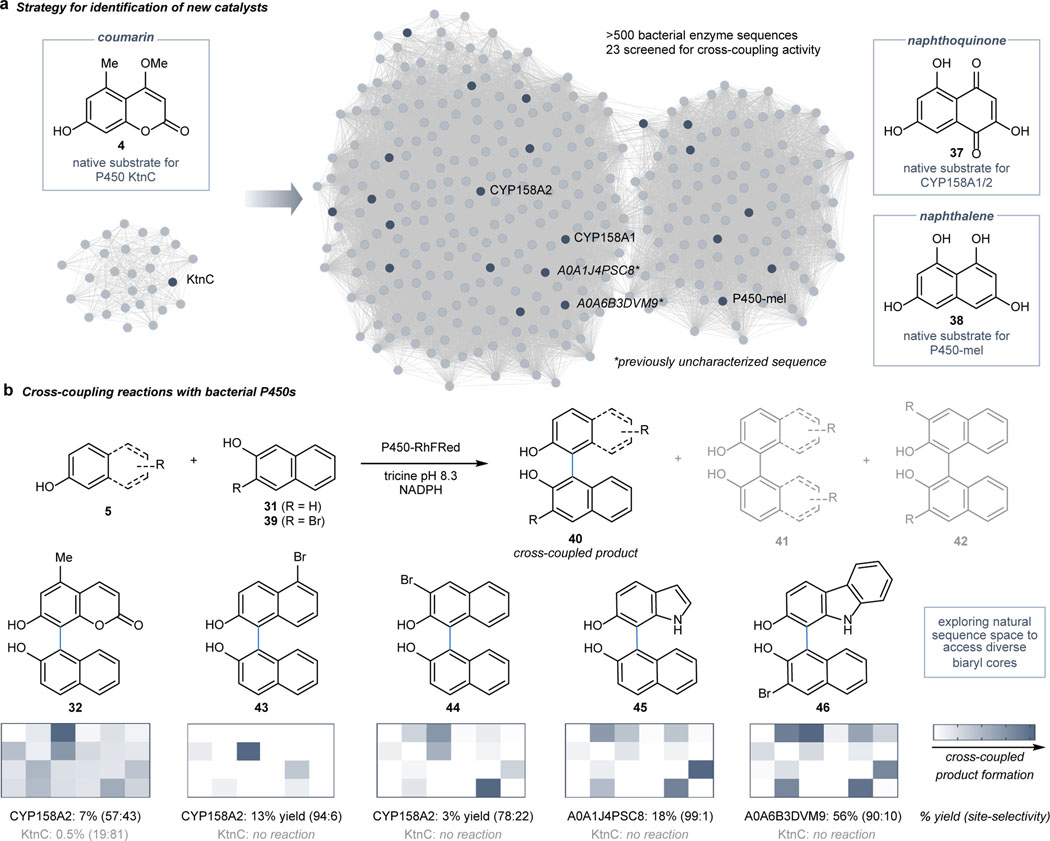
In parallel to employing a protein engineering strategy, we hypothesized that alternative wild-type enzymes could provide complementary catalysts for oxidative cross-coupling reactions. A bioinformatics-guided approach was taken by harnessing SSNs as a tool to visualize the natural sequence space surrounding P450s known to mediate intermolecular oxidative dimerization reactions. An SSN was constructed with the entire P450 protein family (Supplemental Figure S26) and then truncated to edges possessing an alignment score greater than 110 with P450s known to dimerize phenolic substrates (Figure 1d).44–47 Characterized P450s that clustered together (indicating high levels of sequence similarity) dimerized similar natural substrates; in contrast, characterized enzymes that cluster separately typically transformed different substrate scaffolds (Supplemental Figure S27). Based on these trends, the reactivity of a panel of 23 enzymes was screened. This panel included 20 uncharacterized enzymes as well as three characterized P450s known to dimerize substrates with naphthoquinone and naphthalene cores (see 37 and 38) that differed structurally from coumarin 4 (Figure 4a).48,49
Fig. 4 |. Exploring alternative sequence space for catalysts with complementary reactivity.
a, Strategy for selection of alternative catalysts for cross-coupling reactions using SSNs. b, Bacterial fusion enzymes (P450-RhFRed) catalyze cross-coupling reactions with naphthols, demonstrating complementary reactivity to KtnC. The top hits from an initial screen of 23 enzymes were purified for further quantification of reaction yield and selectivity. Reaction conditions: 10 μM purified P450-RhFRed in the presence of 5 mM glucose-6-phosphate, 1 mM NADP+, 1 U/mL glucose-6-phosphate dehydrogenase, and substrates in a 1:1 molar ratio in lysate reactions (1 mM 39 and 1 mM 31 or 40) or 1:3 molar ratio in reactions with purified enzyme (100 μM 39 and 300 μM 31 or 40) in tricine buffer pH 8.3 at 30 ˚C for 16 h; cross-coupling reactions reported as percent yields to major product with cross-coupling site-selectivity in parentheses.
We anticipated that these enzymes would productively catalyze oxidative cross-coupling reactions with naphthols (e.g. 31 and 40) to provide a suite of complementary catalysts to KtnC. Artificial fusion enzymes were engineered by tethering the P450RhF reductase domain (RhFRed) to the C-terminus of each P450 to reconstitute the electron transport required for catalytic activity (Supplemental Figure S29).50 In a series of E. coli lysate screens with the P450-RhFRed library (Supplemental Table S3), cross-coupled products formed across a diverse set of phenolic substrates to a naphthol partner in a 1:1 ratio with surprising promiscuity (Figure 4b). The sequences catalyzing the formation of the highest levels of cross-coupled product were further characterized using purified enzyme. Whereas the cross-coupling yields varied across the diverse set of reactions, the transformations favored the formation of a major cross-coupled product (see 43–46) over competing dimeric product formation (Supplemental Figures S73-S82), thereby minimizing the need for a large excess of one partner over the other and demonstrating both chemo- and site-selectivity. Moreover, two of the outstanding hits were novel sequences with previously uncharacterized function (A0A1J4PSC8 and A0A6B3DVM9). Altogether, these results demonstrate the complementary reactivity achievable with different P450 catalysts and provide biocatalytic access to diverse biaryl scaffolds.
The challenge of overriding innate reactivity and selectivity in oxidative coupling reactions has limited the reliable use of this method for convergent synthetic routes. The presented biocatalytic method overcomes reactivity hurdles by surpassing steric and electronic limitations and redefining substrates that can be efficiently cross-coupled. Furthermore, using a large molecule ligand provides the opportunity to achieve catalyst-controlled site- and atroposelectivity in biaryl coupling reactions. The wealth of natural catalysts available, along with the demonstrated ability to tune enzymes for the desired substrate scope, reactivity, and selectivity, positions this biocatalytic approach to be a powerful tool in the convergent assembly of biaryl molecules.
Supplementary Material
Acknowledgements
This research was supported by funds from the University of Michigan’s Life Sciences Institute and the Chemistry Department, Alfred P. Sloan Foundation, and Research Corporation Cottrell Scholars program. Initial studies on the selectivity of dimerization reactions were supported by the NIH R35 GM124880, and protein engineering (rounds 4–7) and bioinformatic-based library generation were supported by generous funds from the Novartis Global Scholars Program. L. E. Z. is grateful to the NIH National Center for Complementary & Integrative Health of the NIH F31AT010973, J. A. Y. thanks the National Science Foundation GRFP, A. L. L. acknowledges the Rackham Graduate School (UM) and the NIH F31 NS111906 for funding. We thank M. Müller for supplying the expression vector containing KtnC and D. Sherman for supplying the expression vector containing RhFRed. We thank E. A. Meucci, E. C. Bornowski, and J. B. Pyser for assistance with the synthesis of substrates and C-H. Chiang for help with CD acquisition.
Footnotes
Competing Interests The authors have no conflicts of interest to report.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/xxxx.
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability All data are available from the corresponding author upon reasonable request.
Online content Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/xxxx.
References
- 1.Yet L Privileged Structures in Drug Discovery: Medicinal Chemistry and Synthesis. First edn, 83–154 (John Wiley & Sons, Inc., 2018). [Google Scholar]
- 2.Yoon TP & Jacobsen EN Privileged chiral catalysts. Science 299, 1691–1693 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Ashenhurst JA Intermolecular oxidative cross-coupling of arenes. Chem. Soc. Rev 39, 540–548 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Kozlowski MC Oxidative coupling in complexity building transforms. Acc. Chem. Res 50, 638–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Lan J & You J Oxidative C–H/C–H coupling reactions between two (hetero)arenes. Chem. Rev 117, 8787–8863 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Hüttel W & Müller M Regio- and stereoselective intermolecular phenol coupling enzymes in secondary metabolite biosynthesis. Nat. Prod. Rep 38, 1011–1043 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Yin & Liebscher J Carbon−carbon coupling reactions catalyzed by heterogeneous palladium catalysts. Chem. Rev 107, 133–173 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Boström J, Brown DG, Young RJ & Keserü GM Expanding the medicinal chemistry synthetic toolbox. Nat. Rev. Drug Discov. 17, 709–727 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Yin J, Rainka MP, Zhang X-X & Buchwald SL A highly active Suzuki catalyst for the synthesis of sterically hindered biaryls: novel ligand coordination. J. Am. Chem. Soc. 124, 1162–1163 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Cammidge AN & Crépy KVL Synthesis of chiral binaphthalenes using the asymmetric Suzuki reaction. Tetrahedron 60, 4377–4386 (2004). [Google Scholar]
- 11.Martin R & Buchwald SL Palladium-catalyzed Suzuki−Miyaura cross-coupling reactions employing dialkylbiaryl phosphine ligands. Acc. Chem. Res 41, 1461–1473 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valente C et al. The development of bulky palladium NHC complexes for the most-challenging cross-coupling reactions. Angew. Chem. Int. Ed. 51, 3314–3332 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Patel ND et al. Computationally assisted mechanistic investigation and development of Pd-catalyzed asymmetric Suzuki–Miyaura and Negishi cross-coupling reactions for tetra-ortho-substituted biaryl synthesis. ACS Catal. 8, 10190–10209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ackermann L, Potukuchi HK, Althammer A, Born R & Mayer P Tetra-ortho-substituted biaryls through palladium-catalyzed Suzuki−Miyaura couplings with a diaminochlorophosphine ligand. Org. Lett 12, 1004–1007 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Brown DG & Boström J Analysis of past and present synthetic methodologies on medicinal chemistry: where have all the new reactions gone? J. Med. Chem 59, 4443–4458 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Lee YE, Cao T, Torruellas C & Kozlowski MC Selective oxidative homo- and cross-coupling of phenols with aerobic catalysts. J. Am. Chem. Soc 136, 6782–6785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nieves-Quinones Y et al. Chromium-salen catalyzed cross-coupling of phenols: mechanism and origin of the selectivity. J. Am. Chem. Soc 141, 10016–10032 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalit H, Dyadyuk A & Pappo D Selective oxidative phenol coupling by iron catalysis. J. Org. Chem 84, 1677–1686 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Reiss H et al. Cobalt(II)[salen]-catalyzed selective aerobic oxidative cross-coupling between electron-rich phenols and 2-naphthols. J. Org. Chem 84, 7950–7960 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Röckl JL, Schollmeyer D, Franke R & Waldvogel SR Dehydrogenative anodic C−C coupling of phenols bearing electron-withdrawing groups. Angew. Chem. Int. Ed 59, 315–319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang H et al. Enantioselective vanadium-catalyzed oxidative coupling: development and mechanistic insights. J. Org. Chem 83, 14362–14384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Libman A et al. Synthetic and predictive approach to unsymmetrical biphenols by iron-catalyzed chelated radical–anion oxidative coupling. J. Am. Chem. Soc 137, 11453–11460 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Morimoto K, Sakamoto K, Ohshika T, Dohi T & Kita Y Organo-iodine(III)-catalyzed oxidative phenol-arene and phenol-phenol cross-coupling reaction. Angew. Chem. Int. Ed 55, 3652–3656 (2016). [DOI] [PubMed] [Google Scholar]
- 24.More NY & Jeganmohan M Oxidative cross-coupling of two different phenols: an efficient route to unsymmetrical biphenols. Org. Lett 17, 3042–3045 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Egami H & Katsuki T Iron-catalyzed asymmetric aerobic oxidation: oxidative coupling of 2-naphthols. J. Am. Chem. Soc 131, 6082–6083 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Hovorka M, Günterova J & Zavada J Highly selective oxidative cross-coupling of substituted 2-naphthols: a convenient approach to unsymmetrical 1, 1′-binaphthalene-2, 2′-diols. Tet. Lett 31, 413–416 (1990). [Google Scholar]
- 27.Li X, Hewgley JB, Mulrooney CA, Yang J & Kozlowski MC Enantioselective oxidative biaryl coupling reactions catalyzed by 1, 5-diazadecalin metal complexes: efficient formation of chiral functionalized BINOL derivatives. J. Org. Chem 68, 5500–5511 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Tian J-M et al. Copper-complex-catalyzed asymmetric aerobic oxidative cross-coupling of 2-naphthols: enantioselective synthesis of 3,3′-substituted C1-symmetric BINOLs. Angew. Chem. Int. Ed 58, 11023–11027 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Bringmann G et al. Atroposelective synthesis of axially chiral biaryl compounds. Angew. Chem. Int. Ed 44, 5384–5427 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Kočovský P, Vyskočil Š & Smrčina M Non-symmetrically substituted 1,1’-binaphthyls in enantioselective catalysis. Chem. Rev 103, 3213–3246 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Kozlowski MC, Morgan BJ & Linton EC Total synthesis of chiral biaryl natural products by asymmetric biaryl coupling. Chem. Soc. Rev 38, 3193–3207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bringmann G, Gulder T, Gulder TAM & Breuning M Atroposelective total synthesis of axially chiral biaryl natural products. Chem. Rev 111, 563–639 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Aldemir H, Richarz R & Gulder TA The biocatalytic repertoire of natural biaryl formation. Angew. Chem. Int. Ed 53, 8286–8293 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Mate DM & Alcalde M Laccase: a multi-purpose biocatalyst at the forefront of biotechnology. Microb. Biotechnol 10, 1457–1467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagui F et al. Laccase-catalyzed coupling of catharanthine and vindoline: an efficient approach to the bisindole alkaloid anhydrovinblastine. Tetrahedron 65, 312–317 (2009). [Google Scholar]
- 36.Obermaier S, Thiele W, Fürtges L & Müller M Enantioselective phenol coupling by laccases in the biosynthesis of fungal dimeric naphthopyrones. Angew. Chem. Int. Ed 58, 9125–9128 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Fasan R Tuning P450 enzymes as oxidation catalysts. ACS Catal. 2, 647–666 (2012). [Google Scholar]
- 38.Gil Girol C et al. Regio-and stereoselective oxidative phenol coupling in Aspergillus niger. Angew. Chem. Int. Ed 51, 9788–9791 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Mazzaferro LS, Huttel W, Fries A & Müller M Cytochrome P450-catalyzed regio- and stereoselective phenol coupling of fungal natural products. J. Am. Chem. Soc 137, 12289–12295 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Chakrabarty S, Wang Y, Perkins JC & Narayan ARH Scalable biocatalytic C–H oxyfunctionalization reactions. Chem. Soc. Rev 49, 8137–8155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noji M, Nakajima M & Koga K A new catalytic system for aerobic oxidative coupling of 2-naphthol derivatives by the use of CuCl-amine complex: a practical synthesis of binaphthol derivatives. Tet. Lett 35, 7983–7984 (1994). [Google Scholar]
- 42.Nakajima M Synthesis and application of novel biaryl compounds with axial chirality as catalysts in enantioselective reactions. Yakugaku Zasshi 120, 68–75 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Langeslay RR et al. Catalytic applications of vanadium: a mechanistic perspective. Chem. Rev 119, 2128–2191 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Shannon P et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerlt JA et al. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037, doi: 10.1016/j.bbapap.2015.04.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zallot R, Oberg NO & Gerlt JA ‘Democratized’ genomic enzymology web tools for functional assignment. Curr. Opin. Chem. Biol 47, 77–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zallot R, Oberg N & Gerlt JA The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 58, 4169–4182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Funa N, Funabashi M, Ohnishi Y & Horinouchi S Biosynthesis of hexahydroxyperylenequinone melanin via oxidative aryl coupling by cytochrome P-450 in Streptomyces griseus. J. Bacteriol 187, 8149–8155 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao B et al. Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2. J. Biol. Chem 280, 11599–11607 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Li S, Podust LM & Sherman DH Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain. J. Am. Chem. Soc 129, 12940–12941 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.