Abstract
Haloacid dehalogenases are enzymes that catalyze the hydrolytic removal of halogens from haloalkanoic acids. Dehalogenase IVa (DehIVa) from Burkholderia cepacia MBA4 and dehalogenase CI (DehCI) from Pseudomonas sp. strain CBS3 exhibit 68% identity. Despite their similarity DehIVa is a dimeric enzyme while DehCI is a monomer. In this work, we describe the identification of the domain that confers the dimerization function of DehIVa. Recombinant DNA molecules were constructed by fusion of the respective dehalogenase genes hdlIVa and dehCI. When amino acids 73 to 89 of DehCI were replaced by amino acids 74 to 90 of DehIVa, the recombinant molecule migrated like that of DehIVa in a nondenaturing activity-stained gel. Similarly, when residues 73 to 89 of DehIVa were replaced by the corresponding residues of DehCI, the chimera migrated as a monomer. These 17 amino acid changes were able to determine the aggregation states of the molecules. The retention of the catalytic function in these chimeras indicated that the overall folding of these proteins was not affected. Site-directed mutagenesis on hdlIVa however indicated that amino acids Phe58, Thr65, Leu78, and Phe92 of DehIVa are also important for the aggregation state of the protein. This indicates that the 17 residues are not sufficient for the dimerization of the protein.
Dehalogenases are enzymes that remove halogen from the carbon moiety (6). Several dehalogenases have been purified and characterized since their first detection in bacteria. These dehalogenases were categorized according to their substrate specificities (28) and recently according to their phylogenetic relationships (8). Much attention has been drawn to the 2-haloacid dehalogenases or halidohydrolases. These are hydrolytic enzymes that cleave the halogen-carbon bond(s) in halogenated aliphatic acids, yielding hydroxy- or oxoalkanoic acids from a substrate with a mono- or disubstitution, respectively (7, 28). At least 11 2-haloacid dehalogenase genes have been isolated and sequenced. Comparative study on the amino acid sequences of these enzymes has exhibited 37 to 67% homology (17, 32). Among these enzymes three conserved motifs have been identified. These include residues 4 to 18 in motif 1, residues 105 to 123 in motif 2, and residues 139 to 194 in motif 3 (1). Motif 1 contains a highly conserved aspartate and a threonine, motif 2 contains a highly conserved hydroxy residue (serine or threonine), and motif 3 contains a highly conserved lysine and a pair of aspartates. These conserved motifs were expected to convey functions essential for catalysis. Site-directed mutagenesis had confirmed the role of these motifs in the activity of dehalogenase l-DEX-YL and dehalogenase IVa (DehIVa) (18; B. C. M. Pang and J. S. H. Tsang, unpublished data).
Most dehalogenases were identified from microorganisms isolated from enrichment cultures using specific halogenated substrates (3, 27). These microorganisms are capable of utilizing the substrates as sole carbon and energy sources. Burkholderia cepacia MBA4, isolated with monobromoacetate (MBA), produces a single dehalogenase (DehIVa) under batch culture conditions (31). The structural gene of DehIVa (hdlIVa) has been isolated and characterized and encodes 231 amino acids (22, 30). DehIVa exists as a ∼45-kDa molecule, composed of two identical subunits of ∼26 kDa each (22, 31). Another dehalogenase, DehCI, isolated from the chlorobenzoate-utilizing pseudomonad strain CBS3 (15, 26), exhibits 64% identity in nucleotide sequence and 68% identity in amino acid sequence to DehIVa. This enzyme, however, is a monomer of 26 kDa. It is of interest to find out what causes the dimerization of DehIVa and not DehCI.
The modular replacement technique (2) has been used to study the structure-function relationships, including subunit association, substrate recognition, and enzyme activity, of several dioxygenases (4, 12). The dehalogenases characterized so far range from 26 to 79 kDa (8). The native forms of these enzymes exist as monomer, dimer, trimer, or tetramer (13, 21, 26, 31). In this paper, we described the use of modular replacement technique in identifying the dimerization domain of DehIVa. Chimeric constructs were produced by swapping nucleotide sequences between hdlIVa and dehCI. The recombinant molecules produced in vitro and/or in vivo were characterized by gel electrophoresis, gel filtration, and/or activity assay. This is the first work to address functionally the dimerization domain of 2-haloacid dehalogenases by a molecular analysis approach.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
B. cepacia MBA4 and Pseudomonas sp. strain CBS3 were used as the sources for wild-type dehalogenases. Plasmids pHKU201 (B. C. M. Pang, unpublished data) and pUK1035 (25) contain the structural genes for hdlIVa and dehCI. Escherichia coli BL21(DE3) cells were used for in vivo expression of the recombinant dehalogenases. Plasmid pET19b (Novagen) was used as an expression vector for in vivo and in vitro synthesis of proteins. B. cepacia MBA4 and Pseudomonas sp. strain CBS3 were grown at 30°C in Luria broth with monochloroacetate (MCA) for induction of the dehalogenases. E. coli transformants were grown at 37°C in Luria broth supplemented with ampicillin (100 μg/ml).
Enzymes and chemicals.
Restriction endonucleases were obtained from Gibco-BRL or New England Biolabs. Alkaline phosphatase was purchased from Boehringer-Mannheim. A T7 sequencing kit, [α-S35]methionine, and IPTG (isopropyl-β-d-thiogalactopyranoside) were from Amersham Pharmacia Biotech. Monochloroacetate (MCA) was from Sigma. UITma DNA polymerase was from Perkin-Elmer; T4 DNA ligase and T7 S30 E. coli extract were from Promega.
In vivo and in vitro synthesis of protein.
For in vivo protein expression, 4 ml of overnight culture was inoculated into 100 ml of fresh medium and grown until the optical density at 600 nm was 0.8 to 1. IPTG (1 mM) was then added, and the culture was allowed to grow for another 3 to 12 h before total protein extract was prepared. For in vitro synthesis, about 4 μg of DNA was incubated with T7 S30 E. coli extract (Promega) with 1 μCi of [α-S35]methionine at 37°C for about 3 h.
Plasmid isolation and DNA sequencing.
For preparative purpose, plasmid DNAs were obtained using a Qiagen spin column or a Qiagen tip-20 device. For analytical purpose, plasmid DNAs were isolated by the boiling method (10). Sequencing reactions were performed using the T7 Sequenase kit with Cy-5-labeled nucleotides. The samples were resolved by an ALFexpress automated sequencer (Amersham Pharmacia Biotech).
PCR.
PCR was carried out using a Peltier thermal cycler (PTC-200; MJ Research). Reaction buffer (100 μl; 10 mM Tris-HCl [pH 8.8], 10 mM KCl, 0.002% Tween 20 [vol/vol]) was mixed with 2 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1 μg of each primer, 1 μg of template plasmid DNA, and 3 U of UITma DNA polymerase (Perkin-Elmer). The amplification of the fragments were carried out for 30 cycles with denaturation at 94°C for 2 min, annealing at 72°C for 2 min, and extension at 76°C for 2 min. The PCR products were analyzed on a 1% agarose gel and purified by GeneClean (Bio 101).
Cloning of structural genes for hdlIVa and dehCI.
Full-length DehIVa and DehCI were used as controls. Two pairs of oligonucleotide primers designed with reference to the published DNA sequences (22, 25) were used. NdeI and BamHI restriction sites were introduced in the forward and reverse primers respectively. The primers for hdlIVa were 5′-ATT-ATT-ATT-AAG-CTT-(CAT-ATG)NdeI- ATG-GTG-GAT-TCA-CTC-CGC-3′ and 5′-ATT-ATT-ATT-CCC-GGG- (GGA-TCC)BamHI-TCA-GGC-AGC-TTT-TGT-AAC-3′, and the primers for dehCI were 5′-ATT-(CAT-ATG)NdeI-ATG-GAC-CCA-ATT-CGC-GCT-TGC-3′ and 5′-ATT-(GGA-TCC)BamHI-TTA-CTG-CGT-GAG-TCG-CAG-CAA-3′ (restriction sites are shown as superscripts; parentheses indicate the recognition sequences). The first codon, GTG, of dehCI was replaced by ATG (underlined). PCR products containing the structural genes for hdlIVa and dehCI were digested with NdeI and BamHI and cloned into the corresponding sites of pET19b to form pHKU221 and pHKU252, respectively. Transformation into E. coli BL21(DE3) was by the standard calcium chloride method (20). The sequences of all the constructs were verified by DNA sequencing.
Construction of pHKU271, pHKU272, pHKU273, pHKU275, and pHKU281.
hdlIVa and dehCI were isolated from pHKU221 and pHKU252 as NdeI/BamHI fragments, respectively. These fragments were then cut with SspI, MspI, or BsiEI for construction of other derivatives. pHKU271 contains an NdeI/SspI fragment of hdlIVa and an SspI/BamHI fragment of dehCI, while pHKU272 contains an NdeI/SspI fragment of dehCI and an SspI/BamHI fragment of hdlIVa. pHKU273 contains an NdeI/MspI fragment of hdlIVa and an MspI/BamHI fragment of dehCI. pHKU275 contains an NdeI/BsiEI fragment of hdlIVa and an BsiEI/BamHI fragment of dehCI. pHKU281 contains an NdeI/SspI fragment of pHKU252 and an SspI/BamHI fragment of pHKU273. These ligated fragments were all cloned into the corresponding NdeI/BamHI sites of pET19b.
Construction of pHKU284 and pHKU285.
PCR and linker ligation were used for the construction of tripartite molecules. The primer pair IVa-3n and IVa-3c, having the sequences 5′-GCG-TAC-AAG-(GAG-CTC)SacI-AGT-GCA-TAC-CCT-3′ and 5′-ATT-ATT-ATT-(GGA-TCC)BamHI-CCC-GGG-TCA-GGC-AGC-TTT-TGT-AAC-GTT-3′, was used for amplifying nucleotides 274 to 693 of hdlIVa. The PCR product contains nucleotides encoding amino acids 91 to 231 of DehIVa. This fragment was cut with SacI/BamHI and ligated to the NdeI/BamHI sites of pET19b with an NdeI/SacI linker to form pHKU282. The linker was formed by annealing of the following two oligonucleotides: 5′-TAT-GTA-ACT-CGA-GCC-CGG-GAA-TAT-TAA-GCT-TGA-GCT-3′ and 5′-CAA- GCT-TAA-TAT-TCC-CGG-GCT-CGA-GTT-ACA-3′. The primer pair CI-2n and CI-2c, having the sequences 5′-TTC-GCG-CTC-GA(A-AGC-TT)HindIII C-GGC-CTG-3′ and 5′-GGG-GTA-AGC-GCT-(GAG-CTC)SacI-CTT-ATA-GGC-3′, was used for amplifying nucleotides 220 to 267 of dehCI. The PCR product contains nucleotides encoding amino acids 73-89 of DehCI. The primer pair IVa-1n and IVa-1c, having the sequences 5′-ATT-ATT- ATT-(CAT-ATG)NdeI-AAG-CTT-ATG-GTG-GAT-TCA-CTC-CGC-GCG-3′ and 5′-CTC-GAG-ATG-G(AA-GCT-T)HindIIITC-CAA-CGC-GAA-TGT-3′ was used for amplifying nucleotides 1 to 219 of hdlIVa. The PCR product contains nucleotides encoding amino acids 1 to 73 of DehIVa. The PCR products obtained with the primer pairs IVa-1n and IVa-1c and CI-2n and CI-2c were cut with NdeI/HindIII and HindIII/SacI, respectively. These fragments were then ligated to the NdeI/SacI sites of pHKU282 to form the tripartite pHKU284 construct.
For construction of pHKU285 a similar strategy was used. The primer pair CI-3n and CI-3c, having the sequences 5′-GCC-TAT-CAT-(GAG-CTC)SacI-AGC-GCT-TAC-CCC-3′ and 5′-ATT-ATT-ATT-(GGA-TCC)BamHI-CCC- GGG-TTA-ACT-GCG-TGA-GTC-GCA-GCA-A-3′, was used for amplifying nucleotides 271 to 681 of dehCI. The PCR product contains nucleotides encoding amino acids 90 to 227 of DehCI. This fragment was cut with SacI/BamHI and ligated to the NdeI and BamHI sites of pET19b with the NdeI/SacI linker to form pHKU283. The primer pair CI-1n and CI-1c, having the sequences 5′-ATT-ATT-ATT-(CAT-ATG)NdeI-AAG-CTT-ATG-GAC-CCA-ATT-CGC-GCT-TGC-3′ and 5′-CAG-CAG-GCC-GAA-(CGT-ACG)SunI-GAG-CGC-GAA-ATC-3′, was used for amplifying nucleotides 1 to 216 of dehCI. The PCR product contains nucleotides encoding amino acids 1 to 72 of DehCI. The primer pair IVa-2n and IVa-2c, having the sequences 5′-TTC-GCG-TTG-(CGT-ACG)SunI-TAC-CAT-CTC-3′ and 5′-AGG-GTA-TGC-ACT-(GAG-CTC)SacIATG-GTA- CGC-3′, was used for amplifying nucleotides 223 to 270 of hdlIVa. The PCR product contains nucleotides encoding amino acids 74 to 90 of DehIVa. The PCR products obtained with the primer pairs CI-1 and CI-1c and IVa-2n and IVa-2c were cut with NdeI/SunI and SunI/SacI, respectively. These fragments were then ligated to the NdeI and SacI sites of pHKU283 to form the tripartite pHKU285 construct.
Site-directed mutagenesis of conserved amino acid residues.
Overlap extension PCR (11) was used to generate a mutation in the hdlIVa gene. Plasmid pHKU221 was used as the template. The primers used and the mutations which resulted are shown in Table 1. The primers flanking the 5′ and 3′ ends of hdlIVa are 5′-ATT-ATT-(AAG-CTT)HindIII-CAT-ATG-ATG-GTG-GAT-TCA-CTC- CGC-3′ and 5′-ATT-ATT-(CCC-GGG)SmaI-GGA-TCC-(1529)TCA-GGC- AGC-TTT-TGT-AAC-3′. The amplification of the fragments was carried out for 30 cycles with denaturation at 94°C for 2 min, annealing at 55°C for 2 min, and extension at 68°C for 2 min. The PCR products were gel purified and subjected to HindIII and SmaI digestion. Purified HindIII/SmaI PCR products were cloned into the HindIII/SmaI site of pFLAG-MAC (Sigma).
TABLE 1.
Primers used for generation of mutations in the hdlIVa gene
| Primer sequencesa | Mutation | Construct |
|---|---|---|
| 5′-G-ATG-AAT(CAT)-CAG-TAT-GCG-G-3′ | H56N | pHKU356 |
| 5′-G-ATG-CAT-CAG-TTT(TAT)-GCG-G-3′ | Y58F | pHKU357 |
| 5′-AT-GCG-GAT-TAT(TTT)-TGG-C-3′ | F61Y | pHKU358 |
| 5′-GG-CAG-TTG-GCC(ACC)-GAT-G-3′ | T65A | pHKU359 |
| 5′-G-ACC-GAT-GAG-ACG(GCG)-CTG-AC-3′ | A68T | pHKU360 |
| 5′-G-ACC-GAT-GAG-GCG-ATG(CTG)-AC-3′ | L69M | pHKU361 |
| 5′-AC-CAT-ATG(CTC)-GAG-GAT-CGC-3′ | L78M | pHKU362 |
| 5′-G-ATG-AGT-GCG-TTC(TAC)-AAG-G-3′ | Y92F | pHKU363 |
| 5′-C-GAA-AAG-ATG(TTG)-AAA-TC-3′ | L108M | pHKU364 |
The mutated nucleotide(s) is shown in boldface type, and the wild-type sequence is shown in parentheses.
Western blot analysis.
Molecules with the FLAG peptide were visualized by Western blot analysis. Cell extracts (CE) were resolved by denaturing or nondenaturing polyacrylamide gel electrophoresis (PAGE). Proteins were blotted onto Hybond-ECL membrane (Amersham). The membrane was blocked in Tris-buffered saline (10 mM Tris-HCl [pH 7.5], 150 mM NaCl) with 5% nonfat dried milk for 1 h and incubated with M2 antibody (Sigma) for 1 h in Tris-buffered saline with 0.05% Tween 20. After washing with the same buffer, the membrane was incubated with peroxidase-conjugated goat anti-mouse immunoglobulin G (Amersham) in Tris-buffered saline with 0.5% Tween 20 for 1 h. The unbound antibody was removed by washing three times for 10 min each in the same buffer. Western blot ECL reagents (Amersham) were added, and the chemiluminescence was detected on BioMax XR film (Kodak).
Preparation of CE and determination of dehalogenase activity.
CE of the bacteria were prepared through two passages of the cell in a French press (Amicon) at 2,500 bar. The lysates were centrifuged at 48,400 × g for 45 min to remove insoluble materials and cell debris. Certain amount of the CE was incubated in 100 mM Tris-SO4, pH 7.9, with 100 mM MCA at 37°C. The amount of chloride ion released was determined using a Corning 925 chloride counter.
Determination of molecular weight.
The relative molecular weights of the chimeras were determined by gel filtration and sodium dodecyl sulfate (SDS)-PAGE. Gel filtration was carried out on a Sephacryl S-200 HR column equilibrated with 20 mM Tris-SO4 (pH 7.9)–0.15 M NaCl at room temperature. Proteins were eluted with the same buffer at a flow rate of 0.5 ml per min. Fractions of 0.25 to 1 ml were collected. Elution volumes were determined by A280 reading or by enzyme activity. The void volume of the column was determined with blue dextran, and the column was calibrated with a low-molecular-weight standard (Amersham Pharmacia Biotech). The protein standards used were albumin, 67,000 (67K); ovalbumin, 43K; chymotrypsinogen A, 25K; and ribonuclease A, 13.7K. Standard SDS-PAGE was used (20). The denatured protein standard used was prestained high-molecular-weight Rainbow marker (Amersham Pharmacia Biotech), which included lysozyme, 14.3K; trypsin inhibitor, 21.5K; carbonic anhydrase, 30K; ovalbumin, 46K; bovine serum albumin, 66K; phosphorylase b, 97.4K; and myosin, 220K.
Activity-stained PAGE.
CE were electrophoresed in a nondenaturing acrylamide gel. After electrophoresis, the gel was incubated in 20 mM Tris-SO4, pH 7.9, supplemented with 100 mM MCA for about 30 min at 37°C; the gel was then incubated with 0.1 M AgNO3. White precipitate was formed at the position where the dehalogenase released the chloride ion from the MCA.
RESULTS
The first 55 residues of DehIVa are not essential for dimerization.
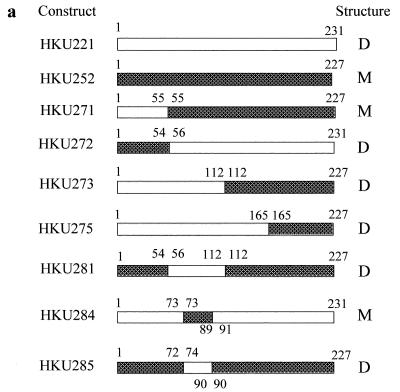
Analysis of the DNA sequences of hdlIVa and dehCI revealed that restriction recognition sites for SspI provide a convenient site for swapping of the N-terminal 54 to 55 amino acids between DehIVa and DehCI. Constructs HKU271 and HKU272 were created (Fig. 1a). Molecule HKU271 contains amino acids 1 to 55 of DehIVa followed by amino acids 55 to 227 of DehCI. This molecule was produced in vitro (Fig. 1b), but dehalogenase activity was not detected in the activity-stained gel (Fig. 1c, lane 6). However, when this protein was made in vivo in E. coli, dehalogenation of the substrate MCA was detected (data not shown). This molecule migrated as a monomer like DehCI in nondenaturing gel (Fig. 1d, lane 6). Molecule HKU272 contains amino acids 1 to 54 of DehCI followed by amino acids 56 to 231 of DehIVa. This molecule was produced in vitro and was found to be active (Fig. 1b and c). This molecule migrated as a dimer like DehIVa in nondenaturing gel (Fig. 1d, lane 5). These results indicated that the dimerization domain of DehIVa resides in amino acids 56 to 231.
FIG. 1.
Various chimeric dehalogenases and their electrophoretic properties. (a) Schematic representation of various constructs. The open box indicates the DehIVa portion, while the shaded box represents the DehCI region. The numbers represent the numbering of the amino acids of the corresponding enzyme. The last column indicates the dimeric (D) or monomeric (M) nature of the chimera. (b) Autoradiograph of an SDS-PAGE gel analyzing the in vitro-produced proteins. The proteins were synthesized in the presence of [35S]methionine and analyzed on a 12.5% denaturing gel. (c) Nondenaturing activity-stained gel of various native and in vitro-produced proteins. The activities of the dehalogenases and derivatives were visualized using MCA as the substrate. (d) Autoradiograph of the activity-stained gel shown in panel c. (b to d) Lanes: 1, CE of B. cepacia MBA4; 2, CE of E. coli producing DehCI; 3 to 9, in vitro-produced HKU221, HKU252, HKU272, HKU271, HKU273, HKU275, and HKU281, respectively; 10, in vitro-produced mock lysate.
Residues 112 to 231 of DehIVa are not essential for dimerization.
Analysis of hdlIVa and dehCI also revealed that restriction sites for MspI and BsiEI could provide convenient sites for construction of other chimeras. Molecule HKU275 contains amino acids 1 to 164 of DehIVa followed by amino acids 164 to 227 of DehCI (Fig. 1a). This molecule was found to be inactive (Fig. 1b, lane 8) when produced in vitro but active under in vivo conditions (data not shown). This molecule migrated as a dimer in nondenaturing gel (Fig. 1d, lane 8). Molecule HKU273 contains amino acids 1 to 111 of DehIVa followed by amino acids 111 to 227 of DehCI (Fig. 1a). This molecule also behaved similarly to HKU275 (Fig. 1b to d, lanes 7). An active dimeric molecule of HKU273 was produced effectively in E. coli. These results showed that amino acids 112 to 231 of DehIVa are not essential for subunit interaction.
Residues 74 to 90 of DehIVa are essential for dimerization.
Results from the previous sections implied that the subunit interaction domain of DehIVa must be located between amino acids 56 and 111. In order to confirm that this is the case, molecule 281 was constructed (Fig. 1a). This molecule contains amino acids 56 to 111 of DehIVa flanked by residues 1 to 53 and 111 to 227 of DehCI. This chimera is active and migrates similarly to dimeric DehIVa (Fig. 1c, lane 9). This confirmed that residues 56 to 111 contain a functional dimerization domain.
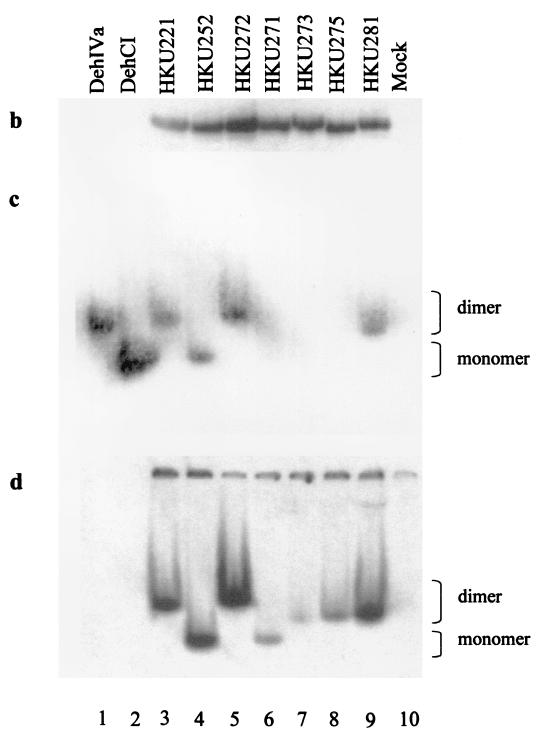
Further analysis of the secondary structure of DehIVa and DehCI by the Garnier-Osguthorpe-Robson prediction in the Wisconsin Genetics Computer Group package revealed that these enzymes are quite alike. There is, however, a region in DehIVa located between amino acids 50 and 110 that exhibits a higher level of hydrophilicity than that of DehCI. These structural prediction results augment our observation that residues 56 to 111 of DehIVa contain a dimerization domain. A closer look showed that residues 74 to 90 of DehIVa are those showing stronger hydrophilic properties. Constructs HKU284 and HKU285 were prepared to confine the interaction domain of the protein. Molecule 284 contains residues 73 to 89 of DehCI flanked by residues 1 to 73 and 91 to 231 of DehIVa (Fig. 1a). This molecule is active and migrated as a monomer like DehCI (Fig. 2, lane 6). Likewise, molecule 285, which contains residues 74 to 90 of DehIVa flanked by residues 1 to 73 and 91 to 227 of DehCI (Fig. 1a), is active and migrated as a dimer like DehIVa (Fig. 2, lane 5). These results showed that amino acids 74 to 90 of DehIVa are able to mediate the dimerization function for the protein.
FIG. 2.
Activity-stained gel of various native and chimeric dehalogenases. Lanes 1, cell extract of B. cepacia MBA4; 2, cell-free extract of E. coli producing DehCI; 3–7, in vitro-produced (IVP) HKU221, HKU252, HKU285, and HKU284, respectively; 8, IVP mock lysate.
The 17 residues are not sufficient for subunit interaction.
It appears that the 17 residues ranging from residues 74 to 90 of DehIVa are able to confer the protein dimerization function. We have, however, obtained evidence suggesting that other regions of the enzymes are also important for the subunit interaction. N-terminal and C-terminal deletion mutants were constructed, and the molecules were analyzed by nondenaturing gel electrophoresis for dimerization ability. Derivatives deleting 3, 6, 8, 9, or 10 residues from the N terminus migrated like the full-length molecule, while removal of 11 amino acids from the N terminus or three residues from the C terminus abolished the dimerization function of the enzyme (data not shown). Although we cannot eliminate the possibility that these 3 or 11 amino acid deletions could affect the overall folding of the protein, these results did suggest that there are other regions critical for protein interaction. This prompted us to investigate the role of some of the individual residues in the protein by means of single amino acid changes.
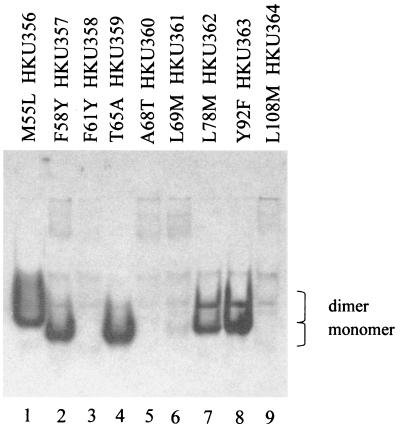
Conserved amino acid residues among various isozymes normally play an important role in certain functions. Amino acid residues 56 to 111 of DehIVa have now been shown to be important for subunit association. Comparison of similar regions among eight 2-haloacid dehalogenases has identified nine conserved residues (17). They are Met55, Tyr58, Phe61, Thr65, Ala68, Leu69, Lys78, Tyr92, and Leu108. Reverse genetics has been used to elucidate the function of important residues in enzymes (19). The nine conserved residues were mutated as follows: Met55Leu, Tyr58Phe, Phe61Tyr, Thr65Ala, Ala68Thr, Leu69Met, Lys78Met, Tyr92Phe, and Leu108Met. These mutants were expressed in vivo in E. coli and analyzed by Western blot analysis. Samples resolved by SDS-PAGE indicated that these DehIVa derivatives were similar in size and were expressed to similar levels (data not shown). However when these samples were analyzed on nondenaturing gel, some of the proteins were denatured and were not admissible into the gel. Figure 3 shows that F61Y, A68T, L69M, and L108M changes affected the conformation of the proteins and the altered molecules were not resolved by native PAGE (lanes 3, 5, 6, and 9). Molecules with Y58F or T65A change were migrated as monomer (lanes 2 and 4). Cells harboring either of these molecules were able to dehalogenate MCA, implying that these protomers were active (data not shown). Molecules with an L78M or Y92F change displayed both monomeric and dimeric products (lanes 7 and 8). Modifications of these two molecules were therefore less detrimental than the other changes, but the conformational stability of the proteins was definitely affected.
FIG. 3.
Western blot analysis of DehIVa derivatives in nondenaturing gel. Each of the recombinant molecules carries a FLAG leader peptide. A 10-μg aliquot of total cellular protein was loaded in each lane. Lanes 1 to 9, in vivo-expressed protein HKU355 (M55L), HKU357 (Y58F), HKU358 (F61Y), HKU359 (T65A), HKU360 (A68T), HKU361 (L69M), HKU362 (L78M), HKU363 (Y92F), and HKU364 (L108M).
The 17 residues affect enzyme activity.
The wild-type DehIVa, DehCI, and chimeras HKU284 and HKU285 have been expressed in vivo in E. coli. The amount of the enzymes produced in each strain corresponded to about 25% of total cellular proteins. This was determined by scanning densitometric analysis using Phoretix 1D standard software (Chinetek Scientific) on Coomassie brilliant blue R250-stained SDS-PAGE gel (data not shown). The similar levels of expression for the four enzymes in the heterologous hosts made it possible to compare their dehalogenase activities without purifying the enzymes. Table 2 reveals the specific activity for these four enzymes. DehCI was the most active one and has a highest activity towards MCA. DehIVa, on the other hand, was most active towards MBA. The monomeric HKU284, which has a DehIVa backbone but a DehCI internal fragment, was similar to DehIVa, except with a decreased activity towards MBA. The dimeric molecule HKU285, which has DehCI backbone and an internal DehIVa fragment, was DehCI like, being active towards dichloroacetate (DCA). It appears that the 17 residues affected the enzyme specificity and activity to some extent.
TABLE 2.
Dehalogenase activities of CE for DehIVa-, DehCI-, HKU284-, and HKU285-producing strains
| Substrate | Sp act (μM halide released/mg/min)a
|
|||
|---|---|---|---|---|
| DehIVa | DehCI | HKU284b | HKU285c | |
| MCA | 24.7 (1) | 53.3 (1) | 28.0 (1) | 32.7 (1) |
| MBA | 34.0 (1.4) | 42.8 (0.8) | 24.0 (0.36) | 27.4 (0.8) |
| L-2MCPA | 5.9 (0.24) | 9.9 (0.19) | 4.1 (0.15) | 7.7 (0.24) |
| DL-2MBPA | 10.3 (0.42) | 16.1 (0.30) | 9.5 (0.34) | 9.7 (0.30) |
| DCA | — | 4.1 (0.08) | — | 4.2 (0.13) |
Activities are averages of two experiments that show no significant deviation. —, no detectable activity. The relative activity with respect to MCA as the 100% was shown in parentheses.
HKU284 is DehIVa1–73–DehCI73–89–DehIVa91–231.
HKU285 is DehCI1–72–DehIVa74–90–DehCI90–227.
DISCUSSION
Isolation and characterization of 2-haloacid dehalogenases have been studied extensively during the last decade. Many of these genes have been cloned and sequenced (5, 13, 22, 23, 25, 29, 32). Arbitrary grouping of these dehalogenases was previously based on electrophoretic mobility and/or substrate specificity (7). Recently the relationships of these enzymes have been studied phylogenetically. These 2-haloacid dehalogenases were grouped into two families, each with four subdivisions (8). DehIVa of MBA4 and DehCI of CBS3 were grouped to the same subdivision of group II; however, the structural relationship of these enzymes has not been examined. DehIVa and DehCI show 68% identity in amino acid sequences, yet the native form of the former is a dimer while the latter is a monomer.
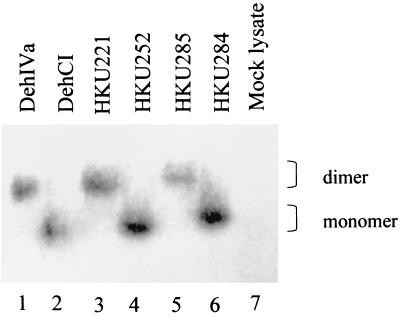
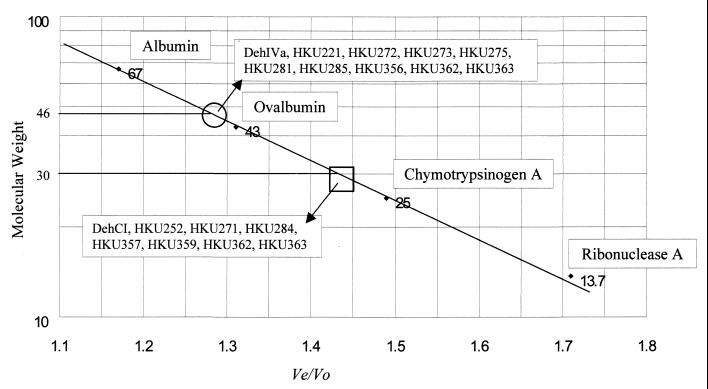
In this work we have identified a region in DehIVa that confers the dimerization ability of the protein. As seen in the activity-stained gel and the autoradiograph, it is reasonable to assume that chimeras with a mobility identical to that of DehCI are monomers and that those migrating similarly to DehIVa are dimers (Fig. 1c and d). However, a previous study has indicated the possibility of the formation of different electrophoretic forms from purified dehalogenase (14) in nondenaturing gel. In order to show that these changes in the mobility of the protein did indeed result from the change in the aggregation state, the molecular weights of the recombinant proteins were analyzed by gel filtration. The chimeras, HKU284 and HKU285, were expressed in vivo in E. coli BL21(DE3), and the CE were analyzed by SDS-PAGE. Both recombinant proteins were the correct size (data not shown). The CE were applied to Sephacryl-S200 HR gel filtration columns, and the dehalogenase active fractions for HKU284 and HKU285 were found to be 30K and 46K, respectively (Fig. 4). These confirmed that the active HKU284 is a monomer and the active HKU285 is a dimer consisting of two identical subunits.
FIG. 4.
Relative molecular weight of the proteins used in this study. A Sephacryl S200 HR column was calibrated with protein standards of known molecular weight. The standard curve was plotted with the molecular weight (in thousands) versus the Ve/Vo ratio. Ve is the elution volume and Vo is the void volume as determined by blue dextran. DehIVa, HKU221, HKU272, HKU273, HKU275, HKU281, HKU285, and HKU356 are dimers and eluted around 46K (within the open circle). DehCI, HKU252, HKU271, HKU284, HKU357, and HKU359 are monomers and eluted around 30K (within the open square). HKU362 and HKU363 contain both dimer and monomer and were found eluted around the 46K and the 30K regions.
The crystal structures of two 2-haloacid dehalogenases are currently known. One of the molecules, l-DEX-YL, exists as a homodimer (9, 18). Amino acid residues distributed along the polypeptide were found to be important for maintenance of the subunit interaction. This molecule was divided into a core domain and a subdomain. The subunit interaction was formed between the subdomain and the carboxyl-terminal half of the core domain. An α-2 helix region runs along the molecular twofold axis and plays an indispensable role in dimerization. This region was proposed to play similar roles in multimerization of other 2-haloacid dehalogenases. The other molecule, DhlB, whose crystal structure is known, has a dimerization interface similar to that of l-DEX-YL (24). However, an additional small subdomain which stretches from residues 193 to 219 and interacts with residues 15 to 93 provides the dimerization interface (24). Dimerization interfaces in l-DEX-YL and DhlB are similar, but DhlB is tighter after dimerization. l-DEX-YL is only 13.5% buried after dimerization, while 19% of the monomer surface is buried in DhlB (9, 24). This suggests that the dimeric interface for these two molecules are different, despite their similarity as group II dehalogenases (8). The α-2 helix of l-DEX-YL stretches from Leu39 to Arg56 and corresponds to Leu40 to His55 of DehIVa. Site-directed mutageneses in this putative α-2 helix region of DehIVa, however, have not produced any monomeric molecule (B. C. M. Pang and J. S. H. Tsang, unpublished data). It is likely that these similar residues could fold into different structures for dimerization.
Dehalogenase activity between DehIVa and DehCI has not been compared previously. In this study it has been shown that the monomeric DehCI has a higher specific activity than the dimeric DehIVa. Moreover, DehCI is active towards DCA while DehIVa is not. Despite their similarity in amino acid sequence, their enzyme activity patterns are different. This is not uncommon, as enzymes having 95% identity while exhibiting different enzymic efficiencies have been found (16). The substrate specificity of the chimeras also shows that there are no general rules for estimating the efficiency of the swapped domains (2). The relative activities suggested that dimeric DehIVa works better on MBA than on MCA while its monomeric counterpart HKU284 works better on MCA than on MBA. On the other hand, both monomeric DehCI and the dimeric derivative HKU285 behave similarly (Table 2). We are now in the process of testing the synergistic interaction of DehIVa protomers under dimeric conditions.
The indication of the presence of active protomer for DehIVa is confirmed by the mutation T65A (data not shown). It is not very surprising that a single subunit is active. Based on the crystal structures of the l-DEX-YL and DhlB, the dimerization domain is not involved in the catalysis of the dehalogenase (18, 24). The monomeric property of the T65A derivative was verified by gel filtration (Fig. 4). However, the relative activity of this in vivo-expressed T65A was only about 1% of the wild-type protein.
Further investigation of the amino acids by site-directed mutagenesis may be able to give a better understanding of not only the dimerization domain but also the substrate specificity and active site of the protein. This information may be important in broadening the substrate range of dehalogenases and their application for biotechnology purposes.
ACKNOWLEDGMENTS
We thank R. Müller for Pseudomonas sp. strain CBS3 and plasmid pUK1035.
This work was supported by a grant from the Hong Kong Research Grants Council. B.C.M.P. thanks the University of Hong Kong for a studentship.
REFERENCES
- 1.Aravind L, Galperin M Y, Koonin E V. The catalytic domain of the P-type ATPase has the haloacid dehalogenase fold. Trends Biol Sci. 1998;23:127–129. doi: 10.1016/s0968-0004(98)01189-x. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong R N. Structure-function relationships in enzymic catalysis. Can chimeric enzymes contribute? Chem Rev. 1990;90:1309–1325. [Google Scholar]
- 3.Barth P T, Bolton L, Thomson J C. Cloning and partial sequencing of an operon encoding two Pseudomonas putida haloalkanoate dehalogenases of opposite stereospecificity. J Bacteriol. 1992;174:2612–2619. doi: 10.1128/jb.174.8.2612-2619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beil S, Mason J R, Timmis K N, Pieper D H. Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J Bacteriol. 1998;180:5520–5528. doi: 10.1128/jb.180.21.5520-5528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brokamp A, Happe B, Schmidt F R J. Cloning and nucleotide sequence of a d,l-haloalkanoic acid dehalogenase encoding gene from Alcaligenes xylosoxidans ssp. denitrificans ABIV. Biodegradation. 1997;7:383–396. doi: 10.1007/BF00056422. [DOI] [PubMed] [Google Scholar]
- 6.Goldman P, Milne G W A, Keister D B. Carbon-halogen bond cleavage. III. Studies on bacterial halidohydrolases. J Biol Chem. 1968;243:428–434. [PubMed] [Google Scholar]
- 7.Hardman D J. Biotransformation of halogenated compounds. Crit Rev Biotechnol. 1991;11:1–40. doi: 10.3109/07388559109069182. [DOI] [PubMed] [Google Scholar]
- 8.Hill K E, Marchesi J R, Weightman A J. Investigation of two evolutionarily unrelated halocarboxylic acid dehalogenase gene families. J Bacteriol. 1999;181:2535–2547. doi: 10.1128/jb.181.8.2535-2547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hisano T, Hata Y, Fujii T, Liu J Q, Kurihara T, Esaki N, Soda K. Crystal structure of L-2-haloacid dehalogenase from Pseudomonas sp. YL. J Biol Chem. 1996;271:20322–20330. doi: 10.1074/jbc.271.34.20322. [DOI] [PubMed] [Google Scholar]
- 10.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 11.Horton R M, Pease L R. Recombination and mutagenesis of DNA sequences using PCR. In: McPherson M J, editor. Directed mutagenesis: a practical approach. Oxford, United Kingdom: IRL Press; 1991. pp. 217–247. [Google Scholar]
- 12.Jiang H, Parales R E, Gibson D T. The α-subunit of toluene dioxygenase from Pseudomonas putida F1 can accept electrons from reduced ferredoxinTOL but is catalytically inactive in the absence of the β-subunit. Appl Environ Microbiol. 1999;65:315–318. doi: 10.1128/aem.65.1.315-318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones D H A, Barth P T, Byrom D, Thomas C M. Nucleotide sequence of the structural gene encoding a 2-haloalkanoic acid dehalogenase of Pseudomonas putida strain AJ1 and purification of the encoded protein. J Gen Microbiol. 1992;138:675–683. doi: 10.1099/00221287-138-4-675. [DOI] [PubMed] [Google Scholar]
- 14.Keuning S, Janssen D B, Witholt B. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol. 1985;163:635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klages U, Krauss S, Lingens F. 2-Haloacid dehalogenase from a 4-chlorobenzoate-degrading Pseudomonas spec. CBS 3. Hoppe-Seyler Z Physiol Chem. 1983;364:529–535. doi: 10.1515/bchm2.1983.364.1.529. [DOI] [PubMed] [Google Scholar]
- 16.Kronbach T, Larabee T M, Johnson E F. Hybrid cytochromes P-450 identify a substrate binding domain in P-450IIC5 and P-450IIC4. Proc Natl Acad Sci USA. 1989;86:8262–8265. doi: 10.1073/pnas.86.21.8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurihara T, Liu J Q, Nardi-Dei V, Koshikawa H, Esaki N, Soda K. Comprehensive site-directed mutagenesis of L-2-haloacid dehalogenase to probe catalytic amino acid residues. J Biochem. 1995;117:1317–1322. doi: 10.1093/oxfordjournals.jbchem.a124861. [DOI] [PubMed] [Google Scholar]
- 18.Li Y F, Hata Y, Fujii T, Hisano T, Nishihara M, Kurihara T, Esaki N. Crystal structures of reaction intermediates of l-2-haloacid dehalogenase and implications for the reaction mechanism. J Biol Chem. 1998;273:15035–15044. doi: 10.1074/jbc.273.24.15035. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Liu D, DeRose E F, Mullen G P. Studies of the dimerization and domain structure of γς resolvase. J Biol Chem. 1993;268:16309–16315. [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 21.Mörsberger F M, Müller R, Otto M K, Lingens F, Kulbe K D. Purification and characterization of 2-halocarboxylic acid dehalogenase II from Pseudomonas spec. CBS3. Biol Chem Hoppe-Seyler. 1991;372:915–922. doi: 10.1515/bchm3.1991.372.2.915. [DOI] [PubMed] [Google Scholar]
- 22.Murdiyatmo U, Asmara W, Tsang J S H, Baines A J, Bull A T, Hardman D J. Molecular biology of the 2-haloacid halidohydrolase IVa from Pseudomonas cepacia MBA4. Biochem J. 1992;284:87–93. doi: 10.1042/bj2840087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nardi-Dei V, Kurihara T, Okamura T, Liu J-Q, Koshikawa H, Ozaki H, Terashima Y, Esaki N, Soda K. Comparative studies of genes encoding thermostable L-2-halo acid dehalogenase from Pseudomonas sp. strain YL, other dehalogenases, and two related hypothetical proteins from Escherichia coli. Appl Environ Microbiol. 1994;60:3375–3380. doi: 10.1128/aem.60.9.3375-3380.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridder I S, Rozeboom H J, Kalk K H, Janssen D B, Dijkstra B W. Three-dimensional structural of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate. J Biol Chem. 1997;272:33015–33022. doi: 10.1074/jbc.272.52.33015. [DOI] [PubMed] [Google Scholar]
- 25.Schneider B, Müller R, Frank R, Lingens F. Complete nucleotide sequences and comparison of the structural genes of two 2-haloalkanoic acid dehalogenases from Pseudomonas sp. strain CBS3. J Bacteriol. 1991;173:1530–1535. doi: 10.1128/jb.173.4.1530-1535.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider B, Müller R, Frank R, Lingens F. Site-directed mutagenesis of the 2-haloalkanoic acid dehalogenase I gene from Pseudomonas sp. strain CBS3 and its effect on catalytic activity. Biol Chem Hoppe-Seyler. 1993;374:489–496. doi: 10.1515/bchm3.1993.374.7-12.489. [DOI] [PubMed] [Google Scholar]
- 27.Schwarze R, Brokamp A, Schmidt F R J. Isolation and characterization of dehalogenases from 2,2-dichloropropionate-degrading soil bacterium. Curr Microbiol. 1997;34:103–109. doi: 10.1007/s002849900152. [DOI] [PubMed] [Google Scholar]
- 28.Slater J H, Bull A T, Hardman D J. Microbial dehalogenation of halogenated alkanoic acids, alcohols and alkanes. Adv Microb Physiol. 1997;38:133–176. doi: 10.1016/s0065-2911(08)60157-5. [DOI] [PubMed] [Google Scholar]
- 29.Smith J M, Harrison K, Colby J. Purification and characterization of D-2-haloacid dehalogenase from Pseudomonas putida strain AJ1/23. J Gen Microbiol. 1990;136:881–886. doi: 10.1099/00221287-136-5-881. [DOI] [PubMed] [Google Scholar]
- 30.Tsang J S H, Hardman D J, Bull A T. Cloning and expression of 2-haloalkanoic acid dehalogenase of Pseudomonas cepacia MBA4 in Escherichia coli and in Pseudomonas putida. In: Chang S T, Chan K-Y, Woo N Y S, editors. Recent advances in biotechnology and applied biology. Hong Kong, Hong Kong: Chinese University Press; 1988. pp. 231–239. [Google Scholar]
- 31.Tsang J S H, Sallis P J, Bull A T, Hardman D J. A monobromoacetate dehalogenase from Pseudomonas cepacia MBA4. Arch Microbiol. 1988;150:441–446. [Google Scholar]
- 32.Tsang J S H, Sam L. Cloning and characterization of a cryptic haloacid dehalogenase from Burkholderia cepacia MBA4. J Bacteriol. 1999;181:6003–6009. doi: 10.1128/jb.181.19.6003-6009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Ploeg J, van Hall G, Janssen D B. Characterization of the haloacid dehalogenase from Xanthobacter autotrophicus GJ10 and sequencing of the dhlB gene. J Bacteriol. 1991;173:7925–7933. doi: 10.1128/jb.173.24.7925-7933.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]