Abstract
Itaconic acid (ITA), an effective alternative fossil fuel, derives from the bypass pathway of the tricarboxylic acid (TCA) cycle. Therefore, the imbalance of metabolic flux between TCA cycle and ITA biosynthetic pathway seriously limits the production of ITA. The optimization of flux distribution between biomass and production has the potential to the productivity of ITA. Based on the previously constructed strain Escherichia coli MG1655 Δ1-SAS-3 (ITA titer: 1.87 g/L), a CRISPRi-mediated self-inducible system (CiMS), which contained a responsive module based on the ITA biosensor YpItcR/Pccl and a regulative CRISPRi-mediated interferential module, was developed to regulate the flux of the TCA cycle and to enhance the capacity of the strain to produce ITA. First, a higher ITA-yielding strain, Δ4-Prmd-SAS-3 (ITA titer: 3.20 g/L), derived from Δ1-SAS-3, was constructed by replacing the promoter PJ23100, for the expression of ITA synthesis genes, with Prmd and knocking out the three bypass genes poxB, pflB, and ldhA. Subsequently, the CiMS was used to inhibit the expression of key genes icd, pykA, and sucCD to dynamically balance the metabolic flux between TCA cycle and ITA biosynthetic pathway during the ITA production stage. The constructed strain Δ4-Prmd-SAS-3 under the dynamic regulation of the CiMS, showed a 23% increase in the ITA titer, which reached 3.93 g/L. This study indicated that CiMS was a practical strategy to dynamically and precisely regulated the metabolic flux in microbial cell factories.
Keywords: CRISPRi, Dynamic regulation, Itaconic acid, Biosensor
1. Introduction
With the rapid development of synthetic biology, conventional chemical and biological extraction technologies to produce valuable compounds are gradually being replaced by microbial fermentation [[1], [2], [3], [4]]. However, owing to the complex metabolic circuits and regulatory networks in living microorganisms, it is difficult to precisely balance the central metabolism for biomass and the metabolite synthesis [[5], [6], [7]]. To accumulate several types of organic acids, it is necessary to inhibit the precursor-consuming pathways, most of which are a part of the basic metabolic pathway, such as glycolysis and the tricarboxylic acid (TCA) cycle. However, the disruption of these pathways would be harmful to cell growth. Therefore, a novel approach is urgently needed to balance the flux of metabolic bypass and product accumulation pathways.
Itaconic acid (ITA) is a C5 unsaturated dicarboxylic acid. As it possesses the methylene group and dicarboxylic acid, ITA has distinct active chemical properties [8]. ITA has garnered much interest, as it can participate in various polymerization reactions to produce promising and renewable polymer materials such as resins, rubbers, and fibers [9,10]. Owing to its wide range of applications, it was considered one of the 12 high-added-value bio-based materials by the US Department of Energy in 2004. ITA has also been found in the metastatic tumor cell line, which indicates that it might also have a role in tumor biology [11]. With its broad utility, researchers have concentrated their efforts on enhancing the production of ITA. Currently, the most predominant way to produce ITA is microbial fermentation. Therefore, the development of metabolic engineering strategies to construct robust microbial cell factories will be critical to improve ITA production.
ITA is synthesized from cis-aconitate, the intermediate of the TCA cycle, by cis-aconitate decarboxylase (CAD). ITA synthesis competes for the cis-aconitate in the TCA cycle, therefore, balancing energy allocation between the TCA cycle and the ITA-producing pathway is important for the high-yielding production of ITA. Recently, a high-yield ITA strain Escherichia coli MG1655 Δ1-SAS-3, based on a sequential self-assembly system that enhances the cascade reaction among citrate synthase (gltA, GA), aconitase (acnA, ACN), and cis-aconitate decarboxylase (cadA, CAD), was reported, and this system could produce 1.87-g/L ITA in a shake flask within 100 h [12]. To further increase the production of ITA, a higher ITA-producing strain Δ6-SAS-3, which could produce 3.06-g/L ITA within 100 h, was engineered by combining downstream gene knock-outs in competitive pathways. However, 4 g/L of α-ketoglutaric acid (α-KG) should be added to the medium to sustain TCA cycle operation, which significantly increased the cost of fermentation. In addition, several strategies have been adopted to improve the ITA productivity of strains. Myung et al. [13] successfully addressed the limitation of the TCA cycle during ITA production in E. coli by constructing a heterologous acetic acid metabolic pathway, and this economic carbon source reduced the cost of ITA production. However, the high concentration of acetic acid lowered the pH of the medium which made it unsuitable for the growth of the strain. Steffen et al. [14] implemented interventions in five metabolic pathways, which increased the production of ITA. In addition, the engineered E. coli strain ita23 could produce 2.27-g/L ITA in minimal medium. Although the effect of pathway interventions was significantly less than that caused by knock-out strategies, the interventions still influenced the growth of ita23. Hence, a novel system design that can dynamically control the metabolic flux during ITA production is required.
Strategies to improve ITA production by dynamically regulating the TCA metabolic flux have been developed. Gene cadA was cloned into the IPTG-inducible vector and CAD expression was induced by adding-IPTG at 20 °C in the stationary growth phase [15]. However, the high cost of the inducer and energy-consuming program limited its industrial application. A temperature sensitive repressor, CI857, activated at 30 °C was also utilized to dynamically regulate the expression of icd in the ITA-accumulation stage, further influencing the metabolic flux of the TCA cycle [16]. However, it is difficult to precisely control the temperature in a large-scale bioreactor. Hence, a novel system design that can dynamically and precisely control the metabolic flux during ITA production is required.
CRISPR interference (CRISPRi), based on a mutant RNA-guided DNA endonuclease derived from the type II CRISPR/Cas9 system, is a tool to precisely regulate gene expression [[17], [18], [19]]. CRISPRi provides several advantages for the design of synthetic circuits. For example, the single guide RNA (sgRNA) is easy to program in silico [20]. It is also possible to target multiple genes or to design multiple targets for a single gene. In addition, short 20 bp sgRNAs impart a low metabolic burden to host cells [21]. Although the knockout of key genes could regulate the metabolic distribution to increase the yield of the metabolite of interest, it would cause severe growth stagnation in host cells [22]. Compared with the hazards of deleting key genes, the dynamic regulation of the expression of target genes via CRISPRi to optimize metabolic distribution is a gentler approach. Recently, CRISPRi has been widely utilized in microbial cell factories for gene regulation, especially in E. coli and Corynebacterium glutamicum, to improve the productivity of organic acids [2,23,24]. However, the distribution of metabolic flux to the biomass and product is a dynamic process, which is difficult to precisely regulate simply through interventions.
In this study, we aimed to address the previously mentioned challenge in the production of ITA through a novel double-module regulation system, the CRISPRi-mediated self-inducible system (CiMS). This system contained an ITA-responding biosensor YpItcR/Pccl to dynamically regulate the expression of dCas9, which could target the desired genes with the corresponding designed sgRNAs to regulate the distributions of metabolic flux between biomass and ITA production. Using the combination of different types of sgRNAs, an optimized sgRNA-array was constructed and verified to be effective in significantly improving ITA production by the E. coli cell factory.
2. Materials and methods
2.1. Bacteria, plasmids, and materials
The bacterial strains and plasmids employed in this study are presented in Supplementary Tables S1 and S2, respectively. Maps of plasmids employed in this study are presented in Supplementary Fig. S1. Escherichia coli DH5α and MG1655 were used for plasmid propagation and CiMS assays, respectively. E. coli MG1655 Δ1-SAS-3, which was constructed in our previous research [12], was adopted as the original host and used for ITA production in this study. The vector pACYCDuet-1 was used to construct the responsive module and regulated module. Antibiotic and standard compounds were purchased from Aladdin (Shanghai, China). Restriction enzymes, DNA polymerase, ligase, and other DNA-modifying enzymes were purchased from TaKaRa (Dalian, China). In addition, the Gibson assembly cloning kit was purchased from Yesen (Shanghai, China).
2.2. Media, culture conditions, and ITA analysis
The transformants of E. coli were cultured in Luria Bertani (LB) broth containing appropriate antibiotics for cell growth and protein expression. Then, the reconstructed strains for ITA production were grown at 30 °C on a rotary shaker at 200 rpm in minimal media (MM) broth [25]. To quantify ITA production, samples were collected from 1-mL of culture and filtered using a 0.45-μm filtration membrane. A high-performance liquid chromatography (Agilent 1200) system equipped with an Agilent ZORBAX SB-Aq C18 column (5 μm, 4.6 mm × 250 mm) and a UV/VIS detector were utilized to quantify ITA levels at a wavelength of 210 nm at 40 °C. The injection volume was 10 μL per sample and the mobile phase was 0.1 M NH4H2PO4 (pH 2.6) at a flow rate of 1.0 mL/min.
2.3. Itaconic acid biosensor construction and functional testing
The sequence of the ITA biosensor, YpItcR/Pccl [26], was synthesized by Tsingke Biotech (Beijing, China) and cloned into pACYCDuet-1, which contains a mrfp gene constructed by the Gibson assembly method, to yield pYpItcR/Pccl-mrfp. The primers used in this study are presented in Supplementary Table S3. To precisely measure the red fluorescence intensity, the MG1655 cells containing pYpItcR/Pccl-mrfp were recultivated in fresh LB medium with a different concentration of ITA at 37 °C. Subsequently, the cells were diluted to an optical density of 600 nm (OD600) of 0.5, and the red fluorescence intensity was measured in vivo using the Ascent (Thermo, Waltham, MA, USA) at excitation and emission wavelengths of 580 nm and 610 nm, respectively [27].
2.4. Regulation module construction
The gene dcas9 was adopted as the template and amplified with pdCas9-Pccl-F/pdCas9-Pccl-R, and then, the amplified fragment was cyclized with the plasmid skeleton pYpItcR/Pccl via the Gibson assembly method to generate pYpItcR/Pccl-dcas9. In addition, the sgRNAs (N20) were designed online (https://zlab.bio/guide-design-resources). The sequences of the sgRNAs are presented in the Supplementary Table S4. A single sgRNA expression cassette was constructed, via the Gibson assembly method, into the Sal Ⅰ/Xba Ⅰ restriction sites of pYpItcR/Pccl-dcas9. sgRNA expression cassettes were amplificated with primers sRn-F/sRn+1-R. The sgRNA arrays were assembled via the Gibson assembly method (Supplementary Fig. S2).
2.5. Real-time quantitative PCR (RT-qPCR) analysis
Cells were grown at 30 °C in MM broth and harvested at 24 and 48 h. The total RNA was extracted from the cells using the Bacterial RNA Kit purchased from Omega Bio-tek (Georgia, USA), according to the manufacturer's instructions. In addition, the 16s rRNA gene was adopted as the housekeeping gene. RT-qPCR was conducted with FastKing RT and SuperReal PreMix Plus kits purchased from Tiangen (Beijing, China) following the manufacturer's instructions. The obtained results were analyzed via the 2−ΔΔCt method.
2.6. Statistical analysis
All quantitative data were measured based on at least three independent experiments. The experimental data were statistically processed using Origin 8.0 software, and the data results were presented as the mean ± standard error (mean ± SE). The data were considered statistically significant at P < 0.05.
3. Results and discussion
3.1. Design of CiMS system
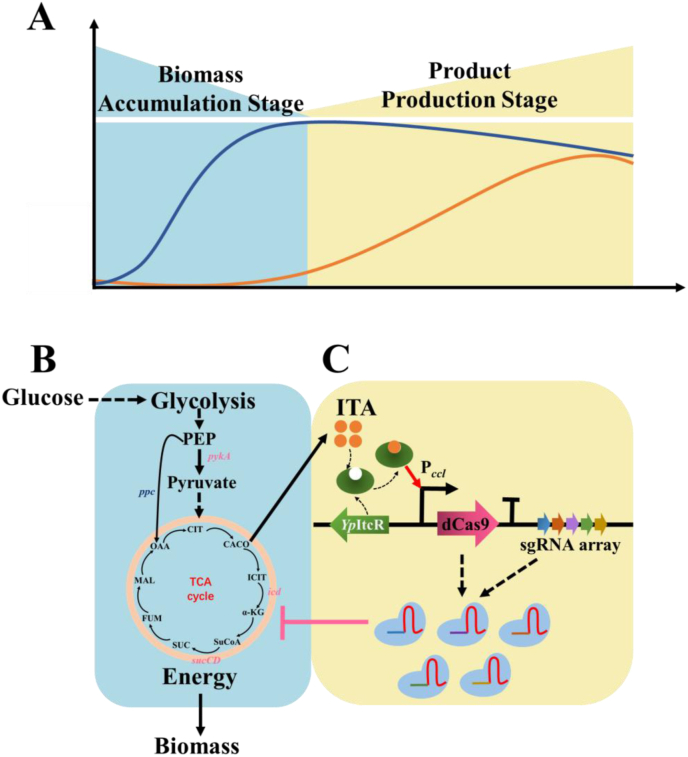
The TCA cycle provides most of the energy for biomass accumulation. When cells enter the stationary phase, the stored energy used for metabolites production is what we expected. ITA synthesis competes for the cis-aconitate in the TCA cycle. Therefore, during the ITA accumulated phase, balancing energy allocation between the TCA cycle and the ITA-producing pathway is essential for the high-yielding production of ITA. Here, we designed a CRISPRi-mediated self-inducible system (CiMS), which functioned as a dynamic regulator to continually optimize the flux distributions between biomass and metabolites production during the fermentation stage. The CiMS contained a regulative module based on the CRISPRi system and a responsive module based on a specific biosensor activated by the target product. (Fig. 1). At the biomass-accumulation stage, ITA was barely accumulated and the CRISPRi system was nearly silent (Fig. 1A and B). Therefore, the metabolic flux could be fully applied for cell growth. At the fermentation stage, ITA rapidly accumulated. The special biosensor was gradually launched which strengthened the CRISPRi to regulate the metabolic flux (Fig. 1A and C). Hence, the metabolic flux transformed from the TCA cycle to the ITA synthesis. To dynamically self-regulate the metabolic flux during the ITA synthesized phase, an efficient ITA-specific biosensor was indispensable. The YpItcR/Pccl, comprising the ITA itaconate-inducible promoter Pccl and its corresponding transcriptional regulator ItcR from Yersinia pseudotuberculosis, has been verified to be a special ITA biosensor in E. coli [26] and was used to construct the responsive module of CiMS.
Fig. 1.
Overview of the CRISPRi-mediated self-inducible system (CiMS). (A) Normal metabolic characteristics of Escherichia coli. The blue line represents the cell growth curve; the orange line represents the product titer. (B) Biomass accumulation via the TCA cycle; little itaconic acid (ITA) could be produced during this stage. The CiMS was almost silent and metabolic flux was turned into biomass. The genes marked with a pink color were regulated via the CiMS. (C) The CiMS was activated via product accumulation. The ITA biosensor YpItcR/Pccl could be activated by sufficient ITA to induce the expression of dCas9. The expression of genes icd, pykA, and sucCD was inhibited by dCas9 with the guide of sgRNA and the metabolic flux was almost converted to ITA production.
To better balance the metabolic flux between the TCA cycle and ITA biosynthetic pathway, several key genes should be dynamically regulated by CiMS to accumulate the precursor of ITA and facilitate ITA production. Knocking out key genes icd and sucCD in the TCA cycle limited the metabolic flux of TCA cycle and accumulated the precursor of ITA, which turned out to be an efficient intervention to increase ITA yield [12]. Instead of gene deletion, inhibition of the expression of these two genes would cause a minor effect on cell growth. Therefore, in this study, the genes icd and sucCD were repressed by the CiMS to control the metabolic flux of the TCA cycle and further accumulate the precursor of ITA.
However, inhibiting the expression of the essential genes icd and sucCD blocked the metabolic flux of the TCA cycle and decreased the production of oxaloacetate (OAA), which was also indispensable for ITA production. Phosphoenolpyruvate (PEP) can be catalyzed by pyruvate kinase Ⅰ (encodes by gene pykA) and pyruvate kinase Ⅱ (encodes by gene pykF) to produce pyruvate, and then to produce acetyl coenzyme A (acetyl CoA). PEP can also be catalyzed by phosphoenolpyruvate carboxylase (encodes by gene ppc) to produce OAA. Acetyl CoA and OAA are the starting substrates for the TCA cycle and the precursors of ITA. Therefore, it is necessary to balance the metabolic flux between the synthesis of acetyl CoA and OAA. Compared with pyruvate kinase Ⅱ, pyruvate kinase Ⅰ contributes significantly low enzymatic activity [28,29]. A study has indicated that deleting pykA gene had a positive effect on ITA production [14]. Therefore, to ensure the proper function of the TCA cycle and to accumulate the precursor of ITA, the expression of gene pykA also needed to be regulated by the CiMS.
3.2. Feasibility analysis of CiMS system
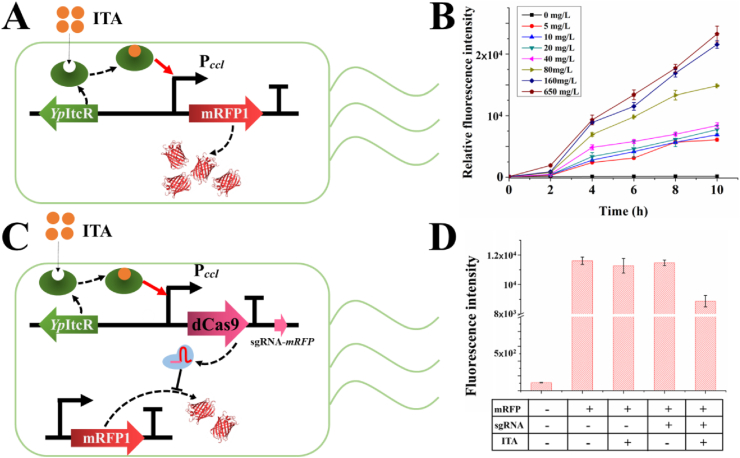
To verify the availability of YpItcR/Pccl for the CiMS, we first constructed the recombinant strain YpItcR/Pccl-mrfp/MG1655, which was utilized to detect the YpItcR/Pccl property to regulate the expression of mRFP (Fig. 2A). To analyze the relationship between the ITA concentration and YpItcR/Pccl strength, different concentrations of ITA were adopted to activate YpItcR/Pccl. The obtained results indicated that the strength of YpItcR/Pccl was extremely weak with 40 mg/L ITA, and significantly increased with >80 mg/L ITA (Fig. 2B). Considering that the ITA titer of the MG1655 Δ1-SAS-3 strain was no more than 50 mg/L at the logarithmic phase, modulating of the TCA cycle via the CiMS would not affect the cell growth.
Fig. 2.
Verification of the feasibility of the regulation module and the response module of the CRISPRi-mediated self-inducible system (CiMS). (A) Construction of the itaconic acid (ITA) biosensor YpItcR/Pccl. (B) Analysis of the RFP fluorescence intensity to verify the expression strength of YpItcR/Pccl with different concentrations of ITA. (C) Construction of the regulative module and response module of the CiMS. (D) Analysis of the RFP fluorescence intensity to verify the efficiency of interference by the CiMS. Error bars show the standard deviation from three independent experiments.
To verify the efficiency of CRISPRi intervention under the regulation of YpItcR/Pccl, the constructed gene circuit YpItcR/Pccl-CRISPRi was applied to regulate the expression of mRFP (Fig. 2C). After adding 80 mg/L ITA, CRISPRi was activated and dCas9 targeted mrfp mRNA, directed by a sgRNA-mrfp. The expression of red fluorescence in strain pYpItcR/Pccl-mrfp/MG1655 was repressed at the fourth hour, and the inhibition efficiency increased significantly over time. The obtained results indicated that the expression of mRFP was decreased by 23% after 10 h under the regulation of YpItcR/Pccl-CRISPRi (Fig. 2D, Supplementary Fig. S3), clearly demonstrating that it was suitable for the modulation of target genes.
3.3. Improving the ITA production of engineered strains
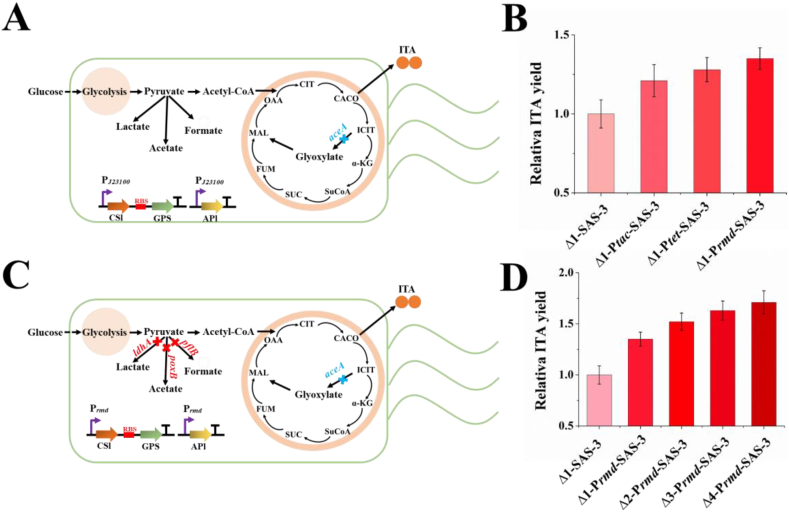
In our previous study, we had knocked out gene aceA in the glyoxylate pathway in the genome of E. coli MG1655 and constructed a self-assembly system that enhanced the cascade reaction among citrate synthase (gltA, GA), aconitase (acnA, ACN), and cis-aconitate decarboxylase (cadA, CAD) to construct strain Δ1-SAS-3 (Fig. 3A). Evidently, the ITA titer of Δ1-SAS-3 (1.87 g/L) was lower than that reported with ITA production strains, such as ita23 (2.27 g/L) [14]. Therefore, before conducting CiMS regulation, we expected to further improve the ITA yield of Δ1-SAS-3 via conventional metabolic engineering strategies. First, three different high-strength promoters, Ptac, Ptet, and Prmd, derived from E. coli were applied to replace PJ23100 for the expression of the self-assembly cascade reaction. The results indicated that the ITA yield of the recombinant strain Δ1-Prmd-SAS-3 was the highest, which could reach 1.35 times that of Δ1-SAS-3 (Fig. 3B). Next, the engineered strain, Δ4-Prmd-SAS-3 (Fig. 3C, Supplementary Fig. S4), derived from Δ1-Prmd-SAS-3 was obtained by knocking out three genes, pflB, poxB, and ldhA, in the pyruvate bypass pathway to accumulate the precursor of ITA and the ITA titer of Δ4-Prmd-SAS-3 (3.2 g/L) reached 1.27 times that of Δ1-Prmd-SAS-3 (Fig. 3D).
Fig. 3.
Construction of engineered strains to improve the itaconic acid (ITA) yield. (A) Overview of Escherichia coli MG1655 Δ1-SAS-3. The gene marked with a blue color represents gene knock-out in the genome of Δ1-SAS-3. CSl (CAD-SH3lig), GPS (GA-PDZ-SH3), and APl (ACN-PDZlig) represent the over-expression cascade reaction for ITA production. (B) Improvement in the ITA yield in engineered strain via promotor optimization. PJ23100 for the expression of the CSl, GPS, and APl was replaced by Ptac, Ptet, and Prmd, respectively. (C) Overview of E. coli MG1655 Δ4-Prmd-SAS-3. The genes marked with blue and red represent the genes knocked out in the genome of Δ4-Prmd-SAS-3. (D) Relative ITA yield of each engineered strain. Error bars show the standard deviation from three independent experiments.
3.4. Effectiveness of CiMS for dynamic regulation of ITA production
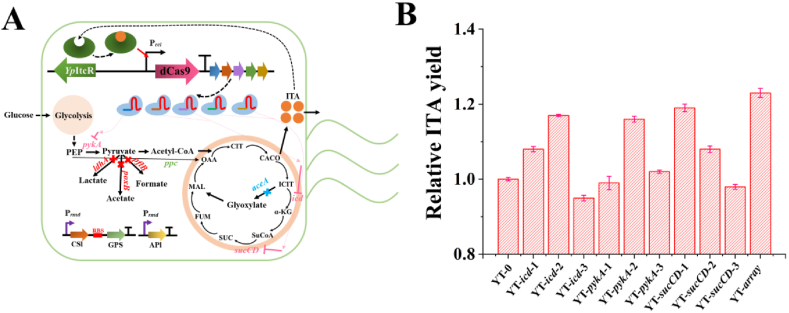
To effectively inhibit the expression of icd, pykA, and sucCD, three different sgRNAs were designed for each gene (Fig. 4A). Consequently, nine regulated strains were constructed to verify the effectiveness of the CiMS to improve ITA production capability (Fig. 4B). Compared with the control strain E. coli MG1655 Δ4-Prmd-SAS-3/sgRNA-0 (Δ4-Prmd-SAS-3 harboring a non-targeting CRISPRi plasmid), ITA production by the five regulated strains with single-targeting intervention (sgRNA-icd-1, sgRNA-icd-2, sgRNA-pykA-1, sgRNA-sucCD-1, sgRNA-sucCD-2) was significantly improved. To verify the superposition of effective inhibited sites, these five sgRNAs were combined into a sgRNA array. The results showed that the regulated strain E. coli MG1655 Δ4-Prmd-SAS-3/sgRNA-array was optimal, which could increase the ITA yield to 1.23 times that of the control strains (Fig. 4B). Moreover, the growth of the engineered strain was similar to that of the control strain but exhibited significantly increased production of ITA at the anaphase of the platform period (100 h, Supplementary Fig. S5), which clearly demonstrated that the CiMS could dynamically and precisely regulate ITA production in the E. coli cell factory.
Fig. 4.
Biosynthesis of itaconic acid (ITA) for the dynamic regulation of metabolic flux by the CRISPRi-mediated self-inducible system (CiMS). (A) Construction of different sgRNAs targeted to essential genes to interfere with the metabolic pathway. CSl (CAD-SH3lig), GPS (GA-PDZ-SH3), and APl (ACN-PDZlig) represent the over-expression cascade reaction for ITA production. The genes marked with blue and red represent the genes knocked out in the genome of Δ4-Prmd-SAS-3. The genes marked with a pink color represent the target genes repressed by the CiMS. After accumulating sufficient ITA, the ITA-bound YpItcR could activate the expression of dCas9 and inhibit the expression of the genes in pink with the help of the sgRNA. (B) Relative ITA yield of the CiMS-regulated strains with different sgRNAs. Escherichia coli MG1655 Δ4-Prmd-SAS-3/sgRNA-0 was used as the control. The sgRNA-array contained five sgRNAs, sgRNA-icd-1, sgRNA-icd-2, sgRNA-pykA-1, sgRNA-sucCD-1, sgRNA-sucCD-2, the inhibition of which could significantly improve the production of ITA. Error bars show the standard deviation from three independent experiments.
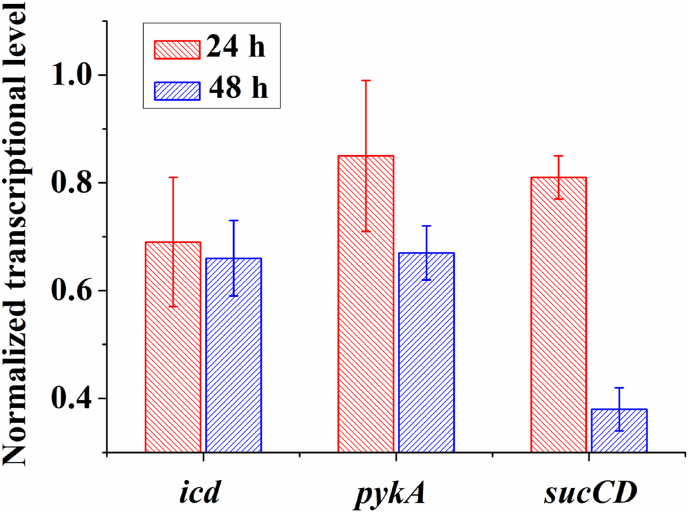
To verify that the ITA yield improvement in the engineered strain was the result of the dynamic regulation via the CiMS, the transcriptional level of each metabolic bypass gene, icd, pykA, and sucCD, was analyzed at 24 and 48 h. The results showed that the transcriptional level of the three genes were slightly decreased at 24 h, which was possibly due to activation of the regulation module by the small amounts of accumulated ITA (Fig. 5). Additionally, the transcriptional levels of these three genes were significantly decreased at 48 h, which respectively reached 66% (icd), 67% (pykA), and 38% (sucCD) of those in the unregulated strain (Fig. 5). These results fully demonstrated that the collateral ITA pathway was dynamically regulated by the CiMS during ITA production.
Fig. 5.
Transcriptional level evaluation of icd, pykA, and sucCD at 24 and 48 h. The strain Escherichia coli MG1655 Δ4-Prmd-SAS-3/sgRNA-0 was used as the control and the strength was set to a value of 1. Error bars show the standard deviation from three independent experiments.
4. Discussion
The biosynthesis of ITA in heterogenous microorganisms has been extensively studied [[30], [31], [32]]. Meanwhile, researchers have also encountered challenges posed by the uneven distributions of the metabolic flux. Isocitrate dehydrogenase, which plays an important role in the TCA cycle, is encoded by the gene icd, and the intervention of icd significantly affects the growth of host cells [33]. In our previous work, the gene icd was completely deleted to accumulate the precursor of ITA, which required the addition of extra expensive α-KG to maintain stability of the TCA cycle [12]. Therefore, it is important to dynamically regulate the metabolic flux between biomass accumulation and metabolite production.
With the development of synthetic biology, novel strategies, drawing from the natural dynamic regulatory mechanisms in microorganisms, have been developed for the dynamic regulation of metabolic pathways. A temperature-sensitive promotor, pH-responsive biosensor, and quorum sensing were applied to dynamically regulate metabolic flux and enhance the production of metabolites [[34], [35], [36]]. However, the artificially engineered Ntr regulon, which senses of acetyl phosphate, was constructed to control the expression of genes pps and ldi in lycopene synthesis in response to flux dynamics [37]. To date, dynamic regulation has made significant progress in the synthesis of valuable products in a low-cost manner. However, the metabolic regulation by a single signal would not be suitable for a complicated metabolic network. Therefore, dynamic regulation, comprised of real-time responses to target metabolites, would probably sustains cellular homeostasis to increase the productivity of metabolites.
Hence, in this study, we constructed a self-inducible system, which contained the biosensor YpItcR/Pccl to specially respond to ITA and a CRISPRi-mediated interferential module regulated by the ITA biosensor. Combined with the designed sgRNAs targeting the vital genes in the critical metabolic pathway, the CiMS was applied to dynamically regulate metabolic flux between biomass accumulation and ITA production (Fig. 1). During the logarithmic growth phase, trace amounts of ITA could be produced and YpItcR/Pccl was almost silent; thus, the CRISPRi system could not be activated to inhibit the TCA cycle and the cells could grow well (Fig. 1A). After the logarithmic growth phase, sufficient ITA accumulation could activate the designed CiMS to inhibit the TCA cycle, while metabolic flux was continuously converted to ITA production (Fig. 1B).
The key role of the CiMS was as a specific biosensor that could respond to the target product. Recently, the ITA biosensor YpItcR/Pccl was obtained, and its effectiveness was verified in E. coli [26]. Therefore, YpItcR/Pccl could function effectively as an important element for the construction of a CiMS to dynamically regulate ITA production in E. coli. In this study, we also utilized YpItcR/Pccl to directly regulate the expression of mRFP (Fig. 2B) and inhibit the transcription of mrfp by combining it with the CRISPRi system (Fig. 2D), which fully confirmed that YpItcR/Pccl was suitable for CiMS application to the dynamic regulation of ITA production. In addition, various biosensors responding to organic acids, such as glutamate and cysteine, have been reported [[38], [39], [40]]. The CiMS exhibited the potential to produce several types of organic acids. Furthermore, YpItcR/Pccl can be reconstructed to respond to other organic acids via protein engineering and directed evolution strategies [26].
However, some issues must be further addressed in future studies. The expression level of dCas9 was barely detected under the regulation of YpItcR/Pccl, such that it was difficult to optimize the expression of dCas9. In addition, the promoter for the transcription of sgRNA should also be optimized, which might further improve the ITA yield of engineered strains. Alternatively, it is worth considering the application of CRISPR/ddCpf1 systems in our further research. Numerous studies have demonstrated that the gene repression mediated by ddCpf1 is highly efficient and has no substantial off-target effects [[41], [42], [43], [44]]. The Cas9-based gene-editing system requires both a crRNA and tracrRNA to mediate interference, whereas the Cpf1 system only needs a pre-crRNA, which remarkably simplifies the construction of sgRNA arrays [45].
5. Conclusions
In conclusion, we designed a CiMS, for dynamic regulation of the TCA cycle and ITA production. This novel system employed the CRISPRi strategy instead of the knock-out strategy to inhibit the expression of vital genes in the TCA cycle. Therefore, extra amounts of expensive organic acids, such as α-KG and l-glutamic acid were not required in the medium, and this would remarkably minimize the costs of fermentation. In addition, the biosensor responding to the target product ITA was employed to regulate the expression of dCas9, and sufficient ITA accumulation to activate the biosensor should be realized during the ITA accumulation period. Therefore, the CiMS exerted a negligible effect on cell growth. Consequently, the CiMS turned out to be an effective method to increase the ITA titer, which provides a novel idea for organic acid production.
CRediT authorship contribution statement
Ming Zhao: designed the research, performed experiments, Formal analysis, analyzed the results, Writing – original draft, wrote the manuscript. Yuting Li: performed experiments, Formal analysis, analyzed the results, Writing – original draft, wrote the manuscript. Fengqing Wang: designed the research, Formal analysis, analyzed the results, Writing – original draft, wrote the manuscript. Yuhong Ren: designed the research, Formal analysis, analyzed the results. Dongzhi Wei: Supervision, supervised the research.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 21778018) and Research Program of State Key Laboratory of Bioreactor Engineering and the grant from the National Key Research and Development Program of China (2021YFC2100300) and the Chinese Plastic Surgery Foundation (No. 2020M671021).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2022.05.008.
Contributor Information
Fengqing Wang, Email: fqwang@ecust.edu.cn.
Yuhong Ren, Email: yhren@ecust.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ji D.N., Li J.H., Xu F.L., Ren Y.H., Wang Y. Improve the biosynthesis of baicalein and scutellarein via manufacturing self-assembly enzyme reactor in vivo. ACS Synth Biol. 2021;1 0(5):1087–1094. doi: 10.1021/acssynbio.0c00606. [DOI] [PubMed] [Google Scholar]
- 2.Miscevic D., Mao J.Y., Kefale T., Abedi D., Moo-Young M., Perry C.C. Strain engineering for high-level 5-aminolevulinic acid production in Escherichia coli. Biotechnol Bioeng. 2021;118(1):30–42. doi: 10.1002/bit.27547. [DOI] [PubMed] [Google Scholar]
- 3.Paddon C.J., Keasling J.D. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol. 2014;12(5):355. doi: 10.1038/nrmicro3240. 67. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y., Daviet L., Schalk M., Siewers V., Nielsen J. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng. 2013;15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Gao C., Hou J., Xu P., Guo L., Chen X., Hu G., et al. Programmable biomolecular switches for rewiring flux in Escherichia coli. Nat Commun. 2019;10(1):3751–3763. doi: 10.1038/s41467-019-11793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keasling J.D. Manufacturing molecules through metabolic engineering. Science. 2010;330(6009):135 5–8. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 7.Kamp A.V., Klamt S. Growth-coupled overproduction is feasible for almost all metabolites in five major production organisms. Nat Commun. 2017;8 doi: 10.1038/ncomms15956. 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuenz A., Krull S. Biotechnological production of itaconic acid-things you have to know. Appl Microbiol Biotechnol. 2018;102(9):3901–3914. doi: 10.1007/s00253-018-8895-7. [DOI] [PubMed] [Google Scholar]
- 9.Nuss P., Gardner K.H. Attributional life cycle assessment (ALCA) of polyitaconic acid production from northeast US softwood biomass. Int J Life Cycle Assess. 2012;18(3):603–612. doi: 10.1007/s11367-012-0511-y. [DOI] [Google Scholar]
- 10.Cordes T., Michelucci A., Hiller K. Itaconic acid: the surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annu Rev Nutr. 2015;35:451–473. doi: 10.1146/annurev-nutr-071714-034243. [DOI] [PubMed] [Google Scholar]
- 11.Strelko C.L., Lu W., Dufort F.J., Seyfried T.N., Chiles T.C., Rabinowitz J.D., et al. Itaconic acid is a mammalian metabolite induced during macrophage activation. J Am Chem Soc. 2011;133(41):163 86–89. doi: 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z.W., Wang H.L., Wang Y.X., Ren Y.H., Wei D.Z. Manufacturing multienzymatic complex reactors in vivo by self-assembly to improve the biosynthesis of itaconic acid in Escherichia coli. ACS Synth Biol. 2018;7(5):1244–1250. doi: 10.1021/acssynbio.8b00086. [DOI] [PubMed] [Google Scholar]
- 13.Noh M.H., Lim H.G., Woo S.H., Song J., Jung G.Y. Production of itaconic acid from acetate by engineering acid-tolerant Escherichia coli W. Biotechnol Bioeng. 2018;115(3):729. doi: 10.1002/bit.26508. 38. [DOI] [PubMed] [Google Scholar]
- 14.Harder B.J., Bettenbrock K., Klamt S. Model-based metabolic engineering enables high yield itaconic acid production by Escherichia coli. Metab Eng. 2016;38:29–37. doi: 10.1016/j.ymben.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Vuoristo K.S., Mars A.E., Sangra J.V., Springer J., Eggink G., Sanders J.P., et al. Metabolic engineering of itaconate production in Escherichia coli. Appl Microbiol Biotechnol. 2015;99(1):221–228. doi: 10.1007/s00253-014-6092-x. [DOI] [PubMed] [Google Scholar]
- 16.Harder B.J., Bettenbrock K., Klamt S. Temperature-dependent dynamic control of the TCA cycle increases volumetric productivity of itaconic acid production by Escherichia coli. Biotechnol Bioeng. 2018;115(1):156–164. doi: 10.1002/bit.26446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81(7):2506. doi: 10.1128/AEM.04023-14. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sander T., Wang C.Y., Glatter T., Link H. CRISPRi-based downregulation of transcriptional feedback improves growth and metabolism of arginine overproducing E. coli. ACS Synth Biol. 2019;8(9):1983–1990. doi: 10.1021/acssynbio.9b00183. [DOI] [PubMed] [Google Scholar]
- 19.Cornuault J.K., Moineau S. Induction and elimination of prophages using CRISPR interference. CRISPR J. 2021;4(4):549–557. doi: 10.1089/crispr.2021.0026. [DOI] [PubMed] [Google Scholar]
- 20.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos-Moreno J., Tasiudi E., Stelling J., Schaerli Y. Multistable and dynamic CRISPRi-based synthetic circuits. Nat Commun. 2020;11:2746–2754. doi: 10.1038/s41467-020-16574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu P., Chen Y., Liu M., Xiao G., Yuan J. Engineering an optogenetic CRISPRi platform for improved chemical production. ACS Synth Biol. 2021;10:125. doi: 10.1021/acssynbio.0c00488. 31. [DOI] [PubMed] [Google Scholar]
- 23.Yi Y.C., Ng I.S. Redirection of metabolic flux in Shewanella oneidensis MR-1 by CRISPRi and modular design for 5-aminolevulinic acid production. Biores Bioprocess. 2021;8:13–24. doi: 10.1186/s40643-021-00366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cleto S., Jensen J.V., Wendisch V.F., Lu T.K. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi) ACS Synth Biol. 2016;5(5):375. doi: 10.1021/acssynbio.5b00216. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto K., Nagata K., Ohara H., Aso Y. Challenges in the production of itaconic acid by metabolically engineered Escherichia coli. Bioengineered. 2015;6(5):303. doi: 10.1080/21655979.2015.1068471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanko E.K.R., Minton N.P., Malys N. A transcription factor-based biosensor for detection of itaconic acid. ACS Synth Biol. 2018;7(5):1436–1446. doi: 10.1021/acssynbio.8b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraut-Cohen J., Afanasieva E., Haim-Vilmovsky L., Slobodin B., Yosef I., Bibi E., et al. Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2013;24(19):3069. doi: 10.1091/mbc.E13-01-0038. 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valentini G., Stoppini M., Iadarolau P., Malcovat M. Divergent binding sites in pyruvate kinases I and II from Escherichia coli. Biol Chem Hoppe Seyler. 1993;374:69–74. doi: 10.1515/bchm3.1993.374.1-6.69. [DOI] [PubMed] [Google Scholar]
- 29.Ponce E., Flores N., Martinez A., Valle F., Bolívar F. Cloning of the two pyruvate kinase isoenzyme structural genes from Escherichia coli: the relative roles of these enzymes in pyruvate biosynthesis. J Bacteriol. 1995;177(19):5719–5722. doi: 10.1128/jb.177.19.5719-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker J., Tehrani H.H., Ernst P., Blank L.M., Wierckx N. An optimized Ustilago maydis for itaconic acid production at maximal theoretical yield. J Basel. 2020;7:20–34. doi: 10.3390/jof7010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao M., Lu X., Zong H., Li J., Zhuge B. Itaconic acid production in microorganisms. Biotechnol Lett. 2018;40(3):455–464. doi: 10.1007/s10529-017-2500-5. [DOI] [PubMed] [Google Scholar]
- 32.Qi H., Du Y., Zhou X., Zheng W., Zhang L., Wen J., et al. Engineering a new metabolic pathway for itaconate production in Pichia stipitis from xylose. Biochem Eng J. 2017;126:101. doi: 10.1016/j.bej.2017.06.011. 8. [DOI] [Google Scholar]
- 33.Goncalves S., Miller S.P., Carrondo M.A., Dean A.M., Matias P.M. Induced fit and the catalytic mechanism of isocitrate dehydrogenase. Biochemistry. 2012;51(36):7098. doi: 10.1021/bi300483w. 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter F., Fonfara I., Bouazza B., Schumacher C.H., Bratovic M., Charpentier E., et al. Engineering of temperature- and light-switchable Cas9 variants. Nucleic Acids Res. 2016;44(20) doi: 10.1093/nar/gkw930. 1 0003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin X., Shin H.D., Li J., Du G., Liu L., Chen J., et al. Pgas, a low-pH-induced promoter, as a tool for dynamic control of gene expression for metabolic engineering of Aspergillus Niger. Appl Environ Microbiol. 2017;83(6) doi: 10.1128/aem.03222-16. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen X., Wang J., Li C., Yuan Q., Yan Y. Dynamic gene expression engineering as a tool in pathway engineering. Curr Opin Biotechnol. 2019;59:122. doi: 10.1016/j.copbio.2019.03.019. 9. [DOI] [PubMed] [Google Scholar]
- 37.Farmer W.R., Liao J.C. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18:533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 38.Zeynaloo E., Yang Y.P., Dikici E., Landgraf R., Bachas L.G., Daunert S. Design of a mediator-free, non-enzymatic electrochemical biosensor for glutamate detection. Nanomedicine. 2021;3 1:102305–102314. doi: 10.1016/j.nano.2020.102305. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Mishra D., Bergman J., Keighron J.D., Skibicka K.P., Cans A.S. Ultrafast glutamate biosensor recordings in brain slices reveal complex single exocytosis transients. ACS Chem Neurosci. 2019;10(3):1744–1752. doi: 10.1021/acschemneuro.8b00624. [DOI] [PubMed] [Google Scholar]
- 40.Chen C., Zhou L., Liu W., Liu W. Coumarinocoumarin-based two-photon fluorescent cysteine biosensor for targeting lysosome. Anal Chem. 2018;90(10):6138. doi: 10.1021/acs.analchem.8b00434. 43. [DOI] [PubMed] [Google Scholar]
- 41.Ji X., Zhao H., Zhu H., Zhu K., Tang S.Y., Lou C. CRISPRi/dCpf1-mediated dynamic metabolic switch to enhance butenoic acid production in Escherichia coli. Appl Microbiol Biotechnol. 2020;104(12):5385–5393. doi: 10.1007/s00253-020-10610-2. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X., Wang J., Cheng Q., Zheng X., Zhao G., Wang J. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017;3:17018–17027. doi: 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M., Mao Y., Lu Y., Tao X., Zhu J.K. Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant. 2017;10(7):1011–1013. doi: 10.1016/j.molp.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Tak Y.E., Kleinstiver B.P., Nunez J.K., Hsu J.Y., Horng J.E., Gong J., et al. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat Methods. 2017;14(12):1163–1166. doi: 10.1038/nmeth.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ungerer J., Pakrasi H.B. Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci Rep. 2016;6:39681–39690. doi: 10.1038/srep39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.