Graphical abstract
Abbreviations: TOCSY, Total Correlation Spectroscopy; KNFST, Kernel Null Foley–Sammon Transform; SVDD, Support Vector Data Description; KDE, Kernel Density Estimation; NMR, Nuclear Magnetic Resonance; ATP, Adenosine Triphosphate; BMRB, Biological Magnetic Resonance Data Bank; HMDB, Human Metabolome Database; ROC, Receiver Operating Characteristic; AUC, Area under Curve
Keywords: Novelty detection, Machine learning, Classification, 2D NMR TOCSY, Metabolic profiling, Breast cancer, Chemometrics, Metabolomics
Highlights
-
•
Automatic novelty detection of metabolites of 2D-TOCSY NMR spectra.
-
•
Metabolic profiling of the dynamics changes in Breast cancer tissue sample.
-
•
Accurate and fast automatic multicomponent peak assignment of 2D NMR spectrum.
-
•
One- and multi- novelty detection of metabolites.
Abstract
Most metabolic profiling approaches focus only on identifying pre-known metabolites on NMR TOCSY spectrum using configured parameters. However, there is a lack of tasks dealing with automating the detection of new metabolites that might appear during the dynamic evolution of biological cells. Novelty detection is a category of machine learning that is used to identify data that emerge during the test phase and were not considered during the training phase. We propose a novelty detection system for detecting novel metabolites in the 2D NMR TOCSY spectrum of a breast cancer-tissue sample. We build one- and multi-class recognition systems using different classifiers such as, Kernel Null Foley-Sammon Transform, Kernel Density Estimation, and Support Vector Data Description. The training models were constructed based on different sizes of training data and are used in the novelty detection procedure. Multiple evaluation measures were applied to test the performance of the novelty detection methods. Depending on the training data size, all classifiers were able to achieve 0% false positive rates and total misclassification error in addition to 100% true positive rates. The median total time for the novelty detection process varies between 1.5 and 20 seconds, depending on the classifier and the amount of training data. The results of our novel metabolic profiling method demonstrate its suitability, robustness and speed in automated metabolic research.
1. Introduction
Metabolic profiling involves the investigation of metabolite concentrations and systematic metabolic variation, caused by new stimuli such as different drugs, dieting, microbiological causes or gene modulation, for the purpose of the characterization of the effects of external interventions [1]. Due to the nature of the biological fluids, cells and tissues, metabolites change to reach a dynamic equilibrium. As a result, any biological process will induce metabolic alteration, which can be related to the diagnosis or prognosis of specific diseases or therapeutic status [2], [3]. Metabolites drive essential cellular functions, like signal transduction, energy production, storage, and apoptosis [4]. ATP, acetyl-CoA, NAD+, and S–adenosyl methionine metabolites can contribute to the regulation of post-translational modifications that affect protein activity[5], [6]. Additionally, metabolite and protein interactions can aid to cellular responses, thus evincing the metabolites role in signal transduction [7], [8].
NMR spectroscopy is one of the most robust tools applied in multicomponent analysis of samples from urine, blood plasma or tissue [2], [9], [10]. Nevertheless, major challenges of NMR spectroscopy include peak overlapping, chemical shift variations, noise and biological matrix effects owing to the continuous change of chemical environments [11]. These challenges can introduce considerable variations in the spectral signature of individual molecules in comparison to its pattern in complex mixtures [11]. Though the identification of metabolites in 2D NMR spectra is simpler than 1D NMR, the straightforward metabolic profiling in 2D NMR is valid only to first order systems with weak coupling [2]. Even in 2D NMR, the identification of metabolites with low concentration or peaks partially or totally overlapping is a complicated task [2]. Consequently, the complexity of experimental measurements, noise, artifacts in addition to phase and baseline distortion, cause peak shifts, misaligned peaks as well as peaks with slight deviation from the expected peak shape, make metabolic profiling a demanding task [11], [12]. Furthermore, the manual analysis of 2D NMR spectra is prone to error and missing assignments in cases of complex mixtures [12].
In biology and medicine, machine learning supports scientists in the prediction, evaluation, uncertainty estimation and model interpretation of medical images, including X-ray, MRI and mammography images in addition to enabling the utilization of data produced from high-throughput omics to identify new molecular biomarkers [13].
NMR spectroscopy and machine learning create a promising interdisciplinary research area that could achieve a notable progress in NMR spectroscopy. Establishing an automatic assignment system that can detect emerging new metabolites or unknown molecule in samples will enhance and support many applications that rely on 2D NMR spectra [12], [14]. In machine learning, novelty detection refers to systems that try to distinguish normal control samples from potentially abnormal variant samples. The concepts normal control and abnormal variant samples are used to differentiate known categories which are consistent with the training model, from new uncommon data that appear in new experiments [15], [16]. Often, due to the complexity of real systems, defining a list of categories that might appear in future samples is inapplicable. Consequently, conventional multi-class classification algorithms are inappropriate for this issue because they will assign a wrong label to the new data sample by employing the predefined categories [15]. Normally, novelty detection is required in two situations. The first is when there are few examples to represent a known class within the training dataset; for instance, a particular category happens rarely, so the classification system does not have enough instances to represent this category. In this case, it is better to consider the rare category as novel or abnormal and test it against the model of normality. The second situation occurs when the training list is incomplete. Although enough instances are available to form a training model, it is expected that new classes will appear in the future [17]. In this article, we introduce the concept of novelty detection of metabolites in 2D NMR TOCSY spectra where one or more metabolites appear in the spectra.
2. Related work
Previous related studies indicate that peak overlap, the absence of reference spectral database of metabolites, and the diversity of metabolites are common challenges when analyzing complex biological mixtures [18]. According to [18], a consistent technique for reporting novel metabolites in NMR is still unavailable. Overall, the identification of metabolites in complex biological mixtures is based only on the most abundant metabolites. The identification of metabolites is done by analyzing 1D and 2D NMR using the literature or online spectral libraries such as HMDB and BMRB [19], [20]. For large-scaled applications, biomarker identification can be boosted through using software tools such as, Chenomx NMRsuite [21], COLMAR query [22] or MetaboMiner [23]. These tools provide a list of possible metabolites based on visual comparison or similarity scores. However, in complex mixtures with crowded and overlapped spectrum, the interpretation of the NMR signal becomes doubtful and unclear. When these methods fail, the most common way to identify novel metabolites is structure elucidation, metabolic purification and isolation in addition to applying further complementary mass spectroscopy techniques [2], [18], [24], [25], [26]. Recently, potential novel biomarkers related to prostate cancer and skin cancer have been identified in 1H NMR employing principal component analysis and least squares discriminant analysis to detect metabolites and outliers [27], [28], [29], [30]. A protocol with multiple workflows to detect known and unknown metabolites using 1D and 2D NMR has been thoroughly discussed [31]. Similarly, it reports principal component analysis, structure discriminant analysis, structure elucidation, and an extensive use of matching biological databases [31]. Nonetheless, these methods are traditional methods that could be time consuming, imperfect and of limited precision [18].
Various approaches have been proposed for breast cancer classification using machine learning. Most of these approaches use magnetic resonance imaging (MRI), mammography and ultrasonography images to detect and classify breast tumors tissue [32], [33], [34], [35], [36]. Machine learning techniques dealing with 1D and 2D NMR spectra that provide peak assignment, chemical shifts prediction and identifying molecular structure have been recently suggested [37], [38], [39], [40], [41], [42], [43], [44]. A recent review listed various approaches that use deep learning in NMR acquisition, spectral reconstruction, de-noising, automated peak picking and chemical shift annotation. However, though they regarded the chemical shift and their corresponding frequencies as the most informative variable in NMR, they reveal that chemical shift is a hard deterministic parameter to be calculated [45]. This study explicitly detects and assigns new metabolites in a crowded 1D NMR spectrum using only the horizontal and vertical frequencies of the 2D TOCSY spectra by employing machine learning.
3. Hypothesis and concept analysis
In our previous published work [44], we established an automated metabolic assignment based on the spectral deconvolution of 2D TOCSY NMR by employing machine learning methods. We customized four semi-supervised learning classifiers and extended them for automatic metabolite assignment of a real breast cancer tissue sample under different training of dataset sizes. The classification results were pooled using the concept of confidence bands, thus, classification results that did not comply with the confidence band values were excluded. Moreover, we constructed a database of metabolites by using a wide range of available 1D NMR metabolic data [46], including the Biological Magnetic Resonance Data Bank (BMRB) [19] and the Human Metabolome Database (HMDB) [20]. The performance of the customized machine learning classifiers was evaluated by comparing the obtained results with those analyzed by NMR specialists. Accordingly, the KNFST and SVM classifiers show better accuracy and smaller mislabeling rates regardless of the sizes of the initially labeled training dataset. On the other hand, under the same settings, cubic and quartic polynomial classifiers were inadequate.
Based on our previous work of employing 2D TOCSY spectra [44], the method is extended to automate the identification of not only known but also potential novel metabolites that might appear due to the dynamicity of living cells.
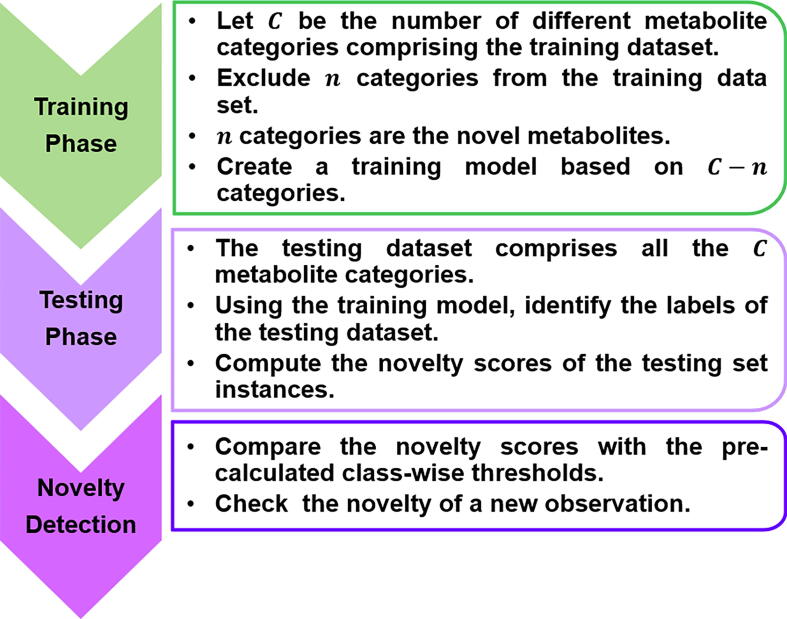
Fig. 1 summarizes the novelty detection protocol: automatic peak picking is performed on the first 2D TOCSY spectra, two characteristic frequencies (F2, F1) are assigned to form the training dataset. The training models will be created based on the KNFST, SVDD and KDE classifiers with different training data volume, observing the classifier performance and the corresponding execution time. The training model will be used in the testing phase to detect novel classes, i.e., novel metabolites in this case. Subsequently, the automatically derived peak picking parameters from the training phase are applied to the second TOCSY. The characteristic frequencies (F2, F1) are studied using the classifiers to identify novel peaks (i.e., metabolites) compared to the reference training models from the previous step. During the testing phase, training models are deployed to assess the novelty of particular metabolites and the success of the learning paradigm.
Fig. 1.
Schematic illustration of the novelty detection procedure in metabolic profiling in a real biological sample based on 2D TOCSY NMR spectra.
4. Experimentations (NMR)
4.1. Breast cancer tissue samples
The breast cancer tissue data used in this work has been previously analyzed and published [30]. The work was part of a comprehensive study in our group focusing on the heterogeneity of cancer tumor tissue. In the study, breast tumor tissue samples from 18 patients were analyzed. After surgery, a specimen for pathological diagnosis was immediately procured and the remaining tissue was snap frozen and stored at –80 °C within 10 min. Six cores each taken from a different patient were analyzed blindly by HR MAS 1H NMR [30], [47].
4.2. NMR data acquisition and processing
As described in [30], [47], 1H NMR measurements were performed using HR MAS probe-head operated by a Bruker Avance III 600 spectrometer at 600.13 MHz for 1H at 276 K. HR MAS spinning frequency was set to 5 kHz, and the magic angle was adjusted typically according to the KBr measurement [30], [47]. The B0 magnetic field shimming was performed manually until the linewidth of the alanine signal at 1.46 ppm adjusted to be within the range of 1.20–1.83 Hz. Metabolites were deduced from the 1H NMR spectrum based on expert knowledge with the assist of 1H, 1H-TOCSY, 13C-1H-HSQS and the Chenomx NMR Analysis Software. Details are reported in [30], [44], [47].
The 1H-1H-TOCSY in this work were specifically recorded with suppressing zero-quantum coherences [48] in order to avoid blurring of the multiple patterns. 1H-1H-TOCSY were measured with a spectral range (SWH) of 7 kHz in both F2 and F1 dimensions. Mixing time and relaxation delay were set to 80 ms and 1 s respectively. Zero filling was performed to 16 K and 128 data points in F2 and F1 dimensions before the 2D Fourier transform applied [30], [44], [47]. The spectral widths in F2 and F1 dimensions were 12.00 ppm, while the spectral width of 9.0 ppm (5600 Hz) is an enlargement of the area of interest in the TOCSY (cross-peaks of the metabolites appeared). The 1D NMR spectral projections on the F2 and F1 axes are external projections from extra 1D NMR measurement using the CPMG pulse sequence with embedded water suppression by excitation sculpting. CPMG was used to suppress protein, lipids and other macromolecules and it was recorded employing 400 echoes with 1 ms echo time. NMR spectra acquisition and processing were achieved by using the TopSpin software package 3.6.
4.3. TOCSY crosspeak picking and de-noising
The cross-peaks entries in F2 and F1 dimensions in ppm and Hz are deduced from the 2D contour lines of the experimental 2D TOCSY NMR spectrum by employing the automatic peak picking function (pp2d) in TopSpin 3.6 provided by Bruker for acquisition and processing. Before applying automatic peak picking, the contour projection magnitude threshold was adjusted for every ppm range in F2 dimension according to the amplitude of the 1D NMR spectrum internal projection on F2 axis to avoid picking artifacts and noise crosspeaks. Afterward, the collected peaks were listed and transferred as text file for data de-noising and artifact cross-peak elimination. In a TOCSY spectrum, every real cross-peak appearing in the upper diagonal (F2, F1) due to the J-coupling should have a mirror (transpose) crosspeak in the lower diagonal (F2, F1) within tolerance threshold of ∼ 30 Hz, based on that we could exclude cross peaks that do not fulfill this criterion. Moreover, most crosspeaks in vicinity of water and solvent signals are associated with t1-noise [49]. Fortunately, t1-noise appears in TOCSY spectrum as random or semi-random spurious streaks along the indirect F1 dimension of a 2D NMR spectrum and they have no transpose (mirror) in the lower diagonal entries (F2, F1). Typically, no metabolite signals in vicinity are taken for assignment, since other characteristic peaks in different F2 and F1 ranges can be considered. It is worth mentioning that, metabolites that have no coupled protons will show singlet signals in 1D NMR and therefore, no crosspeaks in TOCSY. Such signals will only have contour projections in the diagonal. Typically, 2D TOCSY spectra provide information about correlated protons of the same spin system. However, peaks in the diagonal can be used as a part of the data to solve the issue of metabolites with no intrinsic coupling if they are not severely overlapping. A spectroscopic more favorable approach would be correlation measurements between 1H–13C as in HSQC [50], [51].
5. Machine learning and novelty detection
Machine learning is a set of methods used to automatically distinguish between patterns and then uses its knowledge to detect future patterns or to make decisions with some uncertainty without explicit programming [52]. A machine learning system uses three types of datasets: The first data type is the training dataset, which is the labeled training data used to build a generalization model. The second data type is the testing dataset, which is the unlabeled data that is to be learned [26]. To tune the parameters of the classifiers, the validation dataset is used. Importantly, all datasets must belong to the same distribution, but the testing dataset is still unknown to the classifier during the training phase. In cases where new categories of data appear during the testing phase, novelty detection is used. Novelty detection is assorted into distance-, probabilistic-, or domain-based approaches [15]. Distance-based methods learn a distance metric to identify the similarity between different samples. They use the assumption that similar data are located near each other, while novel instances are located far away from known data. Probabilistic methods are based on using density estimation of the data to distinguish normal reference from abnormal unknown instances. Domain-based methods try to describe boundaries that encloses the training data and typically ignore the class density. Depending on the location of the sample with respect to the boundary, the class membership can be determined [15]. The output of the classification algorithms takes the form of a score or a measure that determines the class membership of the test sample. Scores represent the degree of normality or novelty of a data sample. Thresholds are incorporated on the novelty scores as boundaries to differentiate between known and unknown samples [53].
In this work, for the purpose of novelty detection, the following three classifiers are studied:
5.1. Kernel null Foley-Sammon transform
The Kernel Null Foley-Sammon transform (KNFST) is a distance-based method which computes the projection distance in the null space by decreasing the within-scatter between the similar classes while increasing the between-scatter between dissimilar classes. KNFST maps the input feature space with C classes into a low-dimensional embedding, called the null space projection , such that the null space is spanned by null projection directions [54]. KNFST is based on the Fisher discriminant criterion which can be defined as.
| (1) |
where and are the between-class and the within-class scatter, respectively, in a mapped high-dimensional space , i.e., kernel. KNFST tries to achieve the best separation between classes based on the following conditions [54], [55]:
-
•
A zero within-class scatter ;
-
•
A positive between-class scatter .
As a result, using KNFST, samples that belong to the same class are mapped to one point but samples that belong to different classes are mapped to different points.
Following the above conditions, we get [56] and Eq. (1) turns into the maximization problem [54]:
| (2) |
which tries to find the null projection direction matrix of KNFST ensuring the above conditions. KNFST is a joint multi-class model, which is able to achieve classification of all classes at once. The output of KNFST is used as a novelty score, where the larger the novelty score, the more novel is the test sample. A threshold is set to detect novelty borders. KNFST has been used in image classification [54], [57], gesture recognition [58], abnormal event detection in object tracking [59], authentication on mobile devices [60] and fault detection in machinery [61]. In this work, the KNFST code implementation in [54] has been customized.
5.2. Support Vector data Description
Support Vector Data Description (SVDD) is a domain-based method, which employs a hyperplane to represent a boundary based on the training data. This hyperplane tries to maximize the separation between different classes. SVDD was developed by [62] as a one-class classifier that distinguishes a positive (normal) class from all other classes in the dataset and builds its model based on the single positive class [63]. This approach creates a minimum-volume spherically shaped region that encompasses all or most of the training data of a chosen class. The hypersphere acts as a descriptor of normality and a sample is considered an outlier if it falls outside the sphere [63], [64]. The problem of SVDD is an optimization problem that finds the center with minimum radius of the hypersphere that encloses most of the training data. SVDD enables the existence of outliers outside of the hypersphere, but a larger distance from the hypersphere is penalized in Eq. (3).
| (3) |
is a slack variable that permits the existence of outliers, is a parameter that controls the trade-off between the volume of the radius and the number of outliers (set to 1%), and is the high dimensional mapping of [65]. In this work, the binary classification implemented in the Novelty Detection Toolbox (NDtool) [15], [66] is extended to a multi-class approach using one--all classification. SVDD has several applications in image and gesture classification [67], [68], [69], [70], [71], biomarker detection in HSQC NMR spectroscopy [72], and fault detection [73], [74].The novelty threshold of SVDD is defined as the radius of the hypersphere according to [62].
5.3. Kernel density estimation
Kernel density estimation (KDE) is a probability-based method which computes the probability at each point in the data space within a localized neighborhood area of that point. KDE is a non-parametric approach that tries to estimate the probability of unknown distributions. The main assumption of density estimation is that samples reside in low-density areas indicate a low probability of being a known class. Accordingly, this area tends to contain novel data; whereas areas of high probability means the existence of known samples [15]. The probability density function is approximated by estimating the probability density through locating kernels at each point of the dataset, i.e., a kernel is centered at each data point, and then these kernels are summed up. A typical kernel density estimation is the Parzen Window estimator [65]. The Parzen estimator defines a fixed-width region centered at the sample point x and counts the number of neighboring sample points which fall in this region. Parzen estimators can be defined as:
| (4) |
where , is the number of data samples and are the region centers which are sampled from X. The density of is calculated based upon the distance between and and then representing it as a linear combination of the neighboring kernel centers. is a kernel function centered at and has an associated parameter related to the bandwidth parameter of region [75]. A common kernel choice is a multivariate Gaussian kernel function. Combing and Eq. (4) we get
| (5) |
Eq. (5) is the class conditional probability for class. The variable is the dimensionality of the features space and the parameter is the standard deviation of the Gaussian component and can be identified as the Parzen window width. The Parzen width parameter is defined as the mean value of the distances between each and its nearest neighbours. Since the probability must sum up to 1, we normalize the density by where is the number of data points that belong to class [65], [76]. KDE has been employed in tissue segmentation [77], [78], Alzheimer's disease detection in MRI [79], [80] and CT images [81], [82]. In this work, the binary classification implementation in NDtool [15], [66] has been extended to a multi-class approach using one--all classification.
5.4. Threshold setting and novelty detection
Classifiers are designed to assign the already known classes and, consequently, match the novel data sample to one of the known classes. Novelty detection tries to learn a model of normality, which is described by a novelty boundary. Normal instances are expected to be included in the normality model and reside within the novelty boundary, whereas unknown instances are expected to lie outside these boundaries [83]. A validation dataset is used to compute the novelty threshold for each known class in advance by finding the threshold with the minimum error on a validation dataset using grid search. During the testing phase, when classifying a data point, the threshold is compared to the output of the corresponding classifier. If the output does not comply with the pre-computed threshold, the data sample will be classified as novel. KNFST is a distance-based approach, which uses the assumption that similar data are located near each other, while novel instances are located away from known data. Thus, if the distance between the tested samples is larger than the novelty threshold of the class, the test sample is classified as novel, i.e., . This is also valid for SVDD, where the radius of the hypersphere indicates the threshold. For KDE, if the posterior probability is below the threshold , the more probable the test sample is a novel instance, [83], [84].
6. Dataset
1D NMR and 2D TOCSY spectra were acquired according to the settings described in [44]. The metabolite dataset is a two-dimensional dataset which includes the horizontal and vertical chemical shift frequencies of the 2D TOCSY. Data augmentation is used to generate a more comprehensive set of probable data. This improves the size, variety and description of the training datasets [85]. Data augmentation simulates the estimated shifts from the original frequencies, resulting in replications of the samples. Consequently, classifiers will treat the same metabolite in different varieties [86]. Data augmentation is used to generate disjoint datasets of training and validation data sets. Following the procedure described in [44], in the training dataset, white Gaussian noise is added to the standard’s chemical shifts with different signal-to-noise ratios [11]. The validation dataset is created by deviating the chemical shift frequencies by a random shift assuming a chemical shift constraint within 30 Hz (0.049 ppm), which is sufficient to model the chemical shift variability caused by the NMR environmental matrix change [87]. An example of the data augmentation procedure for tyrosine is shown in Table 1.
Table 1.
A subset of the training dataset showing the output of the data augmentation procedure for tyrosine.
| Metabolite | Standard From J-coupling |
Experimental TOCSY |
Augmented Generated |
|||
|---|---|---|---|---|---|---|
|
F2 [Hz] |
F1 [Hz] |
F2 [Hz] |
F1 [Hz] |
F2 [Hz] |
F1 [Hz] |
|
| Tyrosine | 4303 | 4128 | 4316 | 4139 | 4320 | 4139 |
| 2353 | 1914 | 2362 | 1920 | 4317 | 4134 | |
| 4315 | 4140 | |||||
| 2363 | 1922 | |||||
| 2361 | 1921 | |||||
| 2363 | 1919 | |||||
| …… | …… | |||||
7. Novelty detection of metabolites using breast cancer tissue
The classifiers KNFST, SVDD and KDE are customized and tested for novelty detection. The training data of size 29402, where 2940 is the number of independent samples from all classes and 2 is the dimension of the data, representing the horizontal and vertical frequencies, is partitioned into eight portions. These portions are used to test the system using different percentages of training data to observe the relation between the performance and the availability of training data and to examine the minimum size of the training set sufficient to yield a satisfactory performance. The portion of labeled training samples is increased every 50 cycles until all training samples are used in the classification process. In each cycle, different random permutations of training data are applied. The introduction of multiple cycles is vital; this is due to the random selection of the training data before starting the recognition process, which leads to different results for each chosen training dataset. Training portions of sizes 2.5%, 5%, 7.5%, 10%, 25%, 50%, 75% and 100% of the total training dataset size were used. In this experiment, a TOCSY spectrum of a breast cancer tissue sample, which contains the metabolites: Val: Valine; Ile: Isoleucine; Leu: Leucine; Lys: Lysine; Glu: Glutamate; Ala: Alanine; Gln: Glutamine; Asp: Aspartate; GPC: sn‐glycero‐3-phosphocholine; Ser: serine; PE: O-phosphoethanolamine; Asc: ascorbate; mIno: myo-Inositole; Lac: Lactate; Pro: Proline; HB: 3-Hydroxybutyrate; PCho: O-Phosphocholine; Thr: Threonine; GSH: Glutathione; β-Glucose; α-Glucose; Ino: Inosine; Tyr: Tyrosine; Phe, phenylalanine; Tau: Taurine; Ura: Uracil; and Met: methionine is used [44]. The experimentally determined frequencies for the metabolites were added to the supplementary material. Fig. 2 shows the feature space of the metabolites contained in training dataset. It can be observed that the frequency overlaps in the horizontal and vertical axes and cannot be linearly separated.
Fig. 2.
The feature space of the training dataset for 27 metabolites deduced from the TOCSY spectrum of a breast cancer tissue. The insets are selected enlargements of peaks that overlap in (F2, F1) dimensions.
To test novelty detection on the TOCSY spectrum of breast cancer tissue, two scenarios are applied. The first scenario handles the one-class novelty detection case. This experiment is built by excluding one of the metabolites from the training dataset, and afterwards a training model is built based on the remaining 26 metabolites. The testing dataset includes all 27 metabolites, which are the known 26 metabolites plus the excluded metabolite. On the second experiment, multi-class novelty detection is employed by excluding multiple metabolites from the training set, and a training model is built based on the remaining metabolites. Subsequently, during the test phase the novelty scenario is tested based on the known and the excluded metabolites. In both scenarios, the classifiers are expected to detect the excluded metabolites and regard them as novel metabolites. The procedure is illustrated in Fig. 3.
Fig. 3.
Novelty detection procedure by excluding one- and multi-metabolites from the pre-assigned 27 metabolites of the breast cancer tissue.
The assessment of the results is based on the novelty detection metrics used in [88]. The first metric is the percentage of novel metabolites misclassified as known (i.e., missed novelties). The second metric is the percentage of existing instances falsely misclassified as novel (i.e., percentage of wrong detections). The final metric is the percentage of total error, where is the number of novel metabolites misclassified as known metabolites (i.e., false negative). stands for the number of known metabolites misclassified as novel metabolites (i.e., false positives) and denotes the misclassifications within known metabolites, N is the total number of metabolites in the test dataset and is the number of novel metabolites in the test dataset.
7.1. One-class novelty detection
In the scenario of one-class novelty detection, the metabolite entry (tyrosine) is considered novel by excluding it from the list of 27 metabolites. Consequently, the training dataset consists of the remaining metabolites whereas the testing dataset includes the excluded novel metabolite tyrosine in addition to the known training data. Excluding a metabolite during the training process simulates the novelty of the excluded metabolite and ascertains that the training model is only aware of all metabolites excluding the exempted tyrosine. In breast cancer, tyrosine is the most frequent reported metabolic biomarker [89].
Fig. 4(a-c) show the results of the novelty detection procedure of the classifiers using the above assessment matrices for the metabolite tyrosine. Fig. 4a shows that KNFST has a zero rate regardless of the size of the training dataset, which means that tyrosine was correctly identified as novel. However, when using 2.5% of training data, in addition to misclassifying some known classes as novel classes, misclassification between known classes have a median error of 4%. On the other hand, using 2.5% of the training dataset, KDE and SVDD (Fig. 4b and 4c) have a value of around 4% and 50%, respectively, with a relatively high standard deviation. Both classifiers show zero values after using only 5% of the training dataset. In general, for all classifiers it can be seen that the values of and decrease when increasing the size of training samples. All classifiers achieve zero or near-zero values for, and when using 5% of the complete training dataset.
Fig. 4.
The ,and values of breast cancer tissue sample for the classifiers (a) KNFST, (b) KDE and (c) SVDD by applying one-class novelty detection.
To test the overall performance of the system for all possible threshold settings, we use Receiver Operating Characteristic (ROC) curve analysis to show the tradeoff between false positives and true positives. ROC curves and Area under Curve (AUC) provide an assessment of the classification performance without indicating a decision threshold [90]. Fig. 5 shows ROC curves which are generated using the one--all approach for one run. This involves training one class per classifier, considering samples that belong to this particular class as normal samples and all other samples as novel [91]. As mentioned earlier, training portions of sizes 2.5%, 5%, 7.5%, 10%, 25%, 50%, 75% and 100% of the total training dataset size were used, nevertheless, for clarity only portions of sizes 2.5%, 10%, 100% are shown in the ROC curves, novelty scores and thresholds figures. These percentages give an indication of the performance using relatively small, medium and large amounts of training data. In general, it can be seen in Fig. 5(a-c) that the classifiers’ capability to distinguish novel metabolites from known metabolites increases by increasing the size of the training dataset. This can also be observed by the increasing values of the AUC, which implies a high diagnostic accuracy for large training data set sizes. Furthermore, it can be deduced that using 2.5% of the training data results in an inaccurate threshold, and consequently in a low recognition rate. By using 10% of the total training samples, the AUC of ROC curve of the metabolite tyrosine was over 97% for all classifiers. The AUC of the ROC curves are close to 100% for the three classifiers when using 100% of the training data.
Fig. 5.
ROC curves and AUC values showing the accuracy of the novelty threshold for different sizes of training data for the metabolite tyrosine. From left to right, the ROC curve obtained using (a) 2.5%, (b) 10% (b) and (c) 100% of the total training dataset is shown.
Fig. 6 shows the corresponding difference in novelty scores between known and unknown metabolites related to Fig. 5. The decision threshold is calculated using the validation data and plotted as a dotted line. The red, green and blue crosses resemble the unknown test data, known test data and known training data, respectively. It can be seen that the separation between the known and the unknown instances becomes more representative by increasing the training data size.
Fig. 6.
Novelty scores and threshold values of KNFST, KDE and SVDD classifiers using different training dataset sizes in the one-class novelty detection scenario (applied to tyrosine, which has two instances as shown on the supplementary material). The red, green and blue crosses resemble the unknown test data, known test data and known training data, respectively. Subfigures (a) to (c) correspond to the variations of the output of the classifiers when using (a) 2.5%, (b) 10% and (c) 100% of the training dataset. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
7.2. Multi-class novelty detection
Metabolites (leucine, tyrosine, proline and serine) are a subset of the clinically most frequently reported metabolic biomarkers related to breast cancer [89]. Therefore, in the multi-class novelty detection the above-mentioned four metabolites were chosen to be excluded for novelty testing under different conditions. Accordingly, the classifiers were trained on 23 metabolites only. During the test phase, all assigned 27 metabolites of the breast cancer sample were included in the test dataset, likewise the one-class novelty detection.
Fig. 7 shows the,and values in multi-class novelty detection scenario. When using 2.5% of the training data, KNFST and SVDD have similar median values around 16%. The SVDD distribution shows a negative skewness, which means most values are low. Although KDE has a median of zero, KDE and the other classifiers have a high standard deviation. This means a low discrimination capability at extremely low training dataset size. Similarly, the values of and showed unstable standard deviations and median values in all classifiers. Starting from 5% training data size, KNFST showed a negative skewness in values, which implies a progressing discrimination of novel metabolites. On the other hand, KDE and SVDD have zero for and approximately zero value for and . Starting from 50% of the training data size, a median of zero values were reached for KNFST. Using only 25% of the training data, all of the classifiers have reached less than 3% median values for, and values. In addition, already when using only 5% of the training data, all classifiers reached near-zero median values of and , indicating that the classifiers are able to correctly classify known metabolites and detect novel instances.
Fig. 7.
,and values of breast cancer tissue sample for the classifiers (a) KNFST, (b) KDE and (c) SVDD by applying multi-class novelty detection.
Fig. 8(a-c) shows novelty scores of the KNFST, KDE and SVDD classifiers using 2.5%, 10%, and 100% training dataset size by applying the multi-class novelty detection. The red crosses correspond to the six-pattern related to tyrosine, proline, leucine and serine. The validation data are used to compute the threshold for each individual class. For clarity reasons, instead of plotting individual thresholds, the median of the thresholds for each class is plotted as a black dotted line. Comparable to one-class novelty detection, the novelty threshold becomes more accurate and the separation between normal and abnormal instances becomes more distinct when increasing the training dataset size. Remarkably, an acceptable threshold could be calculated even when only 10% of the training data were considered.
Fig. 8.
Novelty scores and threshold values of KNFST, KDE and SVDD classifiers using different training data sizes for multi-class novelty detection. The red, green and blue crosses resemble the unknown test data, known test data and known training data, respectively. The output of the classifiers is shown for (a) 2.5%, (b) 10% and (c) 100% of the training dataset. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Unlike one-class classification, generating ROC curves for multi-class classification tasks is not a straightforward problem. A typical solution is to generate individual ROC curves for each class separately using the one-vs-all method [90].
Fig. 9 shows the mean and standard deviation of the total classification processing time of 50 runs in one- and multi-class novelty detection. The experiments were run on Windows 10 using an Intel Xeon E5 machine with 16 GB memory and 2.8 GHz Quad Core CPU. The computational complexity for KDE is [92], and for KNFST [54] and SVDD [93]. The execution time for KNFST and SVDD grows when increasing the amount of training data. In one-class novelty detection, the execution time for KNFST increases steadily until it exceeds the SVDD execution time. However, rather than increasing, the execution time for one- and multi-class novelty in KDE remains almost constant when increasing the size of the training dataset. This might be due to the fixed Parzen window width of the kernel used by KDE. The estimation of the optimal Parzen window width is the most effecting computational factor [92]. As stated earlier, the Parzen width parameter is defined as the mean distance between the -nearest neighbours and the instances in the training dataset. The number of neighbours in our experiments was two [94]. In SVDD, computational cost is related to tuning the parameters of the kernel, and there is a direct relation between the size of the training dataset and the execution time [95]. This can be seen on SVDD time consumption on multi-class novelty detection where, in comparison to the one-class scenario, more novel samples are encountered. The main computational cost in KNFST comes from computing a joint kernel feature space for all known classes and the eigenvalue decomposition of the kernel matrix [54], [96].
Fig. 9.
Total time from training to classification for (a) one-class novelty detection and (b) multi-class novelty detection.
The confusion matrices of one- and multi-class novelty, in addition to the ROC curves for the multi-class novelty detection algorithm, are presented in the supplementary material of this work. The confusion matrix is used to describe the performance of the classification algorithm in terms of true positive, true negative, false positive and false negative values.
8. Conclusions
In this work, the novelty detection was established based on 2D NMR TOCSY spectra for metabolic profiling associated to dynamics changes in biological systems, where metabolites of real breast-cancer tissue samples were extracted from the TOCSY. The one- and multi-class novelty detection tests were designed to consider peak assignments appearing in the TOCSY spectrum as a reference database. Subsequently, one and four metabolites were excluded from the reference TOCSY to simulate their novelty. The KNFST, KDE and SVDD classifiers were tested to detect the excluded metabolites. The classifiers achieved explicit labelling to metabolites that appear in the TOCSY and additionally detected new metabolites which are unknown to the training model. Despite the observed overlapping in the training dataset resulting from chemical shifts, the implemented methods in this work achieved 0% false positive rates at 100% true positive rate. The resulting classification performance increases with increasing training dataset size. Generally, the execution time also increases when increasing the training dataset size for all classifiers, nevertheless, the execution time is relatively short. The results are supported by confusion matrices and ROC curves in addition to plotting the novelty outputs. The presented machine learning based novelty detection techniques provide promising perspectives for automated assignment of metabolites that evolve in dynamic biological environments and triggers the metabolic pathways. For future strategies, creating a more comprehensive and standardized metabolic database using ppm, horizontal and vertical frequencies designed for different NMR resolution frequency is essential to stimulate an uncomplicated access to diverse NMR data. This perspective is critical due to the heterogeneity of metabolites and the associated variables and implication. Furthermore, a new feature which is related to spin–spin couplings can be added to the two already existing features to increase the discriminative strength. Moreover, additional 2D NMR methods such as HMBC or HSQC can be employed and integrated in the automatic prediction. The output of the classification using different techniques might then be combined as ensemble classification to generate more accurate results on more complex mixtures.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Financial support by the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, the Senatsverwaltung für Wirtschaft, Technologie und Forschung des Landes Berlin, the Bundesministerium für Bildung und Forschung and the German Academic Exchange Service (DAAD Project no. 57587918) is acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.05.050.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References:
- 1.Beckonert O., et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2(11):2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 2.Dona A.C., et al. A guide to the identification of metabolites in NMR-based metabonomics/metabolomics experiments. Comput Struct Biotechnol J. 2016;14:135–153. doi: 10.1016/j.csbj.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puchades-Carrasco L., et al. Bioinformatics tools for the analysis of NMR metabolomics studies focused on the identification of clinically relevant biomarkers. Briefings Bioinf. 2016;17(3):541–552. doi: 10.1093/bib/bbv077. [DOI] [PubMed] [Google Scholar]
- 4.Johnson C.H., et al. Xenobiotic metabolomics: major impact on the metabolome. Annu Rev Pharmacol Toxicol. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wellen K.E., et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakahata Y., et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X., et al. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell. 2010;143(4):639–650. doi: 10.1016/j.cell.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubbard T.D., et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5(1):12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma U., Jagannathan N.R., Using B.C.M., NMR, in NMR-Based Metabolomics: Methods and Protocols, G.A.N. Gowda and D. Raftery, Editors . New York, NY; New York: 2019. Springer; pp. 195–213. [Google Scholar]
- 10.Vignoli A., et al. Precision Oncology via NMR-Based Metabolomics: A Review on Breast Cancer. Int J Mol Sci. 2021;22(9):4687. doi: 10.3390/ijms22094687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao J., et al. Bayesian deconvolution and quantification of metabolites in complex 1D NMR spectra using BATMAN. Nat Protoc. 2014;9(6):1416–1427. doi: 10.1038/nprot.2014.090. [DOI] [PubMed] [Google Scholar]
- 12.Güntert P. Automated structure determination from NMR spectra. Eur Biophys J. 2008;38(2):129. doi: 10.1007/s00249-008-0367-z. [DOI] [PubMed] [Google Scholar]
- 13.Ching T., et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15(141):20170387. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson M.P., Craven C.J. Automated protein structure calculation from NMR data. J Biomol NMR. 2009;43(3):131–143. doi: 10.1007/s10858-008-9295-6. [DOI] [PubMed] [Google Scholar]
- 15.Pimentel M.A.F., et al. A review of novelty detection. Signal Process. 2014;99:215–249. [Google Scholar]
- 16.Miljković D. Review of novelty detection methods. 2010:593–598. [Google Scholar]
- 17.Roberts S.J. Novelty, Detection using Extreme Value Statistics. 1999 [Google Scholar]
- 18.van der Hooft J.J.J., Rankin N. In: Modern Magnetic Resonance. Webb G.A., editor. Springer International Publishing; Cham: 2018. Metabolite Identification in Complex Mixtures Using Nuclear Magnetic Resonance Spectroscopy; pp. 1309–1341. [Google Scholar]
- 19.Ulrich E.L., BioMagResBank, et al. Nucleic Acids Res. 2007;36(Database):D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wishart D.S., et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35(Database):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weljie A.M., et al. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78(13):4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 22.Bingol K., et al. Customized Metabolomics Database for the Analysis of NMR 1H–1H TOCSY and 13C–1H HSQC-TOCSY Spectra of Complex Mixtures. Anal Chem. 2014;86(11):5494–5501. doi: 10.1021/ac500979g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia J., et al. MetaboMiner – semi-automated identification of metabolites from 2D NMR spectra of complex biofluids. BMC Bioinf. 2008;9(1):507. doi: 10.1186/1471-2105-9-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tulpan D., et al. MetaboHunter: an automatic approach for identification of metabolites from 1H-NMR spectra of complex mixtures. BMC Bioinf. 2011;12(1):400. doi: 10.1186/1471-2105-12-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wishart D.S. Advances in metabolite identification. Bioanalysis. 2011;3(15):1769–1782. doi: 10.4155/bio.11.155. [DOI] [PubMed] [Google Scholar]
- 26.Bingol K., Brüschweiler R. Knowns and unknowns in metabolomics identified by multidimensional NMR and hybrid MS/NMR methods. Curr Opin Biotechnol. 2017;43:17–24. doi: 10.1016/j.copbio.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang B., et al. Novel Metabolic Signatures of Prostate Cancer Revealed by 1H-NMR Metabolomics of Urine. Diagnostics. 2021;11(2):149. doi: 10.3390/diagnostics11020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chessa M., et al. Urinary Metabolomics Study of Patients with Bicuspid Aortic Valve Disease. Molecules. 2021;26(14):4220. doi: 10.3390/molecules26144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosmopoulou M., et al. Human Melanoma-Cell Metabolic Profiling: Identification of Novel Biomarkers Indicating Metastasis. Int J Mol Sci. 2020;21(7):2436. doi: 10.3390/ijms21072436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gogiashvili M., et al. Impact of intratumoral heterogeneity of breast cancer tissue on quantitative metabolomics using high-resolution magic angle spinning 1H NMR spectroscopy. NMR Biomed. 2018;31(2) doi: 10.1002/nbm.3862. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Perez I., et al. Identifying unknown metabolites using NMR-based metabolic profiling techniques. Nat Protoc. 2020;15(8):2538–2567. doi: 10.1038/s41596-020-0343-3. [DOI] [PubMed] [Google Scholar]
- 32.Huang Q., et al. On Combining Biclustering Mining and AdaBoost for Breast Tumor Classification. IEEE Trans Knowl Data Eng. 2020;32(4):728–738. [Google Scholar]
- 33.Huang Q., et al. Segmentation of breast ultrasound image with semantic classification of superpixels. Med Image Anal. 2020;61 doi: 10.1016/j.media.2020.101657. [DOI] [PubMed] [Google Scholar]
- 34.Devi, D.H. and D.M.I. Devi. Outlier Detection Algorithm Combined With Decision Tree Classifier For Early Diagnosis Of Breast Cancer R. 2016.
- 35.Khan S., et al. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recogn Lett. 2019;125:1–6. [Google Scholar]
- 36.Xie T., et al. Machine Learning-Based Analysis of MR Multiparametric Radiomics for the Subtype Classification of Breast Cancer. Frontiers. Oncology. 2019;9(505) doi: 10.3389/fonc.2019.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J., et al. NMR-TS: de novo molecule identification from NMR spectra. Sci Technol Adv Mater. 2020;21(1):552–561. doi: 10.1080/14686996.2020.1793382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paruzzo F.M., et al. Chemical shifts in molecular solids by machine learning. Nat Commun. 2018;9(1):4501. doi: 10.1038/s41467-018-06972-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klukowski P., et al. NMRNet: a deep learning approach to automated peak picking of protein NMR spectra. Bioinformatics. 2018;34(15):2590–2597. doi: 10.1093/bioinformatics/bty134. [DOI] [PubMed] [Google Scholar]
- 40.Jonas E., Kuhn S. Rapid prediction of NMR spectral properties with quantified uncertainty. J Cheminf. 2019;11(1):50. doi: 10.1186/s13321-019-0374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng, W.K., Clustering NMR: Machine learning assistive rapid (pseudo) two-dimensional relaxometry mapping. bioRxiv, 2020: p. 2020.04.29.069195.
- 42.Peng W.K., Ng T.-T., Loh T.P. Machine learning assistive rapid, label-free molecular phenotyping of blood with two-dimensional NMR correlational spectroscopy. Communications Biology. 2020;3(1):535. doi: 10.1038/s42003-020-01262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houhou R., Bocklitz T. Trends in artificial intelligence, machine learning, and chemometrics applied to chemical data. Analytical Science Advances. 2021;2(3–4):128–141. doi: 10.1002/ansa.202000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Migdadi L., et al. Automated metabolic assignment: Semi-supervised learning in metabolic analysis employing two dimensional Nuclear Magnetic Resonance (NMR) Comput Struct Biotechnol J. 2021;19:5047–5058. doi: 10.1016/j.csbj.2021.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D., et al. Review and Prospect: Deep Learning in Nuclear Magnetic Resonance Spectroscopy. Chemistry – A. European Journal. 2020;26(46):10391–10401. doi: 10.1002/chem.202000246. [DOI] [PubMed] [Google Scholar]
- 46.Van Q.N., et al. Comparison of 1D and 2D NMR Spectroscopy for Metabolic Profiling. J Proteome Res. 2008;7(2):630–639. doi: 10.1021/pr700594s. [DOI] [PubMed] [Google Scholar]
- 47.Gogiashvili M., et al. HR-MAS NMR Based Quantitative Metabolomics in Breast Cancer. Metabolites. 2019;9(2) doi: 10.3390/metabo9020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thrippleton M.J., Keeler J. Elimination of zero-quantum interference in two-dimensional NMR spectra. Angew Chem Int Ed. 2003;42(33):3938–3941. doi: 10.1002/anie.200351947. [DOI] [PubMed] [Google Scholar]
- 49.Mo H., et al. A simple method for NMR t1 noise suppression. J Magn Reson. 2017;276:43–50. doi: 10.1016/j.jmr.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mavel S., et al. 1H–13C NMR-based urine metabolic profiling in autism spectrum disorders. Talanta. 2013;114:95–102. doi: 10.1016/j.talanta.2013.03.064. [DOI] [PubMed] [Google Scholar]
- 51.Rai R.K., Sinha N. Fast and accurate quantitative metabolic profiling of body fluids by nonlinear sampling of 1H–13C two-dimensional nuclear magnetic resonance spectroscopy. Anal Chem. 2012;84(22):10005–10011. doi: 10.1021/ac302457s. [DOI] [PubMed] [Google Scholar]
- 52.Murphy, K.P., Machine Learning: A Probabilistic Perspective. 2012: MIT Press.
- 53.Clark J., Liu Z. Japkowicz; Adaptive Threshold for Outlier Detection on Data Streams: 2018. and N. [Google Scholar]
- 54.Bodesheim P., et al. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2013. Kernel Null Space Methods for Novelty Detection. [Google Scholar]
- 55.Zheng W., Zhao L., Zou C. Foley-Sammon optimal discriminant vectors using kernel approach. IEEE Trans Neural Networks. 2005;16(1):1–9. doi: 10.1109/TNN.2004.836239. [DOI] [PubMed] [Google Scholar]
- 56.Guo Y.-F., et al. Rapid and brief communication: Null Foley-Sammon transform. Pattern Recogn. 2006;39(11):2248–2251. [Google Scholar]
- 57.Wang J., et al. IEEE; 2016. Evaluating features for person re-identification. [Google Scholar]
- 58.Luo Y., et al. Manifold learning for novelty detection and its application in gesture recognition. Complex & Intelligent Systems. 2022:1–12. [Google Scholar]
- 59.Shi Y., et al. Kernel null-space-based abnormal event detection using hybrid motion information. J Electron Imaging. 2019;28(2) [Google Scholar]
- 60.Oza P., Patel V.M. in 2021 IEEE International Joint Conference on Biometrics (IJCB) 2021. Federated Learning-based Active Authentication on Mobile Devices. [Google Scholar]
- 61.Tian Y., et al. A subspace learning-based feature fusion and open-set fault diagnosis approach for machinery components. Adv Eng Inf. 2018;36:194–206. [Google Scholar]
- 62.Tax D.M.J., Duin R.P.W. Support vector data description. Machine Learning. 2004;54(1):45–66. [Google Scholar]
- 63.Khan S.S., Madden M.G. One-class classification: taxonomy of study and review of techniques. Knowledge Engineering Review. 2014;29(3):345–374. [Google Scholar]
- 64.Khan S.S., Madden M.G., Survey A., of Recent Trends in One Class Classification. in Artificial Intelligence and Cognitive Science . Springer, Berlin Heidelberg; Heidelberg: 2010. Berlin. [Google Scholar]
- 65.Bishop C.M., Recognition Pattern, Learning Machine. Springer-Verlag; Heidelberg: 2006. Berlin. [Google Scholar]
- 66.Clifton, L.A., Multi-channel novelty detection and classifier combination. 2007: The University of Manchester (United Kingdom).
- 67.Chenge D., et al. in 2021 6th International Conference on Intelligent Computing and Signal Processing (ICSP) 2021. Gesture Classification Algorithm Based on SVDD-EPF. [Google Scholar]
- 68.Zhihong Z., et al. in 7th International Conference on Automatic Face and Gesture Recognition (FGR06) 2006. One-class classification for spontaneous facial expression analysis. [Google Scholar]
- 69.Al-Behadili, H., et al., Incremental Class Learning and Novel Class Detection of Gestures Using Ensemble. 2015.
- 70.Yoshida T., Kitamura T., Machines Semi-Hard Margin Support Vector, for Personal Authentication with an Aerial Signature Motion. in Artificial Neural Networks and Machine Learning – ICANN . Springer International Publishing; Cham: 2021. 2021. [Google Scholar]
- 71.Na X.G.J.Y.C. New medical image classification approach based on hypersphere multi-class support vector data description. Journal of Computer Applications. 2013;33(11):3300. [Google Scholar]
- 72.Belghith, A., C. Collet, and J.P. Armspach. Detection of Biomarker in Biopsies Based on Hr-Mas 2D HSQC Spectroscopy Indexation. in 4th International Conference on Biomedical Engineering in Vietnam. 2013. Berlin, Heidelberg: Springer Berlin Heidelberg.
- 73.Zhao Y., Wang S., Xiao F. Pattern recognition-based chillers fault detection method using support vector data description (SVDD) Appl Energy. 2013;112:1041–1048. [Google Scholar]
- 74.Chen M.-C., et al. An efficient ICA-DW-SVDD fault detection and diagnosis method for non-Gaussian processes. Int J Prod Res. 2016;54(17):5208–5218. [Google Scholar]
- 75.Qin, X., et al. Scalable Kernel Density Estimation-based Local Outlier Detection over Large Data Streams. 2019.
- 76.Clifton D.A. University of Oxford; St. Cross Colleg: 2009. Novelty Detection with Extreme Value Theory in Jet Engine Vibration Data. [Google Scholar]
- 77.Wu S., et al. Automated fibroglandular tissue segmentation and volumetric density estimation in breast MRI using an atlas-aided fuzzy C-means method. Med Phys. 2013;40(12) doi: 10.1118/1.4829496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Veluppal A., et al. Automated differentiation of Alzheimer’s condition using Kernel Density Estimation based texture analysis of single slice brain MR images. Current Directions in Biomedical Engineering. 2021;7(2):747–750. [Google Scholar]
- 79.YAN, H., et al. Volumetric magnetic resonance imaging classification for Alzheimer's disease based on kernel density estimation of local features. Chin Med J. 2013;126(09):1654–1660. [PubMed] [Google Scholar]
- 80.Sadhukhan D., et al. LATERAL VENTRICLE TEXTURE ANALYSIS IN ALZHEIMER BRAIN MR IMAGES USING KERNEL DENSITY ESTIMATION. Biomed Sci Instrum. 2021;57:2. [Google Scholar]
- 81.Sarv Ahrabi S., et al. Exploiting probability density function of deep convolutional autoencoders’ latent space for reliable COVID-19 detection on CT scans. The Journal of Supercomputing. 2022:1–22. doi: 10.1007/s11227-022-04349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patel A., et al. Cross Attention Transformers for Multi-modal Unsupervised Whole-Body PET. Anomaly Detection. 2022 [Google Scholar]
- 83.Clifton D.A., et al. Advances of Computational Intelligence in Industrial Systems. Springer; 2008. Automated novelty detection in industrial systems; pp. 269–296. [Google Scholar]
- 84.Bishop, C.M. Neural networks for pattern recognition. 1995.
- 85.Shorten C., Khoshgoftaar T.M. A survey on Image Data Augmentation for Deep Learning. Journal of Big Data. 2019;6(1) doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mikołajczyk A., Grochowski M. in 2018 International Interdisciplinary PhD Workshop (IIPhDW) 2018. Data augmentation for improving deep learning in image classification problem. [Google Scholar]
- 87.Tredwell G.D., et al. Modelling the acid/base 1H NMR chemical shift limits of metabolites in human urine. Metabolomics. 2016;12(10):152. doi: 10.1007/s11306-016-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Masud M., et al. Classification and Novel Class Detection in Concept-Drifting Data Streams under Time Constraints. Knowledge and Data Engineering, IEEE Transactions on. 2011;23:859–874. [Google Scholar]
- 89.Yang L., et al. Application of metabolomics in the diagnosis of breast cancer: a systematic review. Journal of Cancer. 2020;11(9):2540–2551. doi: 10.7150/jca.37604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fawcett T. ROC Graphs: Notes and Practical Considerations for Researchers. Machine Learning. 2004;31:1–38. [Google Scholar]
- 91.Mandrekar J.N. Receiver Operating Characteristic Curve in Diagnostic Test Assessment. Journal of Thoracic Oncology. 2010;5(9):1315–1316. doi: 10.1097/JTO.0b013e3181ec173d. [DOI] [PubMed] [Google Scholar]
- 92.Gramacki, A., Nonparametric kernel density estimation and its computational aspects. Vol. 37. 2018: Springer.
- 93.Peredriy S., Kakde D. The sampling peak criterion method for large data; Chaudhuri, Kernel bandwidth selection for SVDD: 2017. and A; pp. 3540–3549. [Google Scholar]
- 94.Bishop C.M. Novelty detection and neural network validation. IEE Proceedings-Vision, Image and Signal processing. 1994;141(4):217–222. [Google Scholar]
- 95.Chaudhuri A., et al. IEEE; 2018. Sampling Method for Fast Training of Support Vector Data Description. [Google Scholar]
- 96.Dufrenois F., Noyer J.-C. IEEE; 2016. A null space based one class kernel Fisher discriminant. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.