Abstract
Bacteria adapt to utilize the nutrients available in their environment through a sophisticated metabolic system composed of highly specialized enzymes. Although these enzymes can metabolize molecules other than those for which they evolved, their efficiency toward promiscuous substrates is considered too low to be of physiological relevance. Herein, we investigated the possibility that these promiscuous enzymes are actually efficient enough at metabolizing secondary substrates to modify the phenotype of the cell. For example, in the bacterium Acinetobacter baylyi ADP1 (ADP1), panD (coding for l-aspartate decarboxylase) encodes the only protein known to catalyze the synthesis of β-alanine, an obligate intermediate in CoA synthesis. However, we show that the ADP1 ΔpanD mutant could also form this molecule through an unknown metabolic pathway arising from promiscuous enzymes and grow as efficiently as the wildtype strain. Using metabolomic analyses, we identified 1,3-diaminopropane and 3-aminopropanal as intermediates in this novel pathway. We also conducted activity screening and enzyme kinetics to elucidate candidate enzymes involved in this pathway, including 2,4-diaminobutyrate aminotransferase (Dat) and 2,4-diaminobutyrate decarboxylase (Ddc) and validated this pathway in vivo by analyzing the phenotype of mutant bacterial strains. Finally, we experimentally demonstrate that this novel metabolic route is not restricted to ADP1. We propose that the occurrence of conserved genes in hundreds of genomes across many phyla suggests that this previously undescribed pathway is widespread in prokaryotes.
Keywords: CoA, enzyme kinetics, genomics, gene knockout, metabolism, metabolomics, polyamine, β-alanine
Abbreviations: 3AP, 3-aminopropanal; ADP1, Acinetobacter baylyi ADP1; ALD, aldehyde dehydrogenase; DAB, 2,4-diaminobutyrate; DAP, 1,3-diaminopropane; Dat, 2,4-diaminobutyrate aminotransferase; Ddc, 2,4-diaminobutyrate decarboxylase; DOPA, 3,4-dihydroxy-l-phenylalanine; FMOC, 9-fluorenylmethoxycarbonyl chloride; PLP, pyridoxal phosphate
Bacterial genomes can encode up to thousands of enzymes, most of which are habitually considered as highly specific. However, the notion that enzymes are specialized to carry out a single function is untrue (1, 2, 3, 4, 5). Enzymes exhibit promiscuity, that is, activities other than the one for which they have evolved and which are not parts of the physiology of the organism. When the promiscuous substrates are naturally occurring metabolites, “underground reactions” take place. These underground enzyme activities in Escherichia coli are frequently connected to the native metabolic network but remain at too low abundance to be physiologically relevant (6). Cases in which such underground metabolism led to an observable phenotype have been reported (2, 7, 8), although these functional innovations were always linked to structural or regulatory mutations that increased the flux through these novel enzymatic steps to a physiological level (7, 9, 10, 11, 12).
Since a high proportion of underground reactions can be wired into the existing metabolic network (6), it should be possible that operational underground metabolic pathways exist in the absence of mutations that increase metabolic flux. According to this hypothesis, promiscuous enzymes could have been patched together to generate stochastic salvage pathways that provide bypass to genetic lesions. To evaluate this postulate, a straightforward strategy is analyzing incorrect predictions of gene essentiality. These prediction errors offer the possibility to target biological discoveries.
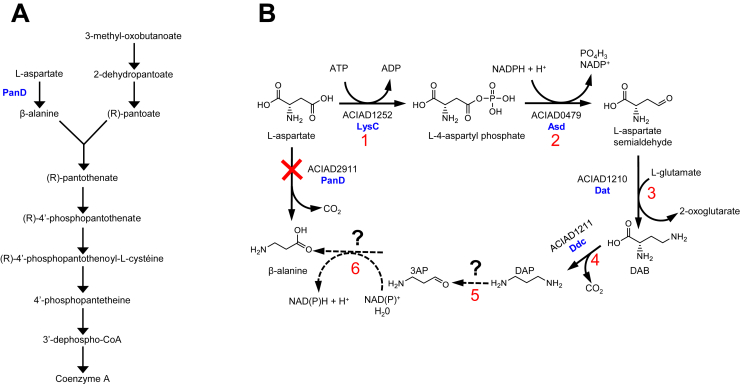
Acinetobacter baylyi ADP1 (ADP1) is a strictly aerobic soil bacterium. Its extraordinary competence for natural transformation and the ease with which it can be genetically engineered (13, 14) make ADP1 a key organism for the study of bacterial metabolism. We have a collection of single-gene deletion mutants for all dispensable genes of this organism (15) and considered in this work the ΔpanD mutant. PanD is an atypical decarboxylase that uses a covalently bound pyruvoyl group (16, 17), which participates in the synthesis of CoA. This enzyme is the only one known to convert l-aspartate into β-alanine, a necessary precursor of pantothenate (Fig. 1A). CoA is an essential cofactor in many metabolic and energy reactions and is a key component of life. The panD gene was thus expected to be essential. Although this is the case in a large number of prokaryotes such as E. coli (18, 19), the ADP1 ΔpanD mutant readily grows on minimal medium (15). This suggested that panD deletion was rescued by an unanticipated reaction or metabolic pathway, as aspartate decarboxylation is not the only way to form β-alanine. At least three other pathways are thought to exist in other organisms, from different precursors (Fig. S1). (1) The polyamine pathway is reported in plants and yeast (20, 21, 22). In plants, spermine and spermidine can be cleaved to form 1,3-diaminopropane (DAP). DAP is converted to 3-aminopropanal (3AP) by polyamine oxidases (23, 24) and further oxidized to β-alanine (25, 26). A similar pathway is reported in yeast (22): the polyamine oxidase FMS1 converts spermine to 3AP and spermidine (27), and the two related aldehyde dehydrogenases ALD2 and ALD3 oxidize 3AP to β-alanine (28). (2) The uracil degradation involves the reduction of uracil to dihydrouracil followed by two hydrolytic steps that eventually yield β-alanine, ammonia, and carbon dioxide. Such a pathway is reported in plants (29) and bacteria (30). (3) The propionate pathway, which is not only well known in plants (31, 32) but also suspected in bacteria (33), proceeds by activation of propionate to propionyl-CoA, which is oxidized to acrylyl-CoA. This latter is in turn hydrated to 3-hydroxypropionyl-CoA, then hydrolyzed to 3-hydroxypropionate, which is further oxidized to malonate semialdehyde. The final step is a transamination that forms β-alanine. None of these pathways is supposed to occur in ADP1. In this work, we show by a metabolomic approach that ADP1 takes advantage of the high DAP content provided by 2,4-diaminobutyrate (DAB) aminotransferase (Dat) and DAB decarboxylase (Ddc) to form 3AP, whose oxidation restores β-alanine synthesis. We selected different candidate enzymes for the two steps of this salvage pathway (from DAP to β-alanine) by a large activity screening assay and further kinetic characterization, and we conducted a growth phenotype analysis of mutants deleted for these candidate genes to evaluate their in vivo contribution. This salvage pathway relies on the two pivotal genes dat and ddc, the last step being performed by different ALDs.
Figure 1.
The synthesis of β-alanine.A, CoA synthesis in prokaryotes. B, hypothetical alternative route for β-alanine synthesis in ADP1. 1: l-aspartate kinase (LysC); 2: l-aspartate semialdehyde dehydrogenase (Asd); 3: l-2,4-diaminobutyrate aminotransferase (Dat); and 4: l-2,4-diaminobutyrate decarboxylase (Ddc). Reactions indicated in dotted lines are unknown. ADP1, Acinetobacter baylyi ADP1; DAB, 2,4-diaminobutyrate; DAP, 1,3-diaminopropane.
The large phylogenetic distribution of this clustered gene pair in bacteria and Euryarchaeota species suggests that this unsuspected salvage pathway for β-alanine is widespread, as supported by its experimental validation in Sinorhizobium meliloti 1021.
Results
A hypothetical alternative pathway for β-alanine synthesis
Since among the many decarboxylases that exist in E. coli none can complement PanD, it seemed unlikely that a promiscuous decarboxylase activity in ADP1 rescued β-alanine synthesis. Indeed, no l-aspartate decarboxylation activity could be detected from the 15 annotated decarboxylases in ADP1 (Table S1). To figure out how ADP1 could synthesize β-alanine, we considered the different pathways listed in Fig. S1, with a particular focus on the polyamine pathway. In this pathway, DAP is formed from spermine or spermidine and oxidized to 3AP, which is eventually oxidized by an NAD(P)-dependent ALD to form β-alanine. Intriguingly, DAP is the major polyamine in Acinetobacter (34, 35). It is used as a chemotaxonomic marker for the genus (36) and was recently shown to be involved in surface-associated motility (37) and siderophore biosynthesis (38, 39). Its abundance prompted us to consider DAP as the starting point for a salvage route for β-alanine synthesis, as illustrated in Figure 1B.
Comparative genomic analysis of wildtype and ΔpanD ADP1 strains
In order to rule out the presence of mutations that would allow bypass for β-alanine synthesis, genomic sequencing of ΔpanD strain was performed and compared with the wildtype sequence (40). The sequencing revealed several events. Most are synonymous mutations in ACIAD1796 (annotated as putative membrane protein), along with a small number of chromosomal mutations, such as SNPs and short deletions/insertions that affected genes and intergenic regions (Table S2). None is located in or close to genes that could be considered as candidates for participating in an alternative β-alanine synthesis route. Together, these data are consistent with a mutation-independent functional salvage pathway.
Targeted metabolomic analysis of the alternative route for β-alanine synthesis
In the alternative route proposed in Figure 1, 3AP is an obligate intermediate. This metabolite is not expected in ADP1 (41), but its presence would strengthen our hypothesis. In this perspective, metabolomes of the wildtype and ΔpanD strains were prepared from cells growing exponentially in a minimal medium devoid of β-alanine containing succinate as the sole carbon source. DAP was detected in both metabolomes, with a slightly lower content in the mutant strain (Fig. 2A). As anticipated, 3AP was observed in ΔpanD, but also in wildtype cells, with a similar concentration (Fig. 2B). These data are in line with the pathway proposed in Figure 1B. The presence of 3AP in the wildtype context supports a constitutive pathway parallel to PanD function. Surprisingly, β-alanine content was dramatically affected in ΔpanD (Fig. 2C). This deficiency was logically reflected in pantothenate (Fig. 2D), the product of the next step of the CoA synthesis pathway (Fig. 1A). In spite of a severe deficiency in intermediates of the CoA biosynthesis pathway (there is 93 times less β-alanine and 60 times less pantothenate), ΔpanD maintains a growth similar to that of the wildtype (15). β-alanine and pantothenate are thus not rate limiting for CoA biogenesis. A similar observation was made for E. coli, in which pantothenate kinase is considered as the primary enzyme regulating the CoA content (42).
Figure 2.
Detection of intermediates of the CoA biosynthesis pathway in wildtype and ΔpanD Acinetobacter baylyi ADP1 strains. Metabolomic analysis of cells grown in minimal medium devoid of β-alanine. LC/MS/MS analyses were conducted on a QTRAP 5500 using a multiple reaction monitoring (MRM)–based approach. DAP and 3AP were analyzed using a C18 BEH column after derivatization with benzoyl chloride and FMOC, respectively. β-alanine and pantothenate were analyzed through a ZIC-pHILIC column. A and B, comparison of DAP and 3AP contents, respectively. C and D, comparison of β-alanine and pantothenate contents, respectively. DAP, 3AP, β-alanine, and pantothenate were identified according to the chromatographic retention time and fragmentation pattern of their corresponding standards (Fig. 3 and Table S3). For DAP and 3AP, data correspond to six and five independent metabolite extractions for each strain, respectively. For β-alanine and pantothenate, data correspond to 10 extractions. The values shown correspond to automatic peak integration for the following mass transitions: DAP, m/z 283.1 → 104.9; 3AP, m/z 296.0 → 179.0; β-alanine: m/z 90.0 → 71.9); and pantothenate, m/z 218.1 → 87.9. Only the values for β-alanine in ΔpanD were close to the detection limit (Fig. S4). Indicated are p values from Welch's t test. 3AP, 3-aminopropanal; DAP, 1,3-diaminopropane; FMOC, 9-fluorenylmethoxycarbonyl chloride.
Identification of genes involved in 3AP oxidation
The most obvious reaction to form β-alanine from 3AP is its oxidation by an ALD (20, 43, 44). To identify the enzymes in ADP1 that may be involved, 237 candidate genes were selected according to their annotation as coding for “dehydrogenases” or “oxidoreductases” by the MicroScope genome annotation platform (https://www.genoscope.cns.fr/agc/microscope/home/index.php). From these, 227 were successfully cloned and overexpressed in E. coli for protein production. The corresponding cell-free extracts were then screened for 3AP dehydrogenation by monitoring NAD(P)H formation (reaction 6, Fig. 1B). Results showed that four proteins exhibited activity (Table 1). Based on these data, ACIAD0960 (GabD-like), ACIAD1009 (BetB), ACIAD2018 (acoD), and ACIAD3339 (AdhA) were purified (see the Experimental procedures section and Fig. S2) and kinetically characterized (Table 2). ACoD and AdhA were specific for NAD+, whereas GabD-like and BetB were active with both nucleotides. GabD-like was slightly more efficient with NADP+ (as indicated by the ratios kcat/Km NADP+ and kcat/Km NAD+: 1.03 103 versus 3.7 102 s−1 M−1, respectively). Conversely, BetB was more efficient with NAD+ (7.4 104 versus 7.2 103 s−1 M−1). Their cofactor preference is in agreement with the presence of a serine at position 159 for GabD-like and a glutamate at position 179 for BetB (45). These enzymes exhibit kcat and kcat/Km values for 3AP in the range of 1 to 4 s−1 and 102 to 104 s−1 M−1, respectively (Table 2). Although they can be considered rather low, these two types of values are similar to those found for a significant proportion of enzymes characterized for their physiological role (46). These data thus suggest that each of these enzymes may contribute in vivo to β-alanine synthesis.
Table 1.
Specific activity of the 3AP dehydrogenases detected from high-throughput enzymatic screening
| Gene ID | Gene name | Predicted function | Specific activity (μM min−1 μg−1) |
|---|---|---|---|
| ACIAD0960 | gabD like | Putative succinate-semialdehyde dehydrogenase [NADP+] (GabD-like) | 5.47 ± 0.08a |
| ACIAD1009 | betB | NAD+-dependent betaine aldehyde dehydrogenase | 1.80 ± 0.02b |
| ACIAD2018 | acoD | Acetaldehyde dehydrogenase II (ACDH-II) | 5.71 ± 0.06b |
| ACIAD3339 | adhA | Alcohol dehydrogenase | 1.99 ± 0.03b |
Activities reported represent the greater value obtained with either 2 mM NADP+ or NAD+.
Value obtained with NADP+.
Value obtained with NAD+. Values are ±SD from two independent experiments conducted in 100 μl 50 mM Tris–HCl, pH 9.0, 200 mM NaCl, and 15% glycerol containing 4 mM 3AP.
Table 2.
Kinetic parameters for the dehydrogenases of the alternative route for β-alanine synthesis
| Enzyme ID | Enzyme name | Substrate | kcat (s−1) | Km (μM) | kcat/Km (s−1 M−1) |
|---|---|---|---|---|---|
| ACIAD0960 | GabD-like | 3APa | 3.86 ± 0.40 | 2934 ± 306 | 1.3 103 |
| NADP+b | 3718 ± 1138 | 1.0 103 | |||
| NAD+b | 1.68 ± 0.04 | 4507 ± 363 | 3.7 102 | ||
| ACIAD1009 | BetB | 3APc | 1.48 ± 0.16 | 82 ± 17 | 1.8 104 |
| NAD+d | 20 ± 4 | 7.4 104 | |||
| NADP+d | 2.43 ± 0.12 | 339 ± 67 | 7.2 103 | ||
| ACIAD2018 | AcoD | 3APe | 1.55 ± 0.35 | 3559 ± 755 | 4.4 102 |
| NAD+f | 124 ± 49 | 1.3 104 | |||
| ACIAD3339 | AdhA | 3APg | 0.93 ± 0.07 | 141 ± 28 | 6.6 102 |
| NAD+h | 1803 ± 305 | 5.2 102 |
Values are the average of two replicates.
NADP+ concentration was 16 mM.
3AP concentration was 15 mM.
NAD+ concentration was 2 mM.
3AP concentration was 2.5 mM.
NAD+ concentration was 2 mM.
3AP concentration was 20 mM.
NAD+ concentration was 9 mM.
3AP concentration was 2 mM.
Identification of genes involved in 3AP formation
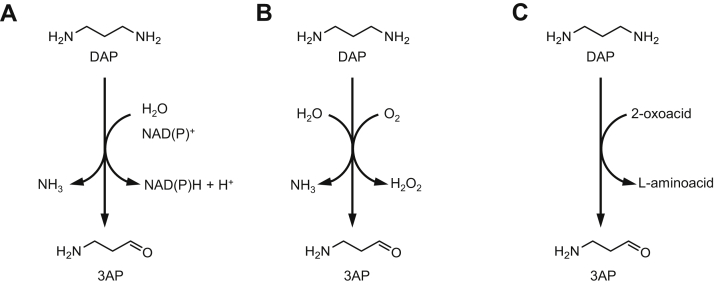
At least three distinct reactions can convert DAP to 3AP (Fig. 3). DAP can undergo a nucleotide-dependent oxidative deamination, in a reaction similar to those reported in the fermentation pathway of lysine or ornithine (47, 48). Alternatively, 3AP can be formed by an O2-dependent amine oxidase, as observed in plants (23, 24, 44) or yeast (22). Last, 3AP can be produced by a polyamine aminotransferase, as described in bacteria (49, 50). We first took advantage of the 227 cell-free extracts containing putative dehydrogenases to search for oxidative deamination of DAP but could not detect activity. Quinohemoprotein amine dehydrogenases were reported to oxidize DAP (51, 52, 53, 54). The activity of these enzymes, which are composed of subunits coded by different genes, could not be screened by our assay. However, it is unlikely that a quinohemoprotein amine dehydrogenase catalyzes 3AP synthesis, since enzymes with known sequences (55, 56) show no homology to any ADP1 protein. We next considered the 29 proteins in ADP1 annotated as “oxidase,” but none was active with DAP (see the Experimental procedures section). This result is consistent with a previous report that indicated that polyamine oxidases were not observed in bacteria (57). Finally, the 27 proteins annotated as “transaminase“ or “aminotransferase“ were evaluated. Briefly, DAP aminotransferase activity was assayed in recombinant cell-free extracts with DAP in the presence of 2-oxoglutarate as the amine acceptor, using an enzymatic assay that monitored l-glutamate formation with l-glutamate dehydrogenase. We detected activity for three ω-transaminases: ACIAD3446 (GabT), ACIAD1284 (AstC), and ACIAD1210 (Dat). Specific activities were 94, 3.4, and 5.9 nmol min−1 mg−1, respectively (see the Experimental procedures section). GabT, AstC, and Dat were purified for further characterization, but the activity of AstC and Dat was too low to determine their kinetic parameters, so only GabT could be studied in more detail (Table 3). The catalytic efficiency of GabT for DAP is low (51 s−1 M−1) but is nonetheless only ∼24-fold lower than that for its physiological substrate (1.2 103 s−1 M−1). With DAP, it is essentially the Km that is affected (one order of magnitude), whereas the kcat remains comparable (0.48 versus 0.22 s−1). This rather weak impact on the kinetic constants for this promiscuous substrate makes GabT unusually efficient as compared with reported promiscuous enzymes (4).
Figure 3.
Enzymatic conversion of DAP to 3AP. A, nucleotide-dependent oxidative deamination. B, oxygen-dependent oxidative deamination. C, transamination. 3AP, 3-aminopropanal; DAP, 1,3-diaminopropane.
Table 3.
Kinetic properties of GabT
| Enzyme | Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) |
|---|---|---|---|---|
| GabT | GABAa | 399 ± 40 | 0.48 ± 0.021 | 1.2 103 |
| 2-Oxoglutarateb | 58 ± 7 | 8.3 103 | ||
| DAPa | 4356 ± 1014 | 0.22 ± 0.016 | 5.1 101 |
Abbreviation: GABA, gamma-aminobutyric acid.
Aminotransferase activities were determined in the presence of 200 μM pyridoxal phosphate, 2 U of l-glutamate dehydrogenase, and 5 mM NAD+ in a spectrophotometric coupled assay by measuring the formation of NADH, as described under the Experimental procedures section. Values are the average of three replicates.
2-Oxoglutarate concentration was 500 μM.
GABA concentration was 4 mM.
Ddc participates in 3AP synthesis
The pathway proposed in Figure 1B is reminiscent of what was observed in a ΔpanD strain of E. coli submitted to laboratory evolution (58). Pontrelli et al. (58) reported a novel pathway—made possible after three mutations in the genome—that also relied on the formation of DAB from aspartate. The authors also considered the pathway we propose here but rejected it because DAP was not detected in their evolved strain. In their work, DAB was directly converted by a decarboxylation-dependent deamination reaction (catalyzed by a mutated form of the pyridoxal phosphate [PLP]–containing ornithine decarboxylase SpeC) into CO2, H2O2, NH4+, and 3AP, which was in turn oxidized into β-alanine by BetB. The capability for PLP-dependent enzymes to catalyze side reactions is documented (59). More specifically, the reaction performed by the mutated SpeC is evocative of the behavior of 3,4-dihydroxy-l-phenylalanine (DOPA) decarboxylase. This latter, in addition to decarboxylating amino acids such as DOPA and 5-hydroxytryptophan, performs oxidative deamination on the decarboxylation products, that is, dopamine and serotonin, respectively, to form the corresponding aldehydes (60, 61). Here, LC/MS/MS analysis indicated that Ddc acted similarly to DOPA decarboxylase as it was able, in addition to forming DAP, to produce 3AP from DAP (along with H2O2; see the Experimental procedures section). The kinetic characterization of Ddc was therefore carried out on both reactions (Table 4). The kinetic constants calculated for the decarboxylation reaction are comparable to those previously reported for an Acinetobacter enzyme (62). The catalytic efficiency of oxidative deamination is less than three orders of magnitude lower than that of decarboxylation (13 versus 7.6 103 s−1 M−1), in agreement with what has been observed for DOPA decarboxylase (61). The difference in the catalytic efficiency between the two Ddc reactions is mainly because of the decrease in kcat. This low kcat/Km value for 3AP formation is however similar to the one calculated for GabT. The two enzymes can therefore potentially have a comparable in vivo contribution to 3AP formation. None of the other ADP1 decarboxylases were active in the formation of DAP and/or 3AP from DAB.
Table 4.
Kinetic properties of Ddc
| Enzyme ID | Enzyme name | Reaction | Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) |
|---|---|---|---|---|---|---|
| ACIAD1211 | Ddc | Decarboxylase | DAB | 1585 ± 257 | 12.1 ± 1.1 | 7.6 103 |
| Amine oxidase | DAP | 710 ± 113 | 0.009 ± 3 10−4 | 13 |
Amine oxidase activity was determined in a spectrophotometric coupled assay by measuring 3AP oxidation in the presence of BetB and 1 mM NAD+, whereas decarboxylase activity was determined by LC/MS/MS, as described under the Experimental procedures section. Values are the average of three replicates.
The growth of ADP1 on minimal medium is impaired in the ΔpanD strains whose genes involved in the alternative β-alanine synthesis route were deleted
As illustrated in Figure 4A, despite the disruption of the canonical β-alanine synthesis pathway, the ΔpanD strain does not show any growth defect. To evaluate the role of each gene of the alternative pathway in vivo, we constructed double mutant strains deleted for panD and for each of the different candidate genes (except ΔpanD/ΔgabD-like, which could not be obtained for technical reasons). We have previously evaluated the impact of single deletions of the different candidate genes on cell growth. Data showed that strains deleted for gabT, adhA, acoD, gabD, and betB have a growth profile globally similar to that of wildtype cells (Fig. 4B). However, Δddc showed a significant growth delay, which was compensated by the addition of DAP in the culture medium but not by β-alanine (Fig. 4C). This delay indicates an important role of this polyamine in ADP1 but not related to β-alanine. It is worthy to note that Δddc has a growth rate of 34% of that of the wildtype (0.150 h−1 versus 0.439 h−1 for the wildtype strain). A similar behavior was observed in an E. coli mutant strain that no longer produced polyamines (63). The growth of the ΔpanD strains that were additionally deleted for any of the candidate genes showed impaired growth (Fig. 4D). These results indicated the relative contribution of each of the candidate genes in vivo in the synthesis of β-alanine. The addition of 10 μM of β-alanine in the culture medium restored a wildtype growth for all mutants (except for ΔpanD/Δddc whose growth is also relying on DAP), thus confirming our hypothesis (Fig. 4E). Specifically, ΔpanD/ΔgabT had a growth rate of 0.129 h−1, which shows GabT contribution. The remaining growth may be due to promiscuous transaminases (such as Dat and astC; see aforementioned) but more likely to the amine oxidase activity of Ddc (Table 4). To better understand the in vivo role of these promiscuous enzymes, we can also rely on transcriptomic data. We previously conducted RNA-Seq analysis on ADP1 grown on the same culture medium used here ((64), Online Resource 2, Table S4) that can help understand the role of the different candidate genes. RNA-Seq is used to determine the set of all expressed transcripts. The number of reads give an estimate of the relative expression levels in the cell. For instance, l-aspartate semialdehyde is a metabolite that must be synthesized in a nonlimiting way, as it is at a metabolic hub that allows for the biosynthesis of l-lysine, l-methionine, and l-isoleucine (65). According to Figure 1B, the formation of l-aspartate semialdehyde is carried out by lysC and asd, which were significantly transcribed (4640 and 43,300 reads, respectively). Similarly, DAP (abundant in ADP1) is formed by dat and ddc, which were also highly expressed (2902 and 5492 reads, respectively). In contrast, GabT, which converts DAP to 3AP in vitro, was poorly expressed (38 reads), especially in comparison with ddc, which also forms 3AP (Table 4). On this basis, considering that the two corresponding enzymes have a similar catalytic efficiency for 3AP formation (51 and 13 s−1 M−1 for GabT and Ddc, respectively), the metabolic contribution of Ddc for 3AP synthesis is expected to be higher than that of GabT. This is in agreement with the lower growth rate observed for panD/ddc, compared with panD/gabT (0.029 versus 0.129 h−1, respectively). GabT still has a role in vivo, which may seem surprising given such low transcription and catalytic efficiency. However, even though its kcat (0.22 s−1) is two orders of magnitude lower than that of an “average enzyme” (46), enzymes that harbor a similar turnover number with their physiological substrate are not rare (46). Its low kcat/Km value for DAP is also because of the high Km value (4.4 mM). DAP content in ADP1 was estimated to 91 μmol/g dry mass (37), which corresponds to ∼35 mM. In these conditions, GabT would function in vivo at DAP saturation and thus have a maximal metabolic contribution. We next assessed the impact of the candidate dehydrogenases for the conversion of 3AP to β-alanine. ΔpanD/ΔadhA was the less affected mutant, with a growth rate of 0.147 h−1. ΔpanD/ΔbetB and ΔpanD/ΔacoD were much more affected with growth rates of 0.076 and 0.064 h−1, respectively, indicating a larger in vivo contribution for BetB and AcoD. Transcriptomic data are more difficult to use here to interpret these results. Since ΔpanD/ΔacoD and ΔpanD/ΔbetB have similar growth rates, it can be assumed that the higher catalytic efficiency of BetB (1.8 104 s−1 M−1) compared with AcoD (4.4 102 s−1 M−1) is counterbalanced by its lower transcript level (405 reads for BetB versus 2885 for AcoD). However, while AcoD and AdhA have a similar catalytic efficiency (4.4 102 and 6.6 102 s−1 M−1, respectively) and a comparable transcriptional level (2885 and 3280 reads, respectively), ΔpanD/ΔacoD growth is much more affected than ΔpanD/ΔadhA (0.147 versus 0.064 h−1, respectively). This shows a weaker involvement of AdhA, which may be due to post-transcriptional events and/or enzymatic regulation. In conclusion, these three dehydrogenases effectively participate in the rescue pathway but in different proportions. Unfortunately, the contribution of the last candidate dehydrogenase, GabD-like (ACIAD0960), could not be evaluated. Finally, the ΔpanD/Δddc growth was barely measurable, with only a doubling of cell numbers in 45 h (Fig. 4D). The residual growth is probably related to the presence of β-alanine in the preculture medium. Another possibility is that a residual enzymatic activity forms β-alanine from another metabolic precursor. It was recently reported that an aminotransferase could complement an E. coli panD mutant by converting malonate semialdehyde to β-alanine (66). However, metabolomic analyses could not detect malonate semialdehyde in ADP1, making this hypothesis unlikely (Supporting information File S2). Therefore, in the absence of PanD, the synthesis of β-alanine seems to be strictly dependent on DAP. This is supported by the fact that DAP could not be detected in the metabolome of Δddc (Fig. 4F). The very low values measured in the mutant are in fact comparable to those obtained with blank solutions (Fig. S4) because DAP remained in the analytical system (carryover). This result is thus in conflict with the work of Skiebe et al. (37), in which DAP content was not affected in a ddc mutant of ADP1. The mutant described by Skiebe et al. is in fact from our own collection of mutant (15). Preliminary experiments in this study with this same mutant confirmed the presence of DAP in the cell, but control PCRs showed that an extra wildtype copy of ddc remained in the chromosome. This phenomenon is caused by a duplication of a part of the genome (which contains the wildtype target gene), when trying to delete a gene essential to the cell (see “Double band” mutants in Ref. (15)). It is noteworthy that during the construction of the collection of single-gene deletion mutants of ADP1, the clones were selected on minimal medium. Here, we constructed a de novo mutant that was selected on a DAP-containing medium. All controls indicated that the ddc mutant described here is valid.
Figure 4.
Relative contribution of the different ADP1 enzymes to the alternative β-alanine biosynthesis pathway.A, comparison of wildtype and ΔpanD growth. B, growth of single-deletion mutants of candidate genes involved in the alternative pathway. C, growth of Δddc complemented with DAP (1 mM) or β-alanine (10 μM). D, growth of the ΔpanD strain additionally deleted for the various candidate genes. E, growth of the double mutants complemented with β-alanine. F, comparison of DAP content in Δddc and WT strains. Independent overnight cultures were grown in minimal medium for Acinetobacter (MA) supplemented with 1 mM DAP and 10 μM β-alanine, harvested, and washed before inoculation in MA minimal medium. Absorbance at 600 nm was recorded by an automated growth curve analysis system (Bioscreen-C; Thermo Fisher Scientific). Values correspond to the average of three replicates. Colors indicate the gene(s) deleted in each strain. LC/MS/MS data for DAP correspond to six independent metabolite extractions for each strain. Indicated values in F correspond to automatic peak integration for the mass transition m/z 283.1 → 104.9. Values of DAP in Δddc were below the detection limit (Fig. S4). ADP1, Acinetobacter baylyi ADP1; DAP, 1,3-diaminopropane.
Phylogenetic repartition of dat and ddc
The alternative β-alanine biosynthetic route builds on the DAP pathway, which is supported by dat and ddc. These genes are linked together on the chromosome with dat always upstream, which makes them easy to identify in databases (67, 68). Comparative analyses conducted on our MicroScope platform (https://www.genoscope.cns.fr/agc/microscope/home/index.php) showed that clusters of genes orthologous to dat–ddc are present in the chromosome of more than 500 organisms (as of March 2022) belonging in phyla as different as Actinobacteria, Cyanobacteria, Firmicutes, Proteobacteria (α-, β-, γ-, and δ-), Spirochaeta, and Euryarchaeota (halobacteria class). These organisms may also form β-alanine using the alternative route. As an example, S. meliloti 1021 has the colocalized Sma2400 (rhba) and Sma2402 (rhbb) genes that are 54 and 37% identical with dat and ddc of ADP1, respectively. Its genome also harbors genes orthologous to those that are involved in the rescue pathway (Fig. 5A). On the other hand, it should be noted that panD is often absent from the genomes of Rhizobiales. In Rhizobium etli CFN42, β-alanine is synthesized from the reductive pyrimidine degradation pathway. Uracil is converted to 3-ureidopropionate, which is hydrolyzed by AmaB (β-alanine synthase) to CO2, NH4+, and β-alanine (69). S. meliloti 1021, which also lacks PanD but harbors AmaB, should therefore produce β-alanine this way.
Figure 5.
Alternative synthesis of β-alanine in Sinorhizobium meliloti 1021.A, conservation of the genes of the alternative β-alanine synthesis pathway in S. meliloti 1021. The gray gene symbol is used for a gene unrelated to β-alanine metabolism. Sma and Smb, plasmidic gene localization; Smc, chromosomal gene localization. Colocalization of genetic loci was observed through the MicroScope platform. B, comparison of wildtype and ΔamaB growth. C–F, detection of DAP and 3AP after derivatization with benzoyl chloride and FMOC, respectively, in wildtype ADP1 and S. meliloti 1021 metabolomes. DAP and 3AP were monitored by LC/MS/MS on a QTRAP 5500 using a multiple reaction monitoring (MRM)–based approach. Extracted ion chromatograms are presented for three specific MRM transitions for both compounds. For DAP: green, 283.1 → 162.0; red, 283.1 → 104.9; and blue, 283.1 → 77.0. For 3AP: blue, 296.0 → 179.0; green, 296.0 → 118.0; and red, 296.0 → 74.0 (Table S3). C, DAP in S. meliloti 1021 metabolome. D, DAP in ADP1 metabolome. E, 3AP in S. meliloti 1021 metabolome. F, 3AP in ADP1 metabolome. LC/MS/MS analyses were conducted in the positive ionization mode. ADP1, Acinetobacter baylyi ADP1; 3AP, 3-aminopropanal; DAP, 1,3-diaminopropane; FMOC, 9-fluorenylmethoxycarbonyl chloride.
Interestingly, in R. etli CFN42, the amaB mutant is auxotrophic for β-alanine (69), a phenotype that can be explained by the absence of dat and ddc. On the opposite, a S. meliloti 1021 mutant disrupted for amaB (Smc01820) is expected to grow in a minimal medium devoid of β-alanine. This could be verified experimentally (Fig. 5B). The formation of β-alanine through the DAP pathway is strengthened by the detection of both DAP and 3AP in the metabolome of the S. meliloti 1021 mutant (Fig. 5, C–F). These results prove that the DAP-dependent β-alanine synthesis pathway is not restricted to ADP1.
Discussion
Substrate ambiguity is considered as a prerequisite for the evolution of new functions. The acquisition of new metabolic activities then comes from mutations that increase the efficiency of a promiscuous activity or increase expression or copy number of the corresponding gene to raise the metabolic flux to a physiological level. These genetic changes can lead to the evolution of a new dedicated and efficient metabolic pathway when an essential metabolite is no longer available and survival depends on the assembly of a new biosynthetic pathway.
In this study, we report an unusual situation, in which in the absence of selection pressure, a serendipitous metabolic pathway parallelizes the formation of a precursor to a key component of life, CoA. We deciphered how ADP1 managed to grow in minimal medium after the deletion of panD. This novel pathway takes advantage of enzyme promiscuity for rerouting the metabolic flux through DAP and forming a novel immediate precursor of β-alanine. These results challenge the accepted assumption that underground reactions occur at rates too low to have essential roles in a wildtype background.
We have shown that in ADP1, two enzymes catalyzing distinct reaction types—transamination (GabT) and oxidative deamination (Ddc)—are both responsible for 3AP synthesis. Furthermore, at least three different dehydrogenases (AcoD, AdhA, and BetB) can handle in vivo this unstable intermediate to form β-alanine. This enzymatic redundancy contributes to both the robustness and efficiency of the pathway. Indeed, in ADP1 and S. meliloti 1021, β-alanine is normally formed by distinct pathways, but in both organisms, this same salvage pathway is remarkably efficient because mutants in the archetypal biosynthetic routes grow as well as the wildtype strains. From these results, we can consider that in most organisms that possess Dat and Ddc, there is also one or more dehydrogenases that ensure the oxidation of 3AP to β-alanine (Ddc being probably sufficient to ensure the formation of 3AP).
Polyamines are found in most cells, although their physiological role is not clearly defined. DAP has been proposed to be the key structural feature for general electrostatic binding to nucleic acids in bacteria (70), an intermediate in a polyamine biosynthetic pathway that is essential for biofilm formation in Vibrio cholera (67), and a precursor in siderophore synthesis (38, 71). We show with this study a novel function for this overlooked metabolite that is part of an unsuspected and likely widespread metabolic pathway in environmental, commensal, and pathogenic bacteria.
Experimental procedures
Chemicals and reagents
l-aspartate, l-DAB, DAP, β-alanine, pantothenate, solvents, and other chemicals were from Sigma–Aldrich. Reagents for molecular biology were from Invitrogen. Oligonucleotides were from Sigma Genosys. 3AP was chemically synthesized starting from 1-amino-3,3-diethoxypropane (Acros Organics) that was incubated in the presence of 3 M HCl overnight at room temperature. The reaction was neutralized with 10 M NaOH, and 3AP was quantified by measuring its complete NAD-dependent oxidation by BetB.
Strains and medium
A. baylyi ADP1 strain (DSM 24193) was provided by Dr Nicholas Ornston (Yale University). ΔpanD was obtained according to de Berardinis et al. (15). S. meliloti 1021 wildtype and ΔamaB (Smc01820 mutant) strains were kindly provided by Pr Anke Becker, LOEWE Center for Synthetic Microbiology, Marburg University, Germany. Briefly, S. meliloti 1021 mutant (SmPI_1021.12.02F11) was constructed by plasmid integration. Information about the S. meliloti mutant library is available at https://synmikro.com/research/chassis-and-genomes/anke-becker/resources/s.meliloti-mutant-libraries.html. All strains were routinely grown on medium for Acinetobacter (minimal medium for Acinetobacter [MA]) consisting of 31 mM Na2HPO4, 25 mM KH2PO4, 18 mM NH4Cl, 41 μM nitrilotriacetic acid, 2 mM MgSO4, 0.45 mM CaCl2, 3 μM FeCl3, 1 μM MnCl2, 1 μM ZnCl2, and 0.3 μM of a mineral ion mix (CrCl3, H3BO3, CoCl2, CuCl2, NiCl2, Na2MoO2, and Na2SeO3) supplemented with 20 mM succinate as the carbon source. S. meliloti cultures were supplemented with 5 μM biotin.
Metabolome preparation
ADP1 wildtype, ΔpanD, Δddc, and Sinorhizobium meliloti 1021 wildtype were grown at 30 °C in 5 ml of MA supplemented with 20 mM succinate. Metabolite extraction was performed in multiwell plates as described previously (72). Dried metabolomes were stored at −80 °C. Before LC/MS analysis, the metabolites were suspended with 120 μl of a mix composed of 80% acetonitrile and 20% 10 mM ammonium carbonate (pH 9.9) or with 250 μl water for derivatization. Samples were finally filtered on 0.22 μm (polytetrafluoroethylene; AcroPrep Advance, Pall).
Derivatization procedures
DAP was labeled with benzoyl chloride for improving LC/MS/MS detection. Before derivatization, DAP standard was prepared at 15 μM in water, and dried metabolomes from 5 ml cultures were suspended in 250 μl water. About 250 μl of sample (including enzymatic reaction products) was basified with 0.5 μl 2 M NaOH, and 250 μl of 4% benzoyl chloride in acetonitrile was added. After 15 min, the mixture was filtered on 0.22 μm (Sterile Filter Unit MF-Millipore MCE Membrane) and diluted to 1/1500 in the mobile phase before LC/MS analysis.
3AP was labeled with 9-fluorenylmethoxycarbonyl chloride (FMOC). Before derivatization, 3AP standard was prepared at 30 μM in water, and dried metabolomes from 5 ml cultures were suspended in 250 μl water. About 20 μl of sample (including enzymatic reaction products) was added to 80 μl of 0.2 M borate buffer (pH 7.7). Then, 100 μl of 15 mM FMOC solution in acetone was added and hand shaken in a 1.5 ml Eppendorf tube for 1 min. The mixture was filtered on 0.22 μm and diluted 1.5-fold in the mobile phase before LC/MS analysis.
LC/MS/MS analysis
The intermediates of the alternative pathway for β-alanine synthesis and pantothenate were detected by LC/MS/MS using a Dionex UltiMate TCC-3000RS chromatographic system (Thermo Fisher Scientific) coupled to a hybrid triple quadrupole linear ion trap mass spectrometer (QTRAP 5500 from ABSciex) equipped with a heated electrospray ionization source. To detect pantothenate and discriminate between β-alanine and l-alanine, chromatographic separation was achieved on a ZIC-pHILIC column (150 × 2.1 mm2; 5 μm; Merck) thermostated at 40 °C. The mobile phase flow rate was set at 0.2 ml/min, and 3 μl was injected. Mobile phase A consisted of 10 mM ammonium carbonate with pH adjusted to 9.9 with NH4OH, and mobile phase B consisted of acetonitrile. The gradient started at 80% B for 0.5 min, followed by a linear gradient to 40% B for 20 min, and remained at 40% B for 8 min. The system returned to the initial solvent composition in 5 min and reequilibrated under these conditions for 15 min.
For the derivatized intermediates, chromatographic separation was achieved on a C18 BEH column (100 × 2.1 mm2; 1.7 μm; Waters) thermostated at 35 °C. The mobile phase flow rate was set at 0.3 ml/min, and 5 μl was injected. A consisted of 10 mM ammonium carbonate with pH adjusted to 9.9 with NH4OH, and B consisted of methanol. The gradient started at 10% B for 1 min, followed by a linear gradient to 70% B for 19 min, and remained at 70% B for 5 min. The system returned to the initial solvent composition in 5 min and reequilibrated under these conditions for 10 min.
Mass spectrometry analyses were conducted with the following parameters: ion source 5500 V (positive mode) or −5500 V (negative mode), curtain gas 20 a.u., temperature 500 °C, gas 1 40 a.u., gas 2 60 a.u., computer-aided design—medium. MS/MS experiments were performed in the triple quadripole mode using multiple reaction monitoring scan type. For each compound, the optimization of following MS parameters (declustering potential, collision energy, and cell exit potential) was performed in order to establish the best intensity transitions (Table S3). Data were processed using Analyst 1.6.2 (ABSciex).
Cloning, expression, and purification of the recombinant enzymes
The laboratory has a collection of approximately 2400 genes coding for proteins annotated as cytosolic enzymes, cloned in a pET22b(+) derivative for recombinant production in E. coli, according to a protocol previously described (73). Dr Véronique de Berardinis has kindly made this collection available for this study. Each expression plasmid was transformed into E. coli BL21 (DE3) strain (Invitrogen). Protein purification was performed using a preparative chromatography system (Äkta Pure; GE Healthcare Life Sciences). A fully automated two-step method was set up for each protein in which a His Trap FF 1 ml (GE Healthcare Life Sciences) column was used in the first purification step. The eluted peak was redirected on a 5 ml HiTrap desalting column (GE Healthcare Life Sciences) and collected in 50 mM Tris–HCl (pH 8.0), 0.15 M NaCl, and 10% glycerol. Decarboxylases were purified by loading the clear crude cell extract from 50 ml cell culture onto a nickel–nitrilotriacetic acid spin column (QIAGEN), according to the manufacturer’s instructions. Proteins were eluted with 50 mM phosphate (pH 7.5), 50 mM NaCl, 250 mM imidazole, and 10% glycerol. For the cell culture and expression of the recombinant proteins for the enzymatic screening (dehydrogenases, amine oxidases, and aminotransferases), all the procedures were conducted in 96-well plates as previously described (74).
Enzymatic assays
Candidate l-aspartate decarboxylase activity was assayed (LC/MS/MS) by monitoring the formation of β-alanine from l-aspartate, using PanD as the positive control. 1 mM l-aspartate was incubated in 150 μl of 100 mM Tris–HCl (pH 8.0) containing 1 mM EDTA, 0.5 mM PLP, and 10 μg of each candidate enzyme for 2 h at 25 °C. A negative control was incubated in the same conditions in the absence of enzyme. Enzymatic reactions were stopped by ultrafiltration and diluted four-fold in 80:20 acetonitrile:10 mM ammonium carbonate (pH 9.9) before LC/MS analysis. Experiments were conducted in duplicates.
Ddc activity was assayed in 150 μl of 100 mM Tris–HCl (pH 8.5) containing 1 mM DAB, 200 μM PLP, 2.5 mM CaCl2, and 10 μg of Ddc for 2 h at 25 °C. A negative control was incubated in the same conditions in the absence of enzyme. The reactions were stopped by ultrafiltration, and after derivatization, the detection of DAP and 3AP was performed in LC/MS as described previously.
The following assays were conducted at 25 °C in 96-well plates using a SpectraMax Plus 384 absorbance microplate reader (Molecular Devices). DAP and 3-AP dehydrogenase activities were assayed in recombinant cell-free extracts by measuring the formation of reduced nucleotide at 340 nm, using 2 to 5 μg total proteins in 200 μl of activity buffer (50 mM Tris–HCl, pH 9.0, 200 mM NaCl, and 15% glycerol) containing either 2 mM NADP+ or NAD+ and 20 mM DAP or 4 mM 3AP. A negative control, measuring the reduction of the cofactor unrelated to the substrate oxidation because of the use of crude cell extracts, was carried out without DAP/3AP to correct the calculated specific activities. The positive control for the oxidative deamination assays was 2,4-diaminopentanoate dehydrogenase using (2R,4S)-diaminopentanoate (47). DAP amine oxidase activity was assayed by measuring H2O2 production using a spectrofluorimetric method based on a peroxidase-coupled assay with Ampliflu Red Kit (Merck, Sigma–Aldrich), using Ddc as the positive control. Experiments were conducted in 150 μl 100 mM Tris–HCl, pH 8.0, in the presence of 10 mM DAP, 100 μM Amplex Red, and 2 U ml−1 horseradish peroxidase. Upon addition of 2 to 5 μg proteins of recombinant cell-free extracts, fluorescence emission was recorded for 15 min. A negative control was carried out in the same conditions without proteins.
DAP aminotransferase activity was assayed in 150 μl 100 mM Tris–HCl (pH 9.0) containing 10 mM DAP, 10 mM 2-oxoglutarate, 2 U of bovine liver l-glutamate dehydrogenase, 5 mM NAD+, 200 μM PLP, and 1 mM KCl. The reactions were initiated by the addition of ∼5 μg proteins of recombinant cell-free extracts; NAD+ reduction was measured continuously at 340 nm. A negative control was carried out in the same conditions without proteins. DAP amine oxidase and aminotransferase assays were conducted using a Xenius XC spectrofluorometer/spectrophotometer (SAFAS).
Kinetic characterizations
Dehydrogenase activity of the four candidate enzymes was determined with 3AP as the substrate and NAD+ or NADP+ as the cofactor. The formation of the reduced nucleotide was monitored continuously at 340 nm. Reactions were carried out in 100 μl of activity buffer.
GabT was characterized in a coupled assay with l-glutamate dehydrogenase in the presence of NAD+, by monitoring the formation of the reduced nucleotide. The assay was conducted in the presence of DAP or gamma-aminobutyric acid using 500 μM 2-oxoglutarate, 2 U of bovine liver l-glutamate dehydrogenase, 200 μM PLP, and 5 mM NAD+ in 100 μl of activity buffer. Reactions were initiated by the addition of the enzyme. Specific activities of GabT, AstC, and Dat were determined in a coupled assay with BetB. The reactions were carried out in activity buffer in the presence of 10 mM DAP, 5 mM 2-oxoglutarate, 200 μM PLP, and 200 μM NAD+. The reactions were initiated by the addition of 35 μg of the enzyme; NAD+ reduction was measured continuously at 340 nm.
Ddc amine oxidase activity was characterized in a coupled assay with BetB in the presence of NAD+ by monitoring the formation of NADH. Assays were conducted in 100 μl of activity buffer containing 1 mM NAD+, 200 μM PLP, 2.5 mM CaCl2, and BetB with various DAP concentrations. The reactions were initiated by the addition of 45 μg of Ddc; NAD+ reduction was measured continuously at 340 nm.
Spectrophotometric assays were performed at 25 °C in a UVmc2 double beam spectrophotometer (SAFAS).
Ddc decarboxylase activity was monitored by LC/MS. Enzymatic reactions were performed at 25 °C by incubating 0.5 μg Ddc in 100 μl of 100 mM Tris–HCl (pH 8.5), 200 μM PLP, and 2.5 mM CaCl2 with varying concentrations of DAB for 0, 10, 20, 40, or 60 min, and stopped with 1 μl of trifluoroacetic acid. Accumulation of DAP with time was linear up to 60 min. Derivatized DAP was quantified by mass spectrometry as described previously.
Kinetic parameters were determined from duplicate experiments by nonlinear analysis of initial rates using SigmaPlot 9.0 (Systat Software, Inc).
Whole genome sequencing and mutational analysis
ΔpanD was grown in 5 ml of MA supplemented with 20 mM succinate. Genomic DNA was purified using ZR-Duet DNA/RNA MiniPrep Plus kit (Zymo Research) and quantified using a Qubit Fluorometer (Thermo Fisher Scientific). For Illumina sequencing, 1500 ng DNA were sonicated to a 500 to 600 bp size range using the E220 Covaris Focused-Ultrasonicator instrument (Covaris, Inc). The fragments were end-repaired, then 3′-adenylated, and NEXTflex HT Barcodes (Bioo Scientific Corporation) were added using NEBNext DNA modules products (New England Biolabs). The ligated products were cleaned up with two consecutive 1× AMPure XP (Beckman Coulter). After library-profile analysis conducted by an Agilent 2100 Bioanalyzer (Agilent Technologies) and quantitative PCR quantification (MxPro; Agilent Technologies), the library was sequenced using a Illumina MiSeq with MiSeq Reagent Kit v2 (2 × 250 bp; Illumina, Inc). A total of 1.8 × 106 paired-end reads were obtained with genome coverage of ∼250. To control the absence of panD in the mutant genome, sequencing reads were first mapped onto the ADP1 strain genome reference (BWA-MEM software, version 0.7.4) (75).
Variant calling
High-throughput sequencing data were analyzed using the PALOMA bioinformatic pipeline implemented in the MicroScope platform (76). First, reads were mapped onto the ADP1 strain reference genome using the SSAHA2 package (version 2.5.1) (77). Only unique matches having an alignment score equal to at least half of their length were retained as seeds for full Smith–Waterman realignment (78) with a region extended on both sides by five nucleotides of the reference genome. Then, all computed alignments were screened for discrepancies between read and reference sequences, and a score based on coverage, allele frequency, quality of bases, and strand bias was computed for each detected event to assess its relevance. Finally the mutation (SNPs and small indels) with a score superior to 0.8 and with at least 10 supporting reads was kept.
Construction of deletion mutants
The construction of single-gene deletion mutants in ADP1 was already described (15). Oligonucleotide primers used are listed in Table S4. Two techniques were considered here to generate strains with a double-gene knockout. First, the kanR cassette in ΔpanD was removed using the tdk negative selection marker to generate unmarked deletions and construct double mutants with the same resistance (14). Second, two different antibiotic resistance cassettes, KanR and AprR, were used, which allowed the selection of the double-gene knockout mutants on minimal medium agar plates supplemented with 30 μg/ml kanamycin and 30 μg/ml apramycin. ΔpanD:betB, ΔpanD:acoD, ΔpanD:adhA, and ΔpanD:gabT were obtained with the first method, whereas ΔpanD:ddc was obtained with the second one. Mutants were then grown on MA supplemented with 20 mM succinate as the carbon source, 10 μM of β-alanine, 1 mM DAP, and the desired antibiotic(s).
Data availability
All data used for the study are presented or cited in the main article or the supporting information. The raw mass spectrometry data are available upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Véronique de Berardinis, Marcel Salanoubat, and the late Annett Kreimeyer for all the fruitful discussions during the conceptualization of this project. We are grateful to Christine Pellé and Peggy Sirvain for excellent technical assistance in protein purification. This work was supported by Commissariat à l’Energie Atomique et aux Energies Alternatives, the CNRS, and the Université Evry-Val-d’Essonne/Université Paris-Saclay.
Author contributions
N. P. and A. P., conceptualization; N. P., C. D., R. M-G., S. D., C. L., D. R., S. F., E. D., and A. P., formal analysis; N. P., C. D., R. M-G., S. D., C. L., investigation; N. P. and A. P., writing–original draft; N.P. and A. P., writing–review & editing.
Edited by Chris Whitfield
Footnotes
Present address for Alain Perret: TBI, Université de Toulouse, CNRS, INRAE, INSA, Toulouse, France.
Supporting information
References
- 1.Copley S.D. Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol. 2003;7:265–272. doi: 10.1016/s1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 2.D'Ari R., Casadesus J. Underground metabolism. Bioessays. 1998;20:181–186. doi: 10.1002/(SICI)1521-1878(199802)20:2<181::AID-BIES10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Jensen R.A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 4.Khersonsky O., Tawfik D.S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 5.Nobeli I., Favia A.D., Thornton J.M. Protein promiscuity and its implications for biotechnology. Nat. Biotechnol. 2009;27:157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- 6.Notebaart R.A., Szappanos B., Kintses B., Pal F., Gyorkei A., Bogos B., et al. Network-level architecture and the evolutionary potential of underground metabolism. Proc. Natl. Acad. Sci. U. S. A. 2014;111:11762–11767. doi: 10.1073/pnas.1406102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J., Kershner J.P., Novikov Y., Shoemaker R.K., Copley S.D. Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5'-phosphate synthesis. Mol. Syst. Biol. 2010;6:436. doi: 10.1038/msb.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patrick W.M., Quandt E.M., Swartzlander D.B., Matsumura I. Multicopy suppression underpins metabolic evolvability. Mol. Biol. Evol. 2007;24:2716–2722. doi: 10.1093/molbev/msm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blank D., Wolf L., Ackermann M., Silander O.K. The predictability of molecular evolution during functional innovation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3044–3049. doi: 10.1073/pnas.1318797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLoughlin S.Y., Copley S.D. A compromise required by gene sharing enables survival: implications for evolution of new enzyme activities. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13497–13502. doi: 10.1073/pnas.0804804105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasvall J., Sun L., Roth J.R., Andersson D.I. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338:384–387. doi: 10.1126/science.1226521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotton C.A., Bernhardsgrutter I., He H., Burgener S., Schulz L., Paczia N., et al. Underground isoleucine biosynthesis pathways in E. coli. Elife. 2020;9 doi: 10.7554/eLife.54207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Berardinis V., Durot M., Weissenbach J., Salanoubat M. Acinetobacter baylyi ADP1 as a model for metabolic system biology. Curr. Opin. Microbiol. 2009;12:568–576. doi: 10.1016/j.mib.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Metzgar D., Bacher J.M., Pezo V., Reader J., Doring V., Schimmel P., et al. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucl. Acids Res. 2004;32:5780–5790. doi: 10.1093/nar/gkh881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Berardinis V., Vallenet D., Castelli V., Besnard M., Pinet A., Cruaud C., et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramjee M.K., Genschel U., Abell C., Smith A.G. Escherichia coli L-aspartate-alpha-decarboxylase: preprotein processing and observation of reaction intermediates by electrospray mass spectrometry. Biochem. J. 1997;323:661–669. doi: 10.1042/bj3230661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williamson J.M., Brown G.M. Purification and properties of L-Aspartate-alpha-decarboxylase, an enzyme that catalyzes the formation of beta-alanine in Escherichia coli. J. Biol. Chem. 1979;254:8074–8082. [PubMed] [Google Scholar]
- 18.Cronan J.E., Jr. Beta-alanine synthesis in Escherichia coli. J. Bacteriol. 1980;141:1291–1297. doi: 10.1128/jb.141.3.1291-1297.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortega M.V., Cardenas A., Ubiera D. panD, a new chromosomal locus of Salmonella typhimurium for the biosynthesis of beta-alanine. Mol. Gen. Genet. 1975;140:159–164. doi: 10.1007/BF00329783. [DOI] [PubMed] [Google Scholar]
- 20.Cona A., Rea G., Angelini R., Federico R., Tavladoraki P. Functions of amine oxidases in plant development and defence. Trends Plant Science. 2006;11:80–88. doi: 10.1016/j.tplants.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Terano S., Suzuki Y. Formation of β-alanine from spermine and spermidine in maize shoots. Phytochemistry. 1978;17:148–149. [Google Scholar]
- 22.White W.H., Gunyuzlu P.L., Toyn J.H. Saccharomyces cerevisiae is capable of de Novo pantothenic acid biosynthesis involving a novel pathway of beta-alanine production from spermine. J. Biol. Chem. 2001;276:10794–10800. doi: 10.1074/jbc.M009804200. [DOI] [PubMed] [Google Scholar]
- 23.Heim W.G., Sykes K.A., Hildreth S.B., Sun J., Lu R.H., Jelesko J.G. Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript. Phytochemistry. 2007;68:454–463. doi: 10.1016/j.phytochem.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki Y. Purification and characterization of diamine oxidase from Triticum aestivum shoots. Phytochemistry. 1996;42:291–293. [Google Scholar]
- 25.Zarei A., Trobacher C.P., Shelp B.J. NAD(+)-aminoaldehyde dehydrogenase candidates for 4-aminobutyrate (GABA) and beta-alanine production during terminal oxidation of polyamines in apple fruit. FEBS Lett. 2015;589:2695–2700. doi: 10.1016/j.febslet.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Zarei A., Trobacher C.P., Shelp B.J. Arabidopsis aldehyde dehydrogenase 10 family members confer salt tolerance through putrescine-derived 4-aminobutyrate (GABA) production. Sci. Rep. 2016;6:35115. doi: 10.1038/srep35115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landry J., Sternglanz R. Yeast Fms1 is a FAD-utilizing polyamine oxidase. Biochem. Biophys. Res. Commun. 2003;303:771–776. doi: 10.1016/s0006-291x(03)00416-9. [DOI] [PubMed] [Google Scholar]
- 28.White W.H., Skatrud P.L., Xue Z., Toyn J.H. Specialization of function among aldehyde dehydrogenases: the ALD2 and ALD3 genes are required for beta-alanine biosynthesis in Saccharomyces cerevisiae. Genetics. 2003;163:69–77. doi: 10.1093/genetics/163.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duhazé C., Gagneul D., Leport L., Robert Larher F., Bouchereau A. Uracil as one of the multiple sources of β-alanine in Limonium latifolium, a halotolerant β-alanine betaine accumulating Plumbaginaceae. Plant Physiol. Biochem. 2003;41:993–998. [Google Scholar]
- 30.Campbell L.L. Reductive degradation of pyrimidines. 5. Enzymatic conversion of N-carbamyl-beta-alanine to beta-alanine, carbon dioxide, and ammonia. J. Biol. Chem. 1960;235:2375–2378. [PubMed] [Google Scholar]
- 31.Parthasarathy A., Savka M.A., Hudson A.O. The synthesis and role of beta-alanine in plants. Front. Plant Sci. 2019;10:921. doi: 10.3389/fpls.2019.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rathinasabapathi B. Propionate, a source of β-alanine, is an inhibitor of β-alanine methylation in Limonium latifolium, Plumbaginaceae. J. Plant Physiol. 2002;159:671–674. [Google Scholar]
- 33.Stadtman E.R. The enzymatic synthesis OF β-ALANYL coenzyme a. J. Am. Chem. Soc. 1955;77:5765–5766. [Google Scholar]
- 34.Hamana K., Matsuzaki S. Diaminopropane occurs ubiquitously in Acinetobacter as the major polyamine. J. Gen. Appl. Microbiol. 1992;38:191–194. [Google Scholar]
- 35.Kämpfer P., Bark K., Busse H.-J., Auling G., Dott W. Numerical and chemotaxonomy of polyphosphate accumulating acinetobacter strains with high polyphosphate: AMP phosphotransferase (PPAT) activity. Syst. Appl. Microbiol. 1992;15:409–419. [Google Scholar]
- 36.Auling G., Pilz F., Busse H.J., Karrasch S., Streichan M., Schon G. Analysis of the polyphosphate-accumulating microflora in phosphorus-eliminating, anaerobic-aerobic activated sludge systems by using diaminopropane as a biomarker for rapid estimation of Acinetobacter spp. Appl. Environ. Microbiol. 1991;57:3585–3592. doi: 10.1128/aem.57.12.3585-3592.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skiebe E., de Berardinis V., Morczinek P., Kerrinnes T., Faber F., Lepka D., et al. Surface-associated motility, a common trait of clinical isolates of Acinetobacter baumannii, depends on 1,3-diaminopropane. Int. J. Med. Microbiol. : IJMM. 2012;302:117–128. doi: 10.1016/j.ijmm.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Funahashi T., Tanabe T., Maki J., Miyamoto K., Tsujibo H., Yamamoto S. Identification and characterization of a cluster of genes involved in biosynthesis and transport of acinetoferrin, a siderophore produced by Acinetobacter haemolyticus ATCC 17906T. Microbiology. 2013;159:678–690. doi: 10.1099/mic.0.065177-0. [DOI] [PubMed] [Google Scholar]
- 39.Okujo N., Sakakibara Y., Yoshida T., Yamamoto S. Structure of acinetoferrin, a new citrate-based dihydroxamate siderophore from Acinetobacter haemolyticus. Biometals. 1994;7:170–176. doi: 10.1007/BF00140488. [DOI] [PubMed] [Google Scholar]
- 40.Barbe V., Vallenet D., Fonknechten N., Kreimeyer A., Oztas S., Labarre L., et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucl. Acids Res. 2004;32:5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durot M., Le Fevre F., de Berardinis V., Kreimeyer A., Vallenet D., Combe C., et al. Iterative reconstruction of a global metabolic model of Acinetobacter baylyi ADP1 using high-throughput growth phenotype and gene essentiality data. BMC Syst. Biol. 2008;2:85. doi: 10.1186/1752-0509-2-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackowski S., Rock C.O. Regulation of coenzyme A biosynthesis. J. Bacteriol. 1981;148:926–932. doi: 10.1128/jb.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Awal H.M.A., Yoshida I., Doe M., Hirasawa E. 3-aminopropionaldehyde dehydrogenase of millet shoots. Phytochemistry. 1995;40:393–395. [Google Scholar]
- 44.Duhazé C., Gouzerh G., Gagneul D., Larher F., Bouchereau A. The conversion of spermidine to putrescine and 1,3-diaminopropane in the roots of Limonium tataricum. Plant Sci. 2002;163:639–646. [Google Scholar]
- 45.Cobessi D., Tete-Favier F., Marchal S., Azza S., Branlant G., Aubry A. Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J. Mol. Biol. 1999;290:161–173. doi: 10.1006/jmbi.1999.2853. [DOI] [PubMed] [Google Scholar]
- 46.Bar-Even A., Noor E., Savir Y., Liebermeister W., Davidi D., Tawfik D.S., et al. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 47.Fonknechten N., Perret A., Perchat N., Tricot S., Lechaplais C., Vallenet D., et al. A conserved gene cluster rules anaerobic oxidative degradation of L-ornithine. J. Bacteriol. 2009;191:3162–3167. doi: 10.1128/JB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kreimeyer A., Perret A., Lechaplais C., Vallenet D., Medigue C., Salanoubat M., et al. Identification of the last unknown genes in the fermentation pathway of lysine. J. Biol. Chem. 2007;282:7191–7197. doi: 10.1074/jbc.M609829200. [DOI] [PubMed] [Google Scholar]
- 49.Yorifuji T., Ishihara T., Naka T., Kondo S., Shimizu E. Purification and characterization of polyamine aminotransferase of Arthrobacter sp. TMP-1. J. Biochem. 1997;122:537–543. doi: 10.1093/oxfordjournals.jbchem.a021786. [DOI] [PubMed] [Google Scholar]
- 50.Yorifuji T., Yonaha K., Shimizu E., Ichikawa H., Matsumoto F., Tabata Y. Diaminopropane aminotransferase, a novel enzyme of a coryneform bacterium. Agric. Biol. Chem. 1991;55:2187–2189. [Google Scholar]
- 51.Durham D.R., Perry J.J. Purification and characterization of a heme-containing amine dehydrogenase from Pseudomonas putida. J. Bacteriol. 1978;134:837–843. doi: 10.1128/jb.134.3.837-843.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eady R.R., Large P.J. Purification and properties of an amine dehydrogenase from Pseudomonas AM1 and its role in growth on methylamine. Biochem. J. 1968;106:245–255. doi: 10.1042/bj1060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Husain M., Davidson V.L. Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans. J. Bacteriol. 1987;169:1712–1717. doi: 10.1128/jb.169.4.1712-1717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kondo T., Kondo E., Maki H., Yasumoto K., Takagi K., Kano K., et al. Purification and characterization of aromatic amine dehydrogenase from Alcaligenes xylosoxidans. Biosci. Biotechnol. Biochem. 2004;68:1921–1928. doi: 10.1271/bbb.68.1921. [DOI] [PubMed] [Google Scholar]
- 55.Datta S., Mori Y., Takagi K., Kawaguchi K., Chen Z.W., Okajima T., et al. Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14268–14273. doi: 10.1073/pnas.241429098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vandenberghe I., Kim J.K., Devreese B., Hacisalihoglu A., Iwabuki H., Okajima T., et al. The covalent structure of the small subunit from Pseudomonas putida amine dehydrogenase reveals the presence of three novel types of internal cross-linkages, all involving cysteine in a thioether bond. J. Biol. Chem. 2001;276:42923–42931. doi: 10.1074/jbc.M107164200. [DOI] [PubMed] [Google Scholar]
- 57.Large P.J. Enzymes and pathways of polyamine breakdown in microorganisms. FEMS Microbiol. Rev. 1992;8:249–262. doi: 10.1111/j.1574-6968.1992.tb04991.x. [DOI] [PubMed] [Google Scholar]
- 58.Pontrelli S., Fricke R.C.B., Teoh S.T., Lavina W.A., Putri S.P., Fitz-Gibbon S., et al. Metabolic repair through emergence of new pathways in Escherichia coli. Nat. Chem. Biol. 2018;14:1005–1009. doi: 10.1038/s41589-018-0149-6. [DOI] [PubMed] [Google Scholar]
- 59.John R.A. Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta. 1995;1248:81–96. doi: 10.1016/0167-4838(95)00025-p. [DOI] [PubMed] [Google Scholar]
- 60.Bertoldi M., Cellini B., Montioli R., Borri Voltattorni C. Insights into the mechanism of oxidative deamination catalyzed by DOPA decarboxylase. Biochemistry. 2008;47:7187–7195. doi: 10.1021/bi800478s. [DOI] [PubMed] [Google Scholar]
- 61.Bertoldi M., Gonsalvi M., Contestabile R., Voltattorni C.B. Mutation of tyrosine 332 to phenylalanine converts dopa decarboxylase into a decarboxylation-dependent oxidative deaminase. J. Biol. Chem. 2002;277:36357–36362. doi: 10.1074/jbc.M204867200. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto S., Tsuzaki Y., Tougou K., Shinoda S. Purification and characterization of L-2,4-diaminobutyrate decarboxylase from Acinetobacter calcoaceticus. J. Gen. Microbiol. 1992;138:1461–1465. doi: 10.1099/00221287-138-7-1461. [DOI] [PubMed] [Google Scholar]
- 63.Chattopadhyay M.K., Tabor C.W., Tabor H. Polyamines are not required for aerobic growth of Escherichia coli: Preparation of a strain with deletions in all of the genes for polyamine biosynthesis. J. Bacteriol. 2009;191:5549–5552. doi: 10.1128/JB.00381-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stuani L., Lechaplais C., Salminen A.V., Segurens B., Durot M., Castelli V., et al. Novel metabolic features in Acinetobacter baylyi ADP1 revealed by a multiomics approach. Metabolomics. 2014;10:1223–1238. doi: 10.1007/s11306-014-0662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lo C.C., Bonner C.A., Xie G., D'Souza M., Jensen R.A. Cohesion group approach for evolutionary analysis of aspartokinase, an enzyme that feeds a branched network of many biochemical pathways. Microbiol. Mol. Biol. Rev. 2009;73:594–651. doi: 10.1128/MMBR.00024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parthasarathy A., Adams L.E., Savka F.C., Hudson A.O. The Arabidopsis thaliana gene annotated by the locus tag At3g08860 encodes alanine aminotransferase. Plant Direct. 2019;3 doi: 10.1002/pld3.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J., Sperandio V., Frantz D.E., Longgood J., Camilli A., Phillips M.A., et al. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J. Biol. Chem. 2009;284:9899–9907. doi: 10.1074/jbc.M900110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michael A.J. Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 2016;473:2315–2329. doi: 10.1042/BCJ20160185. [DOI] [PubMed] [Google Scholar]
- 69.Lopez-Samano M., Beltran L.F.L., Sanchez-Thomas R., Davalos A., Villasenor T., Garcia-Garcia J.D., et al. A novel way to synthesize pantothenate in bacteria involves beta-alanine synthase present in uracil degradation pathway. Microbiologyopen. 2020;9 doi: 10.1002/mbo3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim S.H., Wang Y., Khomutov M., Khomutov A., Fuqua C., Michael A.J. The essential role of spermidine in growth of agrobacterium tumefaciens is determined by the 1,3-diaminopropane moiety. ACS Chem. Biol. 2016;11:491–499. doi: 10.1021/acschembio.5b00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lynch D., O'Brien J., Welch T., Clarke P., Cuiv P.O., Crosa J.H., et al. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 2001;183:2576–2585. doi: 10.1128/JB.183.8.2576-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas M., Stuani L., Darii E., Lechaplais C., Pateau E., Tabet J.C., et al. De novo structure determination of 3-((3-aminopropyl)amino)-4-hydroxybenzoic acid, a novel and abundant metabolite in Acinetobacter baylyi ADP1. Metabolomics. 2019;15:45. doi: 10.1007/s11306-019-1508-3. [DOI] [PubMed] [Google Scholar]
- 73.Vergne-Vaxelaire C., Bordier F., Fossey A., Besnard-Gonnet M., Debard A., Mariage A., et al. Nitrilase activity screening on structurally diverse substrates: providing biocatalytic tools for organic synthesis. Adv. Synth. Catal. 2013;355:1763–1779. [Google Scholar]
- 74.Bastard K., Smith A.A., Vergne-Vaxelaire C., Perret A., Zaparucha A., De Melo-Minardi R., et al. Revealing the hidden functional diversity of an enzyme family. Nat. Chem. Biol. 2014;10:42–49. doi: 10.1038/nchembio.1387. [DOI] [PubMed] [Google Scholar]
- 75.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Quantitative Biol. 2013;0:1–3. [Google Scholar]
- 76.Vallenet D., Calteau A., Dubois M., Amours P., Bazin A., Beuvin M., et al. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucl. Acids Res. 2020;48:D579–D589. doi: 10.1093/nar/gkz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ning Z., Cox A.J., Mullikin J.C. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used for the study are presented or cited in the main article or the supporting information. The raw mass spectrometry data are available upon request.