Abstract
Significant substratum damage can occur when plasticized PVC (pPVC) is colonized by microorganisms. We investigated microbial colonization of pPVC in an in situ, longitudinal study. Pieces of pPVC containing the plasticizers dioctyl phthalate and dioctyl adipate (DOA) were exposed to the atmosphere for up to 2 years. Fungal and bacterial populations were quantified, and colonizing fungi were identified by rRNA gene sequencing and morphological characteristics. Aureobasidium pullulans was the principal colonizing fungus, establishing itself on the pPVC between 25 and 40 weeks of exposure. A group of yeasts and yeast-like fungi, including Rhodotorula aurantiaca and Kluyveromyces spp., established themselves on the pPVC much later (after 80 weeks of exposure). Numerically, these organisms dominated A. pullulans after 95 weeks, with a mean viable count ± standard error of 1,000 ± 200 yeast CFU cm−2, compared to 390 ± 50 A. pullulans CFU cm−2. No bacterial colonization was observed. We also used in vitro tests to characterize the deteriogenic properties of fungi isolated from the pPVC. All strains of A. pullulans tested could grow with the intact pPVC formulation as the sole source of carbon, degrade the plasticizer DOA, produce extracellular esterase, and cause weight loss of the substratum during growth in vitro. In contrast, several yeast isolates could not grow on pPVC or degrade DOA. These results suggest that microbial succession may occur during the colonization of pPVC and that A. pullulans is critical to the establishment of a microbial community on pPVC.
Plasticized PVC (pPVC) is highly susceptible to microbial attack in many different environmental situations. The problem was first identified in U.S. government reports of the deterioration of military equipment (8, 44), and subsequent reports described defacement and deterioration of commercial pPVC products (20, 50). Biodeterioration of pPVC is now known to occur in a wide range of industrial, commercial, and structural applications (18, 19, 22).
The susceptibility of pPVC results from the presence of plasticizers, commonly organic acid esters such as dioctyl phthalate (DOP) and dioctyl adipate (DOA), added to modify physical or mechanical properties of the polymer. Both bacteria (6, 7, 14) and fungi (5, 36) can degrade ester-based plasticizers. Loss of plasticizers from pPVC due to microbial degradation results in brittleness, shrinkage, and ultimately failure of the pPVC in its intended application.
Microbial deterioration of pPVC has been studied extensively in vitro. Many studies have examined the resistance of pPVC formulations incorporating biocides to colonization by test organisms (38, 39, 46). Other research has determined biodegradability by measuring changes in the physical properties of pPVC, such as changes in tensile strength (49), mass (9), or electrical properties (42) during biodegradation. Several international standard test methods for microbiological susceptibility of plastics have been established (1, 2, 26).
Colonization processes occurring on pPVC in the environment have received comparatively little attention. Nothing is known about the temporal sequence of microbial colonization of pPVC in situ. Existing studies have examined fungal defacement of pPVC in tropical or subtropical climates (24, 40). In both studies fungal growth was evaluated with a subjective, visual assessment of defacement of the pPVC. Neither study examined the role of bacteria in the colonization process. Further, unrecognized fungal growth was normally identified only to genus level with basic morphological techniques. Recently, RNA gene (rDNA) sequencing has been used as a rapid and reliable tool for the identification of fungi to the species level (23). This technique has not been used to identify microorganisms that colonize and deteriorate pPVC in the environment.
We examined the microbial colonization of pPVC in situ by exposing pPVC to the atmosphere in a longitudinal experiment. The principal objectives of this work were (i) to investigate colonization processes, (ii) to identify important deteriogenic organisms to the species level using rDNA sequencing, and (iii) to determine if there was a relationship between the microbial colonization sequence observed in situ and the ability of microorganisms to cause biodeterioration of pPVC in laboratory tests.
MATERIALS AND METHODS
Culture media and maintenance.
Fungi and yeasts enriched from pPVC exposed to the atmosphere were maintained on malt extract agar (MEA) (Oxoid, Basingstoke, United Kingdom), a medium used widely for the detection, isolation, and enumeration of fungi. Bacteria were maintained on R2A medium (35) (Difco, Detroit, Mich.), which contains low concentrations of organic nutrients and is used routinely for the enrichment of bacteria from oligotrophic environments. For long term storage at −80°C, fungal spores were harvested from agar plates and frozen in 20% (vol/vol) glycerol. The basal mineral salts medium (MSM) used for determining the deteriogenic properties of organisms contained the following (in grams · liter of distilled H2O−1): K2HPO4, 7; KH2PO4, 3; MgSO4 · 7H2O, 0.1; and (NH4)2SO4, 1. DOA agar, used for the isolation of organisms able to degrade the plasticizer DOA, contained MSM supplemented with 2 ml of DOA liter−1 and 15 g of bacteriological agar (Oxoid) liter−1. For preparation of DOA agar, medium including the DOA was autoclaved at 121°C for 15 min and allowed to cool to approximately 50°C. An emulsion of plasticizer was then created within the medium using a homogenizer at full power for 2 min (260 W, 25,000 rpm; model D-7801; Ystral, Hemel Hempstead, United Kingdom). DOA agar plates were poured immediately after homogenization. DOA liquid medium was prepared as DOA agar except without agar.
Plasticized PVC.
Sheets of pPVC, 0.5 mm thick, were formulated that contained the following components (parts per hundred resin): EP 6779 PVC resin, (European Vinyls Corporation Ltd., Runcorne, United Kingdom), 75; Vinnolit C65V PVC resin (Vinnolit, Cologne, Germany), 25; DOP plasticizer (Exxon Chemicals, Southampton, United Kingdom), 25; DOA plasticizer (Exxon Chemicals), 25; Lankromark LN138 calcium-zinc stabilizer, (Akcros Chemicals, Burnley, United Kingdom), 2; Lankroflex ED63 epoxidized oleate ester (Akcros Chemicals), 3; and titanium dioxide pigment (Tioxide Europe, Grimsby, United Kingdom), 10. Individual pPVC pieces were 4.2 by 7 cm and had two 6-mm-diameter holes in the corners of one of the long edges for attachment to in situ support racks.
Exposure of pPVC in situ.
Three replicate support racks were constructed on-site at Avecia Biocides (Blackley, Manchester, United Kingdom). Each rack consisted of three lines of 8-mm-diameter polypropylene wound rope (Stevecraft, Manchester, United Kingdom), positioned 1.0, 1.5, and 2.0 m above ground level and held between two steel poles set into concrete and at 7 m apart. Nylon cable ties (4-mm width; RS Components, Corby, United Kingdom) were inserted through the rope windings to support pPVC pieces at numbered locations on the rack. Each rack supports up to 300 pPVC pieces which are held free-hanging in order to eliminate fixed-orientation effects. The three racks were positioned in parallel 1 m apart and were orientated at 60° to the horizontal to limit cross contamination of specimens during rainfall. Ten pieces of pPVC were positioned on each of the three racks at locations chosen using a random number table.
At each sampling time, three replicate pPVC pieces were selected at random, one from each rack. Each piece was cut into three 4.2-by-2.5-cm sections, and a separate analysis was carried out on each section.
Viable counts of fungi, bacteria, and DOA-degrading microorganisms.
pPVC sections (4.2 by 2.5 cm) were placed into 25-ml universal tubes (32-mm diameter; 80-mm length) containing 10 ml of sterile distilled H2O. The tubes were shaken vigorously in an automatic side-arm shaker (Gallenkamp, Leicester, United Kingdom) for 1 min. Plasticized PVC samples were transferred to a petri dish containing 5 ml of H2O and scraped heavily three times on both sides using the flat edge of a sterile scalpel blade. The pPVC, H2O, and scalpel blade were then returned to the universal tubes and shaken for a further minute using the sidearm shaker. A dilution series to 10−3 was prepared from each universal tube. Aliquots of 0.2 ml from each dilution were spread onto three replicate plates each of malt extract, R2A, and DOA agar. Viable counts were performed on MEA plates after 5 days of incubation at 25°C, on R2A plates after 7 days of incubation, and on plasticizer agar plates after 14 days of incubation. To investigate whether statistically significant changes in CFU counts occurred between sample times, overall mean viable counts for three replicate pPVC pieces at each time point were compared using analysis of variance.
Viable counts of fungi within the atmosphere.
As a control, to determine whether DOA-degrading organisms could be isolated from the atmosphere, plates of both MEA and DOA agar media were exposed to the atmosphere at a height of 20 m on the roof of the Stopford Building, Manchester, United Kingdom, in March 1997 (during United Kingdom's winter). Three replicate plates of each medium were retrieved after 20 h of exposure to the atmosphere. The number of fungal CFU on each medium was counted after 3, 5, and 7 days of incubation at 25°C.
Scanning electron microscopy (SEM).
pPVC pieces exposed in situ for 95 weeks were rapidly frozen in liquid nitrogen. Specimens were freeze-dried overnight (model B5A; British Oxygen Company Edwards, Crawley, United Kingdom). Specimens were attached to stubs using Electrodag 915 (Acheson Industries, Reading, United Kingdom) and sputter coated (model S150 device; BOC Edwards) with gold before being examined using a Stereoscan 360 scanning electron microscope (Cambridge Instruments, Cambridge, United Kingdom).
Identification of fungi isolated from pPVC.
Fungal isolates were identified by PCR amplification and partial sequencing of the internally transcribed spacer (ITS) regions and the 5.8S rDNA or of the V3 domain of large subunit (28S) rDNA. Initially only the ITS region was sequenced. However, when this sequence was insufficient to establish identity, the V3 region also was sequenced.
For preparation of genomic DNA, fresh mycelia or yeasts cells were harvested in deionized water from overnight plate cultures grown on MEA. The biomass was pelleted by centrifugation at 8,000 × g for 10 min, and the supernatant was discarded. The pellet was frozen by placing the centrifuge tube into liquid nitrogen, and tubes were then stored at −80°C until the pellet was ground in a mortar under liquid N2. DNA was extracted according to the method of Anderson et al. (3). DNA was observed following electrophoresis in 1% agarose in TPE buffer (90 mM Tris-phosphate, 2 mM EDTA) and staining with ethidium bromide (1 μg ml−1 in TPE buffer).
The V3 variable region at the 5′ end of the 28S rDNA was amplified with the fungal universal primers V3-1 (5′ GCATATCAATAAGCGGAGGAAAAG) and V3-2 (5′ GGTCCGTGTTTCAAGACGG) (16). PCR reagent concentrations were 0.2 μM for primers V3-1 and V3-2, 2.5 mM MgCl2, 200 μM for each of the four deoxynucleoside triphosphates, and 1.25 U of Taq DNA polymerase (Roche Diagnostics Ltd., Lewes, United Kingdom) per 50-μl reaction mixture. Amplification was performed for 30 cycles with denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min. ITS regions were amplified using fungal universal primers ITS-1 (5′ TCCGTAGGTGAACCTGCGG) and ITS-4 (5′ TCCTCCGCTTATTGATATGC) (45). PCR reagent concentrations were as for V3-1 and V3-2 with the exception of concentrations of 0.25 μM for primers ITS-1 and ITS-4 and 1.5 mM for MgCl2. Amplification was performed for 35 cycles with denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min. Amplified products were purified using the QIAquick PCR purification kit (Qiagen Ltd., Crawley, United Kingdom).
Both strands of the amplified products were sequenced using the ABI BigDye Dideoxy Terminator Cycle Sequencing kit (Applied Biosystems Inc., Warrington, United Kingdom). Cycle-sequencing conditions were denaturation at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 4 min for 25 cycles, with a final extension at 60°C for 4 min. The annealing temperature was increased to 55°C for sequencing reactions using the V3-1 primer. Forward and reverse sequences were aligned using ABI Autoassembler software (Applied Biosystems Inc.), and the overlapping consensus sequence was compared with sequences in the EMBL fungal DNA database using Fasta3 sequence homology searches. Isolates that were identified using their rDNA sequences were compared with published descriptions of colony and conidial morphology using microscopic examination. A number of isolates, including those for which no rDNA identity was found, were identified using morphological techniques at the International Mycological Institute at Commonwealth Agricultural Bureau International Bioscience, Egham, United Kingdom.
In vitro tests for biodeterioration of pPVC.
We characterized the deteriogenic properties of fungi isolated from pPVC and two additional strains of Aureobasidium pullulans. A. pullulans IMI70103 was obtained from CABI Bioscience, Egham, United Kingdom, and A. pullulans PRAFS8 was provided by Avecia Biocides, Manchester, United Kingdom.
(i) Preparation of inocula.
Spores or yeast cells were harvested from MEA plates in deionized water and filtered through three layers of lens tissue paper (Whatman, Maidstone, United Kingdom). Suspensions were washed three times in deionized water by centrifugation at 12,000 × g for 8 min and then adjusted to approximately 106 spores or yeast cells ml−1 in deionized water using a hemocytometer. Suspensions were checked for purity by streaking onto MEA.
(ii) Biomass and extracellular esterase production with DOA as sole carbon source.
Isolates were grown in DOA liquid medium for 2 weeks at 25°C and shaken at 250 rpm. Each 100-ml flask, containing 40 ml of medium, was inoculated with 0.2 ml of spore or yeast suspension. Three replicate flasks were inoculated with each microorganism. For esterase assays, 1.5-ml samples of culture fluid were removed using a syringe, clarified by filtration through a 0.2-μm-pore-size cellulose-nitrate filter (Sartorius, Epsom, United Kingdom), and then either stored at −20°C or used immediately for assays. Cultures of filamentous fungi were then filtered through preweighed filter paper (Whatman no. 1) and dried at 70°C until a constant weight was reached. Yeast cultures were decanted to preweighed 50-ml centrifuge tubes and centrifuged at 3,600 × g for 5 min to pellet cells. The supernatant was discarded, and tubes were incubated at 70°C until a constant weight was reached, usually 24 to 36 hours.
Nonspecific esterase activity was determined by a spectrophotometric assay with p-nitrophenol butyrate (PNB) (Sigma) as substrate (17). Hydrolysis of PNB yields p-nitrophenol, which absorbs maximally at 400 nm under alkaline conditions. The assay mixture (1 ml volume) contained 2.2 mM PNB in sodium acetate buffer (50 mM; pH 5.5) in cuvettes. Culture fluid (500 μl) was added, and cuvettes were incubated at 25°C for 1 h. The reaction mix was then made alkaline by the addition of 0.75 ml of 0.1 M sodium borate (Sigma) before measurement of absorbance at 400 nm. Esterase activity was determined by reference to a standard curve of p-nitrophenol. A relative extracellular esterase (REE) unit was defined as the enzyme activity that liberates 1 nmol of p-nitrophenol from PNB in 1 h at 25°C at pH 5.5. Three replicate measurements of the extracellular esterase activity of all fungal isolates were made on separate occasions.
(iii) pPVC weight loss.
Preweighed pPVC pieces (4.2 by 2.5 cm) were placed into petri dishes containing 30 ml of MSM liquid supplemented with 0.5 g of yeast extract liter−1. Three replicate petri dishes were inoculated with 0.2 ml of spore or yeast suspension from each organism and incubated at 25°C for 6 weeks. Fungal biomass was removed from pPVC pieces by washing in nonionic detergent (LIP Lipsol, Shipley, United Kingdom), and samples were air-dried at room temperature (21 to 24°C) until a constant weight was reached. Statistically significant differences in percentage weight loss relative to control pPVC pieces incubated in sterile medium were determined using analysis of variance.
(iv) Clear-zone production and colony growth on DOA agar.
Clear-zone production on agar plates containing emulsified DOA as the sole carbon source was used to test the ability of fungi to degrade DOA plasticizer. Plates containing 20 ml of DOA agar were inoculated with 50 μl of spore or yeast suspension placed into 5-mm-diameter wells cut at the center of each plate. Three replicate plates were inoculated for each organism and incubated at 25°C for 14 days. Clear-zone production on DOA-agar was scored according to the following criteria: 0, no clearing; 1, faint clearing below colony; 2, clearing extending beyond colony boundary; 3, intense clearing (agar completely transparent) extending beyond colony boundary. Colony growth was reported as follows: 0, no visible growth; 1, slight growth within inoculation well; 2, colony diameter < 2 cm; 3, colony diameter ≥ 2 cm. Tests for clear-zone production and growth on DOA agar were replicated on three separate occasions.
(v) Growth using pPVC formulation as sole source of carbon.
pPVC pieces (4.2 by 2.5 cm) were placed on 15 ml of solidified MSM agar in a petri dish. A further 10 ml of molten MSM agar, cooled to 45°C, was inoculated with 0.1 ml of spore or yeast suspension and poured over the pPVC. Plates were incubated at 25°C for 4 weeks. The following criteria were used to score growth on the pPVC: 0, no visible growth; 1, slight growth, barely visible; 2, growth clearly visible around the edges of the pPVC; 3, strong growth visible around edges and in the agar above the pPVC.
rDNA sequences.
EMBL accession numbers for rDNA sequences from the 12 fungi identified in this study are as follows (sequences with which matches were made and the percent match for each sequence are shown in parentheses): MZ7, AJ276055 (UO5195, 99.6%); MZ8, AJ276054 (AJ000198, 100%); MZ10, AJ276058 (AIY17066, 99.6%); MZ14, AJ276057 (AJ005674, 94.7%); MZ20, AJ276059 (UO5915, 99.6%); MZ58, AJ276062 (AJ244236, 100%); MZ65, AJ276061 (AJ244236, 99.3%); MZ95, AJ276060 (AF138904, 100%); MZ103, AJ276063 (AF138289, 99.8%); MZ104, AJ276056 (AF033407, 100%); MZ107, AJ276065 (AF050278, 94.8%); MZ109, AJ276064 (U94948, 98.9%).
RESULTS
Viable counts of microorganisms colonizing pPVC in situ.
Numbers of viable fungi, bacteria, and DOA-degrading organisms occurring on pPVC were monitored throughout the in situ trial (Table 1). Fungi were established on the pPVC surface after 40 weeks of exposure (February 1998; during United Kingdom's winter). No significant increase (P = 0.82) in fungal viable counts occurred during the next 15 weeks, but by week 80 (June 1998) the population had more than doubled. A rapid increase to 1,500 ± 220 CFU cm−2 (mean ± standard error) occurred in the last 15 weeks of exposure (December 1988 to March 1989; during United Kingdom's winter). CFU counts of fungi able to produce clear zones on DOA agar varied between 65 and 84% of the MEA fungal count.
TABLE 1.
Viable counts of fungi isolated from pPVC exposed in situ over 95 weeks
| Sample wk (date) | All fungi on MEA (CFU cm−2)a | % DOA-degrading fungib |
|---|---|---|
| 10 (July 1997) | 5 ± 1 | 74 |
| 25 (October 1997) | 2 ± 0 | 65 |
| 40 (February 1998) | 110 ± 27 | 84 |
| 55 (June 1998) | 120 ± 15 | 76 |
| 80 (December 1998) | 260 ± 27 | 71 |
| 95 (March 1999) | 1,500 ± 220 | 80 |
Mean viable counts ± 1 standard error of the mean from three replicate pPVC pieces, one taken from each in situ rack, are shown.
Numbers of fungi recovered on DOA agar expressed as a percentage of all fungi recovered on MEA.
Viable counts of fungi within the atmosphere.
CFU counts were made on MEA and DOA agar plates exposed to the atmosphere for 20 h. The fungal counts (mean ± standard error) on plates of MEA and DOA agar were 40 ± 14 and 2 ± 1 CFU, respectively. Therefore, we estimate the proportion of DOA-degrading fungi deposited onto agar plates from the atmosphere was 5%.
Identification of in situ isolates.
Fungi with 17 morphologically distinct colony types were identified. rDNA sequence lengths ranged from 496 to 576 bp for the 5.8S rDNA and ITS regions and from 518 to 562 bp for 28S rDNA. Representative colonies of 10 fungal morphotypes were identified using partial rDNA sequences, and these identifications were confirmed by microscopic examination and comparison with published descriptions of colony and conidial morphology. Unambiguous matches (98 to 100% sequence identity) were made using ITS sequences from isolates MZ8 (Thanatephorous cucumeris) (13), MZ10 (Alternaria infectoria) (13), MZ58 (A. pullulans) (13), MZ65 (A. pullulans), and MZ103 (Emericella nidulans; anamorph, Aspergillus nidulans) (13) and using the 28S V3 region from isolate MZ109 (Taphrina deformans) (10). The ITS sequence from isolate MZ104 showed 100% identity with Penicillium glabrum and 99.8% identity with Penicillium lapidosum, Penicillium thomii, and Penicillium purpurescens, differing from these species by only a single base pair. However, it was possible to distinguish MZ104 as P. glabrum on the basis of published colony morphology descriptions (33). ITS sequences from isolates MZ7 and MZ20 (both Alternaria alternata) showed the same level of identity (99.63%) with Alternaria lini as well as with A. alternata, and we were unable to differentiate these species following morphological examination. As A. alternata is an extremely common species found in abundance in soil and on other substrates (15), while A. lini is relatively rare (37) and may be contained within A. alternata (31), isolates MZ7 and MZ20 were named A. alternata. The ITS sequence of MZ14 had a relatively low (95%) identity with Petromyces muricatus. However, P. muricatus is a teleomorphic species of the Aspergillus ochraceus group (13, 41), and the colony and conidial morphology of MZ14 were identical to published descriptions of A. ochraceus (34). Thus MZ14 was named Aspergillus ochraceus. The 28S rDNA sequence from isolate MZ107 showed 95% identity with Phaeococcomyces nigricans. As species level identification requires further biochemical characterization, MZ107 was named only to the genus level.
Seven fungal morphotypes, including isolates MZ108 and MZ110 for which no ITS or 28S rDNA match was available, were identified at the International Mycological Institute using morphological techniques. Identification to the species level was possible for the yeasts MZ108 (Kluyveromyces marxianus) and MZ110 (Rhodotorula aurantiaca) and the filamentous fungi MZ111 (Epicoccum nigrum) and MZ112 (Paecilomyces lilacinus). MZ113 was identified only to the genus level (Kluyveromyces sp.), and MZ114, an intensely red yeast, could not be identified.
In situ colonization sequence.
We made viable counts of the 17 fungal morphotypes throughout the colonization period (Table 2). A. pullulans was the primary colonizing isolate and was dominant between 25 and 80 weeks of exposure. After 25 weeks, A. pullulans was isolated from only two of the three in situ racks, but by 40 weeks this fungus was established on all three racks and its frequency had increased significantly (Table 2) (P = 0.02). The A. pullulans mean viable count was stable from 40 to 55 weeks (P = 0.46) but increased significantly from 55 to 80 weeks and again from weeks 80 to 95 (Table 2). After 95 weeks, larger colonies of A. pullulans were visible to the eye as black specks (≤2 mm in diameter) on the pPVC substratum.
TABLE 2.
Frequency of occurrence of fungi with different colony morphologies recovered from pPVC throughout the in situ trial
| Colony morphotype | Mean CFU cm−2 at wka
|
|||||
|---|---|---|---|---|---|---|
| 10 | 25 | 40 | 55 | 80 | 95 | |
| Aureobasidium pullulans | 0 | 1 ± 1 (2) | 93 ± 50 (3) | 120 ± 35 (3)b | 150 ± 20 (3) | 390 ± 53 (3) |
| Epicoccum nigrum | 0 | 0 | 0 | 0 | 0.3 ± 0.7 (1) | 32 ± 13 (3) |
| Phaeococcomyces sp. | 0 | 0 | 0 | 0 | 10 ± 9 (2) | 64 ± 49 (3) |
| Pink-red yeast, unidentified | 0 | 0 | 0 | 0 | 17 ± 11 (3) | 170 ± 110 (3) |
| Rhodotorula aurantiaca | 0 | 0 | 0 | 0 | 0.7 ± 0.7 (3) | 73 ± 69 (3) |
| Taphrina deformans | 0 | 0 | 0 | 0 | 86 ± 55 (3) | 490 ± 240 (3) |
| Kluyveromyces sp. | 0 | 0 | 0 | 0 | 0 | 220 ± 153 (3) |
| Alternaria alternata | 3 ± 1 (3) | 0.3 ± 0.6 (1) | 4 ± 3 (3) | 0 | 0 | 4 ± 7 (1) |
| Alternaria infectoria | 0 | 0 | 0.2 ± 0.4 (1) | 0 | 0.1 ± 0.3 (1) | 0 |
| Aspergillus niger | 0.3 ± 0.5 (1) | 0 | 0 | 0 | 0 | 7 ± 11 (2) |
| Aspergillus ochraceus | 0 | 0 | 3 ± 5 (1) | 0 | 0 | 0 |
| Cladosporium herbarum | 0 | 0 | 0 | 0 | 0 | 0.6 ± 0.7 (2) |
| Emericella nidulans | 2 ± 2 (2) | 0.3 ± 1.2 (1)b | 0 | 0 | 0.1 ± 0.3 (1) | 0 |
| Kluyveromyces marxianus | 0 | 0 | 0 | 0 | 0.6 ± 1.1 (1) | 0 |
| Paecilomyces lilacinus | 0 | 0 | 0 | 0 | 0 | 6 ± 7 (2) |
| Penicillium glabrum | 0.5 ± 1.2 (1) | 0 | 12 ± 22 (1) | 0 | 0 | 0 |
| Thanatephorous cucumeris | 0 | 0 | 0 | 0 | 0.3 ± 0.5 (2) | 3 ± 5 (1) |
The mean viable counts and standard errors from three replicate pPVC pieces, one taken from each in situ rack, are shown. The number of racks on which fungi occurred at each sample time is indicated in parentheses.
No statistically significant change in mean CFU count in comparison with previous sample time (P > 0.05).
A group of yeasts and yeast-like fungi were established after 80 weeks of exposure. These microorganisms had colony morphologies identical to isolates MZ107, MZ108, MZ109, MZ110, and MZ114. A significant increase in the numbers of each of these organisms occurred on all three racks between 80 and 95 weeks of exposure (P ≤ 0.005) (Table 2). The most abundant of these isolates had the same colony morphology as MZ109. E. nigrum, a filamentous fungus, also appeared to be a secondary colonizer. It was initially isolated after 80 weeks from one of the in situ racks, but by 95 weeks this organism was recovered from all three racks (Table 2).
Throughout the in situ trial, several filamentous fungi and the yeast K. marxianus occurred sporadically (0 to 15 CFU cm−2). The filamentous fungi included representatives of the genera Alternaria, Aspergillus, Cladosporium, Emericella, Paecilomyces, Penicillium, and Thanatephorous. Usually these organisms were isolated from only one or two of the in situ racks during sampling, although Alternaria alternata was recovered in low numbers from all three racks after 10 and 40 weeks of exposure (Table 2).
SEM of pPVC samples exposed in situ.
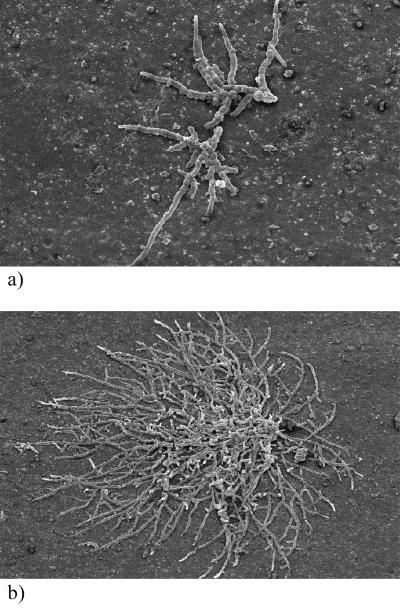
pPVC samples exposed to the atmosphere for 95 weeks were examined under SEM (Fig. 1). Colonies of A. pullulans, 50 to 2,000 μm in diameter, were randomly dispersed across the surface of the pPVC. A. pullulans appeared both as young colonies in the early stages of development (Fig. 1a) and as well-established, circular or oval colonies with extensive hyphal growth (Fig. 1b). Yeast phase growth of A. pullulans was not observed, and mycelia appeared as chains of branching, septate hyphae. There was no evidence of penetration of the pPVC substratum by hyphae of A. pullulans. Very few other microorganisms were observed on the pPVC under SEM. Ovoid, yeast-like cells occurred occasionally either singly or in clumps of two to three cells, both associated with A. pullulans colonies and on uncolonized areas of the plastic. No bacteria were observed.
FIG. 1.
SEM of the surface of pPVC exposed in situ for 95 weeks. (a) A. pullulans colony in early stages of development (magnification, ×250); (b) established A. pullulans colony (magnification, ×125).
In vitro tests for biodeterioration of pPVC.
Organisms with the highest level of extracellular esterase activity (100 to 250 REE ml−1) included all three strains of A. pullulans, MZ10, MZ95, MZ107, and MZ111. Isolates with little or no extracellular esterase activity (≤2.0 REE ml−1) included MZ110, MZ114, MZ103 (E. nidulans), and MZ8 (Table 3).
TABLE 3.
Deteriogenic properties of fungi isolated from pPVC during the in situ triala
| Isolate | Identification | Esterase activity (REEb ml−1) | Biomass in DOA liquid medium (mg/40 ml) | Specific esterase activity (REE mg−1) | Growth on DOA agarc | Clearing on DOA agard | Growth on pPVCe | pPVC weight loss (%)f |
|---|---|---|---|---|---|---|---|---|
| MZ58 | Aureobasidium pullulans | 250 ± 40 | 11 ± 4.1 | 1500 ± 1300 | 1 | 3 | 1 | 3.7 ± 0.7 |
| MZ10 | Alternaria infectoria | 240 ± 17 | 12 ± 0.3 | 800 ± 75 | 3 | 3 | 2 | 3.9 ± 0.5 |
| MZ95 | Aspergillus niger | 190 ± 7 | 13 ± 2.8 | 630 ± 210 | 3 | 0 | 1 | 1.8 ± 0.2 |
| PRA FS8 | Aureobasidium pullulans | 160 ± 6 | 8.8 ± 1.5 | 760 ± 240 | 1 | 2 | 3 | 3.6 ± 0.7 |
| IMI70103 | Aureobasidium pullulans | 150 ± 16 | 5.2 ± 1.5 | 1300 ± 700 | 2 | 2 | 2 | 3.4 ± 0.7 |
| MZ107 | Phaeococcomyces sp. | 120 ± 4 | 6.9 ± 2.4 | 840 ± 370 | 0 | 0 | 3 | 4.0 ± 0.2 |
| MZ111 | Epicoccum nigrum | 100 ± 19 | 23 ± 4.2 | 180 ± 44 | 3 | 2 | 2 | 1.4 ± 0.3 |
| MZ7 | Alternaria alternata | 81 ± 10 | 17 ± 0.4 | 300 ± 120 | 3 | 3 | 2 | 4.1 ± 0.3 |
| MZ109 | Taphrina deformans | 68 ± 10 | 27 ± 9.6 | 160 ± 130 | 1 | 1 | 1 | 1.4 ± 0.4 |
| MZ113 | Kluyveromyces sp. | 51 ± 19 | 18 ± 12 | 260 ± 260 | 1 | 1 | 1 | 1.9 ± 0.7 |
| MZ115 | Cladosporium herbarum | 50 ± 21 | 14 ± 4.2 | 150 ± 78 | 3 | 0 | 3 | 1.7 ± 0.4 |
| MZ112 | Paecilomyces lilacinus | 27 ± 9 | 44 ± 7.0 | 26 ± 11 | 3 | 2 | 3 | 4.0 ± 1.5 |
| MZ108 | Kluyveromyces marxianus | 18 ± 6 | 25 ± 8.7 | 49 ± 51 | 0 | 0 | 0 | 1.8 ± 0.2 |
| MZ14 | Aspergillus ochraceus | 12 ± 5 | 17 ± 1.3 | 29 ± 8.4 | 3 | 2 | 1 | 3.9 ± 0.3 |
| MZ20 | Alternaria alternata | 11 ± 6 | 12 ± 2.3 | 27 ± 14 | 3 | 3 | 2 | 4.2 ± 1.2 |
| MZ104 | Penicillium glabrum | 5 ± 3 | 31 ± 5.3 | 8.1 ± 6.4 | 1 | 0 | 1 | 2.0 ± 0.4 |
| MZ8 | Thanatephorous cucumeris | 2 ± 1 | 11 ± 0.6 | 6.1 ± 2.3 | 2 | 2 | 0 | 1.7 ± 0.1 |
| MZ114 | Unidentified pink-red yeast | 0.4 ± 0.3 | 17 ± 3.6 | 1.3 ± 1.4 | 0 | 0 | 0 | 1.8 ± 0.4 |
| MZ103 | Emericella nidulans | 0.4 ± 0.3 | 7.3 ± 3.2 | 6.5 ± 8.3 | 1 | 1 | 1 | 6.8 ± 0.6 |
| MZ110 | Rhodotorula aurantiaca | 0.3 ± 0.4 | 16 ± 4.5 | 1.0 ± 1.5 | 0 | 0 | 0 | 1.9 ± 0.2 |
Test methods used were measurement of extracellular esterase activity and biomass production during growth using DOA as the sole carbon source, clear-zone production, growth on DOA agar, growth on pPVC, and pPVC weight loss. Isolates are ranked in order of decreasing extracellular esterase activity. Mean values ± 1 standard error of the mean are shown.
An REE is the enzyme activity that liberates 1 nmol of p-nitrophenol from PNB in 1 h at 25°C and at pH 5.5.
Scores for growth on DOA agar: 0, no visible growth; 1, slight growth within inoculation well; 2, colony diameter < 2 cm; 3, colony diameter ≥ 2 cm.
Scores for clear-zone production on DOA agar: 0, no clearing; 1, faint clearing below colony; 2, clearing extending beyond colony boundary; 3, intense clearing (agar completely transparent) extending beyond colony boundary.
Scores for growth on pPVC: 0, no visible growth; 1, slight growth, barely visible; 2, growth clearly visible around the edges of the PVC; 3, strong growth visible around the edges and in the agar above the pPVC.
The initial weight of individual pPVC pieces (100% weight loss value) ranged from 500 to 730 mg. The mean weight loss ± standard error of the mean from sterile pPVC controls was 0.1 ± 0.1%.
Extracellular esterase activity was not correlated with activity in other tests. No significant difference (P = 0.15) occurred between the mean esterase activity of isolates that produced strong DOA clear zones (score 3; MZ7, MZ10, MZ58, and MZ20) and those that demonstrated no clearing (score 0; MZ95, MZ104, MZ107, MZ108, MZ110, MZ114, and MZ115). Similarly, there was no significant difference (P = 0.28) in mean esterase activity between isolates showing strong growth on DOA agar (score 3; MZ7, MZ10, MZ14, MZ20, MZ95, MZ111, MZ112, and MZ115) and those showing no growth (score 0; MZ107, MZ108, MZ110, and MZ114). We also observed differences between strains of the same fungal species. For example, A. alternata strain MZ20 showed poor esterase activity in comparison with A. alternata strain MZ7. Measurements of extracellular esterase activity, clear-zone production, and growth on DOA agar were carried out on three separate occasions, and the same trends among all of the fungal isolates were observed.
The ability of in situ isolates to cause weight loss from pPVC was determined after incubation with pPVC for 6 weeks under MSM supplemented with yeast extract. The net weight loss in all cases was very low (7 to 50 mg), and therefore direct comparisons of the weight loss caused by individual species were not possible. However, all 20 isolates tested caused significant (P ≤ 0.01) weight losses of >1% in comparison to control pPVC pieces incubated in sterile medium (Table 3). The greatest weight reduction, 6.8%, was caused by MZ103.
DISCUSSION
This study is the first detailed, quantitative investigation of the microbial colonization of pPVC. A. pullulans was the principal colonizing fungus. This organism initially colonized the pPVC after 25 weeks of exposure to the atmosphere and was isolated throughout the remainder of the in situ trial. SEM studies demonstrated that A. pullulans colonized pPVC in the absence of other microorganisms and therefore acts as a primary colonizer of pPVC. A. pullulans is increasingly recognized as the major causative agent of defacement of various diverse materials, such as painted surfaces (11, 21, 48) and wood (27), in addition to pPVC exposed to tropical conditions (24, 40). The present study demonstrates that A. pullulans is also an important colonizer of pPVC in temperate climates.
The success of A. pullulans in colonizing pPVC in situ is probably due to a combination of several factors. A. pullulans, which usually colonizes the phylloplane (13), can withstand periods of desiccation and high temperatures and produces highly melanized hyphae that protect against UV exposure (12). A. pullulans also produces extracellular polysaccharides that may facilitate permanent adhesion to surfaces (4), and factors controlling adhesion of A. pullulans to pPVC have recently been characterized (43). Therefore, adaptations for survival within the phylloplane probably confer advantages on A. pullulans for the colonization of painted and plastic surfaces within the environment (51).
A. pullulans also has considerable enzymatic capabilities. All three strains of A. pullulans produced high levels of extracellular esterase and could degrade DOA in vitro. In addition to producing extracellular esterase, A. pullulans also produces significant amounts of cellulase, proteinase, phosphatase, invertase, and maltase (47). The ability of A. pullulans to secrete such a variety of hydrolytic enzymes might enable it to utilize exogenous carbon sources that accumulate on the pPVC during long periods of exposure in situ. Extracellular esterase production is hypothesized to aid in the colonization of pPVC through the hydrolysis of organic-ester plasticizers (5, 28, 32). However, in this study extracellular esterase production did not correlate with DOA clearing or growth using pPVC as the sole source of carbon. These results may be due to differences in the specificity of esterase enzymes towards DOA plasticizers and the PNB synthetic substrate used in esterase assays. Thus, measurement of esterase activity alone is not a reliable indicator of the ability of an organism to degrade plasticizers or colonize pPVC.
A group of yeasts and yeast-like fungi became established on the pPVC much later than A. pullulans, towards the end of the in situ trial. Therefore, these yeasts probably play a secondary role in the colonization of pPVC in the sense that they require additional nutrients, e.g., the metabolites of other fungi or accumulated exogenous nutrients, before they can grow on the pPVC. These hypotheses are consistent with the observation that none of the yeasts, except Kluyveromyces sp., can degrade DOA or grow on pPVC as the sole source of carbon in vitro. Yeasts are not generally considered as important deteriogenic organisms on artificial surfaces, even though they have been recovered from deteriorated rubber and building materials (29) and from deteriorated pPVC during tropical exposure trials in Puerto Rico (24). Neither study provided information on the abundance of the yeasts or their role in the biodeterioration of these materials. Thus, the present study is the first to attribute a significant role to yeasts in the colonization of pPVC.
Interestingly, only a few yeast cells were observed during SEM studies of the pPVC substratum. These results suggest that high yeast CFU counts resulted from a small number of rapidly multiplying yeast colonies and highlight the general problem of using CFU counts to quantify fungi. CFU counts depend on how readily the fungal material on the pPVC breaks into individual propagules during the isolation procedure. For example, for the same amount of biomass, a colony of a budding yeast or a sporulating filamentous fungus may yield many more CFUs than a spreading hyphal mycelium. Thus, while CFU counts are useful in determining which organisms are colonizing and multiplying on the pPVC, they are not usually a reliable indicator of the fungal biomass present on the substratum.
Bacteria were not isolated from the pPVC during the trial, and none were detected during SEM studies or following DAPI (4′,6′-diamidino-2-phenylindole) fluorescence staining of organisms removed from the pPVC substratum (data not shown). Bacterial growth might be inhibited by desiccation, solar irradiation, or in situ acidification of the pPVC following photochemical or thermal degradation (25, 30). Acidification of the pPVC could inhibit bacterial growth, as fungi tolerate lower pHs than do bacteria.
Fungal colonization of pPVC appeared to be influenced by seasonal climatic changes. Major increases in fungal CFU counts occurred only during the British winter. For example, fungal CFU counts increased by 470% during the winter period between November 1998 (80 weeks) and March 1999 (95 weeks), largely due to the establishment of yeasts and yeast-like fungi on the pPVC. In contrast, no increases in fungal CFU counts occurred when pPVC pieces were sampled during the British summer months. Fungal growth or sporulation during the summer period might be inhibited by desiccation and high temperatures caused by long periods of direct solar irradiation. Indeed, Upsher and Roseblade (40) reported that a marked reduction in the amount of fungal growth on pPVC can occur during dry periods. Extended studies of the colonization process are needed to determine whether yeasts that establish on the pPVC during the winter months can survive on the pPVC through the succeeding summer period.
Whether an organism can colonize pPVC also probably depends on its ability to obtain carbon from the pPVC formulation. Evidence supporting this hypothesis comes from the observation that between 64 and 84% of fungi colonizing pPVC could degrade DOA. In contrast, among fungi deposited onto agar plates from the atmosphere, only 5% could degrade DOA at the time of sampling. Although the proportion of DOA-degrading fungi within the atmosphere is likely to be influenced by environmental parameters and probably varies seasonally, these results support the hypotheses that selection for fungi that can degrade DOA occurs on the pPVC substratum and that organisms that can degrade DOA may have a competitive advantage during the colonization of pPVC.
In addition to A. pullulans, many of the recovered fungi, e.g., Alternaria spp., Aspergillus spp., Paecilomyces sp., and Cladosporium sp., have previously been isolated from deteriorated pPVC in tropical exposure trials (24, 36) and are common colonizers of painted surfaces and building materials (for a review, see reference 19). In particular, A. alternata, A. infectoria, and P. lilacinus had high activity in all of the test methods, demonstrating that they are potentially important degraders of pPVC within the environment. Under ideal growth conditions within warm and humid tropical exposure trials, these organisms would probably grow readily on the pPVC substratum. However, these organisms were isolated infrequently and in low numbers in this study, suggesting that environmental factors limited the establishment of these organisms on the pPVC. Thus, while in vitro methods identified fungi potentially capable of causing biodeterioration of pPVC, they were not predictive of the organisms that colonized pPVC in the environment.
We observed discrepancies between the ability of fungi to grow on the intact pPVC formulation as the sole carbon source and their ability to degrade DOA in vitro. For example, Phaeococcomyces sp. grew well on pPVC but could not produce clear zones in DOA agar. It is possible that components of the pPVC formulation other than the plasticizers can support the growth of fungi on pPVC. We used DOA in the present study because it is known to be more susceptible than the other plasticizer, DOP, to microbial attack (5). However, in addition to DOA and DOP the pPVC formulation also contains small quantities of a calcium-zinc stearate stabilizer and an epoxidized oleate ester stabilizer that can be utilized by various fungi as a sole carbon source (36). Thus, while the homogenized plasticizer agar technique is useful to determine if an organism can degrade a plasticizer, it is not necessarily predictive of the organism's ability to grow on a complex pPVC formulation.
We found no evidence that the mechanical properties of the pPVC were altered due to microbial degradation of plasticizers in the environment. Although fungi can increase the tensile strength of pPVC in vitro due to the degradation of plasticizers (39, 46), tensile testing of exposed pPVC pieces showed no significant change in either the tensile strength or the percent elongation at breaking (data not shown). We think that the fungal biomass that accumulated on pPVC pieces during the in situ trial was insufficient to cause a measurable change in the mechanical properties of the pPVC.
In summary, our results suggest that a colonization sequence may occur during colonization of pPVC in situ. A. pullulans is the principal colonizing fungus, and secondary colonizing yeasts establish themselves much later. In vitro biodeterioration tests were not predictive of the ability of fungi to colonize pPVC in the environment, emphasizing the importance of field trials in investigations of the microbial susceptibility of pPVC formulations. Knowledge of the organisms that colonize pPVC and their ecology is essential for the design of novel pPVC formulations and biocides that provide long-term protection against biodeterioration of pPVC in situ.
ACKNOWLEDGMENTS
This work was supported by a BBSRC CASE award in collaboration with Avecia Biocides.
We thank Michael Anderson and Laurence Hall, University of Manchester, Manchester, United Kingdom, for assistance with rDNA sequencing of fungal isolates.
REFERENCES
- 1.American Society for Testing Materials. ASTM standards on materials and environmental microbiology. Philadelphia, Pa: American Society for Testing Materials; 1985. pp. 178–180. [Google Scholar]
- 2.American Society for Testing Materials. ASTM standards on materials and environmental microbiology. Philadelphia, Pa: American Society for Testing Materials; 1985. pp. 174–177. [Google Scholar]
- 3.Anderson M J, Gull K, Denning D W. Molecular typing by random amplification of polymorphic DNA and M13 southern hybridization of related paired isolates of Aspergillus fumigatus. J Clin Microbiol. 1996;34:87–93. doi: 10.1128/jcm.34.1.87-93.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews J H, Harris R F, Spear R N, Gee W L, Nordheim E V. Morphogenesis and adhesion of Aureobasidium pullulans. Can J Microbiol. 1994;40:6–17. [Google Scholar]
- 5.Berk S, Ebert H, Teitell L. Utilization of plasticizers and related organic compounds by fungi. Indust Eng Chem. 1957;49:1115–1124. [Google Scholar]
- 6.Booth G H, Cooper A W, Robb G A. Bacterial degradation of plasticized PVC. J Appl Bacteriol. 1968;31:305–310. [Google Scholar]
- 7.Booth G H, Robb J A. Bacterial degradation of plasticized PVC—effect of some physical properties. J Appl Chem. 1968;18:194. [Google Scholar]
- 8.Brown A E. Problems of fungal growth on synthetic resins, plastics and plasticizers. U.S. Office of Scientific Research and Development, report no. 6067. Washington, D.C.: U.S. Office of Scientific Research and Development; 1946. [Google Scholar]
- 9.Cavett J J, Woodrow M N. A rapid method for determining degradation of plasticized PVC by microorganisms. In: Walters A H, Elphick J J, editors. Biodeterioration of materials—microbiological and allied aspects. Proceedings of the 1st International Biodeterioration Symposium. London, United Kingdom: Elsevier; 1968. pp. 162–169. [Google Scholar]
- 10.Commonwealth Mycological Institute. CMI descriptions of pathogenic fungi and bacteria, set 72 no. 711. Kew, United Kingdom: Commonwealth Mycological Institute; 1981. [Google Scholar]
- 11.Cooke W B. An ecological life history of Aureobasidium pullulans (de Bary) Arnaud. Mycopathol Mycol Appl. 1959;12:1–45. doi: 10.1007/BF02118435. [DOI] [PubMed] [Google Scholar]
- 12.Crang R E, Pechak D G. Aureobasidium pullulans: fine structure and development. J Coatings Technol. 1978;50:36–42. [Google Scholar]
- 13.Domsch K H, Gams W, Anderson T. Compendium of soil fungi. London, United Kingdom: Academic Press; 1980. [Google Scholar]
- 14.Eaton R W, Ribbons D W. Utilization of phthalate esters by micrococci. Arch Microbiol. 1982;132:185–188. doi: 10.1007/BF00508728. [DOI] [PubMed] [Google Scholar]
- 15.Ellis M B, editor. Dematiaceous hyphomycetes. Kew, United Kingdom: Commonwealth Mycological Institute; 1971. [Google Scholar]
- 16.Fell J W. Rapid identification of yeast species using three primers in a polymerase chain reaction. Mol Mar Biol Biotechnol. 1993;1:175–186. [PubMed] [Google Scholar]
- 17.Fett W F, Gerard H C, Moreau R A, Osman S F, Jones L E. Cutinase production by Streptomyces spp. Curr Microbiol. 1992;25:165–171. [Google Scholar]
- 18.Flemming H-C. Relevance of biofilms for the biodeterioration of surfaces of polymeric materials. Polym Degrad Stabil. 1998;59:309–315. [Google Scholar]
- 19.Gaylarde C C, Morton L H G. Deteriogenic biofilms on buildings and their control: a review. Biofouling. 1999;14:59–74. [Google Scholar]
- 20.Girard T A, Koda C F. Staining of PVC. Modern Plastics. 1959;36:148. [Google Scholar]
- 21.Goll M, Coffery G. Mildew of painted surfaces. Paint Oil Chem Rev. 1948;111:14–17. [Google Scholar]
- 22.Griffin G J L, Uribe M. Biodeterioration 6: papers presented at the 6th International Biodeterioration Symposium. Slough, United Kingdom: C.A.B. International; 1984. Biodegradation of plasticized polyvinyl chloride; pp. 648–657. [Google Scholar]
- 23.Guarro J, Gene J, Stchigel A M. Developments in fungal taxonomy. Clin Microbiol Rev. 1999;12:454–501. doi: 10.1128/cmr.12.3.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton N F. Biodeterioration of flexible polyvinyl chloride films by fungal organisms. In: Oxley T A, Barry S, editors. Biodeterioration 5. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1983. pp. 663–678. [Google Scholar]
- 25.Hollande S, Laurent J. Study of discolouring change in PVC, plasticizer and plasticized PVC films. Polym Degrad Stabil. 1997;55:141–145. [Google Scholar]
- 26.International Standards Organisation. Plastics. Determination of behaviour under the action of fungi and bacteria. Evaluation by visual examination or measurement of change in mass or physical properties, ISO 846-978(E). Geneva, Switzerland: International Standards Organization; 1978. [Google Scholar]
- 27.Kaarik A. Sapwood staining fungi. The International Research Group on Wood Preservation. Document no. IRG/WP/125. England: Princes Risborough; 1974. [Google Scholar]
- 28.Klausmeier R E, Jones W A. Microbial degradation of plasticisers. Dev Ind Microbiol. 1961;42:741–743. [Google Scholar]
- 29.Lodder J, Krejer-van Rij N J W. The yeasts: a taxonomic study. Amsterdam, The Netherlands: North Holland Publishing Co.; 1967. [Google Scholar]
- 30.Martinez J G, Rodriguez O S, Sanchez S, Ramirez E, Allen N S. Prediction of photoageing stability of plasticized PVC films containing UV-stabilizers. Polym Degrad Stabil. 1996;54:49–55. [Google Scholar]
- 31.McKay G J, Brown A E, Bjourson A J, Mercer P C. Molecular characterization of Alternaria linicola and its detection in linseed. Eur J Plant Pathol. 1999;105:157–166. [Google Scholar]
- 32.Mills J, Eggins H O. The biodeterioration of certain plasticisers by thermophilic fungi. Int Biodeter Bull. 1974;10:39–44. [Google Scholar]
- 33.Pitt J I, editor. The genus Penicillium and its teleomorphic states. London, United Kingdom: Academic Press; 1979. [Google Scholar]
- 34.Raper K B, Fennel D I. The genus Aspergillus. Baltimore, Md: Williams and Wilkins; 1965. [Google Scholar]
- 35.Reasoner D J, Geldreich E E. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts W T, Davidson P M. Growth characteristics of selected fungi on polyvinyl chloride film. Appl Environ Microbiol. 1986;51:673–676. doi: 10.1128/aem.51.4.673-676.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rotem J, editor. The genus Alternaria: biology, epidemiology and pathogenicity. St. Paul, Minn: American Phytopathological Society; 1994. [Google Scholar]
- 38.Santoro E D, Koestler R J. A methodology for biodeterioration testing of polymers and resins. Int Biodeter and Biodegrad. 1991;28:81–90. [Google Scholar]
- 39.Seal K J, Pantke M. Microbiological testing of plastics: ongoing activities of IBRG plastics project group to improve standard test procedures. Int Biodegrad. 1988;24:313–320. [Google Scholar]
- 40.Upsher F J, Roseblade R J. Assessment by tropical exposure of some fungicides in plasticized PVC. Int Biodet Biodegrad. 1984;20:243–254. [Google Scholar]
- 41.Varga J, Toth B, Kevei E, Palagyi A, Kozakiewicz Z. Analysis of genetic variability within the genus Petromyces. Antonie Leeuwenhoek. 2000;77:83–89. doi: 10.1023/a:1002475816014. [DOI] [PubMed] [Google Scholar]
- 42.Wasserbauer R, Blahnik R. Microbial corrosion of electrical equipment. Analysis of causes and consequences: a review. Int Biodet Bull. 1975;11:9–15. [Google Scholar]
- 43.Webb J S, van der Mei H C, Nixon M, Eastwood I M, Greenhalgh M, Read S J, Robson G D, Handley P S. Plasticizers increase adhesion of the deteriogenic fungus Aureobasidium pullulans to PVC. Appl Environ Microbiol. 1999;65:3575–3581. doi: 10.1128/aem.65.8.3575-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wellman R H, McCallan S E A. Fungus resistance of pesticides. U.S. Office of Scientific Research and Development, report no. 5683. Washington, D.C.: U.S. Office of Scientific Research and Development; 1945. [Google Scholar]
- 45.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal rRNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 46.Whitney P J. A comparison of two methods for testing defined formulations of PVC for resistance to fungal colonization with two methods for the assessment of their biodegradation. Int Biodet Biodegrad. 1996;37:205–213. [Google Scholar]
- 47.Winters H, Guidetti G. Extracellular enzymes from fungal isolates involved in the biodeterioration of paint films. Dev Ind Microbiol. 1976;17:173–176. [Google Scholar]
- 48.Winters H, Isquith I R, Goll M. A study of the ecological succession in biodeterioration of a vinyl acrylic paint film. Dev Ind Microbiol. 1976;17:167–171. [Google Scholar]
- 49.Yabannavar A V, Bartha R. Methods for assessment of biodegradability of plastic films in soil. Appl Environ Microbiol. 1994;60:3608–3614. doi: 10.1128/aem.60.10.3608-3614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeager C C. Pink staining in PVC. Plastics World. 1952;14:15. [Google Scholar]
- 51.Zabel R A, Terracina F. The role of Aureobasidium pullulans in the disfigurement of latex paint films. Dev Ind Microbiol. 1979;21:179–190. [Google Scholar]