Abstract
Natural gas seeps release significant amounts of methane and other gases including ethane and propane contributing to global climate change. In this study, bacterial actively consuming short-chain alkanes were identified by cultivation, whole-genome sequencing, and stable-isotope probing (SIP)-metagenomics using 13C-propane and 13C-ethane from two different natural gas seeps, Pipe Creek and Andreiasu Everlasting Fire. Nearly 100 metagenome-assembled genomes (MAGs) (completeness 70–99%) were recovered from both sites. Among these, 16 MAGs had genes encoding the soluble di-iron monooxygenase (SDIMO). The MAGs were affiliated to Actinobacteria (two MAGs), Alphaproteobacteria (ten MAGs), and Gammaproteobacteria (four MAGs). Additionally, three gaseous-alkane degraders were isolated in pure culture, all of which could grow on ethane, propane, and butane and possessed SDIMO-related genes. Two Rhodoblastus strains (PC2 and PC3) were from Pipe Creek and a Mycolicibacterium strain (ANDR5) from Andreiasu. Strains PC2 and PC3 encoded putative butane monooxygenases (MOs) and strain ANDR5 contained a propane MO. Mycolicibacterium strain ANDR5 and MAG19a, highly abundant in incubations with 13C-ethane, share an amino acid identity (AAI) of 99.3%. We show using a combination of enrichment and isolation, and cultivation-independent techniques, that these natural gas seeps contain a diverse community of active bacteria oxidising gaseous-alkanes, which play an important role in biogeochemical cycling of natural gas.
Subject terms: Metagenomics, Environmental microbiology
Introduction
Approximately 600–900 million tonnes of the potent greenhouse gas methane are released annually into the atmosphere [1] and it has a high global warming potential [2]. The largest source of methane globally is biogenic methane which arises from the activities of methanogenic archaea in anoxic environments [1]. Thermogenic methane of geological origin (often termed “natural gas”) is emitted to the atmosphere as a consequence of anthropogenic fossil fuel extraction, transport and seepage from natural sources, and unintentional release of natural gas due to leaking gas pipelines, coal mining and shale-gas extraction and incidents such as the Deepwater Horizon disaster of 2010 [3]. Natural gas seeps in terrestrial, freshwater, and marine geothermal regions, submarine volcanoes and mud volcanoes, account for the release of ~45 million tonnes of methane per year [1, 4, 5].
Thermogenic natural gas comprises mainly of methane but also contains substantial amounts of other climate-active gases such as ethane and propane. Ethane is a photochemical pollutant while propane is an ozone precursor and together their estimated global emission is around 24–30 million tonnes per annum [6, 7] of which approximately 3–6.4 million tonnes are from natural geologic sources [8]. Some natural gas seeps such as the Eternal Flames in Chestnut Ridge County Park and Pipe Creek, New York State [9–11] release methane containing high concentrations of ethane and propane, and thus provide potential hotspots for microbes that degrade gaseous hydrocarbons.
The major sink for methane and short-chain alkanes once in the atmosphere is photochemical oxidation by hydroxyl radicals [12, 13] but it is estimated that over 50% of methane from both biogenic and thermogenic sources that is released into the biosphere is consumed by methane-oxidising microbes in aerobic and anaerobic environments [14]. Aerobic methane-oxidising bacteria (methanotrophs) [15] grow using either particulate methane monooxygenase (pMMO) or soluble methane monooxygenase (sMMO), a member of the soluble diiron centre (SDIMO) family of oxygenases [16, 17].
Facultative methanotrophs of the genus Methylocella can grow simultaneously on both methane and propane using sMMO and another SDIMO enzyme, propane monooxygenase [18]. This metabolic versatility of Methylocella, which is widespread in the environment [10, 19–21] stimulated the examination of terrestrial natural gas seep environments rich in both methane and propane.
By comparison with methane, the bacterial metabolism of ethane, propane, and butane is less-well understood and the ecology of bacteria using these gases has not been explored in depth at terrestrial natural gas seeps (the subject of this study). The majority of C2–C4 alkane-utilising bacteria belong to the order Actinomycetales and include members of the genera Mycolicibacterium (formerly Mycobacterium [22]), Corynebacterium, Nocardia, and Pseudonocardia [23–25], and Rhodococcus [26, 27]. Gram-negative C2–C4 alkane utilisers include Pseudomonas [28] and Thauera [29] (reviewed by Rojo [30] and Shennan [31]). The initial oxidation of gaseous-alkanes is usually catalysed by an SDIMO enzyme consisting of an oxygenase, a coupling protein, and a reductase. Examination of the evolutionary relationships between different SDIMOs provides a framework for classification of these enzymes into 6 groups containing aromatic/alkene monooxygenases (Group I), phenol hydroxylase (Group II), sMMOs (Group III), and alkane and alkene monooxygenases (Groups IV-VI) [17, 32, 33], although the boundaries of this classification scheme are often blurred. For example, the butane oxidizer Thauera butanivorans [34] contains a butane monooxygenase which is more closely related to sMMO (Group III). Mycolicibacterium chubuense NBB4, a strain isolated on ethene and growing on C2-C4 alkanes, contains four distinct SDIMOs gene clusters from Groups III and VI, and two from Group IV [25, 35]. It also contains a gene cluster encoding a membrane-bound copper-containing monooxygenase, associated with gaseous-alkane utilisation [36, 37]. Gordonia sp. TY-5 oxidises propane via prmABCD from Group V [38] which is closely related to the propane monooxygenases (prmABCD) of Methylocella silvestris BL2 [18], Rhodococcus sp. strain BCP1 [27], and Pseudonocardia sp. strain TY-7 [24]. Another Mycobacterium strain TY-6 contains a prmACDB from Group VI [24]. With the exception of Methylocella, no C2-C4 alkane utilisers can grow on methane (reviewed in Shennan [31]).
The availability of SDIMO sequences predicted to be involved in gaseous-alkane utilisation, and of extant reference bacteria to investigate their metabolism, has provided a robust taxonomic framework to examine the molecular ecology of these bacteria. DNA sequence retrieval of SDIMOs by PCR [32, 39] or via metagenomics [40–43] can reveal the diversity and abundance of key genes encoding putative gaseous-alkane monooxygenases. Combining DNA-SIP [44, 45], using 13C-labelled gases ethane and propane, with a metagenomics analysis of labelled heavy (13C) will assist in assigning the relevant SDIMO gene clusters to their function in gaseous-alkane utilisation. The objective of this study was to use this combination of techniques to investigate the presence of active gaseous-alkane degraders at two contrasting natural gas seeps, Pipe Creek [10, 11] and Andreiasu Everlasting Fire [46], previously examined for the presence and activity of Methylocella [10, 47]. The hypothesis we were testing was that the analysis of metagenome assembled genomes (MAG) retrieved from 13C-labelled DNA would reveal the identity of the bacteria involved. These approaches were combined with cultivation of ethane and propane degraders from these natural gas seeps.
Material and methods
Sample collection
Samples from two natural gas seeps sites, Andreiasu (pH 8.2) in Romania and Pipe Creek (pH 6.0) in the USA, were collected for this study. Samples from the Romanian site were taken from the liquid mud through which the thermogenic gas bubbles, as described by Baciu et al. [46]. Pipe Creek (geology described by Schimmelmann et al. [11]) is a fast-flowing stream with a rocky bed. Samples consisted of water and sediment taken from the vicinity of fissures in the stream bed, through which gas was emerging. Varying characteristics of physical nature, pH and the proportion of methane, ethane and propane in the gas emitted from both sites along with sampling conditions have been previously reported [10].
Enrichment cultures, isolations, and genome sequencing of novel isolates
Fresh samples (~1 g) from natural gas seeps were incubated in 10 ml modified dilute nitrate mineral salts (DNMS) medium [47], supplemented with 5 µM lanthanum, in 120 ml sealed serum vials. A mixture of gases (20%, v/v), comprising of methane (70%, v/v), ethane (10%, v/v), and propane (20%, v/v) was injected as the only supplemental source of C and energy and the vials were incubated in a shaker (150 rpm) at 25 °C for three weeks in the dark. Enrichment cultures were serially diluted and plated onto modified DNMS agar plates (supplemented with 5 µM lanthanum) and incubated, under the same mixture of gases (10%, v/v), in a sealed jar. Colonies appearing on the plates after two weeks of incubation were picked and resuspended in 20 µl of sterile modified DNMS medium. Aliquots of this cell suspension were replica-plated onto two plates of DNMS medium (supplemented with 5 µM lanthanum) and incubated under ethane or propane individually in the headspace (10% v/v) as the only source of carbon and energy. Colonies growing under both conditions were considered as ethane/propane utilisers. Purity of these isolates was achieved by continuously plating of serial dilutions of isolate cultures and confirmed by microscopic observations. After confirmation of purity of the isolates, genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer’s instructions. Genome sequencing of these isolates was performed at MicrobesNG (Birmingham, UK) using HiSeq (Illumina) and Nanopore technology and assembled using SPAdes 3.7 into contigs. Genome sequence annotation, exploration and comparative genomics of the isolates was performed using MicroScope [48] an online platform by GenoScope (France) providing a collection of bioinformatic tools.
Bacterial strains and growth conditions
Modified DNMS medium supplemented with 5 µM lanthanum (LaCl3) in 120 ml serum vials (with 20 ml culture volume) was used for the growth of bacterial strains. Cultures were grown in DNMS medium with ethane or propane (10% v/v) separately as the only carbon and energy source. Optical density of the cultures was measured at 540 nm to monitor the growth of liquid cultures. Concentrations of substrate gases in the cultures were quantified as described by Crombie and Murrell [18].
DNA-SIP incubations
For DNA-SIP incubations, approximately 2 g of soil/sediment suspensions (1:3 sample and ultra-pure water ratio) were incubated in 120 mL sealed serum vials. Substrate gases (12C-ethane, 12C-propane, 13C-ethane, and 13C-propane) were injected separately into the headspace of the vials to a concentration of 1% (v/v). Gas consumption was measured in each vial using gas chromatography [18]. There is a trade-off between leaving the timescale of incubations long enough to ensure enough 13C-labelled DNA is obtained to undertake metagenome sequencing and conversely that the length of incubation is not too long, so that as much as possible, cross-feeding of 13C-label does not occur. The general guidelines of Neufeld et al. (2007) were followed. All incubations were carried out in duplicate for each substrate and for each time point. Time point 1 samples from DNA-SIP incubations were harvested after they had consumed approximately 100 µmol C per g of fresh sample and for time point 2 samples were harvested after they had consumed approximately 200 µmol C per g of fresh sample as measured by gas chromatography (Fig. S1). Molecular analysis of the time point 2 samples showed that sufficient 13C-label was incorporated for a successful SIP experiment, therefore those samples were used for further analysis. After incubations, samples were harvested by centrifugation at 10,000 × g for 15 min, supernatants were discarded, and the pellets were stored at −20 °C. DNA was extracted from both native samples without incubations and SIP-incubated samples (~0.5 g) by using the spin kit for soil (MP Biomedicals) following the manufacturer’s instructions. DNA was quantified using a Qubit 2.0 fluorometer (Invitrogen) and by NanoDrop (Thermo Fisher Scientific).
CsCl density gradient ultracentrifugation (177,000 × g, 40 h, 20 °C, Beckman Vti 65.2 rotor) and fractionation was used for separating labelled and unlabelled DNA from SIP-incubated samples [41, 49] using the refractive index of CsCl fractions (12 fractions per sample) as a proxy for density. Refractive index of each fraction was measured using a refractometer (Reichert AR200, Reichert Analytical Instruments, Buffalo, USA). The buoyant density of each fraction was calculated as described previously [50] and plotted against the quantity of DNA retrieved from corresponding fraction (Fig. S2). Based on the data shown in Figure S2, three to four fractions of each sample containing labelled DNA (buoyant densities 1.7491–1.7296 g/ml) were mixed and designated as the “heavy” DNA fraction, while two to three fractions of each sample containing unlabelled DNA (buoyant densities 1.7216–1.7123 g/ml) were mixed and designated as the “light” DNA fraction.
Sequencing
The microbial community of both native and DNA-SIP incubated samples was characterised by sequencing the 16S rRNA gene as previously described [47]. PCR amplicons using universal primers 341 F and 785 R targeting the V3-V4 regions [51] were obtained from the unfractionated unenriched native DNA samples and the heavy and light DNA fractions from DNA-SIP incubated samples. These amplicons were processed as described previously [47] and sequenced using the MiSeq (Illumina) platform of MR DNA (Shallowater, USA).
The total metagenomic DNA of the native samples, plus DNA from heavy fractions from incubations with 13C-ethane and 13C-propane from both sites (total of six samples) were sequenced on a HiSeq (Illumina) at Novogene (Cambridge, UK). The metagenome was analysed on a high-performance computing cluster supported by the Research and Specialist Computing Support Service at the University of East Anglia (Norwich, UK).
Bioinformatics
MRDNA proprietary analysis pipeline (www.mrdnalab.com) was used to analyse the sequence data from 16S rRNA gene amplicons as described previously [47]. Operational taxonomic units (OTUs) were defined at a sequence identity level of 97%. Final taxonomic classification of OTUs was performed by BLASTn against databases from RDPII and NCBI using a bootstrap confidence threshold of 97% (www.ncbi.nlm.nih.gov, http://rdp.cme.msu.edu). Taxa fulfilling the following criteria were identified as labelled: (1) the relative abundance in the heavy DNA fraction of the 13C-ethane/13C-propane-incubated sample was >1.0%, (2) the abundance in the heavy DNA fraction of the 13C-ethane/13C-propane -incubated sample was higher than the abundance in the light DNA fraction of the 13C-ethane/13C-propane and (3) the difference in the abundance in the compared heavy and light DNA fractions of the 13C-ethane/13C-propane-incubated sample was higher than that of the 12C-ethane/12C-propane-incubated sample.
For metagenomic DNA sequences, reads were checked using FastQC version 0.11.8 [52]. Low-quality reads were discarded using BBDuk version 38.68 [53]. After quality checking, reads were merged into longer contiguous sequences (scaffolds) using de novo assemblers SPAdes (single assembly) and metaSPAdes (co-assembly) version 3.13.1 [54, 55]. Co-assembly was performed for each site (Pipe Creek and Andreiasu) independently. Downstream binning analysis was performed using scaffolds larger than 1000 bp. Metagenomic binning of the assembled scaffolds was carried out with the metaWRAP version 1.2.1 pipeline [56]. Completion and contamination metrics of the extracted bins were estimated using CheckM [57]. The resulting bins were collectively processed to produce consolidated metagenome-assembled genomes (MAGs) using the bin_refinement module (criterion: completeness >70%; contamination <5%). MAGs retrieved from both single- and co-assembly were then dereplicated using dRep [58]. Only the highest scoring MAG from each secondary cluster was retained in the dereplicated set. The abundance of each MAG and genome in the different sites was calculated separately using BLASTn version 2.5.0+ [59, 60], keeping only hits with >95% identity and e-value cutoff <1e−5 for the analysis. A final heatmap of the abundance of the isolates and MAGs, containing genes encoding SDIMO and membrane-bound methane monooxygenase (PmoA), was constructed as described previously [60].
Functional annotation
All the MAGs retrieved in this study and the three isolate genomes were analysed using BLAST separately against databases containing SDIMO-related genes (mmoX, bmoX, prmA, dmpN, tmoA) and pmoA genes. Afterwards, all the genomes of the isolates and only the MAGs that presented hits from the BLAST were annotated using MicroScope platform [48] for downstream analysis. Automatic annotations were validated manually using BLASTp for translated sequences of the genes involved in metabolic pathways of interest, such as those involved in SDIMO (methane monooxygenase, putative butane monooxygenase, propane monooxygenase, toluene monooxygenase and phenol hydroxylase), and membrane-bound methane monooxygenase.
Phylogenomic analysis
For identification of the isolates, the 16S rRNA gene regions of strains PC2, PC3, and ANDR5 were extracted using Barrnap v. 0.9 software (https://github.com/tseemann/barrnap) and compared with the type strains using BLASTn. Amino acid comparisons between the genomes and MAGs retrieved in this study and their closest-relative type strains were calculated based on reciprocal best hits using the Enveomics collection [61]. Average nucleotide identity (ANI) was calculated using the Enveomics platform and confirmed with the outputs from automated multi-locus species tree analysis (autoMLST) [62]. The recommended species cut-off was 95% for ANI [63] and ~70% for amino acid identity (AAI) indices [64] (Ramon Rosselló-Móra, pers. comm.). In addition, JSpeciesWS software (http://jspecies.ribohost.com/jspeciesws/ [65]) was used to determine the correlation indexes of the tetra-nucleotide signatures between the isolates and their closest type strains.
Genomes of isolates and the MAGs were submitted to the TYGS (Type Strain Genome Server) platform (https://tygs.dsmz.de [66]). This platform predicts digital DNA:DNA hybridisation (dDDH) values of the isolates to the most closely related type strains. Additionally, the translated sequences of a set of ten house-keeping genes (rpoB, secA, gyrB, rho, murB, era, recR, dapD, aroC, and nusG), as well as SDIMO genes (i.e. strains PC2 and PC3 (mmoX), and strain ADNR5 (prmA)) were retrieved from both the isolates and their closest type strains for comparisons using BLASTx.
The taxonomic classification of MAGs was performed using the classify_bins module from metaWRAP which relies on the NCBI_nt database. A phylogenomic tree, which included both isolates and MAGs was created using the autoMLST pipeline and conducted with 1000 bootstrap replicates [62]. Phylogenetic analysis of the key SDIMO and membrane-bound methane monooxygenase enzymes was conducted in MEGA X [67] using the Maximum Likelihood method with a JTT matrix-based model and 100 bootstrap replicates.
Results and discussion
Samples from Pipe Creek [11] and Andreiasu [46] were examined for the presence and activity of gaseous-alkane degrading bacteria using cultivation-independent techniques. These sites were chosen because the natural gas emitted at these sites had been previously shown to contain substantial amounts of ethane and propane, as well as methane, and were known to harbour Methylocella that could use methane, ethane, and propane [10, 47]. The goal of this study was to determine which ethane- and propane-utilising bacteria were active at these sites and to investigate the soluble di-iron centre monooxygenases, which enable them to grow on gaseous-alkanes.
Incubation of natural gas seep samples with ethane and propane
To confirm that samples from natural gas seeps were active and to examine the bacterial community consuming ethane and propane, Pipe Creek and Andreiasu samples were incubated with 12C- or 13C-labelled ethane or propane. There were no major differences in the consumption of 13C-substrate gases compared to 12C-substrate gases in samples from Pipe Creek or Andreiasu. However, Pipe Creek samples consumed ethane and propane faster than samples from Andreiasu (Fig. 1, Fig. S1). This is supported by the fact that the natural gas emitted from Pipe Creek seep site contained very high amounts (~5–10x higher) of ethane and propane compared to those present in natural gas emitted from Andreiasu site [10, 11, 46]. This also suggests that potential ethane and propane consumers at Pipe Creek may have been more abundant and better adapted to ethane and propane utilisation as reported previously [47]. These bacteria may serve as a natural biofilter for gases before they are emitted to the atmosphere.
Fig. 1. Consumption rates of propane and ethane by environmental samples from natural gas seep sites.

Bars represent mean values with standard deviations of independent duplicate incubations for each substrate.
SIP was performed in order to examine the diversity of active ethane- and propane-degrading bacteria at both natural gas seep sites. DNA from incubations with both 12C- and 13C-ethane or propane was separated by isopycnic density gradient centrifugation to yield light and heavy DNA fractions (Fig. S2). This yielded a total of 16 DNA samples: (a) light and heavy DNA from Pipe Creek samples incubated with 12C-ethane or 13C-ethane, and three more corresponding sets of four samples from (b) Pipe Creek incubations with 12C-propane or 13C-propane; (c) Andreiasu incubations with 12C-ethane or 13C-ethane; (d) Andreiasu incubations with 12C-propane or 13C-propane.
16S rRNA genes of putative ethane- and propane-utilising bacteria
Profiles of the relative abundance of 16S rRNA genes in the light and heavy DNA fractions from incubations of Pipe Creek samples with 12C-ethane were similar and as expected for a successful DNA-SIP experiment (Fig. S3A). A background of unlabelled DNA is usually present in the heavy fractions in all SIP experiments (Fig. S2). However, heavy fractions were always enriched in labelled DNA compared with the controls (Fig. S2). Focussing on the heavy DNA, the relative abundance of 16S rRNA genes labelled with 13C-ethane revealed that active members of the phylum Alphaproteobacteria including the genera Sphingomonas (having the highest relative abundance of ~27%), Rhodobacter, Xanthobacter and a genus belonging to Gammaproteobacteria (Hydrogenophaga, formerly Betaproteobacteria) were the most abundant in Pipe Creek (Fig. S3A). The most abundant putative ethane-degrading bacteria labelled in Andreiasu samples belonged to the genera Mycolicibacterium (Actinobacteria, 32%) and Micavibrio (Alphaproteobacteria, 18%) (Fig. S3B). Even though high abundances were observed with members of Sphingomonas and Micavibrio, no MAGs containing SDIMO genes from these genera were retrieved after SIP incubations.
The relative abundance of 16S rRNA genes in heavy DNA from incubations with 13C-labelled propane was analysed for both sites. For Pipe Creek samples, the genus with the highest relative abundance was Hydrogenophaga (13%) together with four other abundant genera from the class Alphaproteobacteria (Xanthobacter, Sphingobium, Sphingomonas and Rhodobacter, ~7–8%) (Fig. S4A). Interestingly, these genera featured as some of the most abundant in 13C-ethane DNA-SIP experiments with Pipe Creek samples (Fig. S3A). From Andreiasu samples, the most abundant genera in incubations with 13C-propane were Mycolicibacterium (19%), Hydrogenophaga (9%), as well as Rhizobium and Rhodobacter (both ~5%) (Fig. S4B). Some of these strains contain monooxygenases that are known or predicted to be involved in gaseous hydrocarbon metabolism (e.g. Hydrogenophaga sp. T4 [43], Xanthobacter sp. Py2 [68] and Sphingobium [19]).
Isolation and genome sequencing of gaseous-alkane degraders
The identification by DNA-SIP and screening of 16S rRNA genes from 13C-DNA samples of putative ethane and propane degraders prompted us to attempt to enrich and isolate key members of the gaseous alkane-degrading community at these sites. Two new isolates were obtained from Pipe Creek (PC2 and PC3) and one from Andreiasu (ANDR5). All isolates grew well on ethane and propane (but not on methane) (Fig. 2A, Fig. S5) and could consume approximately 10% (v/v) of these gaseous-alkanes in the headspace of cultures within 200 h (Fig. 2B, Fig. S5). The completeness of these genomes ranged from 97.9–99.8% and contamination was <5%. The 16S rRNA gene sequences of isolates PC2 and PC3 were highly similar to Rhodoblastus acidophilus DSM 137 T (97.6%, PRJNA396223), and isolate ANDR5 was highly similar to Mycolicibacterium litorale F4T (97.6%, PRJNA374925) (Table S1). The genomes of PC2 and PC3 reveal an AAI of 79% to their closest relative, type strain Rhodoblastus acidophilus (formerly Rhodopseudomonas acidophila [69]) and the genome of ANDR5 indicates an AAI of 82% to Mycolicibacterium litorale [70] (Table 1).
Fig. 2. Growth and substrate consumption of the isolates.
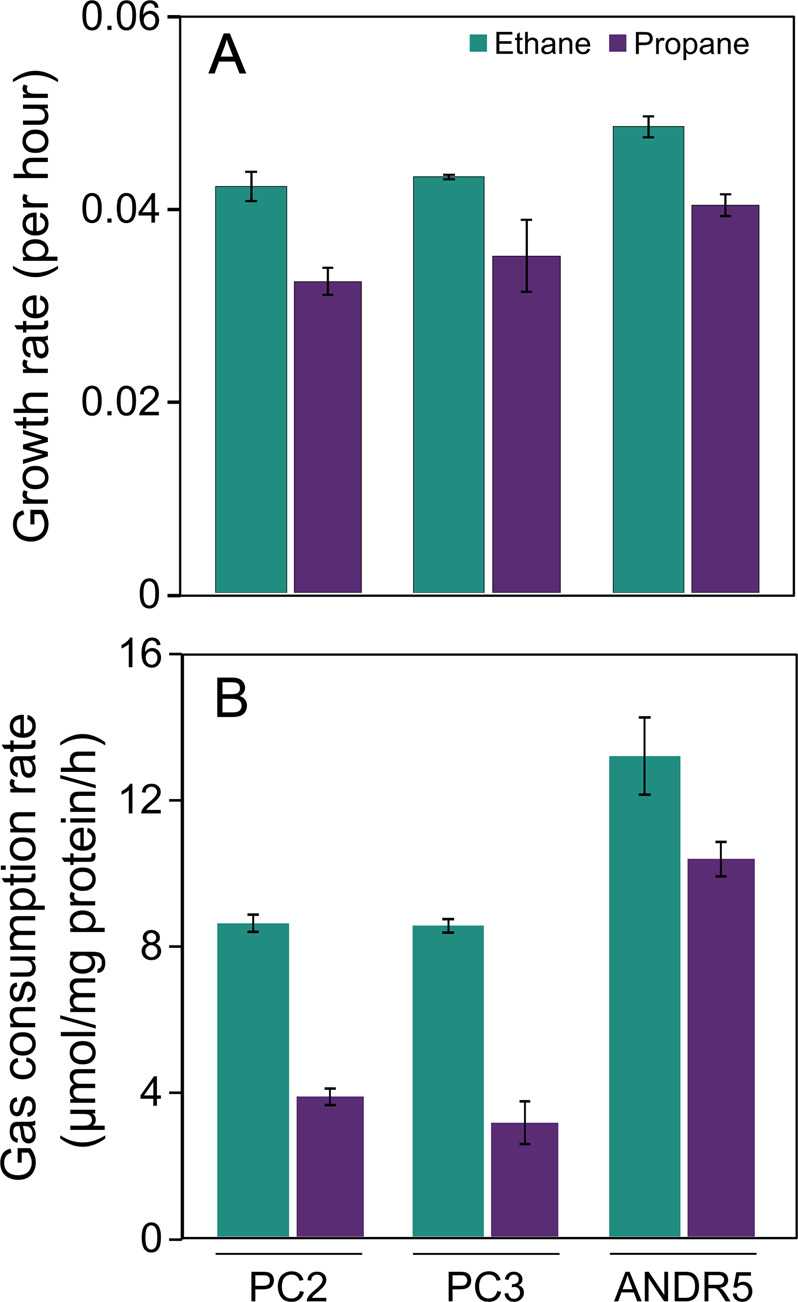
Growth rates (A) and gas consumption (B) of strains PC2, PC3, and ANDR5. Bars represent mean values with standard deviations of independent duplicate incubations for each substrate.
Table 1.
Summary of metagenome-assembled genomes and whole genome sequencing (WGS).
| ID | Origin | MAG retrieved from | Material | Completeness | Contamination | Length | Identification | Reference type strain autoMLST (GenBank accession number) | AAI | ANI |
|---|---|---|---|---|---|---|---|---|---|---|
| (%) | (%) | (bp) | (%) | (%) | ||||||
| MAG12a | Andreaisu | single assembly - ethane incubations | DNA-seq | 86.9 | 4.8 | 2729405 | Methylovulum | Methylovulum miyakonense HT12 (GCF_000384075) | 69.7 | 76.2 |
| MAG19a | Andreaisu | co-assembly | DNA-seq | 99.4 | 1.3 | 3815928 | Mycolicibacterium | Mycolicibacterium litorale F4 (GCA_002007745) | 79.5 | 82.7 |
| MAG22a | Andreaisu | single assembly - propane incubations | DNA-seq | 78.4 | 3.8 | 3341519 | Dechloromonas | Dechloromonas denitrificans ATCC BAA-841 (GCF_001551835) | 78.2 | 82.6 |
| MAG26a | Andreaisu | single assembly - ethane incubations | DNA-seq | 98.9 | 0.9 | 4339169 | Hydrogenophaga | Hydrogenophaga pseudoflava NBRC 102511 (GCF_001592285) | 77.4 | 85.4 |
| MAG27a | Andreaisu | single assembly - propane incubations | DNA-seq | 78.5 | 4.1 | 3487664 | Hydrogenophaga | Hydrogenophaga palleronii NBRC 102513 (GCF_001571225) | 77.2 | 83.8 |
| MAG32a | Andreaisu | co-assembly | DNA-seq | 97.1 | 1.4 | 3163398 | Rhizobiales | Rhodomicrobium vannielii ATCC 17100 (GCF_000166055) | 62.0 | 72.6 |
| MAG33a | Andreaisu | single assembly - ethane incubations | DNA-seq | 82.5 | 1.3 | 4651566 | Rhizobiales | Starkeya novella DSM 506 (GCF_000092925) | 52.9 | 75.5 |
| MAG2p | Pipe Creek | co-assembly | DNA-seq | 89.8 | 1.9 | 4093978 | Mycolicibacterium | Mycolicibacterium vanbaalenii PYR-1 (GCF_000015305) | 70.6 | 78.3 |
| MAG14pa | Pipe Creek | single assembly-original sample | DNA-seq | 98.7 | 0.2 | 3486745 | Methylocapsa | Methylocapsa aurea KYG (GCF_000746085) | 77.4 | 81.5 |
| MAG15p | Pipe Creek | single assembly - ethane incubations | DNA-seq | 85.9 | 2.2 | 2934464 | Rhodoblastus | Rhodoblastus acidophilus DSM 137 (GCF_900187365) | 68.8 | 80.8 |
| MAG16p | Pipe Creek | single assembly - ethane incubations | DNA-seq | 94.9 | 2.2 | 3860394 | Rhodoblastus | Rhodoblastus acidophilus DSM 137 (GCF_900187365) | 68.6 | 78.6 |
| MAG17p | Pipe Creek | single assembly - ethane incubations | DNA-seq | 82.4 | 0.5 | 2824381 | Methylocella | Methylocella silvestris BL2 (GCF_000021745) | 73.7 | 80.7 |
| MAG18p | Pipe Creek | co-assembly | DNA-seq | 96.7 | 0.9 | 3990415 | Methylocella | Methylocella silvestris BL2 (GCF_000021745) | 74.9 | 80.2 |
| MAG38p | Pipe Creek | co-assembly | DNA-seq | 87.7 | 2.4 | 3944496 | Rhodoferax | Rhodoferax fermentans JCM 7819 (GCF_002017865) | 79.6 | 81.6 |
| MAG39p | Pipe Creek | single assembly - ethane incubations | DNA-seq | 91.0 | 2.7 | 3229017 | Xanthobacter | Xanthobacter autotrophicus Py2 (GCF_000017645) | 74.7 | 84.6 |
| MAG47p | Pipe Creek | co-assembly | DNA-seq | 95.7 | 1.1 | 3555802 | Rhodobacteraceae | Loktanella pyoseonensis DSM 21424 (GCF_900102015) | 56.2 | 77.2 |
| MAG49p | Pipe Creek | co-assembly | DNA-seq | 70.7 | 0.0 | 3567447 | Rhodobacteraceae | Loktanella pyoseonensis DSM 21424 (GCF_900102015) | 58.7 | 79.0 |
| PC2 | Pipe Creek | n.a. | WGS | 98.3 | 0.8 | 4345403 | Rhodoblastus | Rhodoblastus acidophilus DSM 137 (GCF_900187365) | 68.6 | 79.3 |
| PC3 | Pipe Creek | n.a. | WGS | 97.9 | 1.7 | 4316956 | Rhodoblastus | Rhodoblastus acidophilus DSM 137 (GCF_900187365) | 68.4 | 79.3 |
| ANDR5 | Andreaisu | n.a. | WGS | 99.8 | 4.1 | 4050246 | Mycolicibacterium | Mycolicibacterium litorale F4 (GCA_002007745) | 79.5 | 82.6 |
AAI Amino acid identity (Enveomics), ANI Average nucleotide identity (autoMLST), n.a. Not applicable.
aMAG contains genes encoding pMMO only.
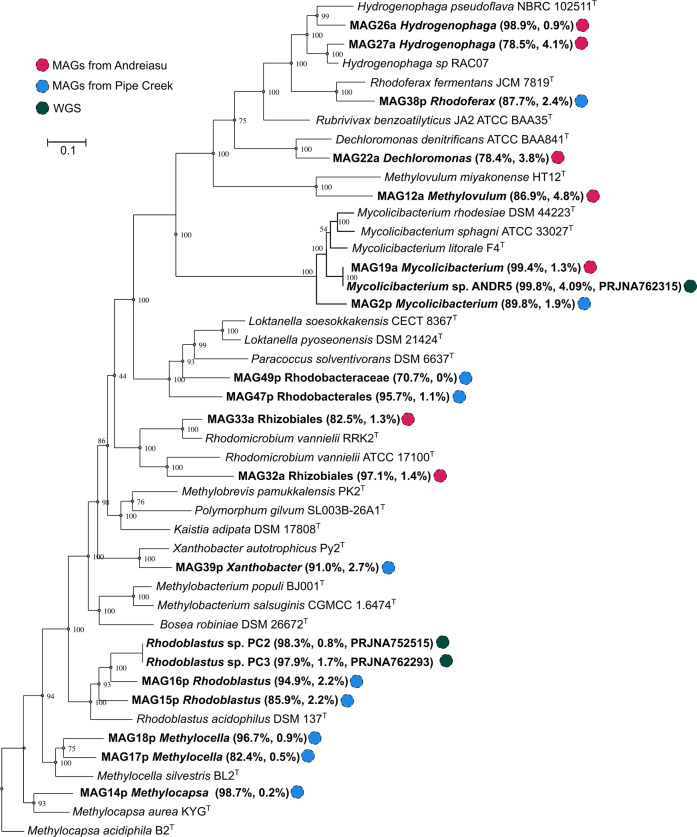
Isolates PC2 and PC3, and two MAGs (MAG15p and MAG16p), all grouped within Rhodoblastus acidophilus (Fig. 3). The AAIs among the isolates and these MAGs are 79.4–80.8%, indicating that the MAGs and the isolates are different species. Isolates PC2 and PC3 have a dDDH of 99.9% and AAI of 99.9% which indicates that they are probably the same strain of R. acidophilus. Interestingly the genome of Mycolicibacterium strain ANDR5 is closely related to MAG19a (Table 1). The multi-locus phylogeny with the genomes of ANDR5 and MAG19a is virtually identical (Fig. 3).
Fig. 3. Multi-locus phylogenetic tree of MAGs and strains PC2, PC3, and ANDR5 using autoMLST.
Bootstrap confidence levels are indicated at internodes. Ten conserved housekeeping genes (infC, hemH, yajC, lipA, frr, nusB, fpg, era, truB and PF00380.15 ribosomal protein S9/S16) were used for the analyses. MAGs and isolates are indicated in bold together with their respective completeness, contamination, and accession number (for isolates). The scale bar represents 10% sequence divergence.
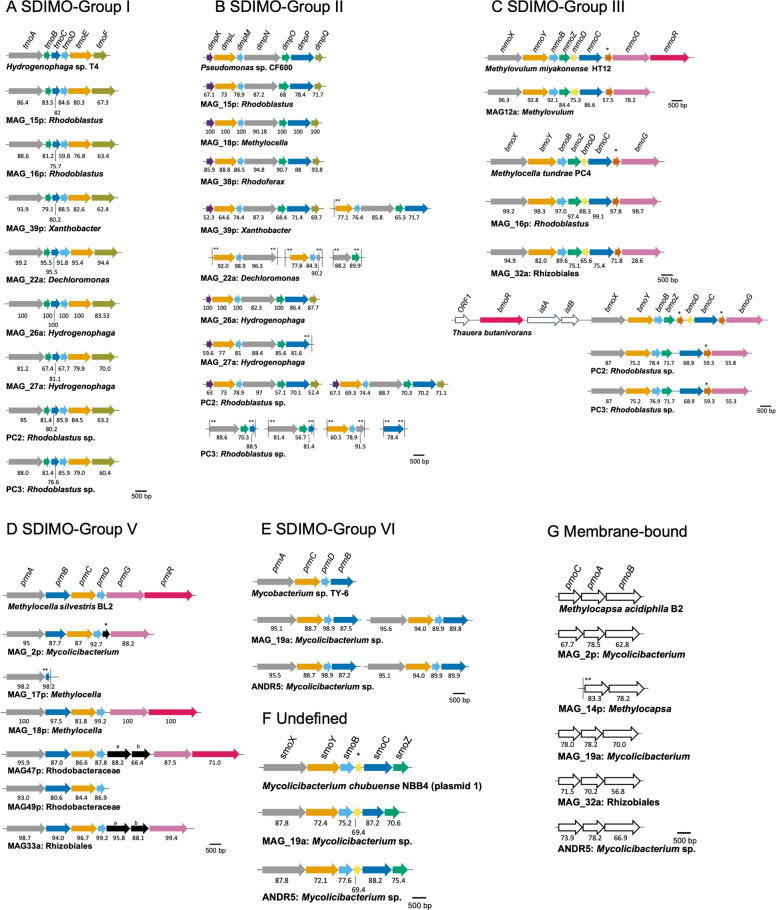
The organisation of the SDIMO gene clusters from PC2, PC3, and ANDR5 was also analysed (Fig. 4). Isolates PC2 and PC3 contained a Group I SDIMO gene cluster (nomenclature of Holmes and Coleman, [17]), which includes toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 [71] and the alkene monooxygenase of Xanthobacter Py2 [68] (Fig. 5A), an SDIMO gene cluster of Group II (Fig. 5B), and an SDIMO gene cluster of Group III (Fig. 5C), related to butane monooxygenase of Thauera butanivorans [29, 34]. The putative butane monooxygenase of both isolates PC2 and PC3 did not show any “hits” when searched by BLAST against the genome of the type strain of R. acidophilus DSM 137 T (Table S1).
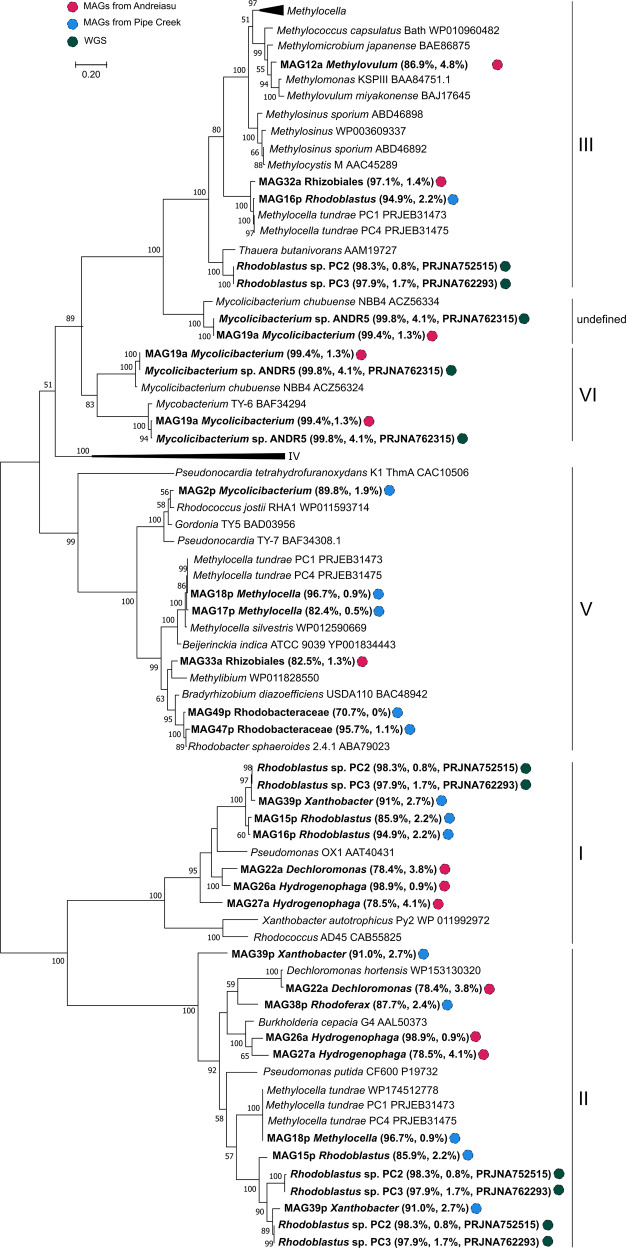
Fig. 4. Phylogenetic tree of the SDIMO large subunit proteins using the Maximum Likelihood method with a JTT matrix-based model.
Bootstrap values (>50) are shown at the nodes. MAGs and isolates are indicated in bold together with their respective completeness, contamination, and accession number (for isolates). The scale bar represents 20% sequence divergence. SDIMO groups analysed in this study: Group-I: toluene, Group-II: phenol, Group-III: methane and butane, Group-IV: ethene, propene, Group-V and Group-VI: propane [17].
Fig. 5. Gene layout of the monooxygenase operons encoded on the MAGs and isolates.
Gene clusters of SDIMO (A–F) and membrane-bound pMMO (G). Names of the genes are given above the reference strain. AAIs with homologous proteins of their closest relative (shown in Table S3) are shown below the arrows. *, unknown function (hypothetical protein); **, end of contig; a, amidohydrolase; b, iron-sulfur cluster assembly protein. The descriptions of SDIMO groups are shown in legend of Fig. 4. Colours are indicated as: large (alpha) subunit (grey), small (beta) subunit (orange), gamma subunit (green), coupling protein (light-blue), unknown function (brown), reductase (blue), ferredoxin (khaki), chaperone (pink), transcriptional regulator (dark pink), and auxiliary protein (purple).
Five MAGs from Pipe Creek and one MAG from Andreiasu contain genes encoding a propane monooxygenase of Group V, but no genes encoding an enzyme from this group were found in the isolates (Fig. 5D). Nevertheless, the isolate Mycolicibacterium strain ANDR5 contains an SDIMO gene cluster of Group VI (also encoding a propane monooxygenase) (Fig. 5E), which was very similar to the corresponding SDIMO from MAG19a (Fig. 4) and most closely related to the propane monooxygenase gene cluster of Mycobacterium sp. TY-6 [24]. The type strain M. litorale F4T only showed 39.9% similarity by BLAST analysis against the propane monooxygenase of isolate ANDR5 (Table S1). Isolate ANDR5 also contains a second undefined SDIMO gene cluster which was again very similar to another SDIMO gene cluster identified in MAG19a (Figs. 4, 5F, Table S2), and which had a high degree of similarity with the SDIMO gene cluster of Mycolicibacterium chubuense NBB4 [25]. Strain ANDR5 also contained a gene cluster, designated pmoCAB, encoding a putative membrane-bound monooxygenase that was also observed in MAG19a (Fig. 5G, Fig. S6). All of this indicates that the MAG19a genome corresponds to a strain related to ANDR5.
Analysis of the metagenomes retrieved from DNA-SIP incubations with 13C-labelled ethane and propane
Since conventional enrichment and isolation only yielded three representative strains of ethane- and propane-utilising bacteria from these natural gas seep samples, the metagenomes of bacteria labelled in SIP incubations with 13C-ethane and 13C-propane were examined. For this, the total metagenomic DNA of the original samples, plus DNA from heavy fractions of incubations with 13C-ethane or 13C-propane from both sites, which is very likely to contain genomes of active hydrocarbon degrading bacteria, were sequenced individually using high-throughput sequencing. In total, 99 MAGs with a completeness >70%, and with a contamination <5% were recovered. 60 of these MAGs originated from Pipe Creek DNA-SIP incubations and 39 MAGs originated from Andreiasu DNA-SIP incubations, and their original samples. Since we aimed to investigate the genomes of putative ethane- and propane-degrading bacteria from both natural gas seep environments, only those MAGs containing gene clusters that resembled either SDIMO gene clusters or membrane-bound monooxygenase gene clusters were selected for detailed analysis. The 17 MAGs selected on the basis of containing putative gaseous hydrocarbon-degrading oxygenases, plus the three genome sequences from the ethane- and propane-degrading isolates, Rhodoblastus strains PC2 and PC3, and Mycolicibacterium strain ANDR5 described above, were further annotated as described in Methods and summarised in Table 1.
For Pipe Creek, one MAG (MAG2p) was affiliated to Actinobacteria (Mycolicibacterium), eight MAGs (MAG14p, MAG15p, MAG16p, MAG17p, MAG18p, MAG39p, MAG47p and MAG49p) belonged to the class Alphaproteobacteria (Methylocella, Rhodoblastus, Methylocapsa, Rhodobacteraceae and Xanthobacter), and one MAG (MAG38p) was affiliated to Gammaproteobacteria (Rhodoferax) (Table 1). MAG17p and MAG18p, affiliated with Methylocella, are interesting because Methylocella is an abundant and active facultative methanotroph in Pipe Creek natural gas seeps [10, 47] and since Methylocella can grow on ethane and propane [18], it is likely that these could have been enriched in ethane and propane SIP incubations.
For Andreiasu, one MAG (MAG19a) belonged to Actinobacteria (Mycolicibacterium), two MAGs (MAG32a and MAG33a) were affiliated to Alphaproteobacteria (Rhizobiales), four MAGs were affiliated to Gammaproteobacteria (MAG22a belonged to Dechloromonas, MAGs 26a and 27a belonged to Hydrogenophaga, and MAG12a belonged to Methylovulum) (Table 1). Of particular note is the estimated high abundance of MAG19a (Mycolicibacterium) in DNA labelled with 13C-ethane and MAG26a (putative Hydrogenophaga) found in DNA labelled with 13C-propane (Fig. 6B). The gaseous alkane-degrading isolate Mycolicibacterium strain ANDR5 and MAG19a representing a Mycolicibacterium strain from Andreiasu samples have a very high AAI of 99.3% suggesting that we have isolated one of the major gaseous-alkane degraders from this natural gas seep site.
Fig. 6. Summary of the presence/absence of SDIMO and membrane-bound MMO gene clusters in each MAG and genome (left panel) and heatmap showing the abundance of those MAGs and isolates in each metagenome (right panel).
A Pipe Creek; (B) Andreiasu. * undefined SDIMO gene cluster. Numbers indicate the abundance of each MAG and genome in the different sites (the analysis was calculated separately using BLASTn, retaining only hits with >95% identity and e-value <1e-5).
Characterisation of SDIMO and membrane-bound copper-containing monooxygenases in MAGs and the genomes of isolated ethane/propane-degrading bacteria
Representative SDIMO gene clusters in MAGs and isolates were found for all groups of SDIMOs except Group IV SDIMOs [17] (Figs. 4, 5A–E, 6, Table S3). In addition, two near identical undefined SDIMO gene clusters (Table S2), found in MAG19a and Mycolicibacterium strain ANDR5 (Fig. 5F), did not cluster within a known SDIMO group. MAG19a and strain ANDR5 also contained a putative pMMO-like alkane monooxygenase (pmoCAB, Fig. 5G, Fig. S6), potentially involved in alkane metabolism [36, 37, 43, 72–76]. Also, the individual genes of the pmoCAB operon have >99% identity between MAG19a and ANDR5 (Table S2). The phylogeny of these pMMO-like, membrane-bound monooxygenases recovered from MAGs and in Mycolicibacterium strain ANDR5 was also analysed. MAG14p (Methylocapsa) PmoA is not surprisingly affiliated with PmoA of other methanotrophs (Fig. S6). The PmoA-like polypeptide sequences derived from Mycolicibacterium MAGs19a and 2p cluster with the corresponding polypeptide found in Mycolicibacterium strain ANDR5 (Fig. S6). Indeed, the PmoA-like polypeptides from strain ANDR5 and MAG19a are virtually identical, again confirming that the isolated ethane/propane degrader from Andreiasu is very representative of a prominent member of the enriched community, retrieved as MAG19a.
Even though their genomes are not identical, both MAGs identified as Rhodoblastus (MAG15p and MAG16p) were grouped in the same cluster as the isolates PC2 and PC3 (Figs. 3, 4). MAG15p and MAG16p, as well as isolates PC2 and PC3, contain genes encoding a toluene monooxygenase from Group I SDIMOs (Figs. 4, 5A). Also, MAG15p and MAG16p, and the two isolates contain genes encoding a phenol hydroxylase (Group II SDIMOs, Fig. 5B), although dmpN (encoding phenol hydroxylase) in MAG16p was truncated (and therefore not used to build the phylogenetic SDIMO tree). Furthermore, MAG16p and MAG32a contain genes encoding sMMO (Group III SDIMOs, Fig. 5C). No SDIMO gene clusters encoding putative isoprene monooxygenases were detected.
Rhodoblastus isolates PC2 and PC3 contained bmoXYBZDCG, encoding a putative butane monooxygenase from Group III SDIMOs (Fig. 5C). Strain ANDR5 also grows on butane to an optical density of >1.0, but no genes encoding butane monooxygenase were found. Strains PC2 and PC3 also contained putative toluene monooxygenase (tmoABCDEF) and phenol hydroxylase (dmpKLMNOP) gene clusters (two copies each) (Fig. 5A, B). Rhodoblastus genus was observed in heavy DNA fractions of 13C-ethane incubations in Pipe Creek samples (Fig. S3A) but only at low abundance (~1%), therefore we assume that this was not a major player in ethane metabolism at Pipe Creek but nevertheless was one of the easiest alkane degraders to isolate.
An estimation of the abundance of MAGs and isolates containing SDIMO and membrane-bound monooxygenases in DNA from both native and enriched samples was carried out (Fig. 6). The MAG with the highest abundance in the original soil from Pipe Creek site was from Methylocapsa (MAG14p, 2.1%). After incubations with 13C-labelled ethane, MAGs mostly from Methylocella (MAG18p, 1.1%), a MAG also from the class Rhizobiales (MAG16p, 1.3%) and Xanthobacter (MAG39p, 1.6%) became enriched. Also, the genomes of Rhodoblastus strains PC2 and PC3 had higher abundance within the metagenomes retrieved from incubations with 13C-labelled ethane than with 13C-labelled propane. After incubations with propane, several alphaproteobacterial MAGs were highly abundant (MAG16p: 1.7%, MAG39p: 1.0%, and MAG47p: 1.2%, Fig. 6A). The MAG with the highest abundance retrieved after incubating Andreiasu samples with labelled ethane belonged to the phylum Actinobacteria (MAG19a, 28.1%). Also, the isolate Mycolicibacterium strain ANDR5 was highly abundant in the raw metagenome dataset from the SIP incubations with 13C-ethane (32.1%). After incubations with 13C-labelled propane, the MAG with the highest abundance belonged to Hydrogenophaga (MAG26a, 37.2%), which was also abundant (12.6%) after incubation with ethane (Fig. 6B).
Description of Candidatus “Mycolicibacterium alkanivorans” sp. nov. and Candidatus “Rhodoblastus alkanivorans” sp. nov
Further genomic comparisons of the three isolates using TETRA, dDDH, ANI, AAI, and a set of ten house-keeping genes, revealed similarities below species-level thresholds when the type strain genome sequences were used as query (Table S1). This indicates that these three isolates are novel. Strain ANDR5 is the representative of a novel species of the genus Mycolicibacterium within the order Corynebacteriales (Phylum Actinobacteria). Strains PC2 and PC3 are novel species of the genus Rhodoblastus within the order Rhizobiales (Phylum Proteobacteria). Since both strains PC2 and PC3 shared an ANI of 99.99% between each other, we are proposing PC2 genome as the type material for this candidate new species. All isolates grew well on ethane, propane, and butane as carbon sources, and possessed SDIMO-related genes. We propose the following new Candidatus species:
Description of Candidatus “Mycolicibacterium alkanivorans” sp.nov
Candidatus M. alkanivorans (al.ka.ni.vo’rans. N.L. neut. n. alkanum, saturated aliphatic hydrocarbon; L. inf. v. vorare, to eat; N.L. part. adj. alkanivorans, alkane-devouring).
Gram-positive rod-shaped aerobic bacterium isolated from natural gas seep (liquid mud) located in Romania. The isolate ANDR5 contains SDIMO-related genes including a propane monooxygenase. The isolate ANDR5 also contained a gene cluster encoding a putative membrane-bound monooxygenase. The genome is characterised by a size of 4.3 Mb and has a G + C content of 63.7 mol%.
The type genome of the species has been deposited at the NCBI under the accession number PRJNA762315.
Description of Candidatus “Rhodoblastus alkanivorans” sp.nov
Candidatus R. alkanivorans (al.ka.ni.vo’rans. N.L. neut. n. alkanum, saturated aliphatic hydrocarbon; L. inf. v. vorare, to eat; N.L. part. adj. alkanivorans, alkane-devouring).
Gram-negative rod-shaped, aerobic bacterium isolated from a natural gas seep (river water and sediment) located in Pipe Creek, NY, USA. Strain PC2 contains SDIMO-related genes including a putative butane monooxygenase, a toluene monooxygenase, and a phenol hydroxylase. The genome is characterised by a size of 4.0 Mb and has a G + C content of 67.3 mol%.
The type genome of the species has been deposited at the NCBI under the accession number PRJNA752515.
Both isolates are available upon request to the corresponding author.
Conclusions
In this study, we isolated gaseous-alkane degraders, characterised their monooxygenase enzymes involved in oxidation of ethane, propane, and butane, and retrieved metagenome-assembled genomes from natural gas seeps after DNA-SIP incubations with 13C-labelled ethane or propane. We demonstrate the relative abundance and importance of SDIMO family enzymes in the consumption of gaseous-alkanes at these natural gas seep sites. Metagenome data from these SIP experiments have revealed the diversity of active alkane-degraders present and the diversity of SDIMOs they contain and provides vital molecular data for screening and targeted isolation of new and potentially novel ethane and propane-degrading bacteria.
Supplementary information
Acknowledgements
We thank Professor Calin Baciu for the Andreiasu (Romania) samples and Scott Ensminger for facilitating sampling of Pipe Creek (USA) gas seeps. We thank Professor Ramon Rosselló-Móra for valuable discussion and his insights into bacterial identification. We thank Dr. Blanca Vera Gargallo and Dr. Pengfei Liu for their help with bioinformatics. The authors are grateful to the valuable comments received by three anonymous reviewers. Work on this project was supported by the Leverhulme Trust (RPG2016-050) and ERC Advanced Grant (694578—IsoMet) to JCM, a Leverhulme Trust Early Career Fellowship (ECF-2016-626) to ATC, and a Royal Society Dorothy Hodgkin Research Fellowship (DHF\R1\211076) to MH.
Author contributions
JCM and ATC conceived the project. MFUH and ATC performed the laboratory experiments. MFUH, ATC, MH and JCM analysed the data. MH carried out bioinformatics analysis. JCM and MH drafted the paper. All authors contributed to its revisions and approved the final version.
Data availability
Sequence data were deposited in the NCBI Sequence Read Archive (SRA) under the bioproject accession numbers: PRJNA765146 for amplicon-sequencing data, PRJNA748243 for metagenomic raw data, and PRJNA748244 for metagenome-assembled genomes. Genomes of the isolates were deposited in the GenBank under the bioproject accession numbers PRJNA752515 (for isolate PC2), PRJNA762293 (for isolate PC3), and PRJNA762315 (for isolate ANDR5).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Muhammad Farhan Ul Haque, Marcela Hernández.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01211-0.
References
- 1.Saunois M, Stavert AR, Poulter B, Bousquet P, Canadell JG, Jackson RB, et al. The global methane budget 2000–2017. Earth Syst Sci Data. 2020;12:1561–623. doi: 10.5194/essd-12-1561-2020. [DOI] [Google Scholar]
- 2.Myhre G, Shindell D, Bréon F-M, Collins W, Fuglestvedt J, Huang J, et al. “Anthropogenic and natural radiative forcing”. In: Stocker TF, et al., editors. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, UK: Cambridge University Press; 2013. [Google Scholar]
- 3.Reddy CM, Arey JS, Seewald JS, Sylva SP, Lemkau KL, Nelson RK, et al. Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20229–34. doi: 10.1073/pnas.1101242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etiope G, Schwietzke S. Global geological methane emissions: an update of top-down and bottom-up estimates. Elementa. 2019;7:47. [Google Scholar]
- 5.Mazzini A, Sciarra A, Etiope G, Sadavarte P, Houweling S, Pandey S, et al. Relevant methane emission to the atmosphere from a geological gas manifestation. Sci Rep. 2021;11:4138. doi: 10.1038/s41598-021-83369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pozzer A, Jöckel P, Tost H, Sander R, Ganzeveld L, Kerkweg A, et al. Simulating organic species with the global atmospheric chemistry general circulation model ECHAM5/MESSy1: a comparison of model results with observations. Atmos Chem Phys. 2007;7:2527–50. doi: 10.5194/acp-7-2527-2007. [DOI] [Google Scholar]
- 7.Dalsøren SB, Myhre G, Hodnebrog Ø, Myhre CL, Stohl A, Pisso I, et al. Discrepancy between simulated and observed ethane and propane levels explained by underestimated fossil emissions. Nat Geosci. 2018;11:178–84. doi: 10.1038/s41561-018-0073-0. [DOI] [Google Scholar]
- 8.Etiope G, Ciccioli P. Earth’s degassing: a missing ethane and propane source. Science. 2009;323:478. doi: 10.1126/science.1165904. [DOI] [PubMed] [Google Scholar]
- 9.Etiope G, Drobniak A, Schimmelmann A. Natural seepage of shale gas and the origin of “eternal flames” in the Northern Appalachian Basin, USA. Mar Pet Geol. 2013;43:178–86. doi: 10.1016/j.marpetgeo.2013.02.009. [DOI] [Google Scholar]
- 10.Farhan Ul Haque M, Crombie AT, Ensminger SA, Baciu C, Murrell JC. Facultative methanotrophs are abundant at terrestrial natural gas seeps. Microbiome. 2018;6:118. doi: 10.1186/s40168-018-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schimmelmann A, Ensminger SA, Drobniak A, Mastalerz M, Etiope G, Jacobi RD, et al. Natural geological seepage of hydrocarbon gas in the Appalachian Basin and Midwest USA in relation to shale tectonic fracturing and past industrial hydrocarbon production. Sci Total Environ. 2018;644:982–93. doi: 10.1016/j.scitotenv.2018.06.374. [DOI] [PubMed] [Google Scholar]
- 12.Lelieveld JOS, Crutzen PJ, Dentener FJ. Changing concentration, lifetime and climate forcing of atmospheric methane. Tellus B Chem Phys Meteorol. 1998;50:128–50. doi: 10.3402/tellusb.v50i2.16030. [DOI] [Google Scholar]
- 13.Hodnebrog Ø, Dalsøren SB, Myhre G. Lifetimes, direct and indirect radiative forcing, and global warming potentials of ethane (C2H6), propane (C3H8), and butane (C4H10) Atmos Sci Lett. 2018;19:e804. doi: 10.1002/asl.804. [DOI] [Google Scholar]
- 14.Reeburgh WS, Global methane biogeochemistry in Treatise on Geochemistry, HD Holland, KK Turekian, Eds. (Elsevier, Amsterdam, 2007), pp. 1-32.
- 15.Trotsenko YA, Murrell JC. Metabolic aspects of aerobic obligate methanotrophy. In: Allen SS, Laskin I, Geoffrey MG, editors. Advances in Applied Microbiology. Academic Press; 2008. [DOI] [PubMed] [Google Scholar]
- 16.Leahy JG, Batchelor PJ, Morcomb SM. Evolution of the soluble diiron monooxygenases. FEMS Microbiol Rev. 2003;27:449–79. doi: 10.1016/S0168-6445(03)00023-8. [DOI] [PubMed] [Google Scholar]
- 17.Holmes AJ, Coleman NV. Evolutionary ecology and multidisciplinary approaches to prospecting for monooxygenases as biocatalysts. Antonie Van Leeuwenhoek. 2008;94:75–84. doi: 10.1007/s10482-008-9227-1. [DOI] [PubMed] [Google Scholar]
- 18.Crombie AT, Murrell JC. Trace-gas metabolic versatility of the facultative methanotroph Methylocella silvestris. Nature. 2014;510:148–51. doi: 10.1038/nature13192. [DOI] [PubMed] [Google Scholar]
- 19.Farhan Ul Haque M, Xu H-J, Murrell JC, Crombie A. Facultative methanotrophs – diversity, genetics, molecular ecology and biotechnological potential: a mini-review. Microbiology. 2020;166:894–908. doi: 10.1099/mic.0.000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahman MT, Crombie A, Chen Y, Stralis-Pavese N, Bodrossy L, Meir P, et al. Environmental distribution and abundance of the facultative methanotroph Methylocella. ISME J. 2011;5:1061–6. doi: 10.1038/ismej.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dedysh SN. Exploring methanotroph diversity in acidic northern wetlands: molecular and cultivation-based studies. Microbiology. 2009;78:655–69. doi: 10.1134/S0026261709060010. [DOI] [Google Scholar]
- 22.Gupta RS, Lo B, Son J. Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol. 2018;9:67. doi: 10.3389/fmicb.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamamura N, Yeager CM, Arp DJ. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl Environ Microbiol. 2001;67:4992–8. doi: 10.1128/AEM.67.11.4992-4998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotani T, Kawashima Y, Yurimoto H, Kato N, Sakai Y. Gene structure and regulation of alkane monooxygenases in propane-utilizing Mycobacterium sp. TY-6 and Pseudonocardia sp. TY-7. J Biosci Bioeng. 2006;102:184–92. doi: 10.1263/jbb.102.184. [DOI] [PubMed] [Google Scholar]
- 25.Coleman NV, Yau S, Wilson NL, Nolan LM, Migocki MD, Ly M, et al. Untangling the multiple monooxygenases of Mycobacterium chubuense strain NBB4, a versatile hydrocarbon degrader. Environ Microbiol Rep. 2011;3:297–307. doi: 10.1111/j.1758-2229.2010.00225.x. [DOI] [PubMed] [Google Scholar]
- 26.Ashraf W, Mihdhir A, Murrell JC. Bacterial oxidation of propane. FEMS Microbiol Lett. 1994;122:1–6. doi: 10.1111/j.1574-6968.1994.tb07134.x. [DOI] [PubMed] [Google Scholar]
- 27.Cappelletti M, Presentato A, Milazzo G, Turner RJ, Fedi S, Frascari D, et al. Growth of Rhodococcus sp. strain BCP1 on gaseous n-alkanes: new metabolic insights and transcriptional analysis of two soluble di-iron monooxygenase genes. Front Microbiol. 2015;6:393. doi: 10.3389/fmicb.2015.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson EL, Hyman MR. Propane and n-butane oxidation by Pseudomonas putida GPo1. Appl Environ Microbiol. 2006;72:950–2. doi: 10.1128/AEM.72.1.950-952.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubbels BL, Sayavedra-Soto LA, Bottomley PJ, Arp DJ. Thauera butanivorans sp. nov., a C2–C9 alkane-oxidizing bacterium previously referred to as ‘Pseudomonas butanovora’. Int J Syst Evol Microbiol. 2009;59:1576–8. doi: 10.1099/ijs.0.000638-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rojo F. Degradation of alkanes by bacteria. Environ Microbiol. 2009;11:2477–90. doi: 10.1111/j.1462-2920.2009.01948.x. [DOI] [PubMed] [Google Scholar]
- 31.Shennan JL. Utilisation of C2–C4 gaseous hydrocarbons and isoprene by microorganisms. J Chem Technol Biotechnol. 2006;81:237–56. doi: 10.1002/jctb.1388. [DOI] [Google Scholar]
- 32.Coleman NV, Bui NB, Holmes AJ. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ Microbiol. 2006;8:1228–39. doi: 10.1111/j.1462-2920.2006.01015.x. [DOI] [PubMed] [Google Scholar]
- 33.Osborne CD, Haritos VS. Beneath the surface: evolution of methane activity in the bacterial multicomponent monooxygenases. Mol Phylogenet Evol. 2019;139:106527. doi: 10.1016/j.ympev.2019.106527. [DOI] [PubMed] [Google Scholar]
- 34.Sluis MK, Sayavedra-Soto LA, Arp DJ. Molecular analysis of the soluble butane monooxygenase from ‘Pseudomonas butanovora’. Microbiology. 2002;148:3617–29. doi: 10.1099/00221287-148-11-3617. [DOI] [PubMed] [Google Scholar]
- 35.Martin KE, Ozsvar J, Coleman NV. SmoXYB1C1Z of Mycobacterium sp. strain NBB4: a soluble methane monooxygenase (sMMO)-like enzyme, active on C2 to C4 alkanes and alkenes. Appl Environ Microbiol. 2014;80:5801–6. doi: 10.1128/AEM.01338-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coleman NV, Le NB, Ly MA, Ogawa HE, McCarl V, Wilson NL, et al. Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME J. 2012;6:171–82. doi: 10.1038/ismej.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khadka R, Clothier L, Wang L, Lim CK, Klotz MG, Dunfield PF. Evolutionary history of copper membrane monooxygenases. Front Microbiol. 2018;9:2493. doi: 10.3389/fmicb.2018.02493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotani T, Yamamoto T, Yurimoto H, Sakai Y, Kato N. Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J Bacteriol. 2003;185:7120–8. doi: 10.1128/JB.185.24.7120-7128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redmond MC, Valentine DL, Sessions AL. Identification of novel methane-, ethane-, and propane-oxidizing bacteria at marine hydrocarbon seeps by stable isotope probing. Appl Environ Microbiol. 2010;76:6412–22. doi: 10.1128/AEM.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M, Jain S, Baker BJ, Taylor C, Dick GJ. Novel hydrocarbon monooxygenase genes in the metatranscriptome of a natural deep-sea hydrocarbon plume. Environ Microbiol. 2014;16:60–71. doi: 10.1111/1462-2920.12182. [DOI] [PubMed] [Google Scholar]
- 41.Crombie AT, Larke-Mejia NL, Emery H, Dawson R, Pratscher J, Murphy GP, et al. Poplar phyllosphere harbors disparate isoprene-degrading bacteria. Proc Natl Acad Sci USA. 2018;115:13081–6. doi: 10.1073/pnas.1812668115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi M, Huang H, Zhang Y, Wang H, Li H, Lu Z. Novel tetrahydrofuran (THF) degradation-associated genes and cooperation patterns of a THF-degrading microbial community as revealed by metagenomic. Chemosphere. 2019;231:173–83. doi: 10.1016/j.chemosphere.2019.05.137. [DOI] [PubMed] [Google Scholar]
- 43.Rochman FF, Kwon M, Khadka R, Tamas I, Lopez-Jauregui AA, Sheremet A, et al. Novel copper-containing membrane monooxygenases (CuMMOs) encoded by alkane-utilizing Betaproteobacteria. ISME J. 2020;14:714–26. doi: 10.1038/s41396-019-0561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumont MG, Murrell JC. Stable isotope probing - linking microbial identity to function. Nat Rev Microbiol. 2005;3:499–504. doi: 10.1038/nrmicro1162. [DOI] [PubMed] [Google Scholar]
- 45.Dumont MG, Hernández García M, editors. Stable isotope probing; methods and protocols. Totowa, NJ, US: Humana Press; 2019. [Google Scholar]
- 46.Baciu C, Ionescu A, Etiope G. Hydrocarbon seeps in Romania: gas origin and release to the atmosphere. Mar Pet Geol. 2018;89:130–43. doi: 10.1016/j.marpetgeo.2017.06.015. [DOI] [Google Scholar]
- 47.Farhan Ul Haque M, Crombie AT, Murrell JC. Novel facultative Methylocella strains are active methane consumers at terrestrial natural gas seeps. Microbiome. 2019;7:134. doi: 10.1186/s40168-019-0741-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxf) 2009;2009:bap021. doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, et al. DNA stable-isotope probing. Nat Protoc. 2007;2:860–6. doi: 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- 50.Jia Z, Cao W, Hernández García M. DNA-based stable isotope probing. In: Dumont MG, Hernández García M, editors. Stable Isotope Probing: Methods and Protocols. New York, NY: Springer New York; 2019. pp. 17–29. [DOI] [PubMed] [Google Scholar]
- 51.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrews S. (2010) FastQC: a quality control tool for high throughput sequence data.
- 53.Bushnell B, Rood J, Singer E. BBMerge - Accurate paired shotgun read merging via overlap. PloS one. 2017;12:e0185056–e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–34. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olm MR, Brown CT, Brooks B, Banfield JF. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–8. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 60.Hernández M, Vera-Gargallo B, Calabi-Floody M, King GM, Conrad R, Tebbe CC. Reconstructing genomes of carbon monoxide oxidisers in volcanic deposits including members of the class Ktedonobacteria. Microorganisms. 2020;8:1880. doi: 10.3390/microorganisms8121880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-R LM, Konstantinidis KT. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016;4:e1900v1901. [Google Scholar]
- 62.Alanjary M, Steinke K, Ziemert N. AutoMLST: an automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 2019;47:W276–82. doi: 10.1093/nar/gkz282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0906412106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez-R LM, Konstantinidis KT. Bypassing cultivation to identify bacterial species. Microbe. 2014;9:111–8. [Google Scholar]
- 65.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stecher G, Tamura K, Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol. 2020;37:1237–9. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Small FJ, Ensign SA. Alkene monooxygenase from Xanthobacter strain Py2. J Biol Chem. 1997;272:24913–20. doi: 10.1074/jbc.272.40.24913. [DOI] [PubMed] [Google Scholar]
- 69.Imhoff JF. Transfer of Rhodopseudomonas acidophila to the new genus Rhodoblastus as Rhodoblastus acidophilus gen. nov., comb. nov. Int J Syst Evol Microbiol. 2001;51:1863–6. doi: 10.1099/00207713-51-5-1863. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Zhang J, Fang C, Pang H, Fan J. Mycobacterium litorale sp. nov., a rapidly growing mycobacterium from soil. Int J Syst Evol Microbiol. 2012;62:1204–7. doi: 10.1099/ijs.0.033449-0. [DOI] [PubMed] [Google Scholar]
- 71.Bertoni G, Martino M, Galli E, Barbieri P. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl Environ Microbiol. 1998;64:3626–32. doi: 10.1128/AEM.64.10.3626-3632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sayavedra-Soto LA, Hamamura N, Liu C-W, Kimbrel JA, Chang JH, Arp DJ. The membrane-associated monooxygenase in the butane-oxidizing Gram-positive bacterium Nocardioides sp. strain CF8 is a novel member of the AMO/PMO family. Environ Microbiol Rep. 2011;3:390–6. doi: 10.1111/j.1758-2229.2010.00239.x. [DOI] [PubMed] [Google Scholar]
- 73.Tavormina PL, Orphan VJ, Kalyuzhnaya MG, Jetten MSM, Klotz MG. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep. 2011;3:91–100. doi: 10.1111/j.1758-2229.2010.00192.x. [DOI] [PubMed] [Google Scholar]
- 74.Picone N, Mohammadi SS, Waajen AC, van Alen TA, Jetten MSM, Pol A, et al. More than a methanotroph: a broader substrate spectrum for Methylacidiphilum fumariolicum SolV. Front Microbiol. 2020;11:604485. doi: 10.3389/fmicb.2020.604485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Awala SI, Gwak J-H, Kim Y-M, Kim S-J, Strazzulli A, Dunfield PF, et al. Verrucomicrobial methanotrophs grow on diverse C3 compounds and use a homolog of particulate methane monooxygenase to oxidize acetone. ISME J. 2021 doi: 10.1038/s41396-021-01037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zou B, Huang Y, Zhang P-P, Ding X-M, Camp HJMOD, Quan Z-X, et al. Horizontal gene transfer of genes encoding copper-containing membrane-bound monooxygenase (CuMMO) and soluble di-iron monooxygenase (SDIMO) in ethane- and propane-oxidizing Rhodococcus bacteria. Appl Environ Microbiol. 2021;87:e00227–00221. doi: 10.1128/AEM.00227-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data were deposited in the NCBI Sequence Read Archive (SRA) under the bioproject accession numbers: PRJNA765146 for amplicon-sequencing data, PRJNA748243 for metagenomic raw data, and PRJNA748244 for metagenome-assembled genomes. Genomes of the isolates were deposited in the GenBank under the bioproject accession numbers PRJNA752515 (for isolate PC2), PRJNA762293 (for isolate PC3), and PRJNA762315 (for isolate ANDR5).