Abstract
We recently described the use of PCR to identify the environmental source of Mycobacterium ulcerans during an outbreak of ulcerative disease that occurred in a localized region of southeast Australia. The PCR used was based on amplification of the M. ulcerans-specific insertion sequence, IS2404. In this study we developed a new test that is a substantial improvement over the original PCR method in terms of sensitivity, reliability, and ease of use. In the new method magnetic bead sequence capture-PCR is used to detect two M. ulcerans sequences (IS2404 and IS2606) and total mycobacterial 16S ribosomal DNA. We used sequence capture-PCR to test water and plant material collected over a 12-month period during 1998 and 1999 from sites near the centers of two distinct foci of M. ulcerans infections. A golf course irrigation system in one area and a small shallow lake in another area repeatedly were PCR positive for M. ulcerans. Nearby sites and sites unrelated to the endemic areas were negative. Based on the PCR data, a most-probable-number method was used to estimate the concentration of M. ulcerans cells in positive samples from both regions. This procedure resulted in average concentrations of 0.5 cell per 100 ml of water and 40 cells per 100 g of detritus. Loss of the PCR signal coincided with a decrease in ulcerative disease in each area. These results provide further evidence that M. ulcerans may be transmitted from a point environmental source and demonstrate the utility of magnetic bead sequence capture-PCR for identification of nonculturable microbial pathogens in the environment.
Mycobacterium ulcerans infection causes progressive destructive skin ulceration in otherwise healthy humans. Established, small, endemic foci of disease exist in the Gippsland and Westernport regions of southeast Australia (9) and in far north Queensland (28). However, in world terms, the prevalence of M. ulcerans infection is greatest in rural West Africa, where the disease is known as the Buruli ulcer. In some countries in this region, the incidence of this disease has risen dramatically (16, 17). While Buruli ulcer is rarely fatal, the social and economic burden of advanced disease on local communities is severe as the only effective treatment is surgical excision of the infected tissue, which is often followed by skin grafting. This leads to lengthy hospital stays or alternatively, when either surgical treatment or appropriate rehabilitation is not available, the prospect of life-long functional deformities. The significance of M. ulcerans as an emerging pathogen was recently recognized by the World Health Organization when it established the Global Buruli Ulcer Initiative (2).
Buruli ulcer endemic foci are characteristically small (this area is often just a few square kilometers) and near swamps or slowly flowing rivers (10, 11, 21). Transmission of M. ulcerans is thought to be mediated via contact with the environment which results in inoculation of the organism through minor cuts and abrasions. Local environmental changes may be associated with the emergence of new endemic foci (10).
It has been very difficult to test any of these hypotheses because of the inability to culture M. ulcerans from the environment (20, 25). Recently, we developed a highly specific PCR based on IS2404 detection (26) and used it to confirm that a golf course irrigation system and a small nearby swamp were the likely sources of M. ulcerans during an outbreak of ulcerative disease on Phillip Island in southeast Australia (25). In that investigation, a gel chromatography procedure was used to reduce problems of PCR inhibition caused by coconcentrated environmental contaminants. However, while this approach was effective, it was cumbersome and labor-intensive. Using paramagnetic beads to selectively isolate DNA or RNA is a straightforward and efficient process that has been used successfully to isolate multiple microbial pathogen target sequences in PCR-inhibitory clinical and environmental samples (12, 15, 29). In the present study we used a nucleic acid extraction and purification method based on magnetic bead sequence capture (15). This technique was initially developed for isolating specific Mycobacterium tuberculosis DNA sequences from clinical specimens that contained a large amount of background DNA and PCR-inhibitory material. It is an indirect capture method in which biotinylated oligonucleotides are hybridized with target DNA in solution. The hybridization complex is then immobilized by adding streptavidin-coated paramagnetic beads, and the bound DNA is then washed to remove inhibitors, resuspended in a small volume of water, and added directly to a PCR mixture.
The primary aims of this study were to build on the initial findings obtained at Phillip Island and to begin to assemble some basic knowledge regarding the prevalence of M. ulcerans in the environment and its correlation with disease. We used sequence capture-PCR to search for evidence of M. ulcerans in environmental samples collected from another endemic focus in southeast Australia that was not related to the Phillip Island outbreak. We then monitored the persistence of the organism and the disease over time in that region and at Phillip Island. We also adapted an M. ulcerans immunomagnetic separation (IMS) method (24) to try to culture M. ulcerans from the PCR-positive samples by selective capture of intact cells. We reasoned that this approach was more likely to succeed than previous efforts to culture M. ulcerans from the environment as we targeted samples that were PCR positive for the organism. Also, by using IMS, we could potentially overcome the problem of culture overgrowth by reducing the concentration of other, faster-growing mycobacteria in a sample.
MATERIALS AND METHODS
Oligonucleotide probes and PCR primers.
The sequences of all of the oligonucleotides used in this study and their functions are listed in Table 1. Development of the PCR primers used for detection of IS2404, IS2606, and mycobacterial 16S ribosomal DNA (rDNA) has been described previously (6, 30). The 5′-biotinylated oligonucleotide capture probes were synthesized by Beckman Instruments, Gladesville, Australia. These probes were reverse-phase cartridge purified to ensure that they were full length and to remove unincorporated biotin molecules. Capture probe MUCAP1 was designed to hybridize with IS2404-containing DNA fragments; MUCAP3 was designed to hybridize with IS2606-containing fragments; and MYC16SCAP was designed to hybridize with mycobacterial 16S rDNA fragments (Table 1).
TABLE 1.
PCR primers and oligonucleotide probes used in this study
| Oligonucleotide | Sequence (5′-3′) | Position in gene | Function | Reference |
|---|---|---|---|---|
| MUCAP1 | CAGTGTTGACGCATCCAGGTCATGATCGCA | 981–952 | 5′ BIOTIN-LABELED CAPTURE PROBE FOR IS2404 | THIS STUDY |
| MUCAP3 | AGGACCTGTCCGAGCGCCTTGGATATGACC | 270–299 | 5′ BIOTIN-LABELED CAPTURE PROBE FOR IS2606 | THIS STUDY |
| MYC16SCAP | ATGCACCACCTGCACACAGGCCACAAGGGA | 1056–1027 | 5′ BIOTIN-LABELED CAPTURE PROBE FOR MYCOBACTERIAL 16S RDNA | THIS STUDY |
| MU5 | AGCGACCCCAGTGGATTGGT | 392–411 | MU5 AND MU6; 492-BP PCR PRODUCT FROM IS2404 | 30 |
| MU6 | CGGTGATCAAGCGTTCACGA | 862–881 | ||
| MU7 | GGCCTGGCGGATTGCTCAAGG | 213–233 | MU7 AND MU8: 332-BP PCR PRODUCT FROM IS2606 | 30 |
| MU8 | CGTAGATGTGGGCGAAATGG | 542–523 | ||
| MUNEST2 | TGATCGTCAAGTCCAACCAA | 664–683 | MU5 AND MUNEST2: 206-BP SEMINESTED PCR PRODUCT FROM IS2404 | THIS STUDY |
| MUNEST3 | CGGTGCTCACTGATGGGTGC | 357–376 | MU7 AND MUNEST3: 200-BP SEMINESTED PCR PRODUCT FROM IS2606 | THIS STUDY |
| MYCGENF | AGAGTTTGATCCTGGCTCAG | 16–35 | MYCGENF AND MYCGENR: 1,030-BP PCR PRODUCT FROM 16S RRNA IN ALL MYCOBACTERIA | 6 |
| MYCGENR | TGCACACAGGCCACAAGGGA | 1046–1027 | ||
| UPS-IPCF | GCGGTCCCAAAAGGGTCAGTATGACAGCCACTCCT | IPCF AND IPCR: 331-BP PRODUCT FROM IPC DNA | 12 | |
| UPS-IPCR | GCGGTCCCAAAAGGGTCAGTATGTCAGTTGTGACCACGAA |
Preparation of environmental samples.
Water samples (500, 1000, or 2,000 ml) were collected in sterile plastic containers by dip sampling from approximately 20 cm below the surface of the water. Bacteria were then concentrated by membrane filtration (pore size, 0.45 μm) or by centrifugation as previously described (25). Sediment samples were collected in sterile plastic jars, and total DNA was extracted from 50-mg (wet weight) subsamples as described below without further concentration. Samples of detritus were prepared by stomaching 100-g portions of each specimen in 200 ml of phosphate-buffered saline supplemented with 0.01% Tween 80 for 10 min by using a Lab-Blender 400 stomacher (Seward Laboratory, London, England). Ten-centimeter portions of a water reed stem (Triglochin spp.), collected below the water surface, were also taken from some sites and stomached as described above. After stomaching, the cells in the wash solutions obtained from either detritus or reed samples were concentrated by centrifugation. All concentrates prepared from water, detritus, and plant samples were resuspended in 10 or 20 ml of nutrient broth (Unipath, Basingstoke, England) containing 15% (vol/vol) sterile glycerol. One-milliliter portions were analyzed by sequence capture-PCR as described below, and the remainder of each sample was stored at −70°C. Sediment samples were stored at 4°C for 1 week.
DNA extraction from environmental samples.
Total DNA was extracted from environmental sample concentrates by using a modification of the method described by Mangan et al. (14). A 1-ml portion of sample concentrate was centrifuged at 17 000 × g for 2 min. The pellet was then resuspended in 300 μl of water and added to a 2-ml screw-cap tube containing 300 μl of 100-μm-diameter, washed, glass beads, 500 μl of lysis buffer (containing [per 100 ml] 9.6 ml of liquid Pyroneg glass-washing detergent [Diversey Ltd., Sydney, Australia], 24 ml of 500 mM sodium acetate, and 66.4 ml of water; pH adjusted to 8.0 with 10 M HCl), 500 μl of pH 7.0 equilibrated phenol, and 100 μl of chloroform-isoamyl alcohol (24:1). Mycobacterial cells were disrupted by one 40-s treatment at a speed setting of 6 with a model FP120 FastPrep cell disruptor (Savant Instruments Inc., Holbrook, N.Y.). After cell disruption, the tubes were cooled briefly on ice, and then the preparations were centrifuged at 17 000 × g for 20 min. In each case the aqueous phase was retained and reextracted with 500 μl of chloroform-isoamyl alcohol (24:1). The DNA in the aqueous phase was then precipitated with 500 μl of isopropanol in the presence of 0.3 M sodium acetate (pH 5.2). All traces of isopropanol were removed, and the DNA-containing pellets were solubilized in 510 μl of buffer (100 mM NaCl, 100 mM Tris, 50 mM EDTA; pH 8.0). The crude DNA preparations were stored at −20°C prior to sequence capture-PCR.
Sequence capture.
Sequence capture was performed essentially as described previously (15) but with some modifications to optimize the sensitivity for this application. In a 1.5-ml screw-cap microtube, the crude DNA extracted from an environmental sample homogenate as described above was heated at 100°C for 10 min and then quenched on ice to produce sheared, single-stranded molecules. To this solution were added 2.5 pmol of each biotinylated capture probe (Table 1) and 200 μl of 3.75 M NaCl (total volume, 750 μl). The probes were allowed to hybridize with potential target DNA at 43°C with rotation for 1.5 h. Twenty micrograms of washed, streptavidin-coated paramagnetic beads (Dynal, Oslo, Norway) was then added, and incubation was continued for an additional 1.5 h at 43°C. The beads were immobilized against the wall of the tube by using a magnetic particle concentrator (model MPC-M; Dynal) and were washed once with 500 μl of wash buffer (10 mM Tris-HCl [pH 8.0], 0.15 M LiCl, 1.0 mM EDTA, 0.1% lithium dodecyl sulfate) and once with 500 μl of 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl). The beads, bound probes, and target DNA were finally resuspended in 30 μl of pyrocarbonic acid diethyl ester-treated H2O. Occasionally, inhibition of the PCR by the magnetic beads was observed, so a dissociation step was included. This involved heating the sample at 95°C for 5 min, cooling it rapidly on ice, and then centrifuging it at 17 000 × g for 20 s. A 5-μl portion of the supernatant was then added to each of three PCR mixtures (IS2404, IS2606, and 16S rRNA mixtures) along with the internal positive control (IPC). The remaining 15 μl was stored at −20°C. The IPC was produced as previously described by PCR amplification of nonmycobacterial DNA with primers UPS-IPCF and UPS-IPCR (Table 1). It was diluted to a concentration that would just support amplification, as determined by agarose gel electrophoresis.
PCR.
The reaction conditions used for PCR detection of IS2404 and IS2606 have been described previously (30). PCR detection of mycobacterial 16S rDNA and the IPC was performed with a 30-μl (total volume) reaction mixture which contained 1× PCR buffer II (10× PCR buffer II contained 500 mM KCl and 100 mM Tris-HCl [pH 8.3]), 2.5 mM MgCl2, 1 mM dATP, 1 mM dTTP, 1 mM dCTP, 1 mM dGTP, 0.5 μM primer MYCGENF, 0.5 μM primer MYCGENR, 0.5 μM primer UPS-IPCF, 0.5 μM primer UPS-IPCR, 1 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer), 10 fg of IPC DNA, and 5 μl of pyrocarbonic acid diethyl ester-treated water containing DNA. A hot-start PCR was performed with a model FTS-960 thermal sequencer (Corbett Research, Sydney, Australia) by using the thermal cycling conditions that were used for IS2404 and IS2606 detection. PCR products were kept at 4°C until they were analyzed by 1.5% agarose gel electrophoresis with ethidium bromide staining and Southern blot hybridization. Appropriate negative controls were included with each batch of samples. Reactions which failed to support amplification of the 331-bp IPC fragment were considered to be inhibited and were retested following 1:5 dilution of the template DNA.
Southern blot hybridization analysis.
Southern blot analysis of PCR products was performed by using 3-h capillary alkaline transfer with Hybond N+ nylon membranes (Amersham Corp.). The probes used for confirmation of IS2404 and IS2606 PCR products were prepared by PCR amplification of 50 ng of M. ulcerans DNA with primers MU5 and MUNEST2 and primers MU7 and MUNEST3, respectively. Probes were labeled and DNA was detected with the digoxigenin nonradioactive labeling system and CDP-Star, respectively, by using the protocols of the manufacturer (Roche). All hybridizations were performed at 65°C with high-stringency posthybridization washes.
Mycobacterial cultures from environmental samples.
Mycobacteria were cultured from environmental samples by using a modification of an IMS method described previously for PCR detection of M. ulcerans (24). Environmental sample concentrates (maximum weight, 200 mg [wet weight]), prepared as described above, were incubated with 15 μg of a polyclonal anti-M. ulcerans immunoglobulin G-fractionated antibody in 1 ml of SLA/B buffer (Dynal) at 4°C for 18 h. In each case cells were pelleted, the supernatant was removed, and the pellet was resuspended in 1 ml of SLA/B buffer. Fifteen micrograms of washed sheep anti-mouse-coated paramagnetic beads (Dynal) was added, and the preparation was incubated with rolling at room temperature for 1 h. The magnetic beads and bound cells were immobilized with a magnetic particle concentrator (model MPCM; Dynal), washed once with phosphate-buffered saline (PBS), and then incubated for 15 min in 1 ml of 0.005% cetyl pyridinium chloride to decontaminate the sample. The beads were washed once more with 1 ml of PBS and then resuspended in 100 μl of PBS. For spiking experiments in which PCR was used to compare the detection limit of the IMS method with that of the sequence capture method (see below), total DNA was extracted from the 100-μl sample by cell homogenization as described above. The isopropanol-precipitated DNA was washed once with 70% ethanol, dried briefly, and then resuspended in 30 μl of water. A 5-μl portion was used for PCR. For attempts to culture M. ulcerans by the IMS method, 20 μl of the 100-μl cell preparation was spread directly onto egg yolk agar slopes containing 500 μg of cycloheximide (Sigma) per ml. The slopes were incubated under microaerophilic conditions (19) for 12 weeks at 32°C. Single colonies were subcultured from the slopes and identified as mycobacteria by 16S rRNA nucleotide sequence analysis as previously described (13). Sequences were compiled with Sequencher 3.1.1 software (Gene Codes). All sequences were aligned between nucleotides 119 and 921 (numbering based on the Escherichia coli 16S rRNA gene) with CLUSTALW (31). This region spanned the two hypervariable regions of the mycobacterial 16S rRNA gene. The maximum-parsimony method (8) was used to analyze the phylogenetic relationship between isolates with bootstrapping (500 iterations), and a consensus tree was drawn with Treeview software (18).
Spiked-sample preparation.
To permit comparisons of data obtained by sequence capture-PCR, data obtained by the IMS method, and data obtained in a previous study by gel filtration-PCR, a three-way spiking experiment was conducted to measure the performance of each method. The spiking matrix was prepared by pooling several detritus concentrates that were IS2404 negative in order to produce an approximately 200-ml portion of material from which we obtained a 20-mg (wet weight) pellet from each 300-μl subsample. Six 30-ml aliquots of this concentrate were then spiked with dilutions of mid-log-phase M. ulcerans cell cultures to produce predicted final concentrations of 10,000, 1,000, 100, 10, 1, and 0 cells per 300 μl (20 mg) of detritus. For this experiment, M. ulcerans cells from a mid-log-phase culture were stained with a Live/Dead kit (Molecular Probes, Eugene, Oreg.) and enumerated by epifluorescence microscopy performed with an Aksioskop microscope (Zeiss, Oberkochen, Germany). Cells were cultured in the presence of Tween 80 as previously described (30) to reduce the problem of cell clumping. However, occasional clumps consisting of less than 10 cells were observed; thus, these experiments were used to compare relative method performance rather than to accurately assess the absolute detection limit of each method. The six spiked samples were vortexed for 1 min, and three 300-μl portions were withdrawn from each dilution and tested by the sequence capture-PCR and IMS-PCR methods as described above and by the gel filtration-PCR method as described previously (25).
RESULTS
Comparison of M. ulcerans environmental PCR detection methods.
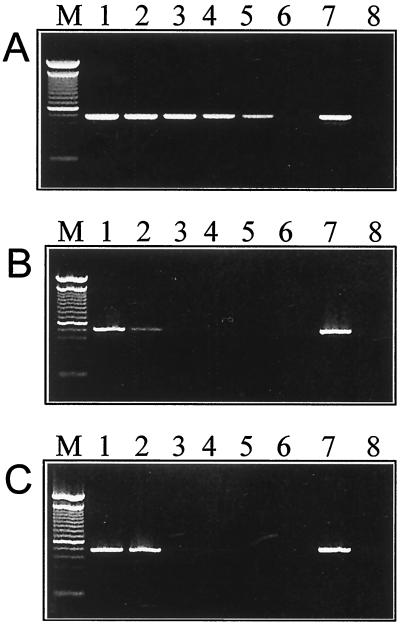
A 5-log unit dilution series of M. ulcerans, spiked into detritus concentrates, was prepared as described above and was tested by the sequence capture-PCR, gel filtration-PCR, and IMS-PCR methods. The results of this analysis are shown in Fig. 1 and indicate that the sequence capture-PCR method resulted in a 1,000-fold improvement compared with both the IMS-PCR and gel filtration-PCR methods.
FIG. 1.
Comparison of detection sensitivities of the sequence capture-PCR (A), gel filtration-PCR (B), and IMS-PCR (C) methods as measured by identification of IS2404 DNA in environmental samples spiked with M. ulcerans. Lane 1, 104 cells; lane 2, 103 cells; lane 3, 102 cells; lane 4, 10 cells; lane 5, 1 cell; lane 6, no cells; lane 7, PCR positive control; lane 8, PCR negative control; lane M, 100-bp ladder size marker.
PCR analysis of samples from the Frankston-Langwarrin focus.
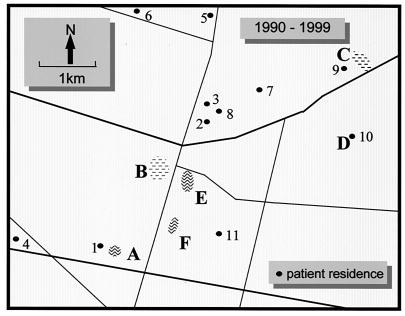
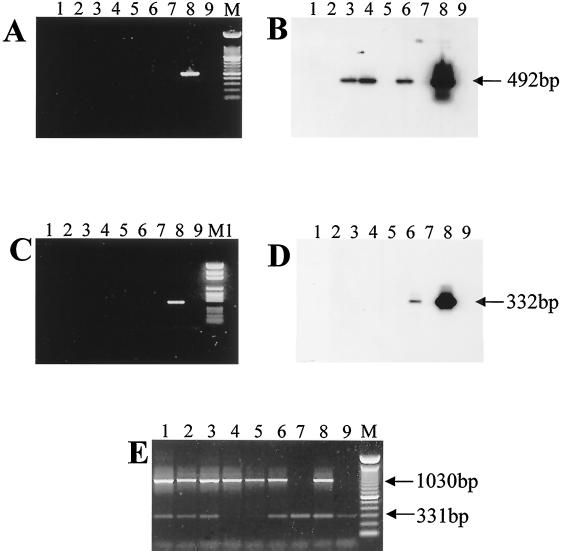
We previously investigated and described a cluster of 29 cases of M. ulcerans disease that occurred at Phillip Island, which is 80 km from the center of Melbourne, Australia, between 1993 and 1995. In investigating other cases, we concentrated on cases that occurred within 70 km of the center of Melbourne between 1990 and 1997, a definition chosen to exclude Phillip Island. Of 25 cases, 10 occurred in three adjacent suburbs (Frankston, Frankston North, and Langwarrin), and one subject visited this area frequently. The remaining cases appeared to be scattered fairly randomly and were not considered further. Langwarrin is an outer suburb of Melbourne with a population of approximately 20,000 people. Sandy soil and remnant, native, sclerophyll woodlands with a growing suburban population characterize the geography of the area. The 11 confirmed or highly likely cases of M. ulcerans disease were clustered in an area approximately 4 by 6 km, suggesting that there was a common source of infection (Fig. 2). Several water sources were identified as potential reservoirs of M. ulcerans in this area. These water sources were a permanent swamp on the site of a disused sand quarry, a farm dam opposite the swamp, and several small ponds or lakes, including a fishpond at a patient's home (Fig. 2). A total of 57 water, sediment, and vegetation samples were collected from the region over a 12-month period and were analyzed by sequence capture-PCR and Southern hybridization for IS2404 and IS2606 and by sequence capture-PCR for mycobacterial 16S rDNA. The results are summarized in Table 2. Water, plant, and detritus samples from two sites (identified as sites B1 and B2) in a swamp near the center of the case cluster area were the only samples that were positive as determined by IS2404 PCR or by both IS2404 and IS2606 PCR. Site B1 was approximately 30 m from site B2, and samples were collected from the littoral zone of the swamp in both areas. Detection of IS2404 in the absence of IS2606 was not unexpected given the higher copy number of IS2404 and the 10-fold difference in PCR detection limits previously reported for these insertion elements (30). However, the two IS2404-positive samples from site B1 were the same two samples that were IS2606 positive (Table 2). All samples obtained from all other sites were negative for both repeated sequences but positive for mycobacterial 16S rDNA and the IPC, indicating that PCR inhibition did not occur and also that mycobacteria were ubiquitous in the environment. An example of results obtained from analysis of some swamp samples is shown in Fig. 3. In this example the 492-bp PCR product for IS2404 was detected in plant, water, and detrital material by Southern hybridization but not by ethidium bromide staining. The 332-bp IS2606 PCR product was detected by Southern hybridization in the detritus sample. The presence of 1,030-bp mycobacterial 16S rRNA PCR products in all samples indicated that other mycobacteria were present (Fig. 3).
FIG. 2.
Map of the Frankston-Langwarrin region showing the distribution of cases of M. ulcerans disease (numbers 1 through 11) and potential environmental sources of M. ulcerans. Site A, Shaxton Lake; site B, swamp on the site of a disused sand quarry; site C, Langwarrin Woodlands; site D, fishpond at patient's home; site E, farm dam opposite site B; site F, flora and fauna reserve.
TABLE 2.
PCR results obtained from the Langwarrin region
| Site | Site description | Sample type | Southern hybridization results (no. of positive samples/total no. of samples examined)a
|
|
|---|---|---|---|---|
| IS2404 | IS2606 | |||
| A | Shaxton Lake | Water | 0/4 | 0/4 |
| B1 | Swamp (area 1)b | Water | 2/14 | 2/3 |
| Detritus | 2/13 | 2/3 | ||
| Sediment | 0/1 | 0/1 | ||
| B2 | Swamp (area 2) | Water | 0/2 | 0/2 |
| Vegetationc | 1/2 | 0/1 | ||
| Sediment | 0/1 | 0/1 | ||
| C | Langwarrin Woodlands | Water | 0/1 | 0/1 |
| D | Fish pond, patient's home | Water | 0/2 | 0/2 |
| Sediment | 0/2 | 0/2 | ||
| E | Dam opposite swamp | Water | 0/1 | 0/1 |
| Sediment | 0/1 | 0/1 | ||
| F | Flora and fauna reserve | Water | 0/11 | 0/2 |
| Detritus | 0/1 | 0/1 | ||
| Sediment | 0/1 | 0/1 | ||
All samples were positive for mycobacterial 16S rRNA genes as determined by PCR.
IMS culture isolate JKD2391 was obtained at this site.
Stem of a water plant (Triglochin sp.) collected below the surface of the water.
FIG. 3.
Identification of M. ulcerans from the Langwarrin swamp (Fig. 2, site B) in the Frankston-Langwarrin region based on detection of the 492-bp IS2404 PCR product (A and B) and the 332-bp IS2606 PCR product (C and D). The PCR products were separated by gel electrophoresis (A and C) and were detected by Southern hybridization (B and D). The total mycobacteria in the samples were identified by detection of the 1,030-bp mycobacterial 16S rDNA PCR product and the 331-bp IPC PCR product (E). Lane 1, water from site B, area 2; lane 2, sediment from site B, area 2; lane 3, plant sample (Triglochin spp.) from site B, area 2; lane 4, water from site B, area 1; lane 5, sediment from site B, area 1; lane 6, detritus from site B, area 1; lane 7, sterile water procedural control; lane 8, PCR positive control; lane 9, PCR negative control; lanes M and M1, molecular weight markers (100-bp ladder [Gibco, BRL] and PGEM size ladder [Promega], respectively).
PCR analysis of samples from the Phillip Island focus.
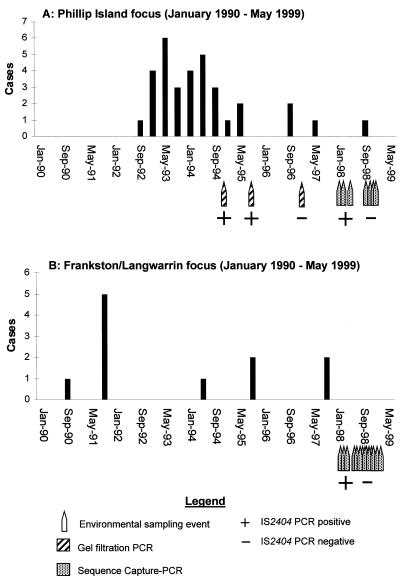
In our previous investigation of the outbreak that occurred between 1993 and 1995 (11), using epidemiological and PCR data, we identified a golf course irrigation system and a nearby swamp as likely sources of infection (25, 32). Modifications of the swamp and the irrigation system were undertaken in 1994 and 1995, respectively, and the epidemic ceased. Only four additional cases of ulcerative disease attributable to this area have been reported since 1995 (Fig. 4). This preliminary evidence that there was an environmental source of M. ulcerans provided an opportunity to explore the ecology of M. ulcerans. A total of 63 water and detritus samples were collected on seven separate occasions between 1998 and 1999 from various sites around the outbreak area on Phillip Island and from other sites on the island that were some distance from this area. A summary of the sites tested and the results of the PCR analysis are presented in Table 3. All 63 samples were screened by the sequence capture-PCR and Southern hybridization methods for IS2404 and by the sequence capture-PCR method alone for total mycobacterial 16S rDNA. Eleven samples were positive for IS2404. Fifty-two of the 63 samples were also tested for IS2606, and 23 were positive. With 10 of these 23 samples, IS2606 was codetected with IS2404, and with 13 IS2606 was detected in the absence of IS2404. Water samples collected from the irrigation system, which included two covered concrete tanks, a pumping system, and a reticulated sprinkler network (system layout details are described in reference 25), were positive for IS2404 on two separate sampling occasions in March 1998 as determined by PCR but were negative on all four subsequent sampling occasions. The covered tanks received water from the local sewage treatment plant, but all samples of water coming to the golf course from the plant were negative for IS2404 as determined by PCR. Detritus and water samples collected from the littoral zone of the dam on the golf course, which was adjacent to the covered tanks, were positive for IS2404 in March and June 1998, but samples from both sites were negative on the four subsequent sampling occasions. The dam was adjacent to the covered tanks and pump system but since 1995 has not been connected to the irrigation network. Samples collected from a shallow bore hole (depth, approximately 3 m) on the golf course were negative, as were control samples collected from the town water supply at the golf course and from the Silverleaves housing estate area. This housing estate is adjacent to the golf course and was the focus of most of the cases that occurred during the outbreak that occurred between 1993 and 1995. Samples collected from the swampy area near a fire access track in the Silverleaves area were negative for IS2404, as were water samples collected from other wetland areas on Phillip Island that were some distance from the outbreak area (Table 3).
FIG. 4.
Epidemic curves for the Phillip Island (A) and Frankston-Langwarrin (B) regions, showing the numbers of cases of human M. ulcerans disease from 1990 to 1999 overlaid with the results obtained from IS2404 PCR analysis of the Phillip Island golf course dam and Langwarrin swamp (site B, area 1), respectively.
TABLE 3.
Summary of PCR results obtained from samples collected from Phillip Island between March 1998 and February 1999
| Region | Site description | Sample type | Southern hybridization results (no. of positive samples/total no. of samples examined)
|
|
|---|---|---|---|---|
| IS2404 | IS2606 | |||
| Golf course | Storage tanksa | Water | 3/12 | 5/8 |
| Irrigation systemb | Water | 3/3 | 2/3 | |
| Dam near storage tanksc | Water | 2/7 | 3/5 | |
| Dam near storage tanks | Detritus | 3/7 | 4/7 | |
| Bore hole | Water | 0/3 | 2/3 | |
| Town water supply at golf coursed | Water | 0/6 | 0/5 | |
| Silverleaves housing estate | Drain under fire access track (east) | Water | 0/4 | 2/3 |
| Coughlan's Road drain (south) | Water | 0/2 | 1/2 | |
| Farm dam | Water | 0/1 | 1/1 | |
| Rain water tank | Water | 0/4 | 0/4 | |
| Town water supply, consumer tapd | Water | 0/5 | 2/4 | |
| Beach | Water | 0/1 | 0/1 | |
| Other sites | Rhyll swamp | Water | 0/3 | 0/1 |
| Cowes Cemetery wetland | Water | 0/1 | 0/1 | |
| Treated sewage effluent, water leaving plant | Water | 0/4 | 1/4 | |
IMS culture isolates JKD2379, JKD2380, JKD2381, JKD2384, JKD2385, and JKD2388 were obtained at this site.
IMS culture isolates JKD2387, JKD2389, and JKD2390 were obtained at this site.
IMS culture isolate JKD2386 was obtained at this site.
Samples from these sites were negative for mycobacterial 16S rRNA genes as determined by PCR. Samples from all other sites were positive for mycobacterial 16S rRNA genes.
Comparisons of M. ulcerans concentrations in water and detritus samples.
Repeat PCR analyses of IS2404-positive samples obtained from each focus were performed to confirm the positive results. These data were then used to estimate the concentrations of M. ulcerans in the original samples by a most-probable-number (MPN) method (5). The results (Table 4) suggested that 0.2 to 1.0 M. ulcerans cell was present in 100 ml of each water sample and 30 to 51 cells were present in 100 g of each detritus sample.
TABLE 4.
MPN estimates for the concentrations of M. ulcerans cells in IS2404 PCR-positive samples collected from the Frankston-Langwarrin and Phillip Island regions
| Site | Collection date (day/mo/yr) | Sample type | Amt of original sample analyzed per replicatea | Results
|
|
|---|---|---|---|---|---|
| No. of positive tubes per dilution | MPN per 100 ml or 100 g (wet wt) of sampleb | ||||
| Langwarrin swamp, site B1 | 28/1/98 | Water | 150 ml (1) | 1 | 0.80 (0.19–3.37) |
| 75 ml (2) | 1 | ||||
| 10/2/98 | Water | 75 ml (3) | 1 | 0.43 (0.06–3.07) | |
| 28/1/98 | Detritus | 1.5 g (4) | 2 | 38 (10–154) | |
| 3/2/98 | Detritus | 1.5 g (2) | 1 | 30 (4–210) | |
| Phillip Island Golf Club | |||||
| Tank | 3/3/98 | Water | 150 ml (2) | 1 | 0.30 (0.04–2.13) |
| Pump shed | 3/3/98 | Water | 150 ml (3) | 1 | 0.21 (0.03–1.54) |
| Irrigation system | 24/3/98 | Water | 300 ml (4) | 3 | 0.50 (0.20–1.25) |
| 75 ml (4) | 2 | ||||
| Dam | 16/6/98 | Water | 75 ml (3) | 2 | 1.0 (0.25–4.22) |
| Dam | 3/3/98 | Detritus | 1.5 g (3) | 2 | 51 (12–211) |
The numbers in parentheses are numbers of replicates.
The numbers in parentheses are 95% confidence intervals.
Correlation of the IS2404 PCR data with the epidemiological data.
The epidemic curves for both regions were overlaid with the environmental PCR data obtained from the dam at the Phillip Island golf course (Fig. 4A) and the swamp at Langwarrin (Fig. 4B). In each area, within 8 weeks the last reported case of disease was accompanied by a loss of the IS2404 PCR-positive signal in the environment. Note that the incubation period for M. ulcerans disease has been estimated to be at least 2 to 4 months (3, 23). Figure 4A also shows the PCR data obtained by the gel filtration method, which was 1,000-fold less sensitive than the sequence capture method, as indicated by the results of the spiking experiments (Fig. 1). In agreement with these results, the gel filtration-PCR test produced negative results in 1997, while the sequence capture-PCR method still detected positive samples early in 1998.
IMS.
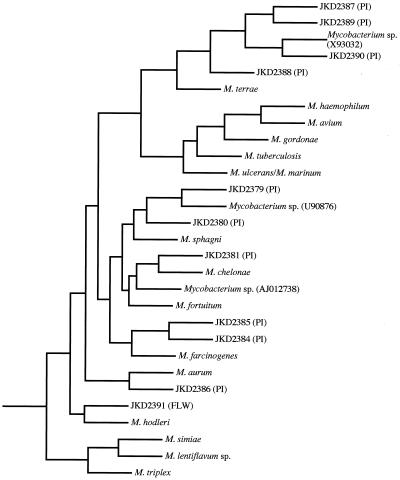
An IMS method was also used in an attempt to culture M. ulcerans from IS2404-positive samples, but this attempt was not successful. However, a diverse range of other mycobacteria were isolated with this technique, presumably because of some cross-reactivity with the polyclonal anti-M. ulcerans antibodies used in the assay. These mycobacteria included fast-growing and slowly growing species. They were characterized by performing a partial sequence analysis of the 16S rRNA gene. A BLASTN analysis (1) was performed to identify closely related species. The results of this analysis and the relationships of the environmental isolates to other high-scoring mycobacteria are shown in Fig. 5. The sequences obtained from these isolates have been deposited in the GenBank database under the following accession numbers: JKD2379, AF221084; JKD2380, AF221085; JKD2381, AF221086; JKD2384, AF221087; JKD2385, AF221088; JKD2386, AF221089; JKD2387, AF221090; JKD2388, AF221091; JKD2389, AF221092; JKD2390, AF221093; and JKD2391, AF221094. The accession numbers for the partial 16S rRNA gene sequences obtained from high-scoring database matches and other relevant mycobacterial species are as follows: Mycobacterium sphagni, X55590; Mycobacterium fortuitum, X52933; Mycobacterium chelonae, U92090; Mycobacterium farcinogenes, AJ012738; Mycobacterium aurum, X55595; Mycobacterium avium, X52918; Mycobacterium haemophilum, L24800; Mycobacterium gordonae, X52923; Mycobacterium tuberculosis, X52917; Mycobacterium ulcerans, Z13990; Mycobacterium marinum, X52920; Mycobacterium terrae, M29568; Mycobacterium hodleri, X93184; Mycobacterium triplex, U57632; Mycobacterium lentiflavum, X93995; Mycobacterium simiae, X52931; and Nocardia asteroides, Z36934. Isolate JKD2391 was obtained from site B1 at the Langwarrin swamp (Table 2). Isolates JKD2379, JKD2380, JKD2381, JKD2384, JKD2385, JKD2387, JKD2388, JKD2389, and JKD2390 were all cultured from the golf course irrigation network, while isolate JKD2386 was obtained from the golf course dam (Table 3). All isolates were negative for IS2404 and IS2606 as determined by PCR (data not shown).
FIG. 5.
Maximum-parsimony tree based on partial rRNA 16S sequences from environmental mycobacterial isolates detected by IMS in IS2404-positive samples, closely related species identified by BLASTN analysis, and other mycobacterial species of clinical significance. The designations for the mycobacteria isolated by the IMS method begin with the prefix JKD, which is followed by a unique four-digit number. The region from which each isolate was cultured is indicated in parentheses, as follows: PI, Phillip Island; FLW, Frankston-Langwarrin. The tree was rooted by using N. asteroides as an outgroup.
DISCUSSION
Based on compelling epidemiological evidence, it has been proposed that M. ulcerans is an environmental organism that occasionally infects humans (21). The mode of transmission is unknown, although primary contact with the environment and infection by inoculation of the organism at a site where there is minor skin trauma are a possible route of infection (10, 21). As a first step towards determining the mode or modes of transmission, it is important to understand the ecology of M. ulcerans. Previously, we have described identification of M. ulcerans in a golf course irrigation system by PCR during an outbreak of M. ulcerans disease (25). This was the first direct evidence that M. ulcerans occurs in the environment and provided an important opportunity to address the question of environmental sources in more detail. To do this, we collected samples from another endemic area in southeast Australia and monitored the occurrence of M. ulcerans at this site and at the site of the original outbreak over a 12-month period.
The gel filtration DNA purification method which was used in the previous study to overcome problems of PCR inhibition was cumbersome, time-consuming, and not suitable for screening large numbers of samples. Sequence capture-PCR was used to overcome these problems. In this study 110 environmental samples were analyzed by this method, and the frequency of inhibition, as determined by the absence of the 331-bp internal control fragment, was only 5.5% (data not shown). In addition, spiking of M. ulcerans cells into environmental material revealed that the detection limit for sequence capture-PCR was low (Fig. 1A). The technique was also found to be 1,000-fold more sensitive than both the gel filtration-PCR method and the IMS-PCR method. The lower detection sensitivity of IMS may in part account for the lack of correlation of the IMS data with the data produced by sequence capture-PCR technique in this study.
IS2606 was used as a second M. ulcerans-specific sequence to support IS2404-positive results and to reduce the likelihood of false-positive results. However, it became apparent during this study that the primer pair used to detect IS2606 was not specific for M. ulcerans. The 332-bp IS2606 PCR product was often detected and identified by Southern hybridization in the absence of IS2404. This lack of species specificity was supported by the discovery of a putative isoform of IS2606 in M. lentiflavum (30). Detection of IS2606 by PCR and Southern hybridization in the town water supply in the absence of mycobacterial 16S rDNA (tested by using PCR alone) (Table 3) was probably a reflection of the different detection limits for the assays rather than the presence of the insertion element in another genus. Taken together, these data suggest that IS2606 (or isoforms of IS2606) may be quite promiscuous, present in several species, and widely distributed in the environment.
The results of this study improved our understanding of the prevalence and distribution of M. ulcerans. The MPN-PCR estimates obtained for water samples collected at the Phillip Island golf course (Table 4) suggest that the average M. ulcerans cell concentration was 0.5 cell per 100 ml of water during 1998. Considering this concentration of cells, the 1,000-fold difference in detection limits between the sequence capture-PCR and gel filtration-PCR methods, and the fact that positive gel filtration-PCR results for 1994 and 1995 were obtained by only Southern hybridization (i.e., at the detection limit of the method), then by extrapolation it seems probable that M. ulcerans was present at a concentration of at least 500 organisms per 100 ml of water at the peak of the Phillip Island outbreak. It is particularly interesting that M. ulcerans was still present in the golf course irrigation system even though the Phillip Island epidemic appeared to have been halted (Fig. 4A). Thus, it is possible that the burden of M. ulcerans in that environment dropped below a critical disease-causing threshold. A dose-response effect is supported by similar data obtained for the Frankston-Langwarrin focus. At this site there have been no cases of disease since November 1997 and no detectable levels of M. ulcerans DNA since January 1998 (Fig. 4B). Also, in both the Phillip Island and Langwarrin foci, the sites that were IS2404 positive were physically close to each cluster of cases (Fig. 2) (25). This is consistent with an airborne method of dispersal, either by aerosolized droplets or perhaps by an insect vector from a point source (10).
The estimates of higher concentrations of M. ulcerans cells in detritus than in water (Table 4) give some insight into the likely habitat of this organism, indicating that the organism may be saprophytic in detrital material. The presence of M. ulcerans in the golf course irrigation network remains unexplained and may represent a transient, opportunistic colonization of this system. Since the golf course receives reuse water from the local sewage facility, we reasoned that this might have been the source of the organism. However, all tests conducted on this water before it entered the golf course system were negative for IS2404, suggesting that local contamination of the irrigation network with M. ulcerans occurred, possibly from the golf course dam.
Now that we have some indication of the environmental sources of M. ulcerans, increases in our understanding of the environmental ecology of this organism may be better achieved by establishing aquatic microenvironments that mimic the conditions at these sites. A method for fluorescent in situ hybridization analysis of mycobacteria in which peptide nucleic acid probes are used has recently been described (7). This method could be readily applied to detailed ecological studies of M. ulcerans by designing peptide nucleic acid probes for either IS2404 or IS2606.
There have been many attempts in the last 30 years to culture M. ulcerans from the environment, and all of them have been confounded by the presence of faster-growing mycobacteria that overgrow the culture media (4, 22, 25). IMS was used in an unsuccessful attempt to selectively isolate and culture M. ulcerans from IS2404-positive samples obtained from both Phillip Island and the Frankston-Langwarrin area. However, a wide variety of other mycobacteria, some of which have not been described previously, were detected by the IMS method. The range of species recovered from the recycled wastewater at the golf course is consistent with previous studies of mycobacterial diversity in treated wastewater (27). The lack of species specificity of the IMS method suggests that the antisera used in this assay contained antibodies that recognized common mycobacterial antigens. In future attempts to culture M. ulcerans from environmental samples, the researchers will need to address the issues of specificity and detection sensitivity.
While this study provided some insights into the habitat of M. ulcerans in southeast Australia, because of climatic and other significant geographic differences these results may not be valid for West Africa or other tropical endemic regions. In these regions, the rates of disease are much higher than the rates in southeast Australia, and direct contact with potentially contaminated water or vegetation is more common. Indeed, in a recent study the researchers identified M. ulcerans DNA in aquatic insects collected in Benin, West Africa (F. Portaels, P. Elsen, A. Guimares-Peres, P. A. Fonteyne, and W. M. Meyers, Letter, Lancet 353:986, 1999). Thus, in tropical regions carefully planned, PCR-based environmental surveys are the most likely means for identifying the sources of M. ulcerans, particularly if the results can be supported by accurate epidemiological data.
ACKNOWLEDGMENTS
We thank Christine Drummond for advice during collation and analysis of the epidemiological data and Brenda Roberts for generously providing the M. ulcerans antisera used in this work. We also thank the Cowes Golf Club and Westernport Water for their cooperation and assistance.
This work was supported in part by funds from the Government of Victoria through the Department of Human Services and also by AWT Victoria.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. The Yammoussoukro declaration on Buruli ulcer. In: Asiedu K, Raviglione M, Scherpbier R, editors. Proceedings of the International Conference on Buruli Ulcer Control and Research. Geneva, Switzerland: World Health Organization; 1998. p. 5. [Google Scholar]
- 3.Anonymous. Epidemiology of Mycobacterium ulcerans infection (Buruli ulcer) at Kinyara, Uganda. Trans R Soc Trop Med Hyg. 1971;65:763–765. doi: 10.1016/0035-9203(71)90090-3. [DOI] [PubMed] [Google Scholar]
- 4.Barker D J. Epidemiology of Mycobacterium ulcerans infection. Trans R Soc Trop Med Hyg. 1973;67:43–50. doi: 10.1016/0035-9203(73)90317-9. [DOI] [PubMed] [Google Scholar]
- 5.Best D J. Optimal determination of most probable number. Int J Food Microbiol. 1990;11:159–166. doi: 10.1016/0168-1605(90)90051-6. [DOI] [PubMed] [Google Scholar]
- 6.Boddinghaus B, Rogall T, Flohr T, Blocker H, Bottger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drobniewski F A, More P G, Harris G S. Differentiation of Mycobacterium tuberculosis complex and nontuberculous mycobacterial liquid cultures by using peptide nucleic acid-fluorescence in situ hybridization probes. J Clin Microbiol. 2000;38:444–447. doi: 10.1128/jcm.38.1.444-447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP (phylogeny inference package), version 3.572c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 9.Hayman J. Mycobacterium ulcerans infection in Victoria: celebration of a golden jubilee? Australas J Dermatol. 1987;28:99–105. doi: 10.1111/j.1440-0960.1987.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 10.Hayman J. Postulated epidemiology of Mycobacterium ulcerans infection. Int J Epidemiol. 1991;20:1093–1098. doi: 10.1093/ije/20.4.1093. [DOI] [PubMed] [Google Scholar]
- 11.Johnson P D, Veitch M G, Leslie D E, Flood P E, Hayman J A. The emergence of Mycobacterium ulcerans infection near Melbourne. Med J Aust. 1996;164:76–78. doi: 10.5694/j.1326-5377.1996.tb101352.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaucner C, Stinear T. Sensitive and rapid detection of viable Giardia cysts and Cryptosporidium parvum oocysts in large-volume water samples with wound fiberglass cartridge filters and reverse transcription-PCR. Appl Environ Microbiol. 1998;64:1743–1749. doi: 10.1128/aem.64.5.1743-1749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschner P, Bottger E C. Species identification of mycobacteria using rDNA sequencing. In: Parish T, Stoker N G, editors. Mycobacteria protocols. Vol. 1. Totowa, N.J: Humana; 1998. pp. 349–362. [DOI] [PubMed] [Google Scholar]
- 14.Mangan J A, Sole K M, Mitchison D A, Butcher P D. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 1997;25:675–676. doi: 10.1093/nar/25.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangiapan G, Vokurka M, Schouls L, Cadranel J, Lecossier D, van Embden J, Hance A J. Sequence capture-PCR improves detection of mycobacterial DNA in clinical specimens. J Clin Microbiol. 1996;34:1209–1215. doi: 10.1128/jcm.34.5.1209-1215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marston B J, Diallo M O, Horsburgh C R, Jr, Diomande I, Saki M Z, Kanga J M, Patrice G, Lipman H B, Ostroff S M, Good R C. Emergence of Buruli ulcer disease in the Daloa region of Cote d'Ivoire. Am J Trop Med Hyg. 1995;52:219–224. doi: 10.4269/ajtmh.1995.52.219. [DOI] [PubMed] [Google Scholar]
- 17.Meyers W M, Tignokpa N, Priuli G B, Portaels F. Mycobacterium ulcerans infection (Buruli ulcer): first reported patients in Togo. Br J Dermatol. 1996;134:1116–1121. [PubMed] [Google Scholar]
- 18.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 19.Palomino J C, Obiang A M, Realini L, Meyers W M, Portaels F. Effect of oxygen on growth of Mycobacterium ulcerans in the BACTEC system. J Clin Microbiol. 1998;36:3420–3422. doi: 10.1128/jcm.36.11.3420-3422.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Portaels F. Epidemiology of ulcers due to Mycobacterium ulcerans. Ann Soc Belg Med Trop. 1989;69:91–103. [PubMed] [Google Scholar]
- 21.Portaels F. Epidemiology of mycobacterial diseases. Clin Dermatol. 1995;13:207–222. doi: 10.1016/0738-081x(95)00004-y. [DOI] [PubMed] [Google Scholar]
- 22.Portaels F. Etude d'Actinomycetales isolées de l'homme et de son environment en Afrique Centrale. Ph.D. thesis. Brussels, Belgium: Faculté des Sciences, Université Libre de Bruxelles; 1978. [Google Scholar]
- 23.Radford A J. Mycobacterium ulcerans. II. Clinical presentation, histopathology, treatment and presentation. Papua New Guinea Med J. 1973;17:134–144. [Google Scholar]
- 24.Roberts B, Hirst R. Immunomagnetic separation and PCR for detection of Mycobacterium ulcerans. J Clin Microbiol. 1997;35:2709–2711. doi: 10.1128/jcm.35.10.2709-2711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross B C, Johnson P D, Oppedisano F, Marino L, Sievers A, Stinear T, Hayman J A, Veitch M G, Robins-Browne R M. Detection of Mycobacterium ulcerans in environmental samples during an outbreak of ulcerative disease. Appl Environ Microbiol. 1997;63:4135–4138. doi: 10.1128/aem.63.10.4135-4138.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross B C, Marino L, Oppedisano F, Edwards R, Robins-Browne R M, Johnson P D. Development of a PCR assay for rapid diagnosis of Mycobacterium ulcerans infection. J Clin Microbiol. 1997;35:1696–1700. doi: 10.1128/jcm.35.7.1696-1700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuppler M, Mertens F, Schon G, Gobel U B. Molecular characterization of nocardioform actinomycetes in activated sludge by 16S rRNA analysis. Microbiology. 1995;141:513–521. doi: 10.1099/13500872-141-2-513. [DOI] [PubMed] [Google Scholar]
- 28.Smith M. Epidemiology of Mycobacterium ulcerans infection in northern Australia. M.Sc. thesis. Townsville, Australia: James Cook University; 1997. [Google Scholar]
- 29.Stinear T, Matusan A, Hines K, Sandery M. Detection of a single viable Cryptosporidium parvum oocyst in environmental water concentrates by reverse transcription-PCR. Appl Environ Microbiol. 1996;62:3385–3390. doi: 10.1128/aem.62.9.3385-3390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stinear T, Ross B C, Davies J K, Marino L, Robins-Browne R M, Oppedisano F, Sievers A, Johnson P D R. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J Clin Microbiol. 1999;37:1018–1023. doi: 10.1128/jcm.37.4.1018-1023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veitch M G, Johnson P D, Flood P E, Leslie D E, Street A C, Hayman J A. A large localized outbreak of Mycobacterium ulcerans infection on a temperate southern Australian island. Epidemiol Infect. 1997;119:313–318. doi: 10.1017/s0950268897008273. [DOI] [PMC free article] [PubMed] [Google Scholar]