Abstract
Soils harbor the most diverse naturally evolved antibiotic resistance genes (ARGs) on Earth, with implications for human health and ecosystem functioning. How ARGs evolve as soils develop over centuries, to millennia (i.e., pedogenesis), remains poorly understood, which introduces uncertainty in predictions of the dynamics of ARGs under changing environmental conditions. Here we investigated changes in the soil resistome by analyzing 16 globally distributed soil chronosequences, from centuries to millennia, spanning a wide range of ecosystem types and substrate age ranges. We show that ARG abundance and diversity decline only after millions of years of soil development as observed in very old chronosequences. Moreover, our data show increases in soil organic carbon content and microbial biomass as soil develops that were negatively correlated with the abundance and diversity of soil ARGs. This work reveals natural dynamics of soil ARGs during pedogenesis and suggests that such ecological patterns are predictable, which together advances our understanding of the environmental drivers of ARGs in terrestrial environments.
Subject terms: Microbial ecology, Biogeography
Soils harbour ancient evolutionary origins of antibiotic resistance and have been proposed as dominant reservoirs for antibiotic resistance genes (ARGs) and antibiotic resistant bacteria (ARB) [1, 2]. Bacteria can develop antibiotic resistance through acquisition of ARGs via mutation or horizontal gene transfer under natural selection, which is a common strategy used by bacteria to withstand the negative effects of antibiotics [3, 4]. Several soil-dwelling bacteria, such as Streptomyces, produce antibiotics that confer an evolutionary advantage to outcompete other soil microorganisms for resources and that must be suppressed by the presence of antibiotic resistance mechanisms [5]. There are still major unknowns related to long-term dynamics of soil antibiotic resistance and we lack a conceptual understanding for how and why antibiotic resistance changes during soil formation (pedogenesis). Improving understanding of how soil ARGs evolve as soil develops, from centuries to millennia, informs important ecological mechanisms, such as microbial competition, which can influence global terrestrial biodiversity and ecosystem services.
Soil chronosequences and associated space-for-time substitutions have been proposed as a model-system to investigate long-term dynamics in soil elements, plant succession, and ecosystem processes [6–9]. As soil develops from hundreds of thousands to millions of years, vegetation develops and organic carbon accumulates. These build-up conditions support a larger number of microorganisms and a higher load capacity of microbial biomass [10]. Using this system model, we proposed that younger soils are likely to support higher abundances of ARGs than older soils because microbial competition for resources is expected to be more significant when resources are low. The majority of antibiotic resistance mechanisms are associated with a fitness cost that is typically observed as a reduced bacterial growth rate [11]. We predict that, as soil develops and resource availability increases, microbial competition will be relieved and, therefore, the need for ARGs may be reduced.
To test this hypothesis, we conducted a global field survey including 435 composite soil samples from 87 global locations in 16 soil chronosequences. These soil chronosequences included soils ranging in age from hundreds to thousands and millions of years (Fig. S1 and Table S1), and encompassed a wide range of ecosystem types (grasslands, shrublands, forests, and croplands), chronosequence origins (volcanic, sedimentary, sand dunes, and glaciers), and climatic conditions (tropical, temperate, continental, polar, and arid). We examined changes in the soil resistome during soil formation across global biomes and identified factors regulating soil antibiotic resistance, which is a microbial trait associated with competition. We used high throughput quantitative PCR (HT-qPCR) [12] to characterize the diversity (Shannon index) and abundance of 285 ARGs encoding resistance to all major known antibiotics that are commonly used in human, animal, and agriculture settings (Table S2). Although this PCR-based method is limited by primer design and selection, and it is possible that novel ARGs will be missed with this approach, this method was validated previously [13, 14] and is more comprehensive than conventional qPCR that targets relatively few ARGs. Soil bacterial and fungal abundances were estimated by phospholipid fatty acid analysis and soil nutrient availability and stoichiometry were assessed (see Supplementary Methods). Using this approach, we produced high quality data for 93% of all soil samples (i.e., 403 out of 435 samples).
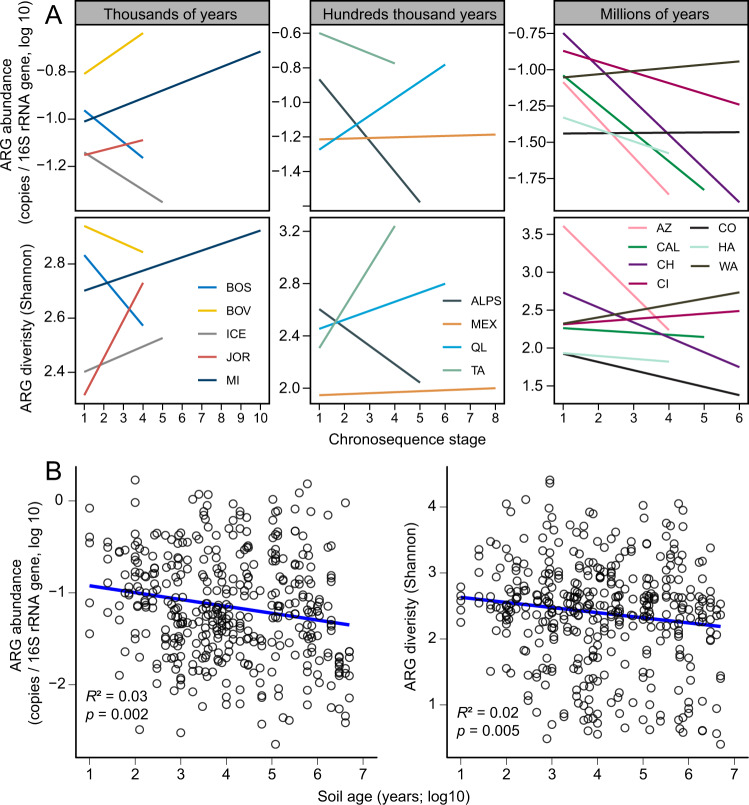
Pedogenesis-related changes in soil bacteria, fungi, and protist biodiversity are relatively well documented [15, 16]. However, changes to the soil resistome, as ecosystems grow older, are less well understood. Our analyses show that young (i.e., thousands of years; five sites) and intermediate (i.e., hundreds of thousands of years; four sites) chronosequences support contrasting patterns in resistome development. In general, over 50% of the chronosequences support increases and decreases in the abundance and diversity of ARGs in young and intermediate chronosequences, respectively (Fig. 1A). A previous study focused on relatively young soils and found that changes in soil ARGs followed a unimodal pattern, with an increasing trend at early stages (i.e., 0–8 years), and with no significant change at later stages (i.e., 17–50 years) [17]. However, our study includes a very different temporal scale. In young chronosequences, increases in the abundance and diversity of ARGs might be associated with increasing competition for soil resources in ecosystems with poorly developed soils. However, in very old chronosequences (i.e., after millions of years of soil development; seven sites), we found a negative relationship between the abundance and diversity of ARGs with chronosequence stage in most soil chronosequences (Fig. 1A). Similar results were found when working with the most abundant type of ARGs (i.e., aminoglycoside, β-lactams, and multidrug; Figs. S2 and S3). The ordinary least-squares (OLS) model further shows that both abundance (p = 0.002) and diversity (p = 0.005) of ARGs were significantly negatively correlated with soil age, when considering all soil chronosequences (Fig. 1B). In addition, a similar pattern was observed for the composition (NMDS1) of ARGs and soil age (p = 0.002) (Fig. S4).
Fig. 1. Changes in ARG abundance and diversity during pedogenesis.
A The relationships between chronosequence stage and soil ARG abundance and diversity across 16 globally distributed soil chronosequences. B The relationships between soil age and ARG abundance and diversity across all soil chronosequences. The chronosequence stage means a set of soil sites that share similar attributes; higher numbers are older stages.
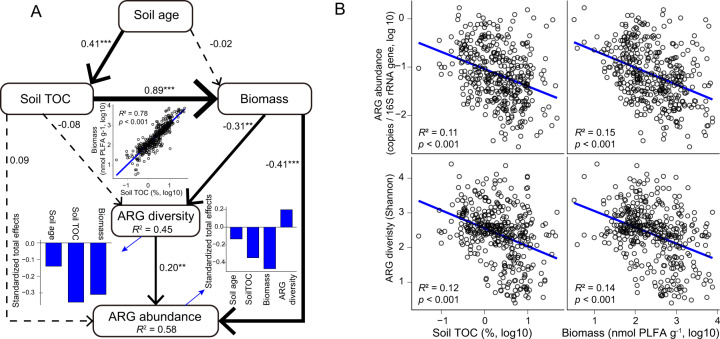
Our results indicate that soil age can have a pivotal role in shaping the resistome during pedogenesis. To better understand how soil age drives the soil resistome, we used structural equation modeling to generate a system-level and global understanding of the associations between soil age and the diversity and abundance of ARGs (Fig. 2A). The optimized model suggests that the increases in soil total organic carbon (TOC) and microbial biomass have negative and significant direct associations with both the diversity and abundance of soil ARGs. The OLS model further shows a significant positive relationship between soil TOC and microbial biomass. Previous research demonstrates that soil nutrient availability, including TOC, is a major determinant of microbial growth and community composition [18, 19]. Our results further suggest that the content of soil TOC and microbial biomass is the most significant controller of the soil antibiotic resistance during soil development (Figs. 2B and S5). Our data indicate that as resources become more abundant in soils after millions of years of ecosystem development, microbial competition decreases, reducing the need for a large production of ARGs and, therefore, the abundance and diversity of ARGs in soil.
Fig. 2. Drivers of the abundance and diversity of ARGs during pedogenesis.
A Structural equation model showing the direct and indirect effects of changes in soil properties on the abundance and richness of ARGs. χ2 = 1.83 (p = 0.40), Degrees of freedom (df) = 2, Root mean square error of approximation (RMSEA) = 0.00. Goodness-of-fit index (GFI) = 0.998. *p < 0.05, **p < 0.01, ***p < 0.001. R2 denotes the proportion of variance explained. B Relationships between the abundance and diversity of soil ARGs, total organic carbon, and microbial biomass.
In summary, our findings show that the abundance and diversity of soil ARGs decays during pedogenesis at a global scale. Our results further indicate that changes in soil TOC and microbial mass are the most significant predictors of soil resistome. These outputs provide new insights into the natural history of the soil resistome during pedogenesis and are essential to forecast the evolution and dissemination of antibiotic resistance under the changing environment and to manage existing and future antibiotic resources.
Supplementary information
Acknowledgements
QL-C and HW-H thank the Australia Research Council (DP210100332, DE210100271) for the projects. We thank the researchers involved in the CLIMIFUN project for the help with soil sampling.
Author contributions
QL-C, HW-H and MD-B conceived the idea of this study. MDB and HW-H conducted soil samplings and processing. QL-C conducted statistical analysis and modeling. The manuscript was written by QL-C, edited by HW-H and MD-B, and all co-authors contributed revisions.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 702057. MD-B is supported by a project from the Spanish Ministry of Science and Innovation (PID2020-115813RA-I00), and a project PAIDI 2020 from the Junta de Andalucía (P20_00879).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hang-Wei Hu, Email: hang-wei.hu@unimelb.edu.au.
Manuel Delgado-Baquerizo, Email: m.delgadobaquerizo@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01225-8.
References
- 1.Van Goethem MW, Pierneef R, Bezuidt OKI, Van De Peer Y, Cowan DA, Makhalanyane TP. A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome. 2018;6:40. doi: 10.1186/s40168-018-0424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–7. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 3.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–9. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 4.Martinez JL, Coque TM, Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol. 2015;13:116–23. doi: 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- 5.Genilloud O. Actinomycetes: still a source of novel antibiotics. Nat Prod Rep. 2017;34:1203–32. doi: 10.1039/C7NP00026J. [DOI] [PubMed] [Google Scholar]
- 6.Ochoa-Hueso R, Plaza C, Moreno-Jimenez E, Delgado-Baquerizo M. Soil element coupling is driven by ecological context and atomic mass. Ecol Lett. 2021;24:319–26. doi: 10.1111/ele.13648. [DOI] [PubMed] [Google Scholar]
- 7.Wardle DA, Walker LR, Bardgett RD. Ecosystem properties and forest decline in contrasting long-term chronosequences. Science. 2004;305:509–13. doi: 10.1126/science.1098778. [DOI] [PubMed] [Google Scholar]
- 8.Crews TE, Kitayama K, Fownes JH, Riley RH, Herbert DA, Mueller-Dombois D, et al. Changes in soil phosphorus fractions and ecosystem dynamics across a long chronosequence in Hawaii. Ecology. 1995;76:1407–24. doi: 10.2307/1938144. [DOI] [Google Scholar]
- 9.Walker LR, Wardle DA, Bardgett RD, Clarkson BD. The use of chronosequences in studies of ecological succession and soil development. J Ecol. 2010;98:725–36. doi: 10.1111/j.1365-2745.2010.01664.x. [DOI] [Google Scholar]
- 10.Delgado-Baquerizo M, Reich PB, Bardgett RD, Eldridge DJ, Lambers H, Wardle DA, et al. The influence of soil age on ecosystem structure and function across biomes. Nat Commun. 2020;11:4721. doi: 10.1038/s41467-020-18451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–71. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 12.Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci USA. 2013;110:3435–40. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu YG, Zhao Y, Li B, Huang CL, Zhang SY, Yu S, et al. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat Microbiol. 2017;2:16270. doi: 10.1038/nmicrobiol.2016.270. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Cao J, Zhu YG, Chen QL, Shen F, Wu Y, et al. Global survey of antibiotic resistance genes in air. Environ Sci Technol. 2018;52:10975–84. doi: 10.1021/acs.est.8b02204. [DOI] [PubMed] [Google Scholar]
- 15.Delgado-Baquerizo M, Bardgett RD, Vitousek PM, Maestre FT, Williams MA, Eldridge DJ, et al. Changes in belowground biodiversity during ecosystem development. Proc Natl Acad Sci USA. 2019;116:6891–6. doi: 10.1073/pnas.1818400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortiz-Álvarez R, Fierer N, de Los Ríos A, Casamayor EO, Barberán A. Consistent changes in the taxonomic structure and functional attributes of bacterial communities during primary succession. ISME J. 2018;12:1658–67. doi: 10.1038/s41396-018-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen J, Li Z-M, Hu H, Zeng J, Zhang L-M, He J, et al. Distribution and succession feature of antibiotic resistance genes along a soil development chronosequence in Urumqi No. 1 Glacier of China. Front Microbiol. 2019;10:1569. doi: 10.3389/fmicb.2019.01569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drenovsky RE, Vo D, Graham KJ, Scow KM. Soil water content and organic carbon availability are major determinants of soil microbial community composition. Micro Ecol. 2004;48:424–30. doi: 10.1007/s00248-003-1063-2. [DOI] [PubMed] [Google Scholar]
- 19.Bastida F, Eldridge DJ, Garcia C, Kenny Png G, Bardgett RD, Delgado-Baquerizo M. Soil microbial diversity-biomass relationships are driven by soil carbon content across global biomes. ISME J. 2021;15:2081–91. doi: 10.1038/s41396-021-00906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.