Abstract
Plant-associated microbes have evolved the ability to independently produce gibberellin (GA) phytohormones as a mechanism to influence their host. Indeed, GA was first discovered as a metabolite from the fungal rice pathogen Gibberella fujikuroi, which uses it as a virulence factor. Though some bacterial plant pathogens similarly use GA to promote infection, symbiotic nitrogen-fixing bacteria (rhizobia), which inhabit the root nodules of legumes, also can produce GA, suggesting a role in symbiosis. The bacterial GA biosynthetic operon has been identified, but in rhizobia this typically no longer encodes the final metabolic gene (cyp115), so that these symbionts can only produce the penultimate intermediate GA9. Here, we demonstrate that soybean (Glycine max) expresses functional GA 3-oxidases (GA3ox) within its nodules, which have the capability to convert GA9 produced by the enclosed rhizobial symbiont Bradyrhizobium diazoefficiens to bioactive GA4. This rhizobia-derived GA is demonstrated to cause an increase in nodule size and decrease in the number of nodules. The increase in individual nodule size correlates to greater numbers of bacterial progeny within a nodule, thereby providing a selective advantage to rhizobia that produce GA during the rhizobia-legume symbiosis. The expression of GA3ox in nodules and resultant nodulation effects of the GA product suggests that soybean has co-opted control of bioactive GA production, and thus nodule size, for its own benefit. Thus, our results suggest rhizobial GA biosynthesis has coevolved with host plant metabolism for cooperative production of a phytohormone that influences nodulation in a mutually beneficial manner.
Subject terms: Symbiosis, Molecular evolution
Introduction
Bacterial fixation of atmospheric nitrogen (N2) is the major natural means by which nitrogen is assimilated into the biological environment [1]. Perhaps most relevant to agriculture is the reduction of N2 by rhizobia, which form symbiotic relationships with legumes, including important crops like soybean (Glycine max), cowpea (Vigna unguiculata), and common bean (Phaseolus vulgaris), resulting in around 200 million tons of fixed nitrogen each year [2]. In this symbiosis, rhizobia reside in organs attached to the plant root called nodules, within which they are provided with a carbon source and a largely competition-free niche in exchange for their production of reduced nitrogen [3].
When inside the nodule, the rhizobia cells assume specialized, nitrogen-fixing forms referred to as bacteroids, and are located within plant nodule cells in membrane-bound symbiosomes [4]. Although the rhizobia-legume symbiosis can be considered to be mutually beneficial for each organism, both the plant and the rhizobium are working to optimize their own cost/benefit ratio within this interaction [5]. This includes rhizobia trying to “cheat” by fixing nitrogen at a sub-optimal level, which is in turn countered by host plant sanctions on these cheaters that affect rhizobial proliferation in the nodule [6]. Additionally, certain legumes can induce the differentiation of symbiotic bacteroids into larger, branched cells. Though this seems to result in a higher symbiotic efficiency for the plant [7], it also results in a decreased ability for the rhizobial bacteroids to revert to a free-living form upon release into the soil [8]. Overall, the active competition between rhizobia and legumes to optimize their own fitness within this intimate relationship incentivizes the development of evolutionary innovations to influence the biology of their symbiotic partner. These strategies must not only specifically modulate the other organism, but also do so without inducing effects that overwhelm the advantage gained through symbiosis.
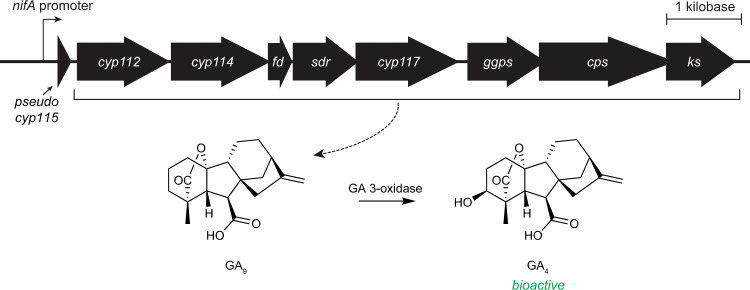
A prominent mechanism by which microorganisms commonly effect changes in plants is through the production of phytohormones [9]. Indeed, rhizobia have long been reported to produce the plant hormone gibberellin (GA) [10], and this metabolic capacity has been confirmed through characterization of GA biosynthesis in these organisms [11–15]. The ability to produce GA is imparted by a cytochrome P450 (CYP)-rich biosynthetic gene cluster referred to as the GA operon (Fig. 1), which is widely distributed in rhizobia, but not universally present [16–18]. Transcriptomic and proteomic studies from several rhizobia species have shown that the GA operon is expressed specifically during symbiosis [19–28]. Symbiotic expression is further evident due to the presence of NifA and RpoN binding sites upstream of the operon [17, 21], both of which are involved in transcription of symbiosis-related genes in rhizobia [29], and seem to control expression of the GA operon [21]. The conditional production of GA, as well as the crucial role of GA as a plant signaling molecule, strongly suggests that bacterial GA is playing a role within the rhizobia-legume symbiosis. Plant-synthesized GA has been demonstrated to be crucial for nodule organogenesis and development, but this hormone activity is critically dependent on timing, location, and concentration [30–34]. For example, both high or low levels of GA can result in decreased nodulation and aberrant nodule morphology [30], and thus an obvious role for rhizobial GA in this symbiosis was not immediately apparent. Previous work has shown that bioactive GA acts as virulence factor for not only the fungal rice pathogen Gibberella fujikuroi (anamorph Fusarium fujikuroi) [35, 36], but also the rice bacterial pathogen Xanthomonas oryzae where the bacterial GA operon also is present [37]. By contrast, the GA operons in rhizobia typically (although not invariably), no longer contain a full-length copy of the final gene (cyp115) required for production of bioactive GA4 [15, 18], such that these only produce the penultimate precursor GA9 [12, 13]. Fragments of the cyp115 gene that lack a full CYP catalytic domain can be found at the 5′ end of some rhizobial GA operons, such as the operon found in B. diazoefficiens (Fig. 1), which suggests that this gene has been selectively lost from the operon in many species of rhizobia [18]. Although GA9 has not been found to exhibit the archetypical GA hormonal activity [38–40], the apparent production of this GA by most rhizobia suggests that it may nevertheless directly affect nodulation.
Fig. 1. GA biosynthetic operon in B. diazoefficiens USDA 110.
The product of the GA operon (blr2143-blr2150) is non-bioactive GA9. However, a single hydroxylation of this compound produces GA4, a bioactive phytohormone.
Initial analysis of the GA operon from the USDA 110 strain of Bradyrhizobium diazoefficiens (previously B. japonicum) in symbiosis with soybean found no differences in soybean height or nodulation phenotypes when plants were inoculated with either wild-type or GA operon knockout strains [41]. Although it was not yet known that this gene cluster was responsible for GA biosynthesis, this negative result suggested that the GA operon did not have an obvious effect on plant growth or nodulation. However, this experiment analyzed soybean growth and nodulation phenotypes relatively early in development (~5 weeks after planting and inoculation with rhizobia). While expression of the GA operon in bacteroids is detectable as early as 3 weeks post-inoculation [19], its expression increases noticeably during the flowering and early pod stages of soybean plant growth, suggesting that GA may have a more prominent effect later in development [12]. Consistently, GA biosynthetic activity could be detected within B. diazoefficiens bacteroids isolated from nodules at the flowering and early pod stage of soybean growth, but not with bacteroids isolated during earlier stages of symbiosis [12]. Collectively, these data suggest that rhizobial GA may not have an effect until relatively late in this symbiotic relationship, as significant amounts of rhizobial GA may not be produced until the flowering stage of the plant host. Therefore a phenotype resulting from this GA production may not be evident until this point in soybean development.
Previous work reported that GA production by Mesorhizobium loti decreases nodule formation by its host legume Lotus japonicus, and further hypothesized that this restriction of nodulation provides a selective advantage by somehow excluding other rhizobia from forming symbiosis with the host plant [14]. However, increases in nodule size were also observed in this study, which could provide a more direct selective advantage, as larger nodules typically accommodate more bacterial symbionts. In addition, M. loti contains a functional copy of cyp115, and thus can produce bioactive GA4 directly [15]. By contrast, the soybean symbiont B. diazoefficiens does not contain cyp115, and thus is only capable of producing GA9. Here, we show that soybean exhibits nodule-specific expression of functional GA 3-oxidase (GA3ox) homologs, which can convert GA9 to GA4, suggesting cooperative production of this bioactive GA phytohormone. We further demonstrate that GA produced by B. diazoefficiens in symbiosis with soybean increases nodule size and decreases nodule number. This increase in nodule size results in an increase in total bacteroids per nodule, and presumably a larger number of bacteria released into the soil upon nodule senescence, thereby providing a selective advantage for the GA-producing rhizobia. We hypothesize that plant expression of GA3ox in nodules may have driven the observed loss of cyp115 in most rhizobia, enabling the host legume to optimize nodule size for its own benefit. Thus, it appears that the biosynthesis and phenotypic effects of bacterial GA have been strongly influenced by the intricate coevolution between rhizobia and their leguminous hosts.
Materials and methods
Bacterial growth
See Supplementary Table 1 for all bacterial strains used in this study. For cloning, Escherichia coli strains were grown in NZY (10 g L−1 casein hydrolysate, 5 g L−1 yeast extract, 5 g L−1 NaCl, 1 g L−1 MgSO4·7H2O) media at 37 °C, with 225 RPM shaking for liquid cultures. For general antibiotic use with all bacterial strains, the following concentrations were used unless stated otherwise: chloramphenicol (Cm) 25 µg mL−1, kanamycin (Km) 50 µg mL−1, and carbenicillin (Cb) 50 µg mL−1.
B. diazoefficiens strains were grown using arabinose-gluconate (AG) media (1 g L−1 arabinose, 1 g L−1 sodium gluconate, and 1 g L−1 yeast extract, pH 7) with Cm at 34 μg mL−1. The appropriate antibiotics to select for knockouts and/or complementing plasmids were applied as needed. For liquid cultures of B. diazoefficiens, 10 mL of the following sterilized supplements were added per 930 mL of AG media (making 1 L total) following autoclaving of media: HEPES-MES buffer (13 g L−1 HEPES, 11 g L−1 MES, pH 6.6–6.9), 0.67 g L−1 FeCl3, 18.0 g L−1 MgSO4·7H2O, 1.3 g L−1 CaCl2·2H2O, 25.0 g L−1 NaSO4, 32.0 g L−1 NH4Cl, and 12.5 g L−1 Na2HPO4.
Rhizobia knockout strains
To generate a GA operon insertional knockout strain (B. diazoefficiens KB2011, ga−), a 1946 bp DNA fragment corresponding to the cyp112 gene and portions of the flanking genes (pseudo-cyp115 and cyp114, Supplementary Fig. 1) was PCR amplified from B. diazoefficiens USDA 110 genomic DNA (gDNA) using primers Bd-cyp112-PmeI-F and Bd-cyp112-PmeI-R (see Supplementary Table 2 for primer sequences). The resulting fragment, which corresponded to the B. diazoefficiens USDA 110 genome coordinates 2317624–2319579, was ligated into the commercial pJET-cloning vector for further manipulation (see Supplementary Table 3 for a list of plasmids used). The cyp112 gene within this fragment was interrupted using an intrinsic EcoRI site (located 739 bp downstream of the cyp112 start codon) and an EcoRI fragment from pHP45Ω [42] containing the aadA gene (encoding streptomycin (Sm) resistance) flanked by strong terminators. This ligation resulted in a 4025 bp GA operon polar knockout cassette, which was inserted into the PmeI restriction sites of the mating/SacB counterselection vector pLO1 [43]. The resulting pLOBJ3 vector was transformed into the E. coli mating strain S17-1 λ pir for subsequent delivery to B. diazoefficiens.
Conjugation reactions between wild type B. diazoefficiens USDA 110 grown in HM medium [44] (0.125 g L−1 Na2HPO4, 0.25 g L−1 Na2SO4, 0.32 g L−1 NH4Cl, 0.18 g L−1 MgSO4·7H2O, 0.004 g L−1 FeCl3, 0.013 g L−1 CaCl2·2H2O, 1.3 g L−1 HEPES, 1.1 g L−1 MES; pH 6.6) supplemented with 0.1% (w/v) arabinose (no other carbon source or yeast extract) and S17-1 λ pir /pLOBJ3 were set up as follows: 1 ml of each culture at an OD600 of ~0.7 were mixed, pelleted, washed two times with HM medium to remove antibiotics, resuspended in 25 µL of HM, and spotted on a 0.45 µm filter placed on a HM/0.1% arabinose plate. Additional carbon sources and yeast extract were not added to promote mating between B. diazoefficiens and S17-1 λ pir as the nutrient limitation prevented exopolysaccharide production by B. diazoefficiens. After 3 days of incubation at 30 °C, the mating reactions were resuspended from the filter into 1 mL of HM and dilutions were plated on YEM-HM medium [45] (HM medium with 0.5% [w/v] mannitol, 0.025% [w/v] yeast extract, and 0.05% [w/v] L-arabinose) containing 30 µg mL−1 Cm to select against the donor S17-1 λ pir (as B. diazoefficiens USDA 110 is naturally resistant), 100 µg mL−1 Sm to select for the aadA interruption cassette, and 100 µg mL−1 Km to select for chromosomal insertion of pLOBJ3 (pLOBJ3 cannot be stably maintained in B. diazoefficiens).
Transconjugants that appeared after 4–5 days were screened for integration of the pLOBJ3 vector into the chromosomal cyp112 region via PCR targeting chromosomal regions outside of the insertion site and regions within the unstable vector. Positive recombinants were grown up in liquid YEM-HM medium with 30 µg mL−1 Cm and 100 µg mL−1 Sm for 3 days to promote removal of the remaining vector (encoding SacB) from the chromosome through a second recombination event between the wild type cyp112 region and pLOBJ3.
The second recombination event and thus final GA operon knockout strain was counter-selected for using Modified Bergersens4 (MB) medium (0.23 g L−1 K2HPO4, 1.1 g L−1 sodium glutamate, 0.1 g L−1 MgSO4·7H2O, 1 mL ultrapure trace element stock, 1 mL vitamin stock, and 4 mL glycerol) containing 5% (w/v) sucrose and 100 µg mL−1 of Sm, as strains still containing the integrated plasmid expressed SacB making them sensitive to sucrose. Sucrose resistant colonies appearing after 6-10 days were also screened for sensitivity to Km to verify loss of the pLOBJ3 plasmid. Interruption of the GA operon was verified in sucrose sensitive, Km sensitive, and Sm resistant colonies by Southern blots hybridized with probes corresponding to the cyp112, aadA, and blr2150 (BdKS; outside of the interrupted GA operon) genes (Supplementary Fig. 2). Compared to the wild type, three selected mutant strains possessed the aadA interruption cassette. The insertion of the aadA interruption cassette resulted in the chromosomal XhoI fragment containing the cyp112 gene to increase in size from 4010 bp (Supplementary Fig. 2c, lane 4) to 6089 bp (Supplementary Fig. 2c, lanes 1–3). Hybridization with a control probe corresponding to the downstream gene cyp114, which is located on a separate XhoI fragment, indicated no differences between the mutant and wild type strains (Supplementary Fig. 2d). Further PCR analysis targeting the region spanning the pseudo cyp115 and cyp114 genes confirmed the interruption of the GA operon as the expected product size increased from the 2195 bp observed in the wild type to the 4275 bp in the mutants (Supplementary Fig. 3a). This 2080 bp increase corresponded to the aadA interruption cassette. The PCR analysis also confirmed the removal of the pLOBJ3 interruption vector through a second recombination event as the plasmid encoded sacB gene was not detected in the mutants (Supplementary Fig. 3b). In total, three GA operon knockout strain were confirmed and named B. diazoefficiens KB903, KB904, and KB2011. Initial PCR confirmation of the knockout insertion was more consistent in B. diazoefficiens KB2011, and thus this strain was selected for further experimentation.
Deletion strains for the individual CYPs within the GA operon for B. diazoefficiens (BdΔcyp117, BdΔcyp114) were previously developed for characterization of GA biosynthesis in rhizobia [13]. These strains were grown in AG media under aerobic growth conditions to determine if knockout of the GA operon significantly affects rhizobial growth. This demonstrated that the deletion of cyp117 and cyp114 did not affect aerobic rhizobial growth, while a small decrease in growth rate was observed with B. diazoefficiens KB2011 (Supplementary Fig. 4), which is presumably due to the constitutive expression of the Sm resistance gene. Collectively, these results suggest that the GA operon does not have an effect on general rhizobial growth.
Plant growth, inoculation, measurements, and nodule harvest
Soybean (Glycine max cv. Williams ’82) seeds were surface sterilized for 10 min. with 20% Clorox (6% [w/v] sodium hypochlorite) followed by five washes with sterile water. Seeds were then immersed in 0.01 M HCl for 10 minutes followed by an additional five washes with sterile water, and then germinated on sterile, moist paper towels in the dark for 3 days. After germination, seeds were planted into a 3:1 mixture (v/v) of sterile vermiculite and perlite within 4″ diameter × 3.5″ tall pots and were grown in a growth chamber with 16 h of light at 27 °C and 8 h of darks at 18 °C. Plants were watered as needed with sterile, deionized water, unless otherwise noted. Once per week, the plants were supplemented with Murashige and Skoog nutrient solution (pH 5.8–6.0) without nitrogen (bioWORLD).
Plants were inoculated with rhizobia one week after planting. To do so, B. diazoefficiens liquid cultures (wild-type or knockout) were grown until late log phase (7–10 days), at which point cultures were centrifuged at 5000 × g for 10 min to pellet cells. Pellets were resuspended in an original culture volume of sterile water to wash away media and antibiotics, then centrifuged again at 5000 × g for 10 min. Pellets were then resuspended in sterile water to a final OD600 of 0.100 ± 0.003. For each plant, 10 mL of the appropriate rhizobial suspension was pipetted into the vermiculite/perlite soil substrate.
Vegetative stage (3 weeks post inoculation), flowering stage (~5–6 weeks post inoculation), early pod stage (~8 weeks post inoculation), full pod stage (>10 weeks post inoculation). Upon harvest, vermiculite and perlite attached to the root system was removed with light shaking and gentle washing with water. Roots were then patted dry with a paper towel. Root and green mass measurements were taken immediately following harvest. Height measurements were taken from the cotyledon scar up to the base of the apical meristem. Note that growth was limited in large part to the height of the growth chamber in use, and thus plant height measurements beyond ~6–7 weeks of growth (at which point plants have grown up to the top of the growth chamber) are likely not representative of normal plant growth. However, the vine-like nature of G. max Williams ’82 allowed for these plants to continue growing laterally within the growth chamber. Nodules were removed from the roots by hand, counted, and the collective total mass of the nodules was measured. Mass per nodule was calculated by dividing the total nodule mass per plant by the number of nodules. For direct comparisons between plants nodulated with GA+ or ga− rhizobia, statistical tests were carried out using Student’s t-test within Microsoft Excel, while multiple comparisons were performed in JMP Pro 13.
Chemical complementation
For chemical complementation experiments, soybean plants were germinated, planted, and inoculated in the same manner as described above (see Plant growth, measurements, and harvest). After 3 weeks, at which point the GA operon has been implicated to be expressed [19, 23], plants were watered twice per week with 50 mL of either a GA solution or sterile water. The GA9 solutions were made by dissolving GA9 (provided by Dr. Peter Hedden, Rothamsted Research, Harpenden, UK) in methanol at stock concentration of 0.1 M. An appropriate amount of this stock (or a serial dilution of the stock) was added to sterile water to make 100, 10, and 1 nM solutions. Preliminary experiments with GA9 suggested that concentrations at or below 100 nM did not have any significant effects on nodulation. As such, follow up experimentation with GA9 supplementation at 1 µM was performed. The GA3 solutions were made in the same method as those for GA9, with the exception that for the 0.1 M stock solution, GA3 (Sigma-Aldrich) was dissolved in ethanol. Statistical analyses for chemical complementation experiments were carried out using JMP Pro 13.
While previous studies have shown that GA3 concentrations of 1 µM are inhibitory to nodule formation, they have also observed that aberrant plant growth phenotypes such as increased height and decreased root formation are associated with concentrations greater than or equal to 10 nM [46]. A subtle, though non-significant increase in height was observed in this experiment from 10 nM GA3, but application with concentrations of 100 nM resulted in extreme increases in height as early as one-week after such treatment was begun. Additionally, a significant reduction in root mass was observed at the higher concentrations (10 and 100 nM), consistent with an inhibitory effect of GA3 on root growth. Because rhizobial GA production does not affect the overall height and mass phenotypes of the plant, it can be reasoned that rhizobia are producing GA at levels lower than those applied in these experiments. While a higher concentration of GA9 was needed to observe a similar effect to lower levels of GA3 (1 µM vs. 10 nM, respectively), we predict that this is due to the difference in solubility between GA9 and GA3 (predicted logP of 2.76 and 0.01, respectively), along with the presumed need for GA9 to be metabolized to a bioactive form.
Bacteroid isolation and counting
Prior to homogenization, soybean nodules were washed five times with 100 µL of sterile water to remove contaminating microbes. B. diazoefficiens bacteroids were extracted from root nodules via grinding in ascorbate buffer (100 mM KH2PO4, 200 mM sodium ascorbate, 2% [w/v] polyvinylpyrrolidone, and 34 µg mL−1 Cm, pH 7.5). Plant material was removed with centrifugation at 100 × g for 10 min. The supernatant was removed, and this fraction was then centrifuged at 5000 × g for 10 min. to pellet the bacteroids. Bacteroid pellets were then resuspended in 1 mL of sterile 0.85% (w/v) NaCl solution.
To determine the number of viable bacteroids, serial dilutions were created for three samples from each time point and/or condition, and 10 µL of these dilutions were plated on AG plates in triplicate. After 6–7 days of incubation at 30 °C, colony forming units (CFUs) were counted, and total number of viable bacteria per nodule mass were calculated.
Total numbers of bacteroids extracted from nodules was measured with flow cytometry. Aliquots of isolated bacteroids from three samples per time point and/or condition were stained using the LIVE/DEAD BacLight Bacterial Viability kit (Thermo Fisher Scientific), which contains SYTO9 dye for staining live cells and propidium iodine (PI) for staining dead cells, and this was done as per the manufacturer instructions. Additionally, counting beads provided in this kit (with a known concentration) were added to each sample in order to facilitate quantification of the total number of cells in the sample. Samples were run and counted using a BD FACSAria III flow cytometer (BD Biosciences). Linear regressions for this data were plotted in Microsoft Excel, and statistical analysis was performed using JMP Pro 13.
Soybean GA 3-oxidase cloning and heterologous expression
Soybean contains eight GA 3-oxidase isoforms (GmGA3ox1-8) that are all annotated as belonging to the Fe(II)/2-oxoglutarate dependent dioxygenase (2ODD) family [47], which was expected based upon previous characterization of GA 3-oxidases. PCR screening for six of these GA3ox (specifically, those previously described in the literature [47]) with cDNA synthesized from nodule RNA revealed that GmGA3ox4 (gene ID: 100780857; locus tag: Glyma14g16060) and GmGA3ox6 (gene ID: 100808546; locus tag: Glyma17g30800) were the only isoforms noticeably expressed in the nodule at 8 weeks post inoculation with B. diazoefficiens. Based on this information, E. coli codon-optimized synthetic clones of the coding sequences of these genes (sGmGA3ox4 and sGmGA3ox6) were synthesized via GeneArt Strings synthesis (Thermo Fisher Scientific). Cloned genes were amplified with primers specific to the 5′ and 3′ of the ORFs, with an additionally CACC added onto the 5′ primer (Supplementary Table 2) to allow for directional cloning into the pET101/D-TOPO (Champion pET101 Directional TOPO Expression Kit; Thermo Fisher Scientific) expression vector. The stop codon of each gene was included on the reverse primer to exclude the C-terminal His-tag encoded within pET101. Following PCR amplification with Accuprime Pfx (Thermo Fisher Scientific) and gel purification, synthetic clones were ligated into pET101/D-TOPO as per the product manual, transformed into chemically competent Top10 cells, and selected on agar plates containing Cb. Positive clones were screened with colony PCR and confirmed with Sanger sequencing. This resulted in the construction of pET101-sGmGA3ox4 and pET101-sGmGA3ox6 expression plasmids. A negative clone of pET101 created during this process was kept and used as an empty vector control.
These plasmids were then transformed into chemically competent E. coli BL21-star for heterologous expression. Positive colonies were used to inoculate 5 mL terrific broth (TB) media (10 g L−1 casein, 24 g L−1 yeast, 0.4% [v/v] glycerol, pH 7) cultures, which were allowed to grow overnight. Five hundred microliters of these cultures was then used to inoculate 50 mL TB cultures containing 5 mL of phosphate buffer (pH 7.5) and appropriate antibiotics, and these cultures were allowed to grow at 37 °C and 225 RPM shaking until reaching an OD600 of 0.6–0.8, at which point they were incubated for an hour at 16 °C with 225 RPM shaking. Following this incubation, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at 1 mM to induce expression. At this time, the 2ODD cofactors 2-oxoglutarate (a.k.a α-ketoglutarate; final concentration of 4 mM), ascorbate (final concentration of 4 mM), and FeSO4 (final concentration of 0.5 mM) were added, along with GA substrate (final concentration of 5 µM). Cultures were then allowed to incubate at 16 °C with 225 RPM shaking for 3–4 days, after which point metabolites were extracted (see “Metabolite extraction and purification”). Through this process, both enzymes were verified as specifically having GA3ox activity, as in addition to C-3β hydroxylation of GA9 to form GA4, heterologously expressed GA3ox4 and GA3ox6 converted GA20 (i.e. 13-hydroxy GA9) into GA1, another common bioactive GA endogenous to plants, via C-3β hydroxylation.
Additionally, GmGA3ox6 function was characterized in vitro within crude cell lysates. Expression conditions were nearly identical to those described earlier, with the exception that NZY media was used instead of TB. To begin, 5 mL NZY cultures of the appropriate transformed E. coli BL21 strain were grown under antibiotic selection overnight, and were used to inoculate 1.0 L cultures of NZY. These were grown at 37 °C with 225 rpm shaking to an OD600 of 0.6–0.8, at which point they were transferred to 16 °C with 225 rpm shaking for 1 h. Cultures were induced with IPTG (1 mM) and allowed to shake overnight at 16 °C. After this incubation, cells were centrifuged at 5000 × g for 10 min to pellet cells, and these pellets were then resuspended in 10 mL Tris buffer (100 mM Tris-HCL, 4 mM dithiothreitol, pH 7.1). Resuspended cells were homogenized with an EmulsiFlex C-5 (Avestin, Canada) or by sonication, and cell debris was pelleted with centrifugation at 17,000 × g for 30 min at 4 °C. Cell lysates were aliquoted and reactions were set up either with or without a 2ODD supplement mixture (final concentrations of 4 mM 2-oxoglutarate, 4 mM ascorbate, 0.5 mM FeSO4, 2 mg mL−1 bovine serum albumin, and 0.1 mg mL−1). GA substrate (at 5 µM) was added to the mixtures, and these were incubated at 30 °C for 12 h, after which they were extracted as described below. These experiments confirmed the nature of these enzymes as 2ODDs, as activity was only detected in the presence of the necessary 2ODD cofactors α-ketoglutarate, ascorbate, and iron.
Metabolite extraction and purification
GA incubation assays (both whole cell and cell lysates) were first acidified to pH 3 with 5 M HCl in order to neutralize free carboxylates. Each incubation was then extracted three times with an equivalent volume of ethyl acetate, and these extracts were pooled in a round bottom flask and dried using a rotary evaporator. Dried extracts were washed 3 times with 3 mL fractions of ethyl acetate, which were pooled in glass tubes and dried under a gentle stream of N2. Each extract was then purified over a silica column. This was done by first resuspending the sample in 1 mL of hexane and adding the sample to a silica column pre-washed with hexane. The column was then washed with successive 1 mL solutions of ethyl acetate in hexane, starting with 100% hexane, and increasing the ethyl acetate proportion with each wash. Collected fractions were derivatized with diazomethane at room temperature for 1 h in order to methylate free carboxylic acids. These were dried under a gentle stream of N2, then resuspended in either BSTFA + TMCS [N,O-Bis(trimethylsilyl)trifluoroacetamide + Trimethylchlorosilane; 99:1 v/v] or MSTFA [N-Methyl-N-(trimethylsilyl)trifluoroacetamide] and incubated at 80 °C for 30 min in order to silylate free alcohols to the trimethylsilyl ether. Derivatized samples were then dried under a gentle stream of N2, and resuspended in n-hexane for gas chromatography-mass spectrometry (GC-MS) analysis.
Gas chromatography-mass spectrometry (GC-MS) analysis
Samples were run on a Varian 3900 GC equipped with an HP-5MS column (Agilent) paired with a Saturn 2100 T MS detector (Varian). For each sample, 1 µL was injected under splitless mode with an initial injector temperature of 250 °C, column flow rate of 1.2 mL min−1, and an initial column oven temperature of 50 °C, which was held for 3 min. The column oven temperature was then increased at a rate of 15 °C min−1 until a final temperature of 300 °C, at which point this temperature was held for 3 min. Electron ionization was used to ionize and fragment compounds, and mass spectra data with a range of 90–650 m/z was collected starting at 13 min and continued until the end of the run. Potential products were compared to previously published mass spectra [48], and were further confirmed via comparison to authentic standards.
RNA isolation, cDNA synthesis, and RT-qPCR
Plant tissue (root nodules and roots, each in triplicate) collected from plants harvested at 3–15 weeks post rhizobial inoculation were ground in liquid nitrogen with a mortar and pestle, and RNA was purified from equivalent amounts of homogenized tissue using the RNeasy Plant Mini Kit (Qiagen) as per the manufacturer’s instructions. Contaminating DNA was digested and removed using the Turbo DNA-free kit (Life Technologies), and cDNA was synthesized using the iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturer’s instructions. PCR amplification using primers specific to the genes of interest was performed using RNA, DNase-treated RNA, and cDNA as templates to confirm that downstream PCR analysis was only detecting cDNA.
Quantitative reverse transcription PCR (RT-qPCR) was performed to test expression of the cyp112 (NCBI gene ID: 1055403; locus tag: blr2144) and ks (NCBI gene ID: 1055399; locus tag: blr2150) genes from the GA operon in B. diazoefficiens USDA110, along with two GA 3-oxidase homologs (GmGA3ox4 and GmGA3ox6) from soybean. For B. diazoefficiens genes, hisS (histidyl-tRNA synthetase; NCBI gene ID: 1054697; locus tag: bll7457) was used as the internal reference gene as previously described [20, 49]. For soybean genes, cons7 (NCBI gene ID: 100804856; locus tag: Glyma_03g137100), which has been identified to be expressed stably under a number of conditions, including nodule development [50], was used as the reference gene (see Supplementary Table 4 for the sequences of RT-qPCR primers used). RT-qPCR samples were prepared with PowerUp SYBR Green Master Mix (Life Technologies) according to the recommended manufacturer conditions with primers at a final concentration of 500 nM. Reactions were run and measured on a StepOnePlus thermocycler (Applied Biosystems). The following program was used for each primer set: 50 °C for 2 min followed by 95 °C for 2 min, then 45 cycles of 95 °C for 15 s and 60 °C for 1 min. After these cycles, a melt curve stage was performed to confirm specificity of the RT-qPCR reaction as per the instructions of the aforementioned RT-qPCR kit. Primer efficiencies were determined for each primer set by using serial dilutions of PCR amplified cDNA fragments generated from soybean cDNA (or fragments generated from gDNA for B. diazoefficiens) as template, and this analysis confirmed the reported primer sets to have between 90 and 110% efficiency (Supplementary Fig. 5).
Results
To confirm expression of the GA operon in B. diazoefficiens during its symbiosis with soybean, nodules were harvested over a 12-week span of soybean development (covering the vegetative to full-pod stages) [51], and quantitative reverse transcription PCR (RT-qPCR) was used to measure the relative expression of two rhizobial GA operon genes (cyp112 and ks). This analysis confirmed that the operon is expressed early in symbiosis (3 weeks post inoculation), and that expression continues throughout the flowering and early pod stages of plant development (Supplementary Fig. 6). Because previous studies have not found any differences in plant phenotypes related to rhizobial GA during early stages of soybean development [41], and because bacteroid GA biosynthetic enzyme activities are not observed until around the soybean flowering stage [12], it was reasoned that studies of such metabolism and its effect should be focused on this stage of host plant development.
Unlike M. loti, the soybean symbiont B. diazoefficiens does not contain a functional copy of cyp115 [13]. Thus, like most other rhizobia containing the GA operon, this species can only produce GA9. Given that GA9 is considered to be the penultimate precursor and does not exhibit hormonal activity on its own, as it does not elicit a response in growth assays [38] and does not bind effectively to the appropriate GA receptor [39, 40], we hypothesized that soybean might express functional GA3ox to convert the rhizobia-produced GA9 to bioactive GA4 in its nodules. To investigate this possibility, PCR with cDNA generated from flowering stage nodule RNA was used to probe the six GA3ox isoforms predicted within the soybean genome [47]. It was possible to detect expression of two isoforms, GmGA3ox4 (gene ID: 100780857) and GmGA3ox6 (gene ID: 100808546), suggesting that these may represent GA3ox enzymes specific to the nodule (Supplementary Fig. 7). Subsequent RT-qPCR analysis demonstrated that transcripts for both GmGA3ox4 and GmGA3ox6 are expressed at significantly higher levels in nodules than elsewhere in the roots, and are present in the nodule throughout all stages of plant development that were assessed here (Supplementary Fig. 7). Our results differ somewhat from previous RNA-seq analyses of the soybean nodule [52, 53], which did not find enrichment of these genes within the nodule relative to other tissues. It is unclear why this discrepancy was observed, and future investigations into GA3ox homolog expression over the full nodule lifetime may be necessary to fully understand the potential roles of soybean GA3ox genes in this symbiosis. Regardless, our ability to detect enrichment of GA3ox homolog expression within the nodule suggests a potential role for these genes in converting rhizobial GA9 into bioactive GA hormones.
The biochemical function of the enzymes encoded by GmGA3ox4 and GmGA3ox6 were confirmed via heterologous expression in E. coli (Supplementary Figs. 8, 9, and 10). Importantly, their capability to act on the rhizobial GA product was demonstrated, as incubation of GA9 with cells expressing either GmGA3ox4 or GmGA3ox6 resulted in 3β-hydroxylation of GA9, thereby producing GA4 (Supplementary Fig. 8). This confirmed that these putative GA3ox exhibit the predicted ability to convert GA9 into the bioactive GA4. Although these data do not confirm in planta conversion of rhizobial GA9 into GA4, they demonstrate that functional soybean GA3ox enzymes are expressed in the correct tissue and at the appropriate time for them to act on GA9, which is presumably secreted by bacteroids within the nodule, thereby suggesting the potential for cooperative production of bioactive GA4 by these symbiotic partners.
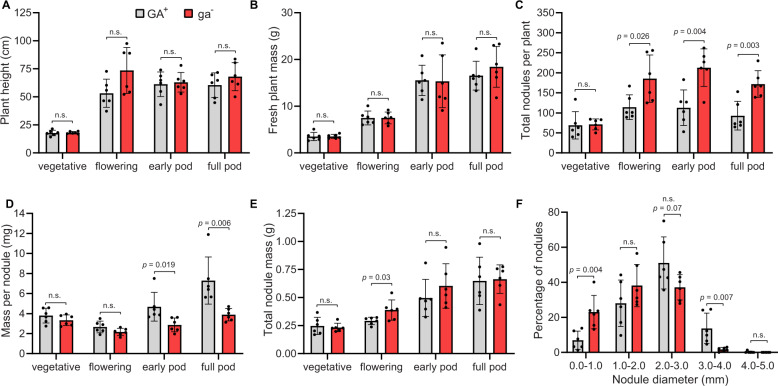
To evaluate the phenotypic role that this rhizobial produced GA may be playing in symbiosis, we generated a GA operon knockout strain from the wild-type strain B. diazoefficiens USDA 110 (GA+). This was accomplished by inserting an antibiotic resistance cassette into cyp112, resulting in the strain B. diazoefficiens KB2011 (ga−), a polar knockout in which transcription of the entire GA operon was essentially eliminated (Supplementary Fig. 11). Plants nodulated with the ga− strain did not have significantly altered height, green tissue mass, or root mass from plants nodulated with the GA+ strain (Fig. 2A, B), which agrees with previous reports showing that the GA operon does not appear to affect major growth phenotypes of the plant [14, 41]. Additionally, much like previous analysis of a GA operon knockout strain of B. diazoefficiens [41], no significant alterations in nodulation were observed early in soybean development (vegetative stage; 3 weeks post inoculation). However, by the soybean flowering stage (7 weeks post-inoculation) there were noticeable differences in nodulation phenotypes. In particular, plants nodulated by the ga− strain had a significant increase in their number of nodules by this stage (Fig. 2C). However, these nodules were on average significantly smaller than those in plants inoculated with the GA+ strain (Fig. 2D). These effects are particularly striking at the full pod stage (18 weeks post-inoculation), as nodules from plants inoculated by GA+ B. diazoefficiens are roughly twice as large as those from plants inoculated by the ga− strain, while the total number of nodules per plant for the ga− strain are essentially twice that of plants inoculated with the GA+ strain. Correspondingly, additional analysis of nodule size distribution at the full-pod time point demonstrated that knockout of the GA operon resulted in significantly fewer large nodules and an increased number of smaller nodules (Fig. 2F).
Fig. 2. Effect of GA operon knockout in the Bradyrhizobium-soybean symbiosis.
Soybean plants were nodulated with either wild-type, GA-producing B. diazoefficiens USDA 110 (GA+) or the GA operon-knockout B. diazoefficiens KB2011 (ga−). Representative plants (n = 6) were harvested at four time points (vegetative, flowering, early pod, and full pod stages) to measure the following phenotypes: A average plant height, B plant fresh mass, C the average number of nodules per plant, D the average mass per nodule for each plant, and E average total nodule mass per plant. F Nodule size distribution of soybean nodulated with GA+ or ga− B. diazoefficiens. The diameters of individual nodules collected at the soybean full pod stage were measured and binned into defined size groups. The distribution of nodule sizes are represented as the percentage of the total nodule number of the plant. n = 6 plants per condition at each time point. For all bar graphs, the mean is shown with ± standard deviation (SD). Statistical significance was assessed at each time point using an unpaired, two-tailed t-test; n.s. = not significant (p > 0.05). Individual p values are shown when ≤0.1.
Overall, this disparity in nodule size and number did not significantly alter the total nodule mass per plant by the early pod and full pod stages (Fig. 2E). The similarity in total nodule mass, as well as total plant mass, suggests that knockout of the GA operon does not restrict nitrogen assimilation by the plant. However, because plants nodulated by the ga− strain form significantly more nodules, it seems likely that the plant needs to compensate for smaller average nodule size, and correspondingly, the nitrogen fixing-capacity of each nodule. A similar effect on nodulation (i.e. an increased number of smaller nodules), with no effect on overall plant growth, was also demonstrated with two additional GA operon gene-deletion strains, BdΔcyp117 and BdΔcyp114 [13], confirming that perturbation of the GA operon, and thus rhizobial GA production, is responsible for the knockout phenotypes (Supplementary Fig. 12). While the presence of GA seems to have a clear effect on the size and number of nodules, the overall morphology and appearance of the nodules from plants inoculated with the GA+ or ga− strains were indistinguishable (Supplementary Fig. 13), which suggests that nodule organogenesis occurs normally, and that rhizobial GA only affects the nodules after their establishment.
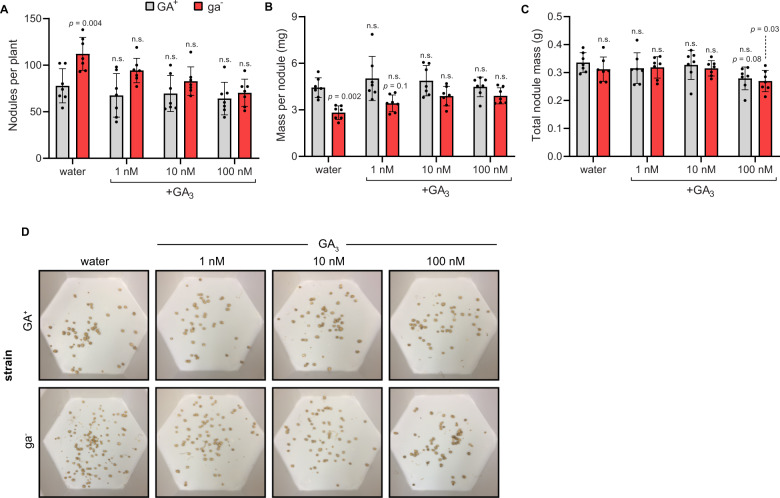
In agreement with bioactive GA being the signaling molecule associated with the observed phenotypes, application of GA3 (a readily available bioactive GA) to the roots of plants inoculated by the ga− strain restored wild-type levels of nodule number and average nodule size (Fig. 3). This was evident for GA3 concentrations as low as 1 nM, and at this lowest concentration only minimal effects on overall plant growth were observed (Supplementary Fig. 14). By contrast, chemical complementation with GA9, the apparent final product of GA biosynthesis by B. diazoefficiens [12, 13], required significantly higher concentrations (1 µM) to restore average nodule mass and nodule numbers to wild-type (GA+) levels for plants in symbiosis with the ga− strain (Supplementary Fig. 15), and significant changes in average plant height and masses were observed at this concentration (Supplementary Fig. 16). Due to the high concentrations of GA9 required to alter the nodule size and numbers, it is unclear whether this chemical treatment is truly rescuing the GA+ phenotype, or if this effect is simply an artifact of GA signaling throughout the roots, which could lead to a restriction in nodulation [30]. Nevertheless, the significantly lower concentrations of bioactive GA required to effect changes in nodule phenotypes for soybean inoculated with ga− rhizobia is consistent with the phenotype being due to low levels of bioactive GA, which would require host conversion of GA9 into GA4.
Fig. 3. Chemical complementation with GA3 restores nodulation phenotypes to GA knockout.
The soil substrate of soybeans nodulated by GA+ or ga− B. diazoefficiens was treated weekly with 1, 10, or 100 nM GA3 in water, or GA-free water as a negative control. The following phenotypes were measured at the early pod stage: A number of nodules per plant B average mass per nodule, and C total nodule mass. D Representative root nodules from each experimental group. Each image shows nodules isolated from one plant. n = 7 for each treatment. Shown for each experimental group is the mean ± SD. Statistical significance was assessed using a two-way ANOVA and Dunnett’s multiple comparison test with the GA+ water treatment as the control group. n.s. indicates p > 0.05. Individual p values are shown when ≤0.1.
Based upon these results we hypothesized that rhizobial GA provides a selective advantage to rhizobia through increasing nodule size. GA phytohormones are known to promote individual cell expansion during plant growth [54], and thus could signal to increase cell size within the nodule, resulting in enlarged nodules. Because larger nodules typically contain greater numbers of bacteroids [55, 56], we hypothesized that GA production confers a selective advantage to rhizobia by increasing nodule size and therefore the number of bacteroids released into the soil upon nodule senescence. To confirm the relationship between nodule size and bacteroid numbers, bacteroids were isolated from mature nodules of varying sizes and counted with flow cytometry to determine the total number of bacteroids, and by plating for CFUs to determine the number of viable cells. These analyses confirmed a positive correlation between viable bacteroid cells and larger nodules (Supplemental Fig. 17), supporting a selective advantage for larger nodules resulting from rhizobial GA production.
Discussion
Because cyp115 has been lost from the GA operon found in most rhizobia [15, 18], thereby restricting their ability to directly make bioactive GA, it might have been expected that the resulting production of GA9, which is not bioactive, would not affect their symbiotic relationship with legumes. However, knockout of the GA operon clearly indicates a phenotypic change with late-stage nodules, thereby indicating that rhizobial production of GA9 plays a role in such symbiosis. The co-expression of functional soybean GA3ox in the nodules suggested that cooperative production of bioactive GA4 by these symbiotic partners is possible, and the corresponding ability for low concentrations of bioactive GA3 to chemically complement the nodulation phenotype exhibited by ga− knock-out strains further suggests that such cooperative biosynthesis may be occurring. Though additional experiments will be necessary to confirm such in planta conversion of rhizobial GA9 into GA4, including analysis of how soybean GA3ox knockouts affect late-stage nodulation phenotypes and more direct observation of GA9 conversion into GA4 within the plant, our data provide circumstantial evidence that GA9 is being supplied by rhizobia to the plant as a precursor for bioactive GA biosynthesis and subsequent signaling that impacts nodulation.
While knocking-out the GA operon affects both nodule numbers and size, consistent with the usual growth-promoting activity associated with GA it is hypothesized here that the primary effect is on size. Accordingly, the increased number of nodules in plants inoculated with ga− strains presumably results from the need for plants to compensate for the smaller nodule size in order to meet fixed nitrogen requirements. Importantly, increased nodule size provides a direct advantage to the rhizobia that initiated the nodulation event by enabling a larger number of descendants (Fig. 4). Although it was previously proposed through study of the Mesorhizobium loti-Lotus japonicus symbiosis that rhizobial GA acts through suppression of nodulation [14], an increase in nodule size for L. japonicus plants inoculated by the GA+ strains of M. loti also was observed. More critically, the competition assay reported in this previous study demonstrated that co-inoculation of L. japonicus with equal amounts of GA+ and ga− M. loti strains led to greater numbers of the GA+ strain in the resulting nodules. Because the GA operon is only expressed after nodulation [28], co-inoculated GA+ and ga− strains should have an equal probability of forming the initial nodules on the legume host. Thus, the observed enrichment in the GA+ strain from this competition assay indicates that GA production acts locally to increase nodule size rather than exerting a systemic effect restricting nodule formation, particularly as it is unclear how this latter effect would differentiate between strains with or without the ability to produce GA. By contrast, local activity of rhizobial GA to increase the size of the individual host nodule would provide a direct advantage to the producing strain (i.e., increased number of descendants), regardless of the strains inhabiting other nodules. However, further experimentation will be necessary to understand the mechanism by which rhizobial GA brings about this change in nodule phenotype, not least due to the relatively late-stage effects on nodulation.
Fig. 4. Proposed model for the function of rhizobial GA in symbiosis.
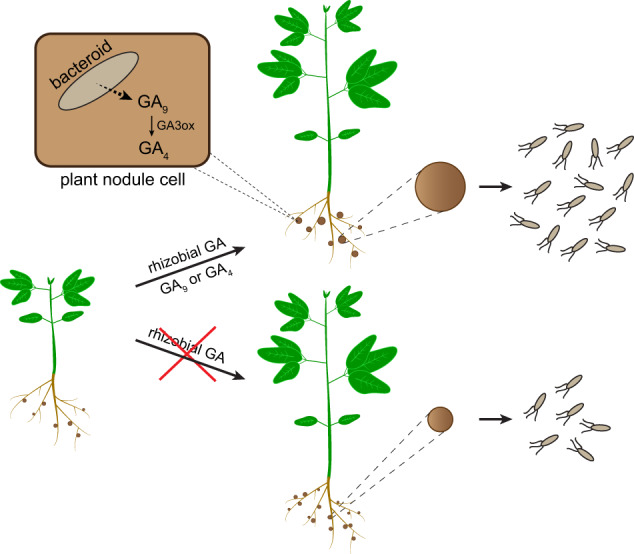
Produce either non-bioactive GA9 or bioactive GA4 during symbiosis, depending on whether the rhizobial species has cyp115. If GA9 is the final product, we propose that host plant GA 3-oxidases convert GA9 into bioactive GA4. GA signaling then leads to an increase in nodule size, which increases bacteroid numbers within that nodule and more rhizobia released into the soil upon nodule senescence. Conversely, an absence of rhizobial GA results in decreased nodule size and thus fewer bacteria being released per nodule.
The observed increase in nodule size from rhizobial GA may also provide a selective advantage for the host plants, thereby explaining why they enable such manipulation by their rhizobial symbiont (e.g., by expression of the necessary receptor to permit a response to GA). First, as larger nodules have been shown to have a higher ratio of nitrogen-fixing tissue to overall nodule volume than smaller nodules [57–59], this presumably provides nitrogen more efficiently for the host plant. In addition, given that each nodulation event represents opening of the plant interior to microbial invaders [60], the resulting reduced number of nodules may provide an additional advantage in this respect as well. Nevertheless, given the use of bioactive GA as a virulence factor by certain plant pathogens, where it acts to suppress the defense signaling molecule jasmonic acid [37, 61], enabling the nodule to respond to GA may also represent a vulnerability in the host plant defense response. This trade-off may then underlie the observed scattered distribution of the GA operon in rhizobia [18], as only those that partner with legumes whose nodules are open to such manipulation would acquire a selective advantage from such biosynthetic capacity.
If cooperative production of bioactive GA by rhizobia and their host plant is indeed occurring, this phenomenon also must have been driven by the legumes. In particular, it would be beneficial for the host plant to control production of bioactive GA, and thus limit nodule size to provide only the necessary amount of nitrogen, as well as to attenuate GA production if a microbial defense response, which can be inhibited by GA [61], becomes necessary. This control over GA signaling would presumably underlie the host plant expression of GA3ox genes in nodules, which would remove selective pressure for maintenance of the functionally analogous bacterial cyp115, which is almost invariably lost from the GA operon in rhizobia [15, 18]. However, because cyp115 is maintained as a separate genetic locus undergoing independent horizontal gene transfer within some GA-operon containing rhizobia [15, 18], host plant expression of GA3ox in late stage nodules is presumably not universal, even among those legumes that enable GA signaling in these organs.
Collectively, the results reported here suggest intricate coevolution between GA-producing rhizobia and their legume hosts that may provide selective advantages for both symbionts. Beyond enabling increased nodule growth in response to rhizobial GA, with obvious implications for population growth of GA-producing rhizobia, our data hint at the possibility that legumes may act cooperatively with rhizobia to produce bioactive GA, potentially to balance optimization of nitrogen supply with the microbial defense response.
Supplementary information
Acknowledgements
This work was supported by grants to RJP from the Iowa Soybean Association and NIH (GM131885).
Author contributions
RSN carried out experimental work and wrote the paper, KSB carried out experimental work, RJP conceived and directed the described work, and helped write the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01236-5.
References
- 1.Canfield DE, Glazer AN, Falkowski PG. The evolution and future of Earth’s nitrogen cycle. Science. 2010;330:192–6. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson BJ, Indrasumunar A. Soybean nodulation and nitrogen fixation. In: Hendricks BP, editor. Agricultural research updates, Volume 1. Nova Science Publishers, Inc.; Hauppauge, NY USA; 2011. p. 1–16.
- 3.Udvardi M, Poole PS. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol. 2013;64:781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 4.Oldroyd GED, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–44. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 5.Sachs JL, Quides KW, Wendlandt CE. Legumes versus rhizobia: a model for ongoing conflict in symbiosis. N Phytol. 2018;219:1199–206. doi: 10.1111/nph.15222. [DOI] [PubMed] [Google Scholar]
- 6.Oono R, Denison RF, Kiers ET. Controlling the reproductive fate of rhizobia: how universal are legume sanctions? N Phytol. 2009;183:967–79. doi: 10.1111/j.1469-8137.2009.02941.x. [DOI] [PubMed] [Google Scholar]
- 7.Oono R, Denison RF. Comparing symbiotic efficiency between swollen versus nonswollen rhizobial bacteroids. Plant Physiol. 2010;154:1541–8. doi: 10.1104/pp.110.163436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kereszt A, Mergaert P, Kondorosi E. Bacteroid development in legume nodules: evolution of mutual benefit or of sacrificial victims? Mol Plant-Microbe Interact. 2011;24:1300–9. doi: 10.1094/MPMI-06-11-0152. [DOI] [PubMed] [Google Scholar]
- 9.Eichmann R, Richards L, Schäfer P. as go-betweens in plant microbiome assembly. Plant J. 2021;105:518–41. doi: 10.1111/tpj.15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacMillan J. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J Plant Growth Regul. 2002;20:387–442. doi: 10.1007/s003440010038. [DOI] [PubMed] [Google Scholar]
- 11.Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, et al. Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 2009;583:475–80. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- 12.Méndez C, Baginsky C, Hedden P, Gong F, Carú M, Rojas MC. Gibberellin oxidase activities in Bradyrhizobium japonicum bacteroids. Phytochemistry. 2014;98:101–9. doi: 10.1016/j.phytochem.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Nett RS, Montanares M, Marcassa A, Lu X, Nagel R, Charles TC, et al. Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat Chem Biol. 2017;13:69–74. doi: 10.1038/nchembio.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatsukami Y, Ueda M. Rhizobial gibberellin negatively regulates host nodule number. Sci Rep. 2016;6:27998. doi: 10.1038/srep27998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nett RS, Contreras T, Peters RJ. Characterization of CYP115 as a gibberellin 3-oxidase indicates that certain rhizobia can produce bioactive gibberellin A4. ACS Chem Biol. 2017;12:912–7. doi: 10.1021/acschembio.6b01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keister DL, Tully RE, Van Berkum P. A cytochrome P450 gene cluster in the Rhizobiaceae. J Gen Appl Microbiol. 1999;45:301–3. doi: 10.2323/jgam.45.301. [DOI] [PubMed] [Google Scholar]
- 17.Hershey DM, Lu X, Zi J, Peters RJ. Functional conservation of the capacity for ent-kaurene biosynthesis and an associated operon in certain rhizobia. J Bacteriol. 2014;196:100–6. doi: 10.1128/JB.01031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nett RS, Nguyen H, Nagel R, Marcassa A, Charles TC, Friedberg I, et al. Unraveling a tangled skein: evolutionary analysis of the bacterial gibberellin biosynthetic operon. mSphere. 2020;5:1–15. doi: 10.1128/mSphere.00292-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pessi G, Ahrens CH, Rehrauer H, Lindemann A, Hauser F, Fischer H-M, et al. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol Plant Microbe Interact. 2007;20:1353–63. doi: 10.1094/MPMI-20-11-1353. [DOI] [PubMed] [Google Scholar]
- 20.Chang W-S, Franck WL, Cytryn E, Jeong S, Joshi T, Emerich DW, et al. An oligonucleotide microarray resource for transcriptional profiling of Bradyrhizobium japonicum. Mol Plant Microbe Interact. 2007;20:1298–307. doi: 10.1094/MPMI-20-10-1298. [DOI] [PubMed] [Google Scholar]
- 21.Hauser F, Pessi G, Friberg M, Weber C, Rusca N, Lindemann A, et al. Dissection of the Bradyrhizobium japonicum NifA+σ54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol Genet Genomics. 2007;278:255–71. doi: 10.1007/s00438-007-0246-9. [DOI] [PubMed] [Google Scholar]
- 22.Perret X, Freiberg C, Rosenthal A, Broughton WJ, Fellay R. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol Microbiol. 1999;32:415–25. doi: 10.1046/j.1365-2958.1999.01361.x. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Tian CF, Chen WF, Wang L, Sui XH, Chen WX. High-resolution transcriptomic analyses of Sinorhizobium sp. NGR234 bacteroids in determinate nodules of Vigna unguiculata and indeterminate nodules of Leucaena leucocephala. PLoS ONE. 2013;8:e70531. doi: 10.1371/journal.pone.0070531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salazar E, Javier Díaz-Mejía J, Moreno-Hagelsieb G, Martínez-Batallar G, Mora Y, Mora J, et al. Characterization of the NifA-RpoN regulon in Rhizobium etli in free life and in symbiosis with Phaseolus vulgaris. Appl Environ Microbiol. 2010;76:4510–20. doi: 10.1128/AEM.02007-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan JT, Brown SD, Ronson CW. The NifA-RpoN regulon of Mesorhizobium loti strain R7A and its symbiotic activation by a novel LacI/GalR-family regulator. PLoS ONE. 2013;8:e53762. doi: 10.1371/journal.pone.0053762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchiumi T, Ohwada T, Itakura M, Mitsui H, Nukui N, Dawadi P, et al. Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J Bacteriol. 2004;186:2439–48. doi: 10.1128/JB.186.8.2439-2448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarma AD, Emerich DW. Global protein expression pattern of Bradyrhizobium japonicum bacteroids: a prelude to functional proteomics. Proteomics. 2005;5:4170–84. doi: 10.1002/pmic.200401296. [DOI] [PubMed] [Google Scholar]
- 28.Tatsukami Y, Nambu M, Morisaka H, Kuroda K, Ueda M. Disclosure of the differences of Mesorhizobium loti under the free-living and symbiotic conditions by comparative proteome analysis without bacteroid isolation. BMC Microbiol. 2013;13:180. doi: 10.1186/1471-2180-13-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Argudo I, Little R, Shearer N, Johnson P, Dixon R. The NifL-NifA system: a multidomain transcriptional regulatory complex that integrates environmental signals. J Bacteriol. 2004;186:601–10. doi: 10.1128/JB.186.3.601-610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi S, Gresshoff PM, Ferguson BJ. Mechanistic action of gibberellins in legume nodulation. J Integr Plant Biol. 2014;56:971–8. doi: 10.1111/jipb.12201. [DOI] [PubMed] [Google Scholar]
- 31.Lievens S, Goormachtig S, Den Herder J, Capoen W, Mathis R, Hedden P, et al. Gibberellins are involved in nodulation of Sesbania rostrata. Plant Physiol. 2005;139:1366–79. doi: 10.1104/pp.105.066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferguson BJ, Ross JJ, Reid JB. Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 2005;138:2396–405. doi: 10.1104/pp.105.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson BJ, Foo E, Ross JJ, Reid JB. Relationship between gibberellin, ethylene and nodulation in Pisum sativum. N Phytol. 2011;189:829–42. doi: 10.1111/j.1469-8137.2010.03542.x. [DOI] [PubMed] [Google Scholar]
- 34.McAdam EL, Reid JB, Foo E. Gibberellins promote nodule organogenesis but inhibit the infection stages of nodulation. J Exp Bot. 2018;69:2117–30. doi: 10.1093/jxb/ery046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiemann P, Sieber CMK, von Bargen KW, Studt L, Niehaus EM, Espino JJ, et al. Deciphering the cryptic genome: Genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9:e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malonek S, Bömke C, Bornberg-Bauer E, Rojas MC, Hedden P, Hopkins P, et al. Distribution of gibberellin biosynthetic genes and gibberellin production in the Gibberella fujikuroi species complex. Phytochemistry. 2005;66:1296–311. doi: 10.1016/j.phytochem.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Lu X, Hershey DM, Wang L, Bogdanove AJ, Peters RJ. An ent-kaurene-derived diterpenoid virulence factor from Xanthomonas oryzae pv. oryzicola. N Phytol. 2015;206:295–302. doi: 10.1111/nph.13187. [DOI] [PubMed] [Google Scholar]
- 38.Nishijima T, Koshioka M, Yamazaki H. Use of several gibberellin biosynthesis inhibitors in sensitized rice seedling bioassays. Biosci Biotechnol Biochem. 1994;58:572–3. doi: 10.1271/bbb.58.572. [DOI] [Google Scholar]
- 39.Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–8. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 40.Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006;46:880–9. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- 41.Tully RE, Keister DL. Cloning and mutagenesis of a cytochrome P-450 locus from Bradyrhizobium japonicum that is expressed anaerobically and symbiotically. Appl Environ Microbiol. 1993;59:4136–42. doi: 10.1128/aem.59.12.4136-4142.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prentki P, Krisch HM. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–13. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 43.Lenz O, Schwartz E, Dernedde J. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–93. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole MA, Elkan GH. Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob Agents Chemother. 1973;4:248–53. doi: 10.1128/AAC.4.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu C, Maier RJ. Identification of a locus within the hydrogenase gene cluster involved in intracellular nickel metabolism in Bradyrhizobium japonicum. Appl Environ Microbiol. 1991;57:3502–10. doi: 10.1128/aem.57.12.3502-3510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maekawa T, Maekawa-Yoshikawa M, Takeda N, Imaizumi-Anraku H, Murooka Y, Hayashi M. Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 2009;58:183–94. doi: 10.1111/j.1365-313X.2008.03774.x. [DOI] [PubMed] [Google Scholar]
- 47.Han F, Zhu B. Evolutionary analysis of three gibberellin oxidase genes in rice, Arabidopsis, and soybean. Gene. 2011;473:23–35. doi: 10.1016/j.gene.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Binks R, MacMillan J, Pryce RJ. Combined gas chromatography-mass spectrometry of the methyl esters of gibberellins A1 to A24 and their trimethylsilyl ethers. Phytochemistry. 1969;8:271–84. doi: 10.1016/S0031-9422(00)85825-2. [DOI] [Google Scholar]
- 49.Lee HI, Lee JH, Park KH, Sangurdekar D, Chang WS. Effect of soybean coumestrol on Bradyrhizobium japonicum nodulation ability, biofilm formation, and transcriptional profile. Appl Environ Microbiol. 2012;78:2896–903. doi: 10.1128/AEM.07336-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, et al. Identification of four soybean reference genes for gene expression normalization. Plant Genome J. 2008;1:44. [Google Scholar]
- 51.Ritchie, SW, Hanway, JJ, Thompson HE. How a soybean plant develops. Spec. Rep. No. 53. 1994. Ames, IA. Iowa State University, Cooperative Extension Service.
- 52.Severin AJ, Woody JL, Bolon Y-T, Joseph B, Diers BW, Farmer AD, et al. RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 2010;10:160. doi: 10.1186/1471-2229-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, et al. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010;63:86–99. doi: 10.1111/j.1365-313X.2010.04222.x. [DOI] [PubMed] [Google Scholar]
- 54.Fleet CM, Sun TP. A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr Opin Plant Biol. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 55.Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume-rhizobium mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- 56.Simms EL, Taylor DL, Povich J, Shefferson RP, Sachs JL, Urbina M, et al. An empirical test of partner choice mechanisms in a wild legume-rhizobium interaction. Proc Biol Sci. 2006;273:77–81. doi: 10.1098/rspb.2005.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen HK, Thornton HG. The structure of ‘ineffective’ nodules and its influence on nitrogen fixation. Proc R Soc Lond B. 1940;129:208–29. doi: 10.1098/rspb.1940.0036. [DOI] [Google Scholar]
- 58.Aprison MH, Magee WE, Burris RH. Nitrogen fixation by excised soy bean root nodules. J Biol Chem. 1954;208:29–39. doi: 10.1016/S0021-9258(18)65621-2. [DOI] [PubMed] [Google Scholar]
- 59.Tajima R, Lee ON, Abe J, Lux A, Morita S. Nitrogen-fixing activity of root nodules in relation to their size in peanut (Arachis hypogaea L.) Plant Prod Sci. 2007;10:423–9. doi: 10.1626/pps.10.423. [DOI] [Google Scholar]
- 60.Berrabah F, Ratet P, Gourion B. Legume nodules: massive infection in the absence of defense induction. Mol Plant Microbe Interact. 2019;32:35–44. doi: 10.1094/MPMI-07-18-0205-FI. [DOI] [PubMed] [Google Scholar]
- 61.Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, et al. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008;18:650–5. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.