Abstract
Little is known about the complexity and plasticity of circulating tumor cell (CTC) biology in different compartments of the fluid microenvironment during tumor metastasis. Here we integrated phenomics, genomics, and targeted proteomics to characterize CTC phenotypic and genotypic heterogeneity in paired peripheral blood (PB) and bone marrow aspirate (BMA) from a metastatic prostate cancer patient following the rapid disease progression, using the High-Definition Single Cell Assay 3.0 (HDSCA3.0). Uniquely, we identified a subgroup of genetically clonal CTCs that acquired a mesenchymal-like state and its presence was significantly associated with one subclone that emerged along the clonal lineage. Higher CTC abundance and phenotypic diversity were observed in the BMA than PB and differences in genomic alterations were also identified between the two compartments demonstrating spatial heterogeneity. Single cell copy number profiling further detected clonal heterogeneity within clusters of CTCs (also known as microemboli or aggregates) as well as phenotypic variations by targeted proteomics. Overall, these results identify epithelial and mesenchymal CTCs in the clonal lineage of an aggressive prostate cancer case and also demonstrate a single cell multi-omic approach to deconvolute the heterogeneity and association of CTC phenotype and genotype in multi-medium liquid biopsies of metastatic prostate cancer.
Subject terms: Prostate cancer, Prognostic markers, Translational research, Tumour heterogeneity
Introduction
The mutation, selection, and adaptation of tumor cells along the pathway of disease progression and metastasis results in a spectrum of phenotypic and genomic heterogeneity1–4. Distinguishing phenotypic states of tumor cells with genetically clonal identity and along clonal lineage is important in the investigation of epithelial-mesenchymal transition (EMT) and for understanding the link between genotype and phenotype5,6. Dissecting the genetic mechanism of a phenotypic transition is especially critical in circulating tumor cells (CTCs) with acquired metastatic capability as the key hallmark of cancer7. The underlying concept behind EMT is that a malignant epithelial cell changes its phenotypic state to become more mesenchymal-like and motile as a means to increase metastatic potential. This implies that to clearly identify EMT as a state change, the genomic alterations in the original epithelial cell must be evident in the resultant mesenchymal cell. Current liquid biopsy methods for CTC detection usually rely on one particular phenotypic state, traditionally the epithelial state characterized by cytokeratin and/or EpCAM expression8. Next generation approaches utilize enrichment-free methods such as the third generation of the High-Definition Single Cell Assay (HDSCA3.0) which has the ability to identify epithelial, mesenchymal, and endothelial phenotypic states among circulating rare cells, as well as to identify those cells in the genetically transformed tumor lineage in the liquid biopsy9.
A number of studies have shown that the presence of mesenchymal-like CTCs (mes.CTCs) is associated with worse prognosis10–19. However, the identification of mesenchymal CTCs in those studies was limited to immunostaining11,13–16,19, RT-PCR10,17, or RNA fluorescence in situ hybridization (RNA FISH)12,18 of EMT biomarkers, without further genomic validation of a genetic lineage with cancer cell identity. In our previous study9, of peripheral blood (PB) and bone marrow aspirates (BMA) from 65 metastatic castration-resistant prostate cancer patients in the “cabazitaxel with or without carboplatin” trial (NCT01505868)20, we identified a large number epithelial-like (CK+) cells that exhibited clonal genomic alterations characteristic of prostate cancer cells as well as platelet-coated cells that comprised a biomarker for therapeutic benefit from additional carboplatin. In that study, we also identified a small number of mesenchymal-like (CK+| VIM+) cells, however with the exception of two cases, they were genomically normal and thus may represent cells from the tumor microenvironment (TME) rather than tumor cells that had been transformed through EMT. Thus, dual identification of phenotypic states and genomic alterations on the same single cell is an essential approach to deconvolute EMT heterogeneity of CTCs with cancer cell identity confirmation and lineage tracing.
Among the potential patients for that trial, there was a single patient that underwent pre-enrollment evaluation but did not enter the trial due to rapid disease progression. Blood and bone specimens were taken as part of the pre-enrollment process and were thus available for this study. Here, we utilized the multi-omic capabilities of the HDSCA3.0 workflow to characterize the CTC phenotypes in paired PB and BMA samples from this unusually aggressive case, followed by single cell copy number profiling based on low pass whole genome sequencing or targeted proteomics based on imaging mass cytometry. In contrast to the patients in our previous study9, this index patient had large numbers of genetically clonal epithelial-like (CK+) CTCs and a nearly equal number of mesenchymal-like (CK+| Vim+) CTCs that comprised a genetic subclone of the original genomic alterations. Meanwhile, differences of CTC abundance and phenotypic diversity were observed between PB and BMA as well as genomic variations. The observed significant change in phenotypic state while maintaining clear genetic relationship to the transformed epithelial clone provides the clear evidence of the EMT state change detected in the liquid biopsy and further suggests that a multi-omic approach can provide useful information about patient condition in near real-time to influence treatment decisions.
Results
Patient demographics and HDSCA3.0 workflow
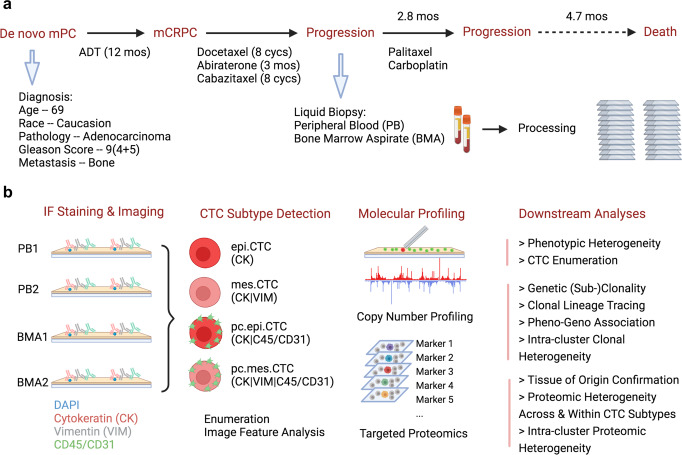
The index patient was diagnosed with de novo mPC at age 69 with high-volume prostatic adenocarcinoma, Gleason Score 9 (4 + 5), PSA 66.4 ng/mL, and bone metastasis. He had acquired castrate resistance after only 12 months of androgen deprivation therapy and was subsequently treated with sequential treatments of docetaxel (8 cycles), abiraterone (3 months), and cabazitaxel (8 cycles). Following PSA progression (from 0.7 to 61.1 ng/mL within 2 months) from cabazitaxel therapy, paired PB and BMA were collected for HDSCA3.0 analysis before the 3rd line chemotherapy paclitaxel and carboplatin which maintained progression-free state for 2.8 months. Unfortunately, the patient passed away in 4.7 months after this progression. (Fig. 1a)
Fig. 1. Patient demographic, pathology, and HDSCA3.0 workflow.
a Patient’s diagnosis, pathology, disease progression, treatment history, and liquid biopsy timepoint. b HDSCA3.0 workflow including immunofluorescence staining and imaging, rare cell detection, CTC classification, molecular profiling, and downstream analyses.
Two slides each from PB and BMA were processed with four-channel staining assay for CTC detection and phenotypic characterization. Furthermore, single cell copy number profiling by whole genome sequencing or targeted proteomics by image mass cytometry were utilized on selected CTCs for genotype and tissue of origin analysis. (Fig. 1b)
CTC enumeration and phenotypical characterization
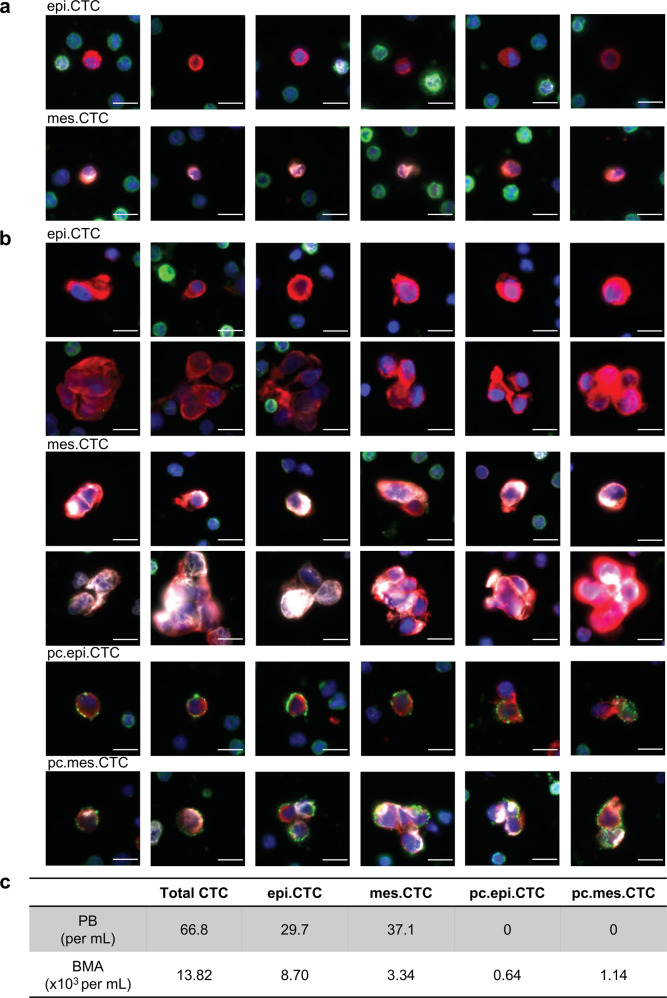
CTC subtypes were classified based on mesenchymal features and platelet attachment status, including epithelial-like CTCs (epi.CTCs), mes.CTCs, platelet-coated epithelial-like CTCs (pc.epi.CTCs), and platelet-coated mesenchymal-like CTCs (pc.mes.CTCs). Consistent with our previous publication6,9,21, the concentration of total CTCs in BMA was ~200-fold higher than in PB (13.82 × 103 vs 66.8 cells per mL). Meanwhile a significant diversity of CTC phenotype was observed, particularly in BMA, with high incidence of CTC clusters and platelet attachment to CTCs compared to the PB (Fig. 2a–b & Supplementary Fig. 1). Within the BMA, 32.4% (4.48 × 103 cells per mL) of CTC expressed VIM and 12.9% (1.78 × 103 cells per mL) of them were coated with platelets. Interestingly, the fraction of VIM positivity was higher in the platelet-coated group, compared to non-coated cells (64.0% vs 27.8%) (Fig. 2c).
Fig. 2. CTC subtype and enumeration in the paired PB and BMA.
a Immunofluorescence images of CTCs from PB sample. Scale bar: 10 μm. b Immunofluorescence images of CTCs from BMA sample. Color coding: DAPI (blue); CK (red); VIM (white); CD45/CD31 (green). Scale bar: 10 μm. c Enumeration of CTC subtypes in each test of PB and BMA.
Based on EBImage-generated features from the four different fluorescence channels, we further evaluated intensity and morphological variations across four CTC subtypes. Following the feature selection pipeline (Supplementary Fig. 2a), we firstly identified ten unique image feature groups and further selected one feature per group as representatives, including four intensity features of DAPI, CK, VIM and CD45/CD31 and six morphological features, i.e., cell and nucleus sizes, cell and nucleus eccentricities, cell/nucleus size ratio, and nucleus location in cell (Supplementary Fig. 2b). As shown in Supplementary Fig. 3, there were minimal differences observed in morphological features, DAPI, or CK intensities among CTC subtypes except for the VIM and CD45/CD31 intensities.
Clonal lineage and its variation between PB and BMA
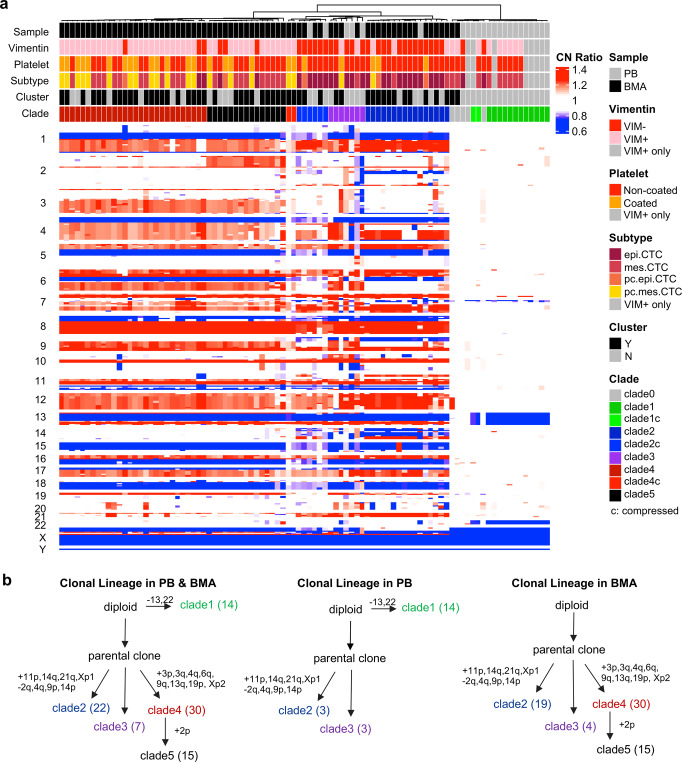
Within PB and BMA samples, 93 cells were sequenced for single cell copy number profiling, including 85 CTCs (27 epi.CTCs, 33 mes.CTCs, 5 pc.epi.CTCs, 20 pc.mes.CTCs) and 8 “VIM + only” rare cells. Of all cells, 88 (94.6%) cells presented clonal alterations and further hierarchical clustering identified one main clone with multiple subclones (clade2–5, 74 cells) and one minor clone (clade1, 14 cells) (Fig. 3a). Unlike common copy number profiling of prostate cancer cells22, the minor clone (clade1) had only chromosome 13 and 22 losses and its breakpoints of chromosome 13 loss were different from those in the main clone (Supplementary Fig. 4c). Clonal lineage tracing indicated that the parental cell of the main clone divided into three different subclones (clade2, clade3, and clade4/5) with the acquisition of different CNAs and clade 5 was generated from clade4 with the additional chromosome 2p gain (Fig. 3bleft) which was further confirmed by phylogenetic tree analysis of individual cells and subclones (Supplementary Fig. 5a–b). Intrasample comparison showed that the subclone (clade4/5) was uniquely detected in the BMA while the emerging minor clone (clade1) was only seen in the PB (Fig. 3bmid & right). As for the fraction of each subclone, 45 of 74 (60.8%) cells were from the clade4-5 (clade4: 30 cells, clade 5:15 cells), 22 cells were from clade2, and 7 cells were from clade3. Due to the small size and high heterogeneity of clade3, we mainly compare the differences between clade2 and clade4-5 and identified 17 distinct CNAs with numerous oncogenes and tumor suppressor genes involved (Supplementary Fig. 4a), e.g., 3q gain (PIK3CA, SOX2), 4q gain (PDGFR), 6q gain (ROS1), and 9q gain (NOTCH1) in the clade4-5 and 2q loss (ERCC3, ZEB2), 9p loss (JAK2, CDKN2A), and 14q gain (HSP90) in the clade2. There was no difference of LSTs observed between clade2 and clade4/5 (Supplementary Fig. 4b).
Fig. 3. Clonal lineage and its variation between PB and BMA.
a Complex heatmap of single cell copy number profiles from paired PB and BMA grouped by hierarchical clustering. The “c” in clade IDs means compressed profile due to normal cell contamination. Red, copy number gain; blue, copy number loss; white, copy number neutral. VIM + only refers to “VIM + only” rare cells. b Clonal lineage in PB and BMA combination and separate. The number in parenthesis is the number of cells analyzed.
Association between genomic subclone and mesenchymal phenotype
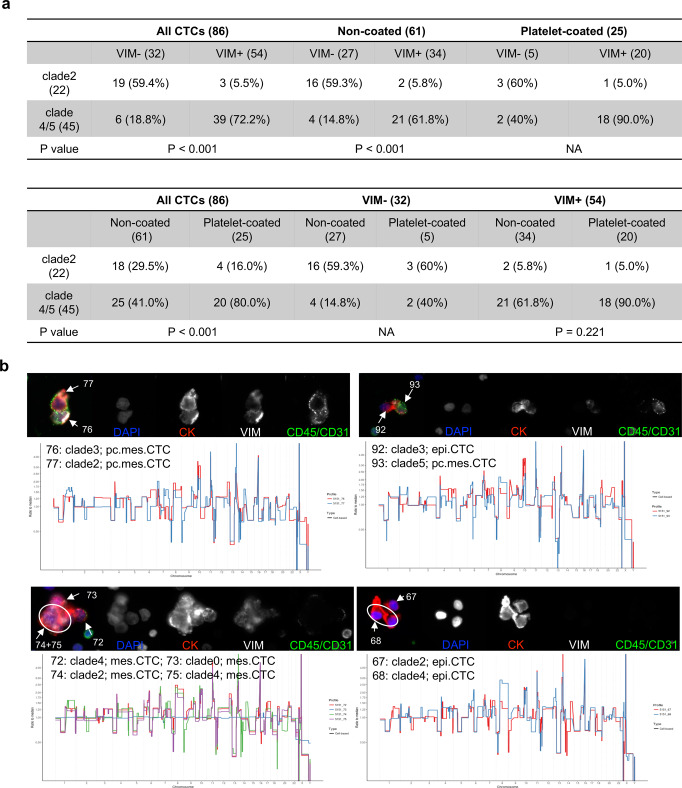
To characterize the relationship between the phenotype and the genotype of the CTCs from the index patient, we firstly compared fluorescence intensity between subclones and observed that the clades4/5 showed higher expression of VIM than clade2 (Supplementary Fig. 6a). To further test if phenotype could be more directly related to the genotype, we initially compared the fractions of clades 2/4/5 between VIM + and VIM− groups in all, non-coated and platelet-coated population. Results showed higher fraction of clade4/5 (39/54, 72.2%) observed in VIM + group while higher fraction of clade2 (19/32, 59.4%) in the VIM- group independent of platelet status, despite the small sample size in the platelet-coated population (Fig. 4a). Meanwhile, we examined the clade fraction difference between non-coated and platelet-coated groups as well and there was no significant difference in the VIM + population while the difference in the CTC population was present due to the sample size imbalance in VIM- population (Fig. 4a). The associations of VIM positivity, platelet attachment, CTC subtypes, and cluster status with genomic clonality were further visualized by t-SNE plots with the input of copy number data (Supplementary Fig. 6b).
Fig. 4. Association between genotype and phenotype.
a Association of genomic subclones with mesenchymal phenotype or platelet attachment in all CTCs or subpopulations. b Immunofluorescence images and single cell copy number profiles of representative heterogeneous CTC clusters in the BMA sample. Images order: composite, DAPI, CK, VIM, and CD45/CD31. The number is the sequencing cell ID.
Meanwhile, we investigated the association between subclones and cluster phenotype to see if genotypes of cells from the same cluster are heterogeneous or homogeneous. Among 14 CTC clusters sequenced from the BMA sample, 10 of them (71.4%) were homogeneous within the same clusters (3 clusters from clade2, 5 clusters from clade4, and 2 clusters from clade 5) and the other 4 (28.6%) clusters were heterogeneous, including various combinations, i.e., clade2 and 4, clade2 and 3, and clade3 and 5 (Fig. 4b & Supplementary Fig. 7a–b).
Proteomic characterization of CTC subtypes
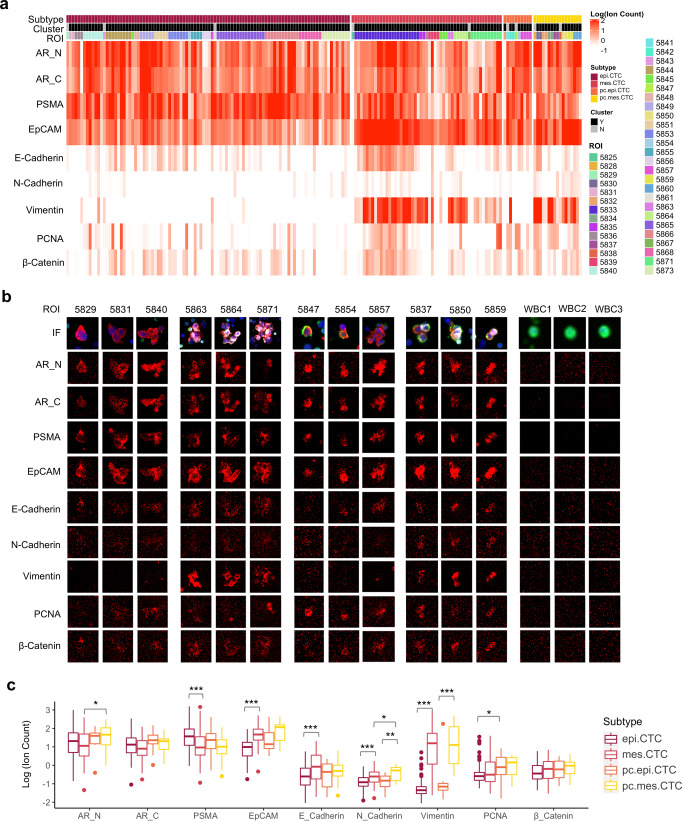
The results of targeted proteomic analysis by imaging mass cytometry are presented as an expression heatmap (Fig. 5a) and CTC images from scanned regions of interest (ROI) (Fig. 5b). The combined results show that EpCAM and prostate-specific biomarkers including AR and PSMA were abundantly expressed in all CTC subtypes while negative in white blood cells thus confirming their tissue of origin. VIM expression was significantly higher in mes.CTCs than epi.CTCs, echoing the results of in the immunofluorescence assay (Fig. 5c). PSMA expression was significantly downregulated while EpCAM, E-Cadherin, and N-Cadherin were significantly upregulated in mes.CTCs compared to epi.CTCs, and similar differences were observed between pc.mes.CTCs and pc.epi.CTCs despite not rising to statistical significance (Fig. 5c). In addition, we observed a trend of increasing AR and PCNA in pc.CTCs compared to CTCs (Fig. 5c). Overall, a spectrum of proteomic heterogeneity was observed across and within CTC subtypes (Fig. 5a–b). Besides, all 31 CTC clusters showed various intra-cluster proteomic heterogeneity. Examples include cluster 5871, where AR, PSMA and PCNA were uniquely expressed in top right two cells of cluster and in cluster 5854, the pc.epi.CTC had higher expression of PSMA, EpCAM, PCNA and beta-catenin compared to the adjacent epi.CTC.
Fig. 5. Targeted proteomics of CTC subtypes.
a Heatmap of proteomic expression in CTC subtypes, including AR-N terminal (AR_N), AR-C terminal (AR_C), PSMA, EpCAM, E-Cadherin, N-Cadherin, Vimentin (VIM), PCNA, and β-catenin. b Immunofluorescence and image mass cytometry images of representative CTC subtypes. c Comparisons of proteomic expressions among CTC subtypes. Center line of box: median; upper/lower hinges of box: 75% or 25% quartile; upper/lower whiskers of box: hinge ± 1.5*IQR (inter-quartile range). *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Phenotypic heterogeneity of tumor cells along disease progression arises from somatic mutations, adaptation to new microenvironments, or resistance against treatments. The purpose of this study was to distinguish different phenotypic states, particularly epithelial-like and mesenchymal-like states, of CTCs with clonal identity and along clonal lineage to analyze genotype-phenotype association and understand CTC biology in mPC. Previous studies from our group6,9 and others23,24 have shown the feasibility of single cell multi-omic approach, e.g., immunostaining, whole genome/exome sequencing, transcriptomic profiling, to simultaneously dissect phenotypic and genotypic heterogeneity of detected or in vitro cultured CTCs. HDSCA3.0, as our most upgraded liquid biopsy workflow, utilizes immunofluorescence (DAPI, CK, VIM, CD45/CD31) to characterize mesenchymal feature and platelet attachment status of CTCs followed with either single cell copy number profiling or targeted proteomics. Here, we applied this approach to paired PB and BMA samples from one prostate cancer patient with bone metastasis and fast progression following multiple lines of treatments including hormonal therapies and chemotherapies. In this index patient, we have identified a subgroup of genetically clonal CTCs that acquired a new phenotypic state, i.e., mesenchymal-like state with the additional expression of VIM, which shows the potential of molecular characterization of EMT on genetically confirmed CTCs in different compartments of fluid microenvironment.
Most importantly, the presence of the mesenchymal-like state in CTCs was significantly associated with a genetic subclone emerged along the clonal lineage. Prior studies have revealed that not only epigenetic events and environmental factors could drive EMT trajectories through transcriptional changes, but also genomic alterations could impact this cell state transition25. For example, CAMK1D (localized at chr.10p13) was highly expressed in basal-like breast cancer due to its gene amplification and exogeneous overexpression could induce EMT26. Similarly, high expression of PLS3 was significantly related to copy number gain of chr.Xq23 which is its genetic locus and promoted EMT through transforming growth factor (TGF)-β signaling in colorectal cancer cells27,28. Oppositely, SLC38A3 (localized at chr.3p21) expression was lower in the tumor tissue than the adjacent normal tissue, linked to its high frequency of gene deletion and further experiments revealed that deletion of SLC38A3 could stimulate EMT in esophageal squamous cell carcinoma29. However, those potential associations were inferred from the observations in bulk tumor samples or the validations in cell lines, while intra-patient genotype-phenotype relationship at single cell level is still unknown, especially in CTCs. Here, our clonal lineage tracing of CTCs has shown that there were at least 3 distinctive subclones emerged under the main clone, include clade2, clade3, and clade4/5, based on the optimal number of clades from hierarchical clustering determined in a mathematical way. More interestingly, the VIM + group had significantly higher percentage of the clade4/5 subclone while the VIM- group was enriched with the clade2 subclone. Further comparison analysis between those two main subclones (clade2 vs clade4/5) had demonstrated a variety of different chromosome or gene level CNAs across the whole genome. We hypothesized the reasons to be that genetic defects of EMT-related genes could cause mesenchymal transformation in clade4/5 subclone or loss of cell state transition ability in clade2 subclone, e.g., copy number gains of PDGFRA, PIK3CA, etc in the clade4/5 subclone while copy number loss of TWIST2, JAK2, etc in the clade2 subclone. In addition to 4 subtypes of CTCs, we also included “VIM + only” classification of cells to represent the full spectrum of rare cells detected and to further investigate the potential of CTCs that completed the EMT and lost epithelial biomarker, e.g., cytokeratin30,31. Single cell copy number profiling showed those cells did not carry the cancer cell genomic architecture (the main clone) which is consistent with our observation in the previous publication9, demonstrating clonality in CK + cell groups in metastatic prostate cancer (mPC). Meanwhile, recent studies have shown hybrid and dynamic phenotypes generated from EMT of CTCs which enhance adaptation ability, treatment resistance, and metastatic potential25,32,33. We acknowledge that the mesenchymal phenotype characterized in our immunostaining has yet to be sufficient to depict the heterogeneity of EMT. We are developing new IF-based assays which will include additional biomarkers to further characterize the plasticity of EMT on CTCs, as well as transcriptomic profiling using single cell RNA sequencing.
Spatial heterogeneity of CTCs during metastasis across multiple liquid compartments has been reported at the transcriptomic level perhaps as an adaptation to different circulation microenvironments34. Here, we investigated the phenotypic and genotypic differences of CTCs from two different compartments of the fluid microenvironment during tumor metastasis. Consistent with phenotypic differences in our previous studies6,21, the abundance and phenotypic diversity of CTCs in the BMA are significantly higher than in the PB which indicates site-specific enrichment and enhanced heterogeneity of CTCs adapting to the bone marrow microenvironment. In terms of genotypic differences, the clade4/5 was uniquely detected in the 66.2% (45/68) of clonal CTCs from BMA as the dominant subclone, while the clade1 minor clone with chromosome 13 and 22 losses was only detected in PB. Interestingly, the cells in the minor clone are all VIM positive with or without CK expression, likely to be circulating TME cells with clonal alterations while the presence of RB1 and BRCA2 losses in the minor clone indicated the probability of the generation of a new cancer clone. To determine if they are TME cells with clonal alteration or CTCs with the new cancer clone, additional biological characterization is warranted. Due to high CTC abundance and additional observation of phenotypic and genotypic heterogeneity in BMA, despite its invasiveness, we suggest that BMA could be a robust supplemental approach, especially when CTC is not detected in PB. Meanwhile, BMA is still less invasive than prostate biopsy and also provides a unique perspective of metastatic cancer cells which usually present aggressive behavior.
CTC cluster or aggregate is another unique phenotype and studies has illustrated its role of indicating worse prognosis21 as well as the epigenetic regulations during its formation35. Here, we explored the genotypic and phenotypic heterogeneity of CTCs from the same clusters. Out of 14 CTC clusters from the BMA, 4 of them had heterogeneous subclones and the rest of them possessed the homogeneous subclones. This observation relates to the hypothesis of CTC cluster formation: (1) the cluster with the same subclone could be formed by cell division from one parental cell or cell aggregation of tumor cells with the same genetic background; (2) the cluster with different subclones could be formed by physical aggregation between 2 or more cells with various genetic backgrounds. It will be interesting to investigate different mechanisms of CTC cluster formation as well as their potential clinical significance.
Overall, this case report demonstrated a robust single cell multi-omic approach to simultaneously deconvolute genotype and phenotype of CTCs in paired liquid biopsy samples from bone mPCs. This approach allows for a complete spatial evolution analysis of cancer metastasis and provides the opportunity to compare multiple compartments of liquid biopsies. The uniqueness of this case includes the confirmation of mes.CTCs (CK + VIM + ) with cancer cell identity, the observation of clonal heterogeneity across subtypes and samples, and, most importantly, morpho-genomic analysis connects a subclonal genotype with an EMT phenotype at the single cell level and elucidates biological heterogeneity and complexity of CTCs.
Methods
Patient selection and sample preparation
The index patient in this study was diagnosed with de novo mPC and following castrate resistance and further disease progression, paired PB and BMA samples were collected and shipped to University of Southern California within 24 hours of the collection for analysis with the HDSCA3.0 workflow9. This patient was registered under the pre-enrollment of “cabazitaxel with or without carboplatin” trial (NCT01505868). The study was approved by the Institutional Review Board and adhered to the principles in the Declaration of Helsinki. The patient provided written informed consent prior to inclusion in the study.
Sample preparation was performed as previously described5,6,21,36. Briefly, upon arrival in the laboratory, both PB and BMA liquid biopsy samples were treated with isotonic ammonium chloride solution for erythrocyte removal and the isolated nucleated cells from centrifuging were plated on the cell-adhesive slides (Marienfeld) and stored at −80 °C.
Immunofluorescent staining and scanning
Two slides with ~3 million nucleated cells each from PB and BMA, respectively, were stained according to the HDSCA3.0 protocol as previously described9. Briefly, slides were thawed for 1 h, fixed with 2% formalin for 20 min, and incubated with 10% goat serum for another 20 min. The following staining steps were conducted on an IntelliPATH FLX autostainer (Biocare Medical LLC) with negative and positive control slides included: (1) a mixture containing an anti-human CD31:Alexa Fluor 647 mouse IgG1 monoclonal Antibody (BioRad; Cat# MCA1738A647; Clone: WM59; Working concentration: 2.5 μg/mL) and an anti-mouse IgG goat monoclonal Fab fragment (Jackson ImmunoResearch; Cat# 115–007–003; Working concentration: 100 μg/mL) for 4 h; (2) 100% cold methanol for permeabilization for 5 min; (3) a mixture consisting of an anti-human cytokeratin (CK) 1,4,5,6,8,10,13,18,19 mouse IgG1/IgG2a monoclonal antibody cocktail (Sigma; Cat# C2562; Clone: C-11, PCK-26, CY-90, KS-1A3, M20, A53-B/A2; Working concentration: 210 μg/mL), an anti-human CK 19 mouse IgG1 monoclonal antibody (Dako; Cat# GA61561–2; Clone: RCK108; Working concentration: 0.2 μg/mL), an anti-human CD45:Alexa Fluor 647 mouse IgG2a monoclonal antibody (AbD Serotec; Cat# MCA87A647; Clone: F10–89–4; Working concentration: 1.2 μg/mL), and an anti-human vimentin (VIM): Alexa Fluor 488 rabbit IgG monoclonal antibody (Cell Signaling Technology; Cat# 9854BC; Clone: D21H; Working concentration: 3.5 μg/mL) for 2 h; (4) a mixture including an anti-mouse IgG1: Alexa Fluor 555 goat IgG polyclonal antibody (Invitrogen; Cat# A21127; Working concentration: 2 μg/mL) and 40,6-diamidino-2-phenylindole (DAPI) for nuclear DNA (Thermo Fisher Scientific; Cat# D1306; Dilution: 1: 50,000) for 1 h. Finally, slides were mounted with a glycerol-based aqueous mounting media followed with adding coverslips. Slides were then scanned, as previously described9. Briefly, slides were scanned using an automated fluorescence scanning microscopy at ×10 objective magnification and generating 2304 frame images for each channel (DAPI: DNA; Alexa Fluor 555: CK; Alexa Fluor 488: VIM; Alexa Fluor 647: CD45/CD31). Exposure time and gain were automatically set up to yield the same background intensity level across slides before auto-scanning. Some cells on the slides were further manually re-imaged using a fluorescence microscopy at ×40 objective magnification for higher resolution images.
Rare cell detection and CTC subtype enumeration
Image analysis was performed as previously reported9. In brief, the EBImage package37 was used to segment DAPI + (cell) events by generating nuclear and/or cytoplasm masks. For each segmented event, 761 quantitative cellular and nuclear features were extracted by “computeFeatures” in the EBImage package and top 350 principal components were further identified from extracted features by principal component analysis. Hierarchical clustering was performed among detected cells with those principal components of features to separate common cells (mainly leukocytes) and rare cells in each frame image.
Manual classification of rare cells into CTC subgroups was also previously reported9. Briefly, epi.CTCs were classified as cells are CK positive, VIM negative and CD45/CD31 negative with distinctive nucleus morphology and mes.CTCs gained VIM expression in addition to CK. Platelet attachment, featured as punctuated CD45/CD31 signals on cell surface, further categorized platelet-coated (pc) epi.CTCs (pc.epi.CTCs) and mes.CTCs (pc.mes.CTCs).
Single cell copy number profiling by low pass whole genome sequencing
Single cell copy number profiling was performed as previously reported5,6,9,21. Briefly, cells of interest were relocated using XY coordinates generated from the scanning and 40× high-quality images were captured before the single cell isolation by micromanipulator. Individual single cells were lysed for whole genome amplification (Sigma-Aldrich; Cat# WGA4) and libraries were constructed using the DNA Ultra Library Prep Kit (New England Biolabs; Cat# E7370) and sequenced by Illumina NextSeq 500 at USC for single-end 50 bp read sequencing. Following genome mapping and PCR duplicate removal, unique reads were seated into ~5000 pre-defined bins and the number of reads per bin was normalized as “ratio to mean” for constructing copy number profile and heatmap and identifying copy number alteration (CNA). The dendrogram on the heatmap was grouped by hierarchical clustering using “cluster-agnes” R package (metric = Manhattan, method = ward). Large scale transition (LST) was a measurement of the number of large scale (>10 Mb) CNAs across the whole genome38. Minimum evolution method39 was used for phylogenetic tree construction of individual cells or clade consensus (the median of cells from the same clade). The contamination of normal cell in CTC could compress the copy number profiles and was manually identified and labeled as “c” standing for “compressed”. The evaluation was based on checking if the “ratio to mean” of CNA was matched with the theoretical ratio between the integer copy number of gain/loss region and the ploidy number of one single cell, e.g., in a diploid cell, the theoretical ratio for one copy loss is 0.5 (1/2) and the value for one copy gain is 1.5 (3/2). In terms of the cell selection criteria for single cell genomics, we sequenced cells across different subtypes including epi.CTC, mes.CTC, pc.epi.CTC, and pc.mes.CTC to observe genotypic heterogeneity and aimed for at least five cells analyzed per subtype per sample in consideration of the number of cells detected and the number of cells successfully isolated and sequenced.
Targeted proteomics by imaging mass cytometry
Cells of interest were subjected to in situ targeted proteomic analyses with the use of the CyTOF Helios imaging mass cytometer (Fluidigm) as previously described6,40. Briefly, sample slides were re-stained with metal-conjugated antibodies (Prostate-specific: AR-N (Cell Signal Technology; Cat# 5153; Clone: D6F11; Dilution: 1:200), AR-C (LS Bio; Cat# LS-C210456-500; Clone: SP242; Dilution: 1:200), PSMA (Novus; Cat# MAB4234; Clone: 460420; Dilution: 1:200); EMT: EpCAM (Fluidigm; Cat# 3144026D; Clone: 9C4; Dilution: 1:200), E-cadherin (Fluidigm; Cat# 3158029D; Clone: 24E10; Dilution: 1:300), Vimentin (Abcam; Cat# ab193555; Clone: EPR3776; Dilution: 1:300), N-cadherin (Abcam; Cat# ab19348; Clone: 8C11; Dilution: 1:200); Cell-proliferation: PCNA (Abcam; Cat# ab18197; Clone: Polyclonal; Dilution: 1:400), β-catenin (Fluidigm; Cat# 3147005 A; Clone: D10A8; Dilution: 1:300) and a DNA intercalator. Antibodies that were not available with Fludigm were sourced from the third-party vendors and were custom conjugated in the lab. Maxpar antibody labeling kits were used to label the antibodies with the metals of choice. Metal labelled antibodies cocktail was applied to the experiment slide during staining process. A region of interest (ROI) of ~400 µm × 400 µm centered on each candidate cell was ablated with a 1 µm diameter pulsed laser, followed by ionization and quantification in the CyTOF Helios instrument. Ion mass data were collected and used for reconstruction of the 1 µm2 ROI spatial resolution, multi-dimensional images of the ROI. Cell segmentation and ion count per cell were generated by IMC segmentation pipeline created by the Bodenmiller Lab41, based on the CellProfiler (version 3.15)42 and Ilastik (version 1.3.3)43 and images with segmented masks could be further displayed by HistoCAT44.
R packages and statistical analysis
For data visualization, we used t-SNE (version 0.15)45 for dimensionality reduction, ggplot2 (version 2.8.0)46 for scatter or bar plots, ape (version 5.5)47 and ggtree (version3.0.4)48 for phylogenetic tree, and Complex Heatmap (version 3.3.5)49 for heatmaps. Chi-square was used for categorical data association analysis and Mann–Whitney U test was used for non-parametric data (i.e., ion count).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors would like to thank the patient who participated in this study, Dr. Jeremy Mason, Dr. Rishvanth Kaliappan, Dr. Chen-Ching Peng, Mike Schmidt, Lisa Welter, Nikki Higa, Libere Ndacayisaba, Sonia Maryam Setayesh, Ryan Storgard, Aidan Plant, and Amanda Hmelar for critical reviewing, comments, initial data analysis, or discussions, Dr. Paul Corn for the registration and supervision of “Cabazitaxel With or Without Carboplatin” trial, Kai-Han Tu for sequencing library preparation, Rafael Nevarez for sequencing data generation and raw data collection, Paul Lanctot for the implementation of IMC analytical pipeline, the Technical team for processing samples, Elvia Nunez and Allison Welsh for project/financial management, and BioRender.com with which we created the Fig. 1. This work is supported fully or partially by the Prostate Cancer Foundation Award 17CHAL01 (A.A., P.K., J.H., S.C.), Breast Cancer Research Foundation Award BCRF-20-089 (J.H., P.K.), NCI’s USC Norris Comprehensive Cancer Center (CORE) Support 5P30CA014089-40 (P.K.), David and Janet Polak Foundation Fellowship in Convergent Science (S.C.), Kalayil & Leela Chacko M.D. Fellowship (S.C.), Vicky Joseph Research Fund, Vassiliadis Research Fund, and Susan Pekarovics.
Author contributions
Assisted with study concept and design: P.K., J.H., S.C., C.R.. Acquired, analyzed, or interpreted the data: S.C., C.R., A.N., M.P., M.S. Provided critical revision of the paper for important intellectual content: L.X., S.S., Calculated statistical analysis: S.C., A.N., Obtained funding: P.K., J.H., A.A., Provided administrative, technical, or material support: N.M., A.K., Collected clinical samples: A.Z., A.A., Supervised study: P.K., J.H.
Data availability
All data discussed in this paper are either included in the main figures or the supplementary files. The single cell sequencing data is available through the Sequence Read Archive with BioProject accession number PRJNA827940. The immunofluorescence image data is available in figshare at 10.6084/m9.figshare.19617717.v1. The image mass cytometry data is available in figshare at 10.6084/m9.figshare.19619007.v1.
Code availability
The code used to analyze the single cell sequencing and image mass cytometry data uses standard third-party open-source libraries and packages in R and/or Python. The code used to identify CTCs in the HDSCA3.0 workflow uses custom code and is proprietary and licensed to Epic Sciences for commercial use.
Competing interests
P.K., J.H., and A.K. hold ownership interest (including patents) in, and P.K. and J.H. hold advisory roles a consultant to Epic Sciences. S.C., C.R-V., N.M., A.K., J.H. and P.K. are royalty recipients on related technology licensed to Epic Sciences for development. No potential competing interests were disclosed by the other authors.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
James Hicks, Email: jameshic@usc.edu.
Peter Kuhn, Email: pkuhn@usc.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-022-00289-1.
References
- 1.Miyamoto DT, Ting DT, Toner M, Maheswaran S, Haber DA. Single-Cell Analysis of Circulating Tumor Cells as a Window into Tumor Heterogeneity. Cold Spring Harb. Symp. Quant. Biol. 2016;81:269–274. doi: 10.1101/sqb.2016.81.031120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller L, Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer. 2019;19:553–567. doi: 10.1038/s41568-019-0180-2. [DOI] [PubMed] [Google Scholar]
- 3.Conteduca V, et al. Circulating tumor cell heterogeneity in neuroendocrine prostate cancer by single cell copy number analysis. NPJ Precis Oncol. 2021;5:76. doi: 10.1038/s41698-021-00211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, et al. Phenotypic Heterogeneity of Circulating Tumor Cells Informs Clinical Decisions between AR Signaling Inhibitors and Taxanes in Metastatic Prostate Cancer. Cancer Res. 2017;77:5687–5698. doi: 10.1158/0008-5472.CAN-17-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dago AE, et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS ONE. 2014;9:e101777. doi: 10.1371/journal.pone.0101777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malihi, P. D. et al. Clonal diversity revealed by morphoproteomic and copy number profiles of single prostate cancer cells at diagnosis. Converg. Sci. Phys. Oncol.4, 10.1088/2057-1739/aaa00b (2018). [DOI] [PMC free article] [PubMed]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.de Bono JS, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 9.Chai, S. et al. Platelet-Coated Circulating Tumor Cells Are a Predictive Biomarker in Patients with Metastatic Castrate-Resistant Prostate Cancer. Mol. Cancer Res. 10.1158/1541-7786.MCR-21-0383 (2021). [DOI] [PubMed]
- 10.Mego M, et al. Prognostic Value of EMT-Circulating Tumor Cells in Metastatic Breast Cancer Patients Undergoing High-Dose Chemotherapy with Autologous Hematopoietic Stem Cell Transplantation. J. Cancer. 2012;3:369–380. doi: 10.7150/jca.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satelli A, et al. Epithelial-mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin. Cancer Res. 2015;21:899–906. doi: 10.1158/1078-0432.CCR-14-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li TT, et al. Evaluation of epithelial-mesenchymal transitioned circulating tumor cells in patients with resectable gastric cancer: Relevance to therapy response. World J. Gastroenterol. 2015;21:13259–13267. doi: 10.3748/wjg.v21.i47.13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindsay CR, et al. Vimentin and Ki67 expression in circulating tumour cells derived from castrate-resistant prostate cancer. BMC Cancer. 2016;16:168. doi: 10.1186/s12885-016-2192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu F, et al. Associations between the Epithelial-Mesenchymal Transition Phenotypes of Circulating Tumor Cells and the Clinicopathological Features of Patients with Colorectal Cancer. Dis. Markers. 2017;2017:9474532. doi: 10.1155/2017/9474532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horimoto Y, et al. Analysis of circulating tumour cell and the epithelial mesenchymal transition (EMT) status during eribulin-based treatment in 22 patients with metastatic breast cancer: a pilot study. J. Transl. Med. 2018;16:287. doi: 10.1186/s12967-018-1663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadaki MA, et al. Circulating Tumor Cells with Stemness and Epithelial-to-Mesenchymal Transition Features Are Chemoresistant and Predictive of Poor Outcome in Metastatic Breast Cancer. Mol. Cancer Ther. 2019;18:437–447. doi: 10.1158/1535-7163.MCT-18-0584. [DOI] [PubMed] [Google Scholar]
- 17.Mego M, et al. Circulating Tumor Cells With Epithelial-to-mesenchymal Transition Phenotypes Associated With Inferior Outcomes in Primary Breast Cancer. Anticancer Res. 2019;39:1829–1837. doi: 10.21873/anticanres.13290. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, et al. Circulating tumor cells undergoing EMT are poorly correlated with clinical stages or predictive of recurrence in hepatocellular carcinoma. Sci. Rep. 2019;9:7084. doi: 10.1038/s41598-019-43572-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batth IS, et al. Cell surface vimentin-positive circulating tumor cell-based relapse prediction in a long-term longitudinal study of postremission neuroblastoma patients. Int J. Cancer. 2020;147:3550–3559. doi: 10.1002/ijc.33140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corn PG, et al. Cabazitaxel plus carboplatin for the treatment of men with metastatic castration-resistant prostate cancers: a randomised, open-label, phase 1-2 trial. Lancet Oncol. 2019;20:1432–1443. doi: 10.1016/S1470-2045(19)30408-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlsson A, et al. Paired High-Content Analysis of Prostate Cancer Cells in Bone Marrow and Blood Characterizes Increased Androgen Receptor Expression in Tumor Cell Clusters. Clin. Cancer Res. 2017;23:1722–1732. doi: 10.1158/1078-0432.CCR-16-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rangel-Pozzo, A. et al. Genomic Analysis of Localized High-Risk Prostate Cancer Circulating Tumor Cells at the Single-Cell Level. Cells9, 10.3390/cells9081863 (2020). [DOI] [PMC free article] [PubMed]
- 24.Soler A, et al. Autologous cell lines from circulating colon cancer cells captured from sequential liquid biopsies as model to study therapy-driven tumor changes. Sci. Rep. 2018;8:15931. doi: 10.1038/s41598-018-34365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genna, A. et al. EMT-Associated Heterogeneity in Circulating Tumor Cells: Sticky Friends on the Road to Metastasis. Cancers (Basel)12, 10.3390/cancers12061632 (2020). [DOI] [PMC free article] [PubMed]
- 26.Bergamaschi A, et al. CAMK1D amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol. Oncol. 2008;2:327–339. doi: 10.1016/j.molonc.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugimachi K, et al. Aberrant expression of plastin-3 via copy number gain induces the epithelial-mesenchymal transition in circulating colorectal cancer cells. Ann. Surg. Oncol. 2014;21:3680–3690. doi: 10.1245/s10434-013-3366-y. [DOI] [PubMed] [Google Scholar]
- 28.Yokobori T, et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 2013;73:2059–2069. doi: 10.1158/0008-5472.CAN-12-0326. [DOI] [PubMed] [Google Scholar]
- 29.Liu R, et al. Defect of SLC38A3 promotes epithelial-mesenchymal transition and predicts poor prognosis in esophageal squamous cell carcinoma. Chin. J. Cancer Res. 2020;32:547–563. doi: 10.21147/j.issn.1000-9604.2020.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang G, et al. Clinical Significance of Mesenchymal Circulating Tumor Cells in Patients With Oligometastatic Hormone-Sensitive Prostate Cancer Who Underwent Cytoreductive Radical Prostatectomy. Front. Oncol. 2021;11:812549. doi: 10.3389/fonc.2021.812549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, et al. Mesenchymal phenotype of circulating tumor cells is associated with distant metastasis in breast cancer patients. Cancer Manag. Res. 2017;9:691–700. doi: 10.2147/CMAR.S149801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassan, S., Blick, T., Thompson, E. W. & Williams, E. D. Diversity of Epithelial-Mesenchymal Phenotypes in Circulating Tumour Cells from Prostate Cancer Patient-Derived Xenograft Models. Cancers (Basel)13, 10.3390/cancers13112750 (2021). [DOI] [PMC free article] [PubMed]
- 33.Balcik-Ercin, P., Cayrefourcq, L., Soundararajan, R., Mani, S. A. & Alix-Panabières, C. Epithelial-to-Mesenchymal Plasticity in Circulating Tumor Cell Lines Sequentially Derived from a Patient with Colorectal Cancer. Cancers (Basel)13, 10.3390/cancers13215408 (2021). [DOI] [PMC free article] [PubMed]
- 34.Sun YF, et al. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat. Commun. 2021;12:4091. doi: 10.1038/s41467-021-24386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gkountela S, et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell. 2019;176:98–112.e114. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marrinucci D, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys. Biol. 2012;9:016003. doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pau G, Fuchs F, Sklyar O, Boutros M, Huber W. EBImage-an R package for image processing with applications to cellular phenotypes. Bioinformatics. 2010;26:979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malihi PD, et al. Single-Cell Circulating Tumor Cell Analysis Reveals Genomic Instability as a Distinctive Feature of Aggressive Prostate Cancer. Clin. Cancer Res. 2020;26:4143–4153. doi: 10.1158/1078-0432.CCR-19-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rzhetsky A, Nei M. Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol. Biol. Evol. 1993;10:1073–1095. doi: 10.1093/oxfordjournals.molbev.a040056. [DOI] [PubMed] [Google Scholar]
- 40.Gerdtsson, E. et al. Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry. Converg. Sci. Phys. Oncol. 4, 10.1088/2057-1739/aaa013 (2018). [DOI] [PMC free article] [PubMed]
- 41.Zanotelli, V. & Bodenmiller, B. ImcSegmentationPipeline: A pixel-classification based multiplexed image segmentation pipeline. Zenodo. 10.5281/zenodo.3841961 (2022).
- 42.McQuin C, et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berg S, et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 44.Schapiro D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods. 2017;14:873–876. doi: 10.1038/nmeth.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Maaten L. Accelerating t-SNE using Tree-Based Algorithms. J. Mach. Learn. Res. 2014;15:3221–3245. [Google Scholar]
- 46.Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York.https://ggplot2.tidyverse.org (2016).
- 47.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 48.Yu G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinforma. 2020;69:e96. doi: 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
- 49.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in this paper are either included in the main figures or the supplementary files. The single cell sequencing data is available through the Sequence Read Archive with BioProject accession number PRJNA827940. The immunofluorescence image data is available in figshare at 10.6084/m9.figshare.19617717.v1. The image mass cytometry data is available in figshare at 10.6084/m9.figshare.19619007.v1.
The code used to analyze the single cell sequencing and image mass cytometry data uses standard third-party open-source libraries and packages in R and/or Python. The code used to identify CTCs in the HDSCA3.0 workflow uses custom code and is proprietary and licensed to Epic Sciences for commercial use.