Abstract
Objective
Type 1 Diabetes (T1D) is characterized by progressive loss of insulin-producing pancreatic β cells as a result of autoimmune destruction. In addition to β cell death, recent work has shown that subpopulations of β cells acquire dysfunction during T1D. We previously reported that β cells undergoing a DNA damage response (DDR) and senescence accumulate during the pathogenesis of T1D. However, the question of how senescence develops in β cells has not been investigated.
Methods
Here, we tested the hypothesis that unrepaired DNA damage in the context of genetic susceptibility triggers β cell senescence using culture models including the mouse NIT1 β cell line derived from the T1D-susceptible nonobese diabetic (NOD) strain, human donor islets and EndoC β cells. DNA damage was chemically induced using etoposide or bleomycin and cells or islets were analyzed by a combination of molecular assays for senescence phenotypes including Western blotting, qRT-PCR, Luminex assays, flow cytometry and histochemical staining. RNA-seq was carried out to profile global transcriptomic changes in human islets undergoing DDR and senescence. Insulin ELISAs were used to quantify glucose-stimulated insulin secretion from chemically-induced senescent human islets, EndoC β cells and mouse β cell lines in culture.
Results
Sub-lethal DNA damage in NIT1 cells led to several classical hallmarks of senescence including sustained DDR activation, growth arrest, enlarged flattened morphology and a senescence-associated secretory phenotype (SASP) resembling what occurs in primary β cells during T1D in NOD mice. These phenotypes differed between NIT1 cells and the MIN6 β cell line derived from a non-T1D susceptible mouse strain. RNA-seq analysis of DNA damage-induced senescence in human islets from two different donors revealed a p53 transcriptional program and upregulation of prosurvival and SASP genes, with inter-donor variability in this response. Inter-donor variability in human islets was also apparent in the extent of persistent DDR activation and SASP at the protein level. Notably, chemically induced DNA damage also led to DDR activation and senescent phenotypes in EndoC-βH5 human β cells, confirming that this response can occur directly in a human β cell line. Finally, DNA damage led to different effects on glucose-stimulated insulin secretion in mouse β cell lines as compared with human islets and EndoC β cells.
Conclusions
Taken together, these findings suggest that some of the phenotypes of senescent β cells that accumulate during the development of T1D in the NOD mouse and humans can be modeled by chemically induced DNA damage to mouse β cell lines, human islets and EndoC β cells in culture. The differences between β cells from different mouse strains and different human islet donors and EndoC β cells highlights species differences and the role for genetic background in modifying the β cell response to DNA damage and its effects on insulin secretion. These culture models will be useful tools to understand some of the mechanisms of β cell senescence in T1D.
Keywords: Pancreatic β cells, Senescence, DNA damage response, Type 1 diabetes
Highlights
-
•
DNA damage elicits senescent phenotypes in mouse β cell lines.
-
•
DNA damage induces a p53 program and senescent phenotypes in human islets and EndoC-βH5 cells.
-
•
DNA damage-induced senescence leads to different effects on insulin secretion depending on the culture model.
1. Introduction
Type 1 Diabetes (T1D) is a disease of chronic insulin deficiency due to progressive autoimmune-mediated destruction of pancreatic β cells [1]. While much of our understanding of T1D stems from investigations of the immune system, recent work has implicated stress responses in β cells as an additional factor driving disease progression and onset [2,3]. For instance, β cells experience unmitigated endoplasmic reticulum (ER) stress and activate the terminal unfolded protein response (UPR) leading to apoptosis during the development of T1D [4]. Small molecule therapeutics that block terminal UPR have shown promise in clinical trials of new onset T1D patients by slowing the decline in C-peptide production after diagnosis [5,6]. Thus, clinical interventions to target β cell stress responses are emerging as a therapeutic strategy for T1D.
In addition to terminal UPR, it was recently shown that a subpopulation of β cells undergo senescence and accumulate during the pathogenesis of T1D [7]. Cellular senescence resulting from unrepairable cell stress or damage is usually activated via the p16Ink4a/Rb and/or p53/p21 pathways, leading to permanent growth arrest and hallmark phenotypes such as an enlarged cell morphology, senescence-associated βgalactosidase (SA-βgal) activity, persistent DNA damage response (DDR), apoptosis resistance and a senescence-associated secretory phenotype (SASP) [8,9].
While healthy β cells are progressively lost in T1D, senescent β cells accumulate during the development of T1D in humans and in the preclinical nonobese diabetic (NOD) mouse model for T1D [7]. Senescent β cells in NOD mice are characterized by upregulation of p21 and p16Ink4a, DDR activation, a Bcl-2-mediated prosurvival phenotype, elevated SA-βgal activity and a SASP involving IL-6, Igfbp3, Serpine1, Mmp2, and Flnb [7]. The presence of persistent DDR, selective Bcl-2 upregulation and contents of SASP generally differentiates stress-induced senescence in β cells that occurs in NOD mice from natural aging and β cell senescence as reported in mouse models of Type 2 Diabetes and maturity onset diabetes of the young (MODY) [[10], [11], [12], [13]]. Notably, in NOD mice, elimination of senescent β cells with Bcl-2 family inhibitors (senolytic compounds), or suppression of SASP with Bromodomain ExtraTerminal domain inhibitors, halts disease progression [7,14]. Therefore, senescence is a novel therapeutic target during T1D progression.
Despite these advances, mechanistic questions around β cell senescence in T1D remain. Senescence progression and phenotypes are variable between cells and are generally cell type- and stress or trigger-dependent [15,16]. In particular, the specific triggers of β cell senescence in T1D are not known and there is currently no β cell culture model to study the phenomenon. In addition, it is unclear the extent to which β cell senescence is modified by genetic background. Different inbred mouse strains used to model diabetes have different genetic background polymorphisms that could influence the development of specific senescence phenotypes. For instance, the NOD mouse strain used to model T1D harbors two polymorphisms in the non-homologous end joining repair factor Xrcc4, lowering the efficiency of DNA double-strand break repair in this strain as compared with C57BL strains [17]. Addressing fundamental mechanistic questions around β cell senescence in T1D using mouse models would greatly benefit from a cell line with the relevant genetically susceptible background.
As with NOD mice, a subset of β cells in human T1D donors express markers of DDR and the p53/p21 senescence pathway [7,18]. Although the source of DNA damage and causes of DDR activation in β cells during T1D are not known, we previously showed that cultured primary adult human islets transcriptionally upregulate p21 (CDKN1A) and can develop a SASP following induction of DNA double-strand breaks with the chemotherapeutic agent bleomycin [7,14]. These findings suggest that sub-lethal DNA damage may be sufficient to elicit β cell senescence during the development of T1D, but a comprehensive analysis of the outcomes of sub-lethal DDR induction in human islets has not been performed and it is unclear whether these phenotypes occur in β cells, as islets are comprised of a diversity of cell types. Similarly, the inter-donor variation in these phenotypes has not been examined.
The objectives of this study were to test the hypothesis that sub-lethal DNA double-strand break damage triggers senescence in β cells of the T1D-susceptible NOD mouse strain and in human β cells. Our results suggest that chemically triggered DNA damage can be used to accurately mimic some of the phenotypes of senescent β cells that progressively accumulate during T1D. Importantly, the extent of senescence phenotypes differed between β cell lines from NOD mice versus C57BL6 mice and between different human donor islet preparations and human EndoC β cells, suggesting an important role of genetic background and other biological variables such as age and sex in modifying outcomes of DDR activation. We also found that chemically induced DDR and senescence had different effects on insulin secretion in mouse β cell lines as compared with human islets and EndoC β cells. These findings have important implications for accurate culture modeling of β cell senescence in diabetes and support a role for unrepaired DNA damage as a mechanism driving senescence in the context of T1D.
2. Materials and methods
2.1. Mouse cell line culture and drug treatments
Mouse insulinoma cell lines MIN6 (RRID:CVCL_0431, sex of cell line is unknown) derived from 13 week C57BL6 mice (gift from Dr. A. Bhushan, UCSF) and NIT1 (RRID: CVCL_3561), derived from 10 week female NOD/LtJ mice (purchased from ATCC) have been previously described [19,20]. Both cell lines were cultured in DMEM high glucose with 2 mM l-glutamine (ThermoFisher) containing 10% FBS (Millipore-Sigma), 20 mM HEPES, 1 mM sodium pyruvate (Gibco), 1X antibiotic-antimycotic (Gibco) and 50 μM 2-mercaptoethanol at 37 °C and 5% CO2. Cell lines were confirmed mycoplasma-free using the PlasmoTest mycoplasma kit (Invivogen). Cell lines were passaged every 4–6 days and MIN6 were detached with 0.25% trypsin-EDTA (Gibco) and seeded at 0.75–1 x 106 cells per 75 cm2 flask, whereas NIT1 were detached with 0.05% trypsin-EDTA and seeded at 2–2.5 × 106 cells per flask. Both MIN6 and NIT1 cells were used between passages 4 and 25 for all experiments. For senescence induction, we modified a previous method that was used to trigger “therapy-induced” senescence in mouse melanoma cell line B16–F10 [21]. MIN6 cells were seeded at a density of 0.7–0.85 × 105 cells/cm2 in 12 or 6-well plates, respectively. Twenty-four hours later, normal growth media was replaced with media containing 2 μM etoposide (Millipore-Sigma) or vehicle (0.01% DMSO) for 2 or 3 days, followed by removal of drug by replacing wells with fresh media. Similarly, NIT1 cells were seeded at a density of 0.85–1.15 × 105 cells/cm2 and 24 h later cultured in 0.5 or 0.25 μM etoposide or vehicle (0.0025 or 0.00125% DMSO) for 3 days before drug removal on day 4 as with MIN6 cells. After drug removal, the media was replaced with fresh media every other day. Vehicle-treated cells often reached confluence before drug washout and required passaging prior to drug removal, so cells were passaged into the same concentration of DMSO-containing media and media was replaced at the same time as the etoposide-treated cells. Cell counting and viability assays were performed with trypan blue staining on an automated cell counter (Bio-Rad TC-20). Cells were cultured for indicated times after drug treatment and harvested as indicated for various assays.
2.2. Human islet culture and EndoC-βH5 cells
Human pancreatic islets from male and female adult donors were provided by the NIDDK-funded Integrated Islet Distribution Program (IIDP) (RRID:SCR_014387) at City of Hope, NIH Grant # 2UC4DK098085 and from the Alberta Diabetes Institute (ADI) IsletCore at the University of Alberta in Edmonton (www.bcell.org/adi-isletcore). Islets from ADI were procured with the assistance of the Human Organ Procurement and Exchange (HOPE) program, Trillium Gift of Life Network (TGLN), and other Canadian organ procurement organizations. Islet isolation was approved by the Human Research Ethics Board at the University of Alberta (Pro00013094). All donors' families gave informed consent for the use of pancreatic tissue in research. Human donor islet information and experiments performed are listed in Supplementary Table 4. We only used donor preparations with >80% purity as determined by dithizone staining. Upon receipt islets were rested in culture for 24 h in RPMI-1640 media containing 10% FBS, 2 mM l-glutamine, 5.5 mM glucose and 1X antibiotic-antimycotic, as previously [7,14]. To induce DDR and senescence, islets were cultured in media containing 50 μM bleomycin (Medchem express) or vehicle (0.2% DMSO) controls for 48 h, followed by removal and culture in drug-free media for 4 or 5 days, as previously [7]. Islets were then harvested for RNA extraction or protein. For conditioned media, islets were put into serum-free islet media on day 4 post-drug washout, for 24 h and the conditioned media collected. To confirm efficacy of the drug treatment, at the day 4 or 5 timepoint we routinely validated DDR activation at the protein level by western blotting for phosphorylated and total ATM and p21 or at the level of SASP by Luminex assay on the conditioned media in all preparations used for experiments reported in this study. To induce ER stress and UPR, islets were cultured in 1 μM Thapsigargin (AdipoGen) for 5 h, before harvest for protein extraction.
The human fetal EndoC β cell lines have been previously described [22]. The latest derivative, EndoC-βH5, were purchased from Human Cell Design. These cells were derived from a female fetus and show the most robust insulin secretory response and β cell identity marker expression as compared with the earlier EndoC-βH3 line, according to the supplier Human Cell Design. βH5 cells [23] were derived from a different donor than the earlier lines and have undergone Cre-mediated excision of the immortalizing SV40 Large T-antigen and hTERT transgenes, similar to the earlier EndoC-βH3 cell line to render the cells mature and non-proliferative. βH5 cells were seeded at 3.75 or 4 × 105 cells per well in 12-well TPP plates treated with gel matrix provided by the supplier (βCOAT) and cultured in a proprietary serum free media (ULTIβ1, supplied by Human Cell Design). After thawing cells, media was changed within 4 h after cells attached. Two days later, growth media was replaced with media containing 0.1% DMSO (vehicle control) or 35 μM bleomycin for 48 or 72 h. Following drug treatment, the media was replaced with fresh media for an additional 2 days of culture or cells were harvested immediately after drug treatment for western blot. In a second experiment cells were cultured in normal growth media for an additional 4 days after drug removal and media was replaced on day 4 to generate conditioned media for 24 h for Luminex assays on day 5 post-drug removal.
2.3. Western blot analysis
Whole cell protein extracts from cells, human islets or mouse tissue were prepared with RIPA buffer (ThermoFisher) in the presence of 1X protease and phosphatase inhibitor cocktail (ThermoFisher) and protein was quantified with the BCA assay (ThermoFisher). Testis tissue from C57BL6/J 3-month-old mice was a gift from Dr. C. Doucette (University of Manitoba). Approximately 3–10 μg of total protein from testis, MIN6 or NIT1 cells, human islets, or EndoC-βH5 cells treated as indicated (equal amounts per sample for each experiment) were electrophoresed on precast 4–12% Bis-Tris Plus Bolt Mini gradient gels and electroblotted onto nitrocellulose membranes with the Mini Bolt-iBlot2 system (ThermoFisher). Membranes were blocked in 5% non-fat milk in Tris buffered saline with 0.1% Tween-20 (TBST) for 1 h at room temperature. Primary antibodies used for westerns and dilutions used are indicated in Supplementary Table 3. Primary antibodies were incubated on membranes in TBST overnight at 4 °C, washed three times for 5 min in TBST and detected with HRP-conjugated secondaries (Jackson Immunoresearch, diluted 1:50,000–1:100,000) after incubation for 1 h at room temperature in 5% non-fat milk in TBST. Membranes were washed three times for 5 min in TBST and were then incubated with SuperSignal West Pico Plus ECL (Thermo Fisher). Membranes were developed after exposure to X-ray film (Fuji Medical) and two or three exposures were collected to ensure signals were not saturated. Densitometry was performed using ImageJ to quantify band intensities relative to β-Actin as a loading control on each blot and samples were normalized to the average of the vehicle-treated controls. To probe different targets of the same molecular weight range on the same blot, membranes were stripped with Restore stripping buffer (ThermoFisher) according to product instructions. Membranes were subsequently washed and reprobed with different primary and secondary antibodies and developed as described above.
2.4. SA-βgal activity assay
SA-βgal activity in MIN6 and NIT1 cells was detected using a commercial kit (Cell Signaling Technology) according to the product instructions. Cells were subsequently washed in PBS and imaged on an EVOS color brightfield imager at 10X magnification.
2.5. EdU labeling assay
EdU incorporation was measured as an assay for DNA synthesis using a commercial kit (Abcam) according to the product instructions modified for adherent cells. Briefly, MIN6 or NIT1 cells were treated with etoposide as indicated above and on the day of drug removal (72 h post-treatment) cells were incubated with media containing 5 μM EdU for 3 h at 37 °C and 5% CO2. Cells were subsequently washed with d-PBS, trypsinized, counted and resuspended at 1–5 x 106 cells/ml in d-PBS. The zombie NIR fixable viability kit (BioLegend) was used according to the manufacturer's instructions prior to fixation in 4% formaldehyde. After fixation, cell pellets were washed twice using Cell Staining Buffer (BioLegend), permeabilized and treated with EdU detection solution. After two more washes in cell staining buffer, cells were filtered through a 70 μm strainer (ThermoFisher Scientific) into 5 mL flow tubes (ThermoFisher Scientific). Flow cytometry was performed on an Attune acoustic focusing cytometer with a gating strategy as follows: FSC-A/SSC-A (cells), FSC-A/FSC-W (single cells), Zombie Near IR-negative (RL3, live cells) and EdU-positive (RL1). Approximately 4–10,000 live cells were scored per sample.
2.6. Luminex assays
Conditioned media (CM) was collected by culturing cells or islets in growth media lacking FBS for 18–24 h. CM was collected, clarified by centrifugation (3000 g for 5 min at room temperature) and either used immediately or stored at 4 °C for up to 2–3 weeks. Luminex assays were carried out on CM from cells treated as indicated using a custom magnetic bead kit that included mouse SASP factors IL-6, Serpine1 and Igfbp3, or human SASP factors CXCL1, CXCL8/IL-8, IGFBP4, GDF-15, or TNFRSF10C (R&D systems). 50 μl of CM was assayed on a Bio-Plex 200 luminex platform (Bio-Rad) as previously [7,14] and secreted amounts in 1–2 ml of CM were normalized to viable cell counts to calculate amount secreted per 100,000 cells or for human islets secretion was normalized to total RNA content. Alternatively, media was spin-concentrated using a 3 kDa MWCO centrifugal unit (Millipore-Sigma) and the concentrated conditioned media was analyzed by Luminex.
2.7. RNA extraction and quantitative RT-PCR
Total RNA was extracted from MIN6, NIT1, human islets or EndoC-βH5 cells using a commercial column-based purification with DNAse I treatment (Direct-Zol RNA MicroPrep with Trizol Reagent, Zymo Research). RNA was quantified by Nanodrop spectrophotometer (ThermoFisher Scientific). Approximately 50–300 ng of total RNA was used for cDNA synthesis with random primers (LunaScript SuperMix Kit, New England Biolabs), depending on the cells used. qPCR was carried out on a Bio-Rad CFX96 using Luna Universal qPCR Master Mix (New England BioLabs). Primers used for qPCR were previously validated from other studies and are indicated in Supplementary Table 3.
2.8. RNA-seq on human islets
Approximately 0.5–1 μg of total RNA from bleomycin and vehicle-treated islets from two different adult female donors (n = 3 biological replicates per group from each donor) at 4 days post-drug removal was submitted for paired-end RNA sequencing on Illumina Nova-Seq 6000 platform by a commercial service provider, LC Sciences. Total RNA was quality checked and ribosomal RNA depleted. Libraries were prepared with Illumina TruSeq-stranded total RNA sample protocol and quality checked by Agilent BioAnalyzer 2100 High Sensitivity DNA Chip. Cutadapt [24] and perl scripts were used to filter adapter contamination and undetermined bases. Sequences were quality checked with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) and mapped to the human genome (version 96) using Bowtie2 [25] and HISAT2 [26]. Mapped reads were assembled for each sample with StringTie [27]. A comprehensive transcriptome was prepared using perl scripts and gffcompare (https://github.com/gpertea/gffcompare/). StringTie and edgeR [28] were used to estimate transcript expression levels. LncRNAs were identified based on filtering transcripts overlapping with known mRNAs, known lncRNAs and transcripts <200 bps. CPC [29] and CNCI [30] were used to predict transcripts coding potential. Transcripts with CPC score <-1 and CNCI score <0 were removed. Remaining transcripts were considered lncRNAs. StringTie was used to perform differential expression level analysis for mRNAs and lncRNAs by calculating FPKM (total exon fragments/mapped reads (millions) x exon length (kb)). A complete list of differentially expressed and all expressed genes is shown in Supplementary Table 1. A complete list of differentially expressed lncRNAs and novel lncRNAs is found in Supplementary Table 2. Differentially expressed mRNAs and lncRNAs were selected with log2 fold-change >1 or < -1 between vehicle control and bleomycin treatment and parametric F-test comparing linear nested models (p < 0.05 and FDR <0.05) by edgeR. GO and KEGG analysis was performed using by hypergeometric distribution fitting (p < 0.05) from gene ontology terms (http://www.geneontology.org/) and KEGG pathways (http://www.kegg.jp/). Hierarchal clustering and heatmap of normalized expression values (FPKM) for differentially expressed SASP genes was generated using pheatmap in R via publicly available software using the Euclidean distance method and plotting row Z-scores (http://www.bioinformatics.com.cn/plot_basic_cluster_heatmap_plot_024_en).
2.9. Static glucose-stimulated insulin secretion assays
Glucose-stimulated insulin secretion was measured from MIN6 and NIT1 cells and human islets according to standard methods with minor changes [[31], [32], [33]]. For mouse β cell lines, cells were seeded as indicated above and cultured with vehicle or etoposide for 72 h to induce senescence. At the 72 h post-treatment timepoint, cells were washed in Krebs-Ringer Bicarbonate (KRB) buffer (128.8 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 2.5 mM CaCl2, 5 mM NaHCO3, 10 mM HEPES) containing 0.1% BSA fraction V (Millipore-Sigma) and incubated for 1 h at 37 °C and 5% CO2. Then cells were incubated for 1 h in fresh KRB buffer with BSA as above containing 0 mM glucose as the low glucose condition, followed by 1 h in KRB with BSA containing 20 mM glucose as the high glucose condition. Total insulin content was collected by lysing cells in an acidified ethanol solution (0.15 M HCl, 95% ethanol). Human islets were treated in n = 5 or 6 biological replicates of vehicle control (DMSO) or 50 μM bleomycin for 48 h to induce senescence, followed by culture in drug-free media for an additional 4 days. At day 5 post-drug removal, islets were washed in KRB + 0.1% BSA containing 2 mM glucose (low glucose) and incubated in the low glucose buffer for 1 h. Islets were then washed into KRB + 0.1% BSA containing 20 mM glucose for 1 h (high glucose buffer) and supernatants of the 2 mM and 20 mM glucose buffers were collected. Total insulin content from human islets was collected by lysing cells in an acidified ethanol solution (0.15 M HCl, 95% ethanol). For EndoC-βH5 cells, GSIS assays were performed according to the protocol provided by the supplier. After induction of senescence by treatment with 35 μM bleomycin or 0.1% DMSO as a control for 72 h as above, on day 2 post-drug removal, cells were cultured for 24 h in starvation buffer ULTI-ST provided by the supplier. The next day, cells were sequentially incubated in βKREBS buffer (provided by supplier) containing low glucose (0 mM) and then βKREBS buffer containing high glucose (20 mM). Insulin content was extracted with TETG solution (20 mM Tris pH 8.0, 137 mM NaCl, 2 mM EGTA, 1% Triton-X-100, and 10% glycerol) containing 1% protease inhibitor cocktail (ThermoFisher). Aliquots of the low and high glucose supernatants and the insulin content extracts were collected and spun at 3000 g for 5 min at 4 °C and stored at −20 °C or used immediately for mouse or human insulin ELISAs (Mercodia) with appropriate dilutions (1:10–1:50 for supernatants, 1:100–3000 for insulin content). Insulin secretion in the supernatants were quantified as a function of total insulin content from each sample.
2.10. Statistical analysis
All statistical analyses were performed using GraphPad Prism version 9.3.1 with a minimum of n = 3 biological replicates per sample group. We considered independently treated wells of islets processed in parallel from a single human donor as biological replicates. Total RNA-seq was carried out on human islets from two different adult female donors. Other human islet experiments were carried out on preparations from 2 or 3 different male or female donors (Supplementary Table 4). Statistical comparisons were performed with two-tailed unpaired T-tests, or two-way ANOVAs where indicated, and were considered significant at p < 0.05.
3. Results
3.1. Sub-lethal DNA damage with etoposide leads to senescent phenotypes in NIT1 cells
To investigate whether direct DNA damage triggers senescent phenotypes in β cells resembling what occurs during T1D, we used the NIT1 β cell line, derived from an insulinoma in the T1D-susceptible NOD/LtJ mouse strain [20]. In parallel we examined the MIN6 β cell line derived from the non-T1D susceptible C57BL6 strain [19] to explore effects of genetic background and as a positive control β cell line known to acquire senescence phenotypes [34]. We induced DNA double-strand breaks with low concentrations of the topoisomerase II (TopoII) poison etoposide [35], as this drug has been used in the clinic to treat insulinomas [36] and leads to an increased burden of unrepaired endogenous DNA double-strand breaks due to inhibited ligation after cleavage by endogenous TopoII enzymes [37]. TopoIIα is typically expressed in proliferating cells, whereas TopoIIβ is expressed in non-proliferating cells [38]. Of the TopoII family members, we found that TopoIIα was expressed in both MIN6 cells and NIT1 cells, while TopoIIβ was expressed only in MIN6 (Supplementary Fig. 1A) confirming that these cells express the target of this drug.
MIN6 or NIT1 cells were cultured in etoposide for 3 days, followed by drug removal to assay early senescent phenotypes or culturing the cells in drug-free media for an additional 5–14 days to assay late senescent phenotypes (Figure 1A). We titrated the doses of etoposide to maintain viability throughout the treatment and culture period. While MIN6 cells tolerated 2 μM with >60% viability for up to 14 days post-drug removal, NIT1 cells could only survive in up to 0.5 μM for 7–9 days post-drug removal (Figure 1B) and similar results were obtained using 0.25 μM etoposide. Viability for both cell lines was maintained at ∼80–90% during 72 h of drug treatment (Figure 1B) but cell counts were significantly lower in the etoposide-treated cells as compared with controls from 24 to 72 h post-treatment (Supplementary Fig. 1B), suggesting that there was a proliferation arrest early during the drug treatment period.
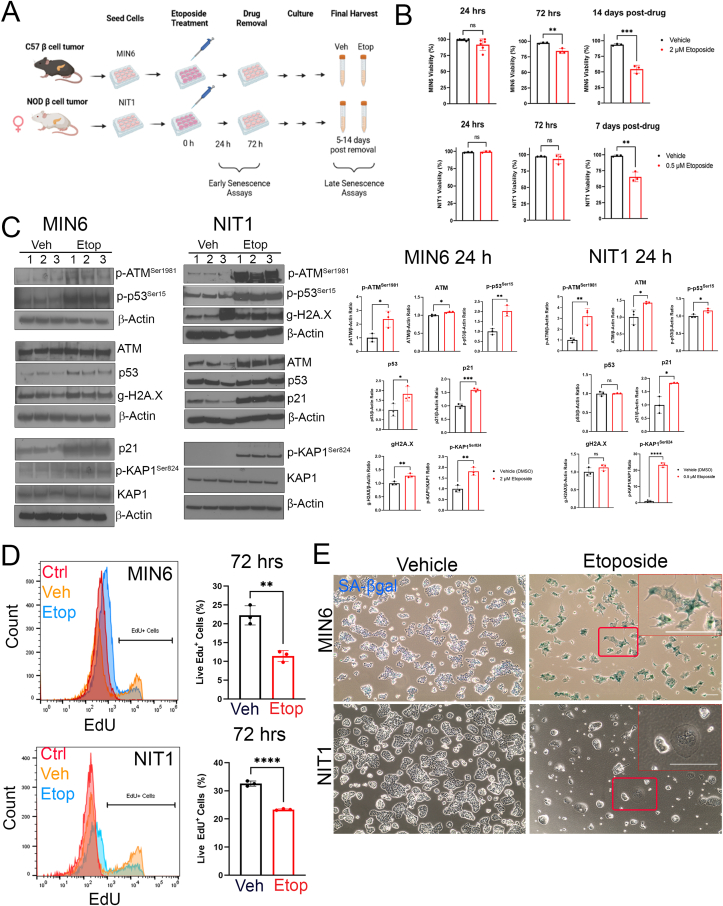
Figure 1.
Sub-lethal DNA damage with etoposide induces DDR, growth arrest and senescent morphology in MIN6 and NIT1 cells. (A) MIN6 or NIT1 cells were seeded and treated with etoposide (2 μM for MIN6, 0.5 μM for NIT1 throughout) or vehicle control (DMSO) for 72 h. Drug was removed by changing cells into fresh media, and cells were cultured an additional 5–14 days before final harvests. Early senescence phenotypes were monitored at 24–72 h post-etoposide treatment and late phenotypes were assayed at 5–14 days post drug removal. B) Cell viability was scored at 24 or 72 h after etoposide treatment and 7 or 14 days post-drug removal. Data are mean ± SD of n = 3–6 biological replicates. C) Western blot analysis and relative quantifications of DDR markers in MIN6 and NIT1 cells at 24 h post etoposide (2 μM or 0.5 μM, respectively) or vehicle treatment. Data are mean ± SD of n = 3 biological replicates. D) EdU labeling flow cytometry assay for DNA replication at 72 h post-etoposide or vehicle control treatment of MIN6 or NIT1 cells. ‘Ctrl’ indicates a no EdU negative control population of cells. Data are mean ± SD of n = 3 biological replicates. E) Representative images of Xgal staining for SA-βgal activity in MIN6 and NIT1 cells at day 5 post-drug removal. Scale bar = 30 μm. Insets show zoomed in regions, scale bars = 60 μm ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.00005, unpaired two-tailed T-tests.
β cells with DNA damage in vivo activate a canonical DDR involving the ATM-p53-21 pathway, phosphorylated KAP1Ser824 and phosphorylated Histone H2A.XSer139 (gamma-H2A.X) [7,18,39,40]. Therefore, we next determined whether the DDR pathway was activated in MIN6 and NIT1 cells 24 h after etoposide treatment (Figure 1C). MIN6 cells exhibited significant increases in phosphorylated ATMSer1981, phosphorylated p53Ser15, total ATM and p53, gamma-H2A.X, phosphorylated KAP1Ser824 and p21 (Figure 1C). Similarly, NIT1 cells had increased phosphorylated and total ATM, phosphorylated p53, phosphorylated KAP1 and p21, but there was no increase in total p53 or gamma-H2A.X (Figure 1C). Total p53 and gamma-H2A.X were already abundant in undamaged NIT1 and MIN6 cells (Figure 1C). Etoposide treatment also reduced the percent of MIN6 and NIT1 cells replicating DNA as indicated by a lower proportion of cells labeled with EdU at 72 h post-drug treatment (Figure 1D) consistent with a growth arrest. Similarly, etoposide treated MIN6 cells developed an enlarged flattened morphology with strong SA-βgal activity by 5 days after drug removal, however, while NIT1 cells showed enlarged flattened morphology, they did not stain positive for SA-βgal activity at the same time point (Figure 1E). Taken together, these data show that NIT1 cells are more sensitive to DNA damage than MIN6 and that sub-lethal DNA double-strand break damage triggers DDR activation, growth arrest and senescence-related morphology changes in NIT1 cells.
3.2. Cdkn1a but not Cdkn2a is upregulated in growth arrested NIT1 cells
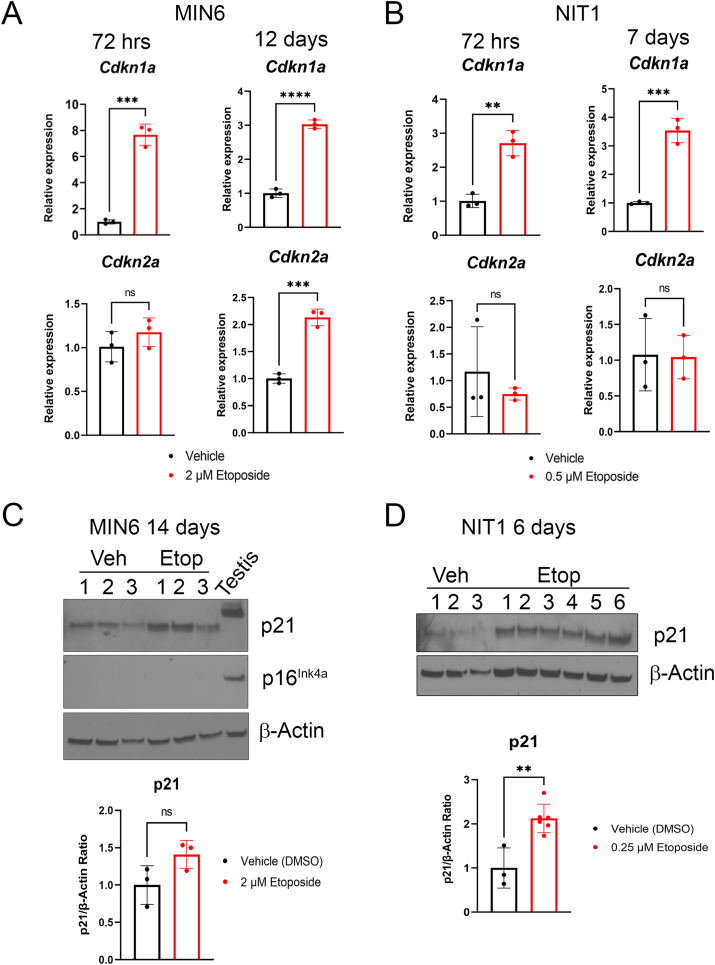
We next assessed whether gene expression levels of CDK inhibitors p21 and/or p16Ink4a were upregulated at later stages of senescence in NIT1 cells (Figure 2). Notably, at the 72 h post-treatment time-point in MIN6 cells, Cdkn1a but not Cdkn2a was upregulated. However, at later stages of senescence by 12 days post-drug removal both Cdkn1a and Cdkn2a were upregulated (Figure 2A). While NIT1 cells also upregulated Cdkn1a at 72 h post-treatment and 7 days post-drug removal, they did not upregulate Cdkn2a at either time-point (Figure 2B). We also analyzed p21 and p16Ink4a protein levels relative to 3-month old mouse testis as a positive control for both proteins [41,42] and in this tissue p21 was expressed as a slower migrating species around 28 kDa (Figure 2C). In late senescent MIN6 cells, p21 levels were more variable and not significantly different from controls at 14 days post-drug removal (Figure 2C). Despite upregulation of p16Ink4a mRNA in MIN6 cells at this late senescence time-point (Figure 2A), the protein was not detected (Figure 2C). In contrast, in late senescent NIT1 cells, p21 levels remained significantly elevated relative to controls, consistent with the transcript levels (Figure 2B, D). We also assessed the expression of Bcl-2, which is selectively upregulated during β cell senescence in NOD mice and confers a prosurvival phenotype [7]. Notably, Bcl-2 was below detection in control and late senescent MIN6 cells, despite robust expression in mouse testis as a positive control [43] (Supplementary Fig. 2) and the same was found in NIT1 cells. Taken together, these data suggest that senescent growth arrest in NIT1 and MIN6 cells involves p21 but not p16Ink4a.
Figure 2.
Induction of Cdkn1a but not Cdkn2a at mRNA and protein level in etoposide treated MIN6 and NIT1 cells. A) qRT-PCR analysis of Cdkn1a and Cdkn2a expression normalized to Gapdh in MIN6 cells at 72 h post-etoposide (2 μM) or vehicle treatment or 14 days post-drug removal (as from Fig. 1). Data are mean ± SD of n = 3 biological replicates. B) qRT-PCR analysis as in B) except on NIT1 cells at 72 h and 7 days post-drug removal. Etoposide was used at 0.5 μM. C) Western blot of p21 and p16Ink4a on whole cell extracts from MIN6 cells treated with 2 μM etoposide or vehicle control 12 days post-drug removal, Actin was a loading control and whole cell extract from 3 month old C57BL6 mouse testes was a positive control for p21 and p16. Data are mean ± SD of n = 3 biological replicates. D) Western blot of p21 on whole cell extracts from NIT1 cells treated with 0.25 μM etoposide or vehicle at 6 days post-drug removal. Data are mean ± SD of n = 3 biological replicates. For all panels, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ns = not significant, two-tailed T-tests.
3.3. Differences in SASP and UPR in MIN6 and NIT1 cells following DNA damage
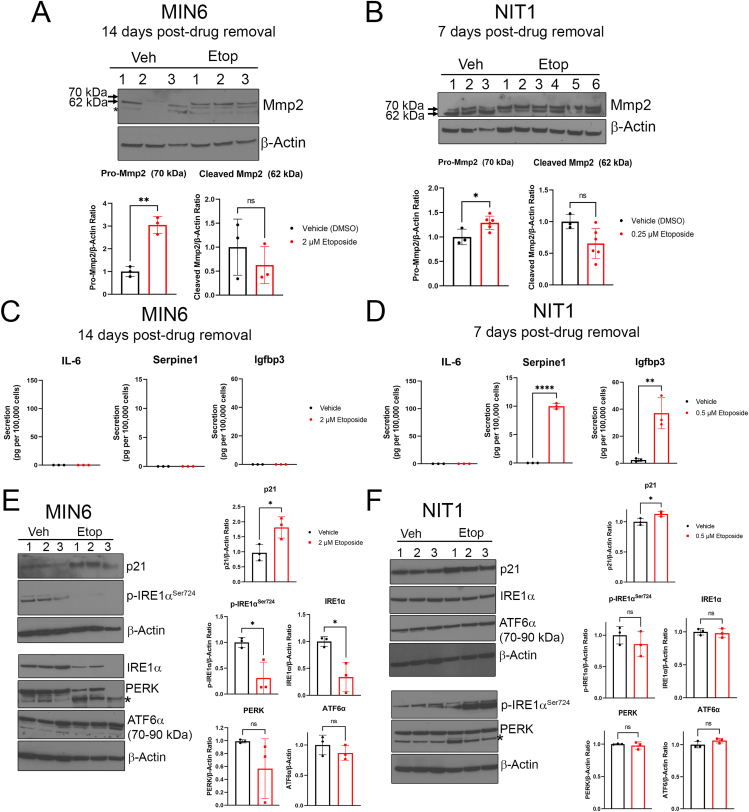
SASPs are cell-type and stressor-dependent, dynamic programs of secreted cytokines, chemokines, matrix proteases, shed receptors, growth factors, microRNAs and extracellular vesicles that are immunogenic and exert paracrine effects within the tissue microenvironment [[44], [45], [46]]. In senescent β cells that accumulate in NOD mice, SASP comprises a wide variety of factors such as interleukins (IL-6), chemokines (Cxcl10), growth factors (Igfbp3), and matrix metalloproteases and protease inhibitors (Mmp2, Serpine1) [7,14]. To determine whether senescence in NIT1 cells led to a SASP resembling primary β cells in NOD mice, we assayed a panel of previously validated SASP factors by a combination of western blot and Luminex secretion assays (Figure 3). Western blot of proenzyme and cleaved active Mmp2 isoforms revealed that etoposide treatment led to increased pro-Mmp2 levels in both MIN6 and NIT1 cells (Figure 3A,B) consistent with increased synthesis of this protein during SASP. Luminex assays for secreted NOD mouse β cell SASP factors [7] on conditioned media showed dramatically increased secretion of Serpine1 and Igfbp3 from senescent NIT1 cells as compared with controls whereas none of these factors were secreted by late-stage senescent MIN6 cells (Figure 3C,D). We also determined whether induction of senescence would impact the UPR, a different β cell stress response implicated in T1D [4]. We validated antibodies for UPR mediators phospho-IRE1αSer724 (activated IRE1α) and total IRE1α, PERK and ATF6α (90 kDa uncleaved form) by western blot on primary human islets treated with 1 μM Thapsigargin for 5 h, which led to increased levels of these UPR factors (Supplementary Fig. 3). Western blot analysis revealed upregulated p21 at 72 h post-etoposide treatment in both lines as expected and MIN6 cells showed reduced phosphorylated and total IRE1α levels and a wide variation in PERK levels (Figure 3E). In contrast, UPR mediators were not affected in early senescent NIT1 cells (Figure 3F). Taken together, these data reveal that senescence induction leads to differences in SASP and UPR proteins in MIN6 and NIT1 cells.
Figure 3.
Differences in SASP and altered UPR in MIN6 and NIT1 cells following etoposide treatment. A) and B) Western blot analysis of Pro-Mmp2 (70 kDa) and cleaved/activated Mmp2 (62 kDa) on whole cell extracts from MIN6 or NIT1 cells treated with vehicle or etoposide as indicated (2 μM or 0.5 μM, respectively). Quantification shows relative Pro-Mmp2 or Cleaved Mmp2 to Actin, mean ± SD of n = 3 biological replicates. C) and D) Luminex assay of secreted SASP factors IL-6, Serpine1 and Igfbp3 in serum-free conditioned media from MIN6 or NIT1 cells treated as indicated at timepoints indicated. Data are mean ± SD from n = 3 biological replicates. E) and F) Western blot analysis of UPR mediators phosphorylated and total IRE1α, total PERK and total ATF6α in MIN6 and NIT1 cells at 72h post-treatment with etoposide (2 μM or 0.5 μM, respectively). p21 levels were used as a positive control and β-Actin was a loading control. Data are mean ± SD of n = 3 biological replicates. Asterisk on the blots indicates a non-specific band for the PERK blots. For all panels, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ns = not significant, two-tailed T-tests.
3.4. RNA-seq analysis of the DNA damage-mediated senescence transcriptional program in human islets
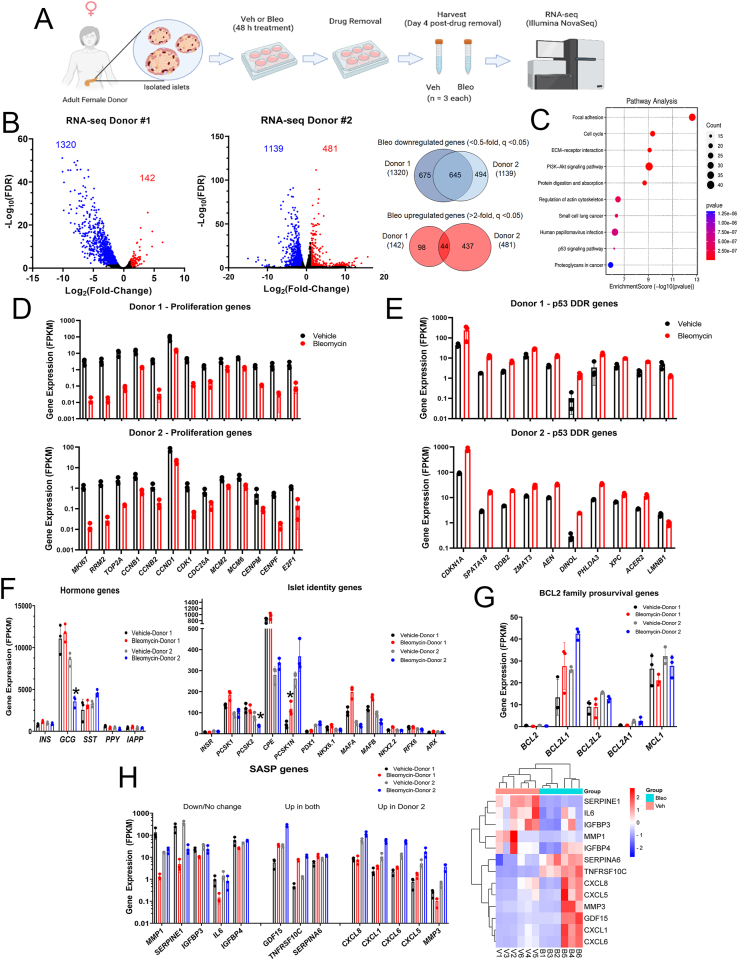
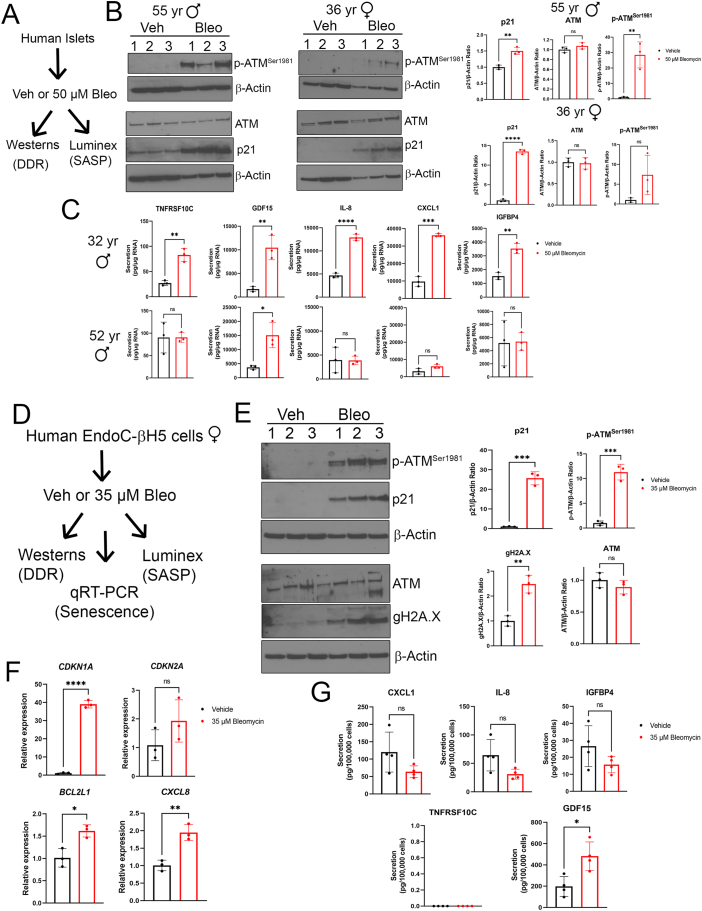
To investigate the transcriptional programs following DDR and senescence in human islets, we next performed RNA-seq on senescent and control human islets from two different donors cultured ex vivo using our previous approach to induce senescence with DNA damage [7,14] (Figure 4). Primary islets isolated from a healthy 44-year-old female donor (Donor 1) or islets from a healthy 50-year-old female donor (Donor 2) (Supplementary Table 4) were cultured with 0.15% DMSO as a vehicle control or 50 μM bleomycin to induce DNA damage for 48 h and total islet RNA from the samples was extracted at 4 days post-drug removal for paired-end RNA-seq (Figure 4A). This timepoint is typically 1 day prior to the detection of SASP secretion in this model [7,14], and we reasoned that transcriptional changes in SASP genes would precede increased protein secretion, as SASP is controlled transcriptionally [14,15]. Differential expression analysis of control and senescent islets from Donor 1 revealed 1320 genes significantly downregulated <0.5-fold and 142 genes significantly upregulated >2-fold, while 1139 genes were downregulated and 481 were upregulated in islets from Donor 2 (FDR <0.05, Figure 4B). In common between the donors, we found 44 genes upregulated and 645 genes downregulated (Figure 4B) suggesting that there was more of a common response in the downregulated genes and more donor variability in the upregulated genes. A complete list of all genes from each donor with expression levels as fragments per kilobase per million reads (FPKM) are listed in Supplementary Table 1. Notably, among the common differentially expressed genes, there was a transcriptome response consistent with senescence induction. Differentially expressed genes in common between the donors were enriched for gene ontology (GO) terms including ‘cell division’ and ‘cell cycle’ and KEGG pathways ‘cell cycle’, ‘p53 signaling pathway’ and ‘PI3K-Akt signaling pathway’ (Figure 4C, Supplementary Fig. 4A). Senescent islets from both donors significantly downregulated genes involved in proliferation and cell cycle progression (Figure 4D) while p53-driven DDR genes [47] were significantly upregulated (Figure 4E). A survey of CDK inhibitor genes showed that only the p53 target gene CDKN1A was consistently upregulated in both donors (∼5-8-fold) (Figure 4E and Supplementary Table 1). Other gene expression changes associated with p53-mediated senescence also occurred, including downregulation of LMNB1 (Figure 4E) [48]. We also noted several small and long noncoding RNAs (lncRNAs) that were differentially expressed (Supplementary Tables 1 and 2). Notably among the lncRNAs, DINOL was found to be upregulated in both donors (Figure 4E), consistent with its established role in amplifying p53-mediated DDR signaling [49]. Comparison of the list of common differentially expressed genes in our dataset with recently generated human islet RNA-seq datasets during natural aging [50] and adult human islets treated with the cytokine IFNα [51] showed that there were only 22/212 (∼10%) age-related genes in common and only 93/1894 (∼5%) IFNα-regulated genes in common, respectively, (Supplementary Fig. 4B, Supplementary Table 1). This suggested that the transcriptional response during islet DNA damage-induced senescence is generally different from natural aging and cytokine exposure. Together, these results demonstrate that bleomycin induces a p53-mediated senescence transcriptional program in human islets.
Figure 4.
RNA-seq analysis reveals activation of the p53 transcriptional program during senescence in human islets. A) Overview of human islet DNA damage-induced senescence model. Islets isolated from a 44-year-old female donor (Donor 1) or from a 50-year-old female donor (Donor 2) were rested overnight and then divided into a total of 6 wells and cultured in the presence of vehicle (DMSO) or 50 μM bleomycin for 48 h (n = 3 biological replicates per group). The islets were then transferred to fresh drug-free media and cultured an additional 4 days prior to harvesting for RNA extraction and paired-end RNA-seq. B) Volcano plots of differentially expressed genes (fold-change cut-off and FDR <0.05) found indicating 1320 genes downregulated <0.5-fold and 142 genes upregulated >2-fold in bleomycin treated islets compared to controls from Donor 1 and 1139 genes downregulated and 480 genes upregulated from Donor 2. 645 genes were downregulated in common and 44 genes were upregulated in common in bleomycin-treated islets from both donors. C) Significant KEGG pathway terms of common differentially expressed genes from Donor 1 and Donor 2. D) Plot of normalized expression levels (FPKM) of selected significantly downregulated proliferation and cell cycle genes from Donor 1 and Donor 2. E) Plot of normalized expression levels of selected significantly upregulated p53 target genes and early senescence gene LMNB1 from Donor 1 and Donor 2. F) Plot of normalized expression levels of selected islet hormone genes, islet cell identity and hormone processing genes from Donor 1 (black and red bars) and Donor 2 (grey and blue bars). Only genes with an asterisk were found to be significantly different in expression. G) Plot of normalized expression levels of BCL-2 family anti-apoptotic genes from Donor 1 and Donor 2. H) Plot of normalized expression of selected significantly downregulated/unchanged SASP genes (Down/No change), SASP genes that were significantly upregulated in both Donors (Up in both), and SASP genes significantly upregulated only in Donor 2 but not Donor 1 (Up in Donor 2). Unsupervised hierarchal clustering heatmap of SASP gene normalized expression values (FPKM). Legend shows Row Z-score values. V1, V2, V3 and B1, B2, B3 were Vehicle-treated or Bleomycin-treated biological replicates from Donor 1. V4, V5, V6 and B4, B5, B6 are Vehicle-treated or Bleomcyin-treated biological replicates from Donor 2. For all barchart plots, data shown are mean ± SD of the n = 3 biological replicates in the RNA-seq datasets per sample group for each donor.
To explore the effect of senescence on islet identity and function and UPR, we next looked at genes encoding endocrine islet cell hormones, processing enzymes, islet cell transcription factors and UPR mediators. The majority of islet hormone, processing or identity genes were not altered in senescent islets from either donor. However there was a ∼50% decrease in GCG expression and a similar decrease in the corresponding glucagon processing enzyme gene PCSK2 [52] in senescent islets from Donor 2 but not Donor 1 (Figure 4F), revealing inter-donor variability. In addition, genes involved in ER stress and UPR [4] were not significantly changed between control and bleomycin treatment (Supplementary Fig. 4C). This suggests that DNA damage and senescence does not substantially impact genes involved in islet cell identity, islet hormones or the UPR.
To assess whether senescent islets developed changes in prosurvival genes and a SASP transcriptional response, we looked at BCL-2 family prosurvival genes and SASP genes. Among the prosurvival family members, BCL2L1, MCL1 and BCL2L2 were the only members expressed at appreciable levels, and of these only BCL2L1 (encoding BCL-XL) was upregulated in both donors (Figure 4G), although it did not reach significance due to the inter-sample variability in Donor 1 and the modest fold-change for Donor 2 (as it was ∼1.6-fold increased, rather than >2-fold). We next looked at SASP genes. There were some canonical proinflammatory SASP factor genes that were either dramatically downregulated or unchanged in bleomycin treated senescent islets relative to controls in both donors, including MMP1, SERPINE1, IGFBP3 and IL6 (Figure 4H, Supplementary Table 1). However, some other SASP genes encoding secreted factors and cell surface receptors were upregulated in both donors, including GDF15 and TNFRSF10C [16,34,44]. Finally, we found inter-donor variability in some SASP genes, as senescent islets from Donor 2 showed significant upregulation of several CXCL family SASP genes CXCL8, CXCL1, CXCL5, CXCL6 along with MMP3 but these were not upregulated in Donor 1 (Figure 4H). Unsupervised hierarchal clustering showed that the expression profiles of SASP genes in bleomycin-induced senescent islets from Donor 1 (B1, B2, B3) and Donor 2 (B4, B5, B6) were more similar to each other than the vehicle controls (V1, V2, V3 and V4, V5, V6) and the variability in SASP gene expression between the donors was clearly apparent (Figure 4H). These results show that bleomycin-induced senescence in human islets has a limited impact on islet identity and hormone gene expression and involves changes in SASP genes.
3.5. Inter-individual variation in senescent phenotypes in human islets
To corroborate the activation of the ATM-p53-p21 pathway during bleomycin-induced senescence in human islets as indicated by RNA-seq and explore the variability of persistent DDR and SASP between islet donors, we carried out western blots for DDR and senescence proteins and Luminex assays for SASP factors at the same time-point (Figure 5A). There was significantly increased expression of phospho-ATMSer1981 and p21 in islets from a 55-year-old male donor at this time-point, but only p21 was significantly increased in the 36-year-old female donor islets, despite the same treatment (Figure 5B). However, the magnitude of increase in p21 varied dramatically between the donors (∼1.5-fold in the 55-year-old male donor versus ∼12-fold in the 36-year-old female donor) (Figure 5B). Luminex assays were carried out on conditioned media at the day 5 post-drug removal timepoint on an additional two different male donor islet preparations. While secretion of previously characterized islet SASP factors CXCL8/IL-8, CXCL1 and IGFPB4 [14] along with newly identified factors TNFRSF10C and GDF15 were all significantly increased from islets of a 32-year-old male donor, islets from a 52-year-old male donor only showed increased GDF15 (Figure 5C). Together, these data confirm the stable activation of DDR and SASP at the protein level in bleomycin-induced senescent human islets and reveal striking inter-donor variation in these phenotypes.
Figure 5.
DDR and SASP activation in senescent human islets and the EndoC-βH5 human β cell line. A) Overview of experiment on human islets. Islets from four different adult donors (ages 55, 36, 32 and 52) were treated as indicated with vehicle (DMSO) or 50 μM bleomycin for 48 h and cultured in drug-free media for an additional 4 or 5 days and harvested for either Western blot (2 donors) or Luminex assays (2 donors). B) Western blot analysis of persistent DDR on vehicle (DMSO) and bleomycin-treated islets from a 55-year-old male and a 36-year-old female at day 4 post-drug removal. Data are means ± SD of n = 3 biological replicates. C) Luminex assays for indicated SASP factors in the conditioned media from islets treated as in (A) from a 32-year-old male donor and 52-year-old male donor at day 5 post-drug removal. Data are normalized to islet RNA content and are means ± SD of n = 3 biological replicates. D) Overview of experiment on human female fetal-derived EndoC-βH5 cells. Cells were treated as indicated with 35 μM bleomycin or vehicle (DMSO) for 48 h, and then harvested for assays or cultured for an additional 2-5 days in drug-free media. Western blotting was carried out on cells after the 48 h after treatment, qRT-PCR was carried out 2 days after drug removal and Luminex assays were carried out 5 days after drug removal. E) Western blot analysis of DDR and early senescence markers in EndoC cells treated as indicated after 48 h in drug-containing media or vehicle media. Data are means ± SD of n = 3 biological replicates. F) qRT-PCR analysis of senescence genes CDKN1A, CDKN2A, prosurvival gene BCL2L1 (encoding BCL-XL) and SASP gene CXCL8 (encoding IL-8) in EndoC cells treated as in (A) at day 2 post-drug removal. Data are means ± SD of n = 3 biological replicates. G) Luminex assays of indicated SASP factors in the conditioned media from EndoC cells treated as in (A) at day 5 post-drug removal. Data are normalized to viable cell counts and are means ± SD of n = 4 biological replicates. For all panels, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, two-tailed T-tests.
3.6. DNA damage triggers senescent phenotypes in a human β cell line
Since islets are a mixed cell population, the contribution of β cells to the observed senescent phenotypes cannot be determined. Therefore, we turned to the EndoC-βH5 human β cell line model to determine whether similar phenotypes could be induced directly in human β cells experiencing DNA damage (Figure 5D). EndoC-βH5 cells [23] are the latest derivative of the widely used EndoC βH fetal β cell lines [22] and exhibit improved maturity characteristics, such as higher insulin secretion and β cell marker expression as compared with earlier EndoC cell lines. The cells were isolated from a female fetus and Cre-mediated excision of the immortalizing transgenes (SV40 Large T antigen and hTERT) renders them non-proliferative, as with the Cre-excised EndoC-βH3 line [53]. After seeding and 2 days in culture, we treated EndoC-βH5 cells for 48 h with vehicle (0.1% DMSO) or 35 μM bleomycin, followed by drug removal and additional culture for 2–5 days to monitor early and late senescence phenotypes as with islets (Figure 5D). Notably, following bleomycin treatment, these cells activated DDR signaling as judged by western blot analysis for phospho-ATMSer1981, gamma-H2A.X and p21 (Figure 5E). qRT-PCR demonstrated a sustained expression of CDKN1A two days after drug removal, but CDKN2A was not significantly affected (Figure 5F). We also found upregulation of SASP and BCL-2 family genes (CXCL8 and BCL2L1, respectively) (Figure 5F) suggesting development of late senescence phenotypes starting at 2 days post-drug removal. To determine whether SASP factor secretion was occurring, we monitored secretion of islet SASP factors CXCL8/IL-8, IGFPB4, CXCL1 and newly identified factors TNFRSF10C and GDF15 by Luminex on conditioned media at day 5 post-drug removal. Of these factors, TNFRSF10C was not detectable and only GDF15 showed significantly increased secretion (Figure 5G). Taken together, these data suggest that DNA damage with bleomcyin induces senescent phenotypes directly in human β cells.
3.7. Effect of senescence on insulin secretion in mouse and human β cell models
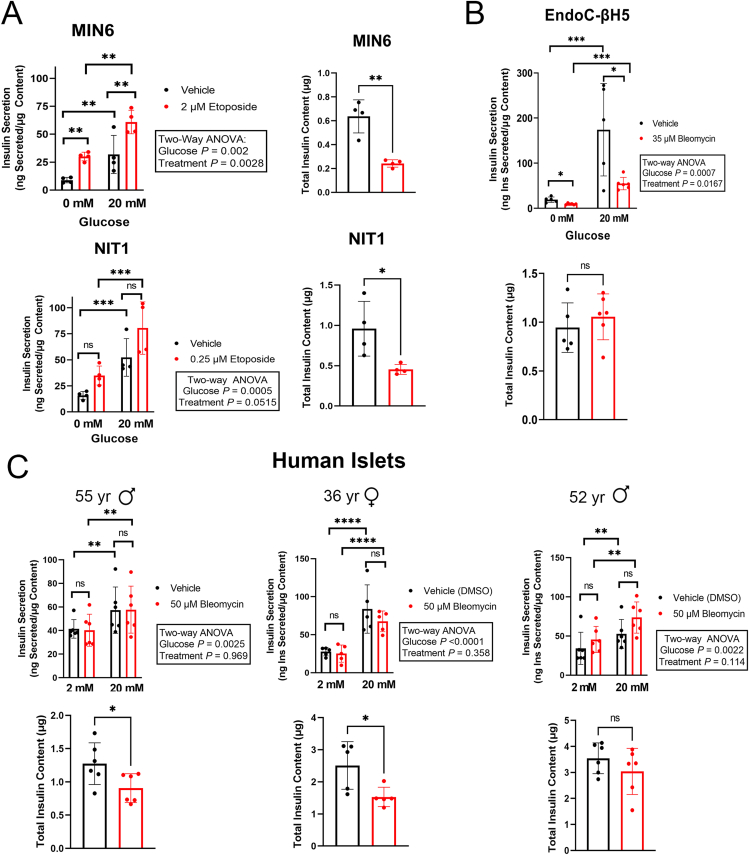
Having validated and characterized our mouse and human β cell culture models for DNA damage-induced senescence, we next used these models to investigate the effect of senescence on insulin secretion (Figure 6). Surprisingly, induction of senescence in MIN6 and NIT1 cells led to abnormally elevated glucose-stimulated insulin secretion (GSIS) from both lines under basal and high glucose conditions at the 72 h post-treatment time point (Figure 6A). Senescent MIN6 and NIT1 cells also showed significantly reduced insulin content (Figure 6A). We next assessed GSIS in human senescent EndoC-βH5 cells. We measured GSIS two days after drug removal, when the cell viability was only modestly reduced and p21 levels were markedly elevated (Supplementary Figs. 5A and 5B). In contrast to the mouse β cell lines, senescent EndoC-βH5 cells had a profound impairment in GSIS with no changes in insulin content (Figure 6B). We also compared GSIS in control and bleomycin-induced senescent human islets. At 5 days post-drug removal, the time-point at which we found that senescent islets had already activated a p53 transcriptional program (Figure 4) and show persistent DDR signaling and SASP factor secretion (Figure 5A,B) there were no significant differences in GSIS as compared to controls in three different human donor islet preparations (Figure 6C). However, two out of three donors had significantly decreased insulin content relative to controls (Figure 6C) similar to the MIN6 and NIT1 cells (Figure 6A). Taken together, these results suggest that DNA damage-induced senescence leads to different effects on GSIS and insulin content in mouse β cell lines, human EndoC-βH5 cells and human islets.
Figure 6.
Impact of DNA damage-induced senescence on GSIS and insulin content in mouse β cell lines, human EndoC-βH5 cells and human islets. A) GSIS assays and total insulin content of MIN6 or NIT1 cells treated with vehicle or etoposide (2 μM or 0.5 μM, respectively) at 72 h post-treatment. Data are means ± SD of n = 4 biological replicates. B) GSIS assays and total insulin content of EndoC-βH5 cells treated with vehicle or bleomycin (35 μM) for 72 h to induce senescence and cultured for 2 days in fresh drug free media before harvest at the day 2 post-drug removal time-point. Data are means ± SD of n = 5 biological replicates for vehicle samples or n = 6 biological replicates for bleomycin samples. C) GSIS assays and total insulin content of human islets from the three indicated donors treated with vehicle or 50 μM bleomycin for 48 h and then cultured in drug free media for 5 days and harvested at day 5 post-drug removal. Data are means ± SD of n = 5 or 6 biological replicates for each donor experiment. For GSIS panels, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, Two-way ANOVAs. For Insulin content panels, ∗p < 0.05, ∗∗p < 0.005, two-tailed T-tests.
4. Discussion
Senescent β cells accumulate during the development of T1D [7], but the triggers/stressors that lead to senescence in this setting are not known and it remains unclear how the outcomes of β cell DDR activation are modified by genetic background and other biological variables. The use of cell culture models has been indispensable for the field of senescence and has been foundational for understanding the mechanisms underpinning this complex process. Senescent β cells in T1D show markers of DNA damage and DDR activation [7,18] although the cause or trigger(s) of the DNA damage is not known. Nevertheless, since persistent unrepaired DNA damage is a potent inducer of senescence in other cell types, we carried out studies to determine the extent to which chemically induced sub-lethal DNA damage in the NIT1 cell line derived from the T1D-susceptible NOD mouse strain, different preparations of human donor islets and EndoC-βH5 cells recapitulates features of senescent β cells that accumulate during the development of T1D. We demonstrated that NIT1 cells responded to DNA double-strand break damage from low concentrations of etoposide by activating the DDR, growth arrest, developing an enlarged flattened morphology and a SASP resembling what is observed in senescent β cells of NOD mice during the development of T1D. There were some notable differences in these phenotypes as compared with MIN6 cells derived from the non-T1D susceptible C57BL6 strain, which did not secrete these SASP factors. Although they were clearly growth arrested and developed an enlarged and flattened morphology, NIT1 cells did not develop SA-βgal activity like MIN6 cells. Both cell lines had sustained upregulation of Cdkn1a expression at the mRNA level but only MIN6 upregulated Cdkn2a, yet p16Ink4a was not detected at the protein level, suggesting that the growth arrest relies on p21 rather than p16Ink4a. Furthermore, Bcl-2 was not detected in NIT1 or MIN6 cells and consistent with a lack of prosurvival phenotype, these lines showed declining viability in culture at later stages (∼60% at days 7–14 post-drug removal). Interestingly, UPR mediator IRE1α was dramatically downregulated in early senescent MIN6 but not NIT1 cells, again highlighting a difference between the strains. Taken as a whole, these results suggest that specific phenotypes of mouse β cell senescence that occur in the NOD mouse model for T1D can be modeled in NIT1 cells treated with sub-lethal concentrations of etoposide and that these phenotypes vary between different genetic backgrounds.
Human islets show substantial differences in architecture, metabolism, and proliferative capacity as compared with rodents [54], necessitating the study of β cell senescence in human models. We previously tested a human islet model for DDR and senescence using the DNA double-strand break agent bleomycin and found that it induces some of the same features observed in β cells expressing senescence markers in human T1D donors [7]. However, a comprehensive investigation of the transcriptional and protein-level response during bleomycin-induced senescence and extent of inter-donor variability in this model was lacking. Whole transcriptome analysis of two different human islet donor preparations by RNA-seq revealed a coordinated p53-mediated transcriptional program during senescence, which was largely distinct from transcriptional changes that occur in human islets during aging [50] or cytokine exposure [51]. Remarkably, islet cell hormone genes, lineage identity genes and UPR genes were largely unaffected in senescent islets although there was inter-donor variability in effects on GCG and PCSK2 expression. Only CDKN1A (p21) was upregulated without any increased expression of CDKN2A (p16Ink4a), consistent with our earlier analysis by qRT-PCR and protein-level changes in senescent β cells of T1D donors [7]. We also found evidence for two other senescence phenotypes by RNA-seq, BCL-2 prosurvival gene upregulation and SASP gene upregulation. While a limitation of our study is that we did not address the specificity of these changes to adult islet β cells versus other islet cells, we also found that bleomycin treatment induced similar senescence phenotypes in a human fetal-derived β cell line model, EndoC-βH5 cells. Together these results showed that DNA damage induces a bona fide senescence transcriptional program in adult donor islets and that human β cells can activate a DNA damage-induced senescence response in a culture model system.
By comparing the persistent DDR and SASP at the protein-level in islets isolated from different human donors we found a wide degree of variability in the extent of these phenotypes, indicating that contributions from genetic background polymorphisms and other biological variables such as age and sex modify the senescence program triggered by bleomycin. This was particularly evident in comparing the extent of p21 induction and the development of SASP. It remains to be determined the extent to which these phenotypes are modified by age and sex. Further studies to correlate donor characteristics with the extent of senescent phenotypes will be important for understanding the biological variables and genetic polymorphisms that modify DNA damage-induced senescence in human islets.
Interestingly, a subset of previously identified SASP genes encoding secreted factors and shed receptors [55] were significantly upregulated at the RNA level, including GDF15 and TNFRSF10C, which were also validated at the protein level in SASP secretion assays. GDF15 was recently identified as a β cell SASP factor in mice in a T2D model [34] and an anti-apoptotic factor in human islets, conferring protection from cytokine-induced apoptosis [56]. GDF15 has also been linked to age-related senescence and was recently suggested as part of a biomarker panel for senescence-related inflammation [44]. Given its potential role as an anti-apoptotic factor, it will be of interest to determine whether GDF15 confers a prosurvival phenotype on senescent β cells. Additionally, we found TNFRSF10C, which is a decoy receptor and also modulates cytotoxic responses [57] to be upregulated both at the RNA and the protein level during senescence induction. Thus, we demonstrated that upregulation of islet SASP components at the mRNA level, such as TNFRSF10C and GDF15, can be linked with increased SASP secretion of these proteins. Inter-donor variability was observed in the extent of SASP at the RNA and protein levels, where some donors showed increased expression or secretion of all tested SASP markers while the others had much lower expression or secretion of fewer SASP factors. During senescence in the EndoC-βH5 human β cell line, we only found GDF15 to be increased in secretion, resembling what we observed in one of the human donor islet preparations. Further investigation will be required to catalogue the similarities and differences between SASP in human islets as compared with EndoC β cell lines and it will be necessary to sort purify β cells from human islets undergoing senescence to more closely quantify differences in gene expression and SASP factor secretion.
Using our mouse β cell lines and human islet/β cell senescence culture models, we addressed how senescence impacts insulin secretion and content. Previous work in mouse models has shown that age-related and transgenic p16-induced senescence improves β cell function [58], whereas senescent β cells that accumulate during MafaS64F MODY in males had impaired function [13]. In euglycemic female NOD mice, treatment with senolytic compounds to remove senescent β cells does not significantly affect glucose tolerance in vivo or static GSIS on isolated islets ex vivo [7]. In contrast, treatment of T2D mouse models with senolytic compounds improved β cell function [12], although the effects of senolytics on senescent cells in other metabolic tissues in T2D models cannot be excluded [59,60]. These studies suggest a variety of potential effects of senescence on β cell insulin secretion, depending on the context and type of senescence and at present it is unclear how DNA damage-induced senescence may impact human β cell function in the context of T1D. In our study, GSIS assays revealed an abnormal hypersecretory response during senescence induction in MIN6 and NIT1 cells, where both basal and stimulated insulin secretion were elevated, concomitant with diminished insulin content. On the other hand, in human islet/β cell senescence models, we found a dramatic decrease in GSIS in EndoC-βH5 cells and no change in GSIS in senescent adult islets. The notable differences in GSIS between EndoC-βH5 cells and adult donor islets may be the result of the substantial differences in age and hence β cell maturation stage. Although EndoC-βH5 cells are non-proliferating and are the most mature EndoC line available, they are nevertheless derived from fetal development and hence not the same maturation stage as adult β cells. Additional contributing factors may also include differences in donor genetic background and/or the presence of other non-β cells in the adult islet. Further studies will be required to investigate the mechanisms underlying these different responses.
In conclusion, our findings demonstrate that chemically induced sub-lethal DNA double-strand break damage in mouse NIT1 cells, human donor islets and EndoC-βH5 cells, accurately models some of the key phenotypes of senescent β cells that accumulate during T1D in NOD mice and humans. These phenotypes are influenced by mouse strain genetic background and show biological variability between human islet donors and EndoC-βH5 cells. Despite their limitations, these models will be useful for dissecting the molecular mechanisms and functional consequences of β cell senescence in the pathogenesis of T1D and other forms of diabetes.
CRediT authorship contribution statement
Gabriel Brawerman: Conceptualization, Methodology, Investigation, Writing - review & editing. Jasmine Pipella: Methodology, Investigation, Peter J. Thompson: Conceptualization, Formal analysis, Methodology, Investigation, Resources, Writing - original draft, Writing - review & editing, Visualization, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Availability of data and materials
The RNA-seq raw data and processed data files described in this study are publicly available on NCBI GEO Accession number GSE176324. Expression data tables are also available in Supplementary Tables 1 and 2
Funding
This study was supported by start-up funds awarded to P.J.T. from the University of Manitoba and the Children's Hospital Research Institute of Manitoba, an operating grant from the Children's Hospital Research Institute of Manitoba (OG-2021-05) and a grant from the University of Manitoba Research Grants Program.
Acknowledgements
The authors thank Dr. Anil Bhushan at University of California, San Francisco supported by NIH R01DK121794 and R01DK118099, for providing IIDP human donor islets for P.J.T. to carry out RNA-seq (Donor 1 islet preparation). Schematic figure panels were created in BioRender.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101524.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Katsarou A., Gudbjornsdottir S., Rawshani A., Dabelea D., Bonifacio E., Anderson B.J., et al. Type 1 diabetes mellitus. Nature Reviews Disease Primers. 2017;3 doi: 10.1038/nrdp.2017.16. [DOI] [PubMed] [Google Scholar]
- 2.Eizirik D.L., Pasquali L., Cnop M. Pancreatic β-cells in type 1 and type 2 diabetes mellitus: different pathways to failure. Nature Reviews Endocrinology. 2020;16(7):349–362. doi: 10.1038/s41574-020-0355-7. [DOI] [PubMed] [Google Scholar]
- 3.Roep B.O., Thomaidou S., Tienhoven R. van., Zaldumbide A. Type 1 diabetes mellitus as a disease of the β- cell (do not blame the immune system?) Nature Reviews Endocrinology. 2020;17(3):150–161. doi: 10.1038/s41574-020-00443-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahin G.S., Lee H., Engin F. An accomplice more than a mere victim: the impact of β-cell ER stress on type 1 diabetes pathogenesis. Molecular Metabolism. 2021:101365. doi: 10.1016/j.molmet.2021.101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ovalle F., Grimes T., Xu G., Patel A.J., Grayson T.B., Thielen L.A., et al. Verapamil and beta cell function in adults with recent-onset type 1 diabetes. Nature Medicine. 2018;24(8):1108–1112. doi: 10.1038/s41591-018-0089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gitelman S.E., Bundy B.N., Ferrannini E., Lim N., Blanchfield J.L., DiMeglio L.A., et al. Imatinib therapy for patients with recent-onset type 1 diabetes: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Diabetes & Endocrinology. 2021;9(8):502–514. doi: 10.1016/S2213-8587(21)00139-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson P.J., Shah A., Ntranos V., Van Gool F., Atkinson M., Bhushan A. Targeted elimination of senescent beta cells prevents type 1 diabetes. Cell Metabolism. 2019;29(5):1045–1060. doi: 10.1016/j.cmet.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 8.He S., Sharpless N.E. Senescence in health and disease. Cell. 2017;169(6):1000–1011. doi: 10.1016/j.cell.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herranz N., Gil J. Mechanisms and functions of cellular senescence. Journal of Clinical Investigation. 2018;128(4):1238–1246. doi: 10.1172/JCI95148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helman A., Avrahami D., Klochendler A., Glaser B., Kaestner K.H., Ben-Porath I., et al. Effects of ageing and senescence on pancreatic β-cell function. Diabetes, Obesity and Metabolism. 2016;18(September):58–62. doi: 10.1111/dom.12719. [DOI] [PubMed] [Google Scholar]
- 11.Brawerman G., Thompson P.J. Beta cell therapies for preventing type 1 diabetes: from bench to bedside. Biomolecules. 2020;10(12):1–20. doi: 10.3390/biom10121681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguayo-Mazzucato C., Andle J., Lee T.B., Midha A., Talemal L., Chipashvili V., et al. Acceleration of β cell aging determines diabetes and senolysis improves disease outcomes. Cell Metabolism. 2019;30(1):129–142. doi: 10.1016/j.cmet.2019.05.006. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker E.M., Cha J., Tong X., Guo M., Liu J.-H., Yu S., et al. Sex-biased islet β cell dysfunction is caused by the MODY MAFA S64F variant by inducing premature aging and senescence in males. Cell Reports. 2021;37(2) doi: 10.1016/j.celrep.2021.109813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson P.J., Shah A., Apostolopolou H., Bhushan A. BET proteins are required for transcriptional activation of the senescent islet cell secretome in type 1 diabetes. International Journal of Molecular Sciences. 2019;20(19):1–13. doi: 10.3390/ijms20194776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez-Segura A., de Jong T.V., Melov S., Guryev V., Campisi J., Demaria M. Unmasking transcriptional heterogeneity in senescent cells. Current Biology. 2017;27(17):2652–2660. doi: 10.1016/j.cub.2017.07.033. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiley C.D., Flynn J.M., Morrissey C., Lebofsky R., Shuga J., Dong X., et al. Analysis of individual cells identifies cell-to-cell variability following induction of cellular senescence. Aging Cell. 2017;16(5):1043–1050. doi: 10.1111/acel.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dooley J., Tian L., Schonefeldt S., Delghingaro-Augusto V., Garcia-Perez J.E., Pasciuto E., et al. Genetic predisposition for beta cell fragility underlies type 1 and type 2 diabetes. Nature Genetics. 2016;48(5):519–527. doi: 10.1038/ng.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horwitz E., Krogvold L., Zhitomirsky S., Swisa A., Fischman M., Lax T., et al. Beta cell DNA damage response promotes islet inflammation in type 1 diabetes. Diabetes. 2018;67(11):2305–2311. doi: 10.2337/db17-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazaki J.I., Araki K., Yamato E., Ikegami H., Asano T., Shibasaki Y., et al. Establishment of a pancreatic β cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127(1):126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 20.Hamaguchi K., Gaskins H.R., Leiter E.H. NIT-1, a pancreatic β-cell line established from a transgenic NOD/Lt mouse. Diabetes. 1991;40(7):842–849. doi: 10.2337/diab.40.7.842. [DOI] [PubMed] [Google Scholar]
- 21.Flor A.C., Wolfgeher D., Wu D., Kron S.J. A signature of enhanced lipid metabolism, lipid peroxidation and aldehyde stress in therapy-induced senescence. Cell Death Discovery. 2017;3(1):1–12. doi: 10.1038/cddiscovery.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scharfmann R., Staels W., Albagli O. The supply chain of human pancreatic β cell lines. Journal of Clinical Investigation. 2019;129(9):3511–3520. doi: 10.1172/JCI129484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szczerbinska I., Tessitore A., Hansson L.K., Agrawal A., Lopez A.R., Helenius M., et al. Large-scale functional genomics screen to identify modulators of human β-cell insulin secretion. Biomedicines. 2022;10(1):1–13. doi: 10.3390/biomedicines10010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet Journal. 2011;17(1):10–12. [Google Scholar]
- 25.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong L., Zhang Y., Ye Z.-Q., Liu X.-Q., Zhao S.-Q., Wei L., et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Research. 2007;35(Web Server issue):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Research. 2013;41(17):e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seshadri N., Jonasson M.E., Hunt K.L., Xiang B., Cooper S., Wheeler M.B., et al. Uncoupling protein 2 regulates daily rhythms of insulin secretion capacity in MIN6 cells and isolated islets from male mice. Molecular Metabolism. 2017;6(7):760–769. doi: 10.1016/j.molmet.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon J., Manning Fox J.E., Spigelman A.F., Kim R., Smith N., O'Gorman D., et al. Research-focused isolation of human islets from donors with and without diabetes at the Alberta diabetes Institute IsletCore. Endocrinology. 2016;157(2):560–569. doi: 10.1210/en.2015-1562. [DOI] [PubMed] [Google Scholar]
- 33.Hardy A.B., Prentice K.J., Froese S., Liu Y., Andrews G.K., Wheeler M.B. Zip4 mediated zinc influx stimulates insulin secretion in pancreatic beta cells. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0119136. e0119136–e0119136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Midha A., Pan H., Abarca C., Andle J., Carapeto P., Bonner-Weir S., et al. Unique human and mouse β-cell senescence-associated secretory phenotype (SASP) reveal conserved signaling pathways and heterogeneous factors. Diabetes. 2021;70(5):1098–1116. doi: 10.2337/db20-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froelich-Ammon S.J., Osheroff N. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. Journal of Biological Chemistry. 1995;270(37):21429–21432. doi: 10.1074/jbc.270.37.21429. [DOI] [PubMed] [Google Scholar]
- 36.Brown E., Watkin D., Evans J., Yip V., Cuthbertson D.J. Multidisciplinary management of refractory insulinomas. Clinical Endocrinology. 2018;88(5):615–624. doi: 10.1111/cen.13528. [DOI] [PubMed] [Google Scholar]
- 37.Morimoto S., Tsuda M., Bunch H., Sasanuma H., Austin C., Takeda S. Type II DNA topoisomerases cause spontaneous double-strand breaks in genomic DNA. Genes. 2019;10(11):1–18. doi: 10.3390/genes10110868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montecucco A., Zanetta F., Biamonti G. Molecular mechanisms of etoposide. EXCLI Journal. 2015;14:95–108. doi: 10.17179/excli2014-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tavana O., Puebla-osorio N., Sang M., Zhu C. Nonhomologous end-joining deficiency leads to a severe diabetic phenotype in mice. Diabetes. 2010;59(January) doi: 10.2337/db09-0792.O.T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chesnokova V., Wong C., Zonis S., Gruszka A., Wawrowsky K. Diminished pancreatic beta-cell mass in securin-null mice is caused by beta-cell apoptosis and senescence. Endocrinology. 2009;150(June 2009):2603–2610. doi: 10.1210/en.2008-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beumer T.L., Gademan I.S., Rutgers D.H. P21 (Cip1/WAF1) expression in the mouse testis before. Molecular Reproduction and Development. 1997;247(January):240–247. doi: 10.1002/(SICI)1098-2795(199707)47:3<240::AID-MRD2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 42.Dai X., Zhang Q., Yu Z., Sun W., Wang R., Miao D. Bmi1 deficient mice exhibit male infertility. International Journal of Biological Sciences. 2018;14(3):358–368. doi: 10.7150/ijbs.23325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W., Guo G., Li J., Ding Z., Sheng J., Li J., et al. Activation of Bcl-2-caspase-9 apoptosis pathway in the testis of asthmatic mice. PLoS One. 2016;11(3):1–14. doi: 10.1371/journal.pone.0149353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basisty N., Kale A., Jeon O.H., Kuehnemann C., Payne T., Rao C., et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biology. 2020;18(1) doi: 10.1371/journal.pbio.3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paez-Ribes M., González-Gualda E., Doherty G.J., Muñoz-Espín D. Targeting senescent cells in translational medicine. EMBO Molecular Medicine. 2019;11(12):1–19. doi: 10.15252/emmm.201810234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prata L.G.P.L., Ovsyannikova I.G., Tchkonia T., Kirkland J.L. Senescent cell clearance by the immune system: emerging therapeutic opportunities. Seminars in Immunology. 2018;40(July 2018) doi: 10.1016/j.smim.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36(28):3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freund A., Laberge R.-M., Demaria M., Campisi J. Lamin B1 loss is a senescence-associated biomarker. Molecular Biology of the Cell. 2012;23(11):2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt A.M., Garcia J.T., Hung T., Flynn R.A., Shen Y., Qu K., et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nature Genetics. 2016;48(11):1370–1376. doi: 10.1038/ng.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seiron P., Stenwall A., Hedin A., Granlund L., Esguerra J.L.S., Volkov P., et al. Transcriptional analysis of islets of Langerhans from organ donors of different ages. PLoS One. 2021;16(3 March):1–13. doi: 10.1371/journal.pone.0247888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colli M.L., Ramos-Rodríguez M., Nakayasu E.S., Alvelos M.I., Lopes M., Hill J.L.E., et al. An integrated multi-omics approach identifies the landscape of interferon-α-mediated responses of human pancreatic beta cells. Nature Communications. 2020;11(1) doi: 10.1038/s41467-020-16327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y.C., Taylor A.J., Verchere C.B. Islet prohormone processing in health and disease. Diabetes, Obesity and Metabolism. 2018;20(May):64–76. doi: 10.1111/dom.13401. [DOI] [PubMed] [Google Scholar]
- 53.Benazra M., Lecomte M.-J., Colace C., Müller A., Machado C., Pechberty S., et al. A human beta cell line with drug inducible excision of immortalizing transgenes. Molecular Metabolism. 2015;4(12):916–925. doi: 10.1016/j.molmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steiner D.J., Kim A., Miller K., Hara M. Pancreatic islet plasticity: interspecies comparison of islet architecture and composition. Islets. 2010;2(3) doi: 10.4161/isl.2.3.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoare M., Ito Y., Kang T.-W., Weekes M.P., Matheson N.J., Patten D.A., et al. NOTCH1 mediates a switch between two distinct secretomes during senescence. Nature Cell Biology. 2016;18(9):979–992. doi: 10.1038/ncb3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakayasu E.S., Syed F., Tersey S.A., Gritsenko M.A., Mitchell H.D., Chan C.Y., et al. Comprehensive proteomics analysis of stressed human islets identifies GDF15 as a target for type 1 diabetes intervention. Cell Metabolism. 2020;31(2):363–374. doi: 10.1016/j.cmet.2019.12.005. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward-Kavanagh L.K., Lin W.W., Šedý J.R., Ware C.F. The TNF receptor superfamily in co-stimulating and co-inhibitory responses. Immunity. 2016;44(5):1005–1019. doi: 10.1016/j.immuni.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helman A., Klochendler A., Azazmeh N., Gabai Y., Horwitz E., Anzi S., et al. p16(Ink4a)-induced senescence of pancreatic beta cells enhances insulin secretion. Nature Medicine. 2016;22(4):412–420. doi: 10.1038/nm.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmer A.K., Xu M., Zhu Y., Pirtskhalava T., Weivoda M.M., Hachfeld C.M., et al. Targeting senescent cells alleviates obesity-induced metabolic dysfunction. Aging Cell. 2019;18(3):1–15. doi: 10.1111/acel.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arora S., Thompson P.J., Wang Y., Bhattacharyya A., Apostolopoulou H., Hatano R., et al. Invariant Natural Killer T cells coordinate clearance of senescent cells. Med. 2021;2(8):938–950. doi: 10.1016/j.medj.2021.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq raw data and processed data files described in this study are publicly available on NCBI GEO Accession number GSE176324. Expression data tables are also available in Supplementary Tables 1 and 2