Abstract
Sphingosine-1-phosphate (S1P) is a sphingolipid metabolite that serves as a potent extracellular signaling molecule. Metabolic regulation of extracellular S1P levels impacts key cellular activities through altered S1P receptor signaling. Although the pathway through which S1P is degraded within the cell and thereby eliminated from reuse has been previously described, the mechanism used for S1P cellular uptake and the subsequent recycling of its sphingoid base into the sphingolipid synthesis pathway is not completely understood. To identify the genes within this S1P uptake and recycling pathway, we performed a genome-wide CRISPR/Cas9 KO screen using a positive-selection scheme with Shiga toxin, which binds a cell-surface glycosphingolipid receptor, globotriaosylceramide (Gb3), and causes lethality upon internalization. The screen was performed in HeLa cells with their sphingolipid de novo pathway disabled so that Gb3 cell-surface expression was dependent on salvage of the sphingoid base of S1P taken up from the medium. The screen identified a suite of genes necessary for S1P uptake and the recycling of its sphingoid base to synthesize Gb3, including two lipid phosphatases, PLPP3 (phospholipid phosphatase 3) and SGPP1 (S1P phosphatase 1). The results delineate a pathway in which plasma membrane–bound PLPP3 dephosphorylates extracellular S1P to sphingosine, which then enters cells and is rephosphorylated to S1P by the sphingosine kinases. This rephosphorylation step is important to regenerate intracellular S1P as a branch-point substrate that can be routed either for dephosphorylation to salvage sphingosine for recycling into complex sphingolipid synthesis or for degradation to remove it from the sphingolipid synthesis pathway.
Supplementary Key words: Sphingolipids, Sphingosine phosphate, Lysophospholipid, Ceramides, CRISPR/Cas9
Abbreviations: CRISPR, clustered regularly interspersed short palindromic repeats; DKD, double knockdown; Gb3, globotriaosylceramide; GeCKO, genome-wide CRISPR/Cas9 KO; KD, knockdown; MAGeCK, model-based analysis of genome-wide CRISPR-Cas9 KO; MOI, multiplicity of infection; S1P, sphingosine-1-phosphate; sgRNA, single guide RNA; TKD, triple knockdown
Sphingolipids, a major lipid class in eukaryotic cells, are characterized by a sphingoid base backbone, often 18-carbon sphingosine. Their biosynthesis occurs through de novo synthesis and via sphingoid base salvage and recycling into the sphingolipid synthesis pathway (1, 2, 3). De novo sphingolipid synthesis takes place in the endoplasmic reticulum (ER) and is initiated by serine palmitoyltransferase. Subsequent enzyme reactions in the ER produce ceramide, composed of a sphingoid base and a fatty acid. Ceramide is modified to generate plasma membrane sphingolipids—sphingomyelin and glycosphingolipids—by the addition of hydrophilic head groups. In the salvage process, sphingoid bases derived from sphingolipids are recycled for the synthesis of ceramide.
Sphingosine-1-phosphate (S1P) is a bioactive sphingolipid metabolite that is transported into the circulation and regulates key physiological functions through interactions with five G protein—coupled receptors (4, 5). Changes in sphingolipid metabolic enzymes and transporters that modify extracellular S1P levels have dramatic physiological effects due to altered S1P receptor signaling (6, 7, 8). The rapid clearance of S1P from blood in vivo suggests that cellular-uptake mechanisms regulate its levels (9, 10, 11). Indeed, a blood S1P clearance pathway mediated by hepatocytes has been described, which leads to its degradation by S1P lyase to form phosphoethanolamine and a long-chain aldehyde and subsequently results in its removal from the sphingolipid synthesis pathway (12). However, the other possible metabolic fate of S1P taken up by cells–sphingoid base salvage for ceramide and complex sphingolipid synthesis–has not been clearly delineated. Here, we use a genome-wide clustered regularly interspersed short palindromic repeats (CRISPR)/Cas9 KO screen in HeLa cells to identify the genes that populate this pathway.
Materials and methods
Reagents
Antibodies and cell lines used are listed in supplemental Table S1. Four-well chamber plates (μ-slide 4 well, 80426) were obtained from Ibidi (Gräfelfing, Germany). Hygromycin B, puromycin, blasticidin, G418, and Lipofectamine 3000 reagent were purchased from Thermo Fisher Scientific (Waltham, MA). Human genome-wide CRISPR/Cas9 KO (GeCKOv2) CRISPR KO pooled library was a gift from Feng Zhang (Addgene, Watertown, MA; Cat# 1000000049), and the lentiviral packaging of the library was accomplished by Vector Builder (Chicago, IL). S1P was purchased from Avanti Polar Lipids (Alabaster, AL), Shiga toxin 2 from Escherichia coli was obtained from List Labs (Campbell, CA), and myriocin was obtained from Cayman Chemical (Ann Arbor, MI).
Genome-wide CRISPR/Cas9 KO library screening and single guide RNA sequence analysis
HeLa cells stably expressing Cas9 (referred as WT HeLa cells in results and figures) were transfected with the pGS-single guide RNA (sgRNA)-Neo plasmid containing the sgRNA sequence of SPTLC1 (supplemental Table S2) using Lipofectamine 3000 reagent and selected with G418 (400 μg/ml) to disrupt the SPTLC1 gene. The SPTLC1 KO HeLa cells expressing Cas9 (9.3 × 107, referred as SPTLC1 KO HeLa cells in results and figures), were transduced with the human GeCKO v2 libraries (13) to achieve a total coverage of ∼50x with a multiplicity of infection (MOI) of 0.25. Puromycin (1 μg/ml) was added 48 h after transduction. Five days after puromycin selection, one-third of the cells were frozen as input, and two-thirds of the cells were replated with 2 μM of S1P, dissolved in DMSO, added to the culture medium. The next day, one half of the cells were exposed to 2.5 ng/ml Shiga toxin for 15 days in culture. After the S1P and toxin treatment, the cells were harvested and the genomic DNA was isolated using a QIAGEN Blood & Cell Culture DNA Maxi kit (Germantown, MD) according to the manufacturer’s protocol. Genomic DNA (400 μg) from each group was used as template DNA to amplify the sgRNA region by PCR. Separate PCR reactions (80 × 100 μl) with 5 μg genomic DNA were set up using NEBNext Ultra TM II Q5 master mix (New England Biolabs, Ipswich, MA; Cat# M0544L) with the following primer set: forward 5′- AATGGACTATCATATGCTT ACCGTAACTTGAAAGTATTTCG -3’; reverse: 5′-CTTT AGTTTGTATGTCTGT TGCTATTATGTCTACTATTCT TTCCA -3’. All PCR reactions were combined and 150 μl of the combined reaction solution was purified with a Zymo-Spin V column (Zymo Research, Irvine, CA) and eluted with 150 μl of the elution buffer. A second-round PCR reaction was performed (13 × 100 μl) using the purified PCR product (10 μl) to amplify and attach Illumina compatible multiplexing sequencing adapters. Finally, PCR products were purified by gel extraction and quantified by Kapa Library Quantification qPCR (Roche, Basel, Switzerland). The libraries were subjected to single-end sequencing by Illumina NextSeq (San Diego, CA) for input, control (+S1P/-Shiga toxin), and treated (+S1P/+Shiga toxin) cells to identify the copy number of sgRNAs. Sequence reads were analyzed using the Mageck Model-based Analysis of Genome-wide CRISPR-Cas9 KO (MAGeCK) computational tool (14). The resultant scores were presented in figures by R script.
Generation of knockdown HeLa cell lines
pLentiGuide-plasmids containing sgRNA sequences of SGPP1, PLPP3 (PPAP2B), SPHK1, SPHK2, or SGPL1 (supplemental Table S2) were used for lentiviral production. The lentivirus packaging was performed by Lenti-X packaging single shots (96-well, VSV-G) and the virus was concentrated using Lenti-X Concentrator according to the manufacturer’s instructions (Takara Bio USA, San Jose, CA). HeLa cells stably expressing Cas9 (3 × 105) were transduced with lentivirus at an MOI of 10 and replated 24 h after transduction into T75 flasks. Puromycin (1 μg/ml) or blasticidin (10 μg/ml) was added 48 h after transduction. After three weeks of antibiotic selection, the loci targeted by sgRNAs were evaluated by DNA sequencing. Protein expression levels in knockown (KD) cells were validated by Western blot except for SGPP1 KD cells as there was no specific detection of SGPP1 protein. SGPP1 KD cells were validated by qPCR using TaqMan gene expression assay for SGPP1 and ACTB as a control (supplemental Table S1) on a Quant Studio 3 (Thermo Fisher Scientific).
Based on Western blot, SPHK1 and SGPL1 KD pools had negligible protein expression and were used for further experiments. For PLPP3, SPHK2, SGPP1 KD, and SPHK1/2 double knockdown (DKD) cell pools, equivalent protein levels were observed between WT and KD cells, therefore, these cells were sorted into 96-well-plates using a BD FACSAria™ III flow cytometer (BD Biosciences, San Jose, CA) for isolation of individual single cell clones. For SPHK1/2, SPTLC1 triple knockdown (TKD) cells, the pGS-sgRNA-Neo plasmid containing the sgRNA sequence of SPTLC1 (supplemental Table S2) was transfected to a SPHK1/2 DKD line using the Lipofectamine 3000 reagent, and individual clones were isolated by cloning cylinders.
Cell culture
For subcellular localization experiment, nongenetically modified HeLa cells were used. These cells were cultured in DMEM containing 10% FBS and penicillin/streptomycin (100 U/ml). For all other experiments, hygromycin B (250 μg/ml) was supplemented for HeLa cells stably expressing Cas9. KD cells were selected and cultured using the antibiotics listed in supplemental Table S2.
Western blot analysis
Protein extracts were prepared from WT and KD HeLa cells using RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific) following the manufacturer’s protocol. Equal amounts of protein (30 μg) were separated on a NuPAGE Novex 4%–12% Bis-Tris gel (Thermo Fisher Scientific) and transferred onto nitrocellulose membranes using the iBlot2 Blotting System (Thermo Fisher Scientific). Membranes were blocked in 5% nonfat dry milk for 1 h at room temperature and incubated overnight at 4°C with antibodies listed in supplemental Table S1. Membranes were then washed in 5% nonfat dry milk and incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (Millipore, Burlington, MA) in 5% nonfat dry milk. Membranes were detected using the ECL prime Western blotting system (Cytiva, Marlborough, MA) and imaged on the Amersham Imager 680 (GE Healthcare Life Sciences, Marlborough, MA). All blots were probed with β-actin (Abcam, Cambridge, UK) as a loading control. The blots were analyzed using the ImageQuant TL 8.2 image software (GE Healthcare Life Sciences).
Measurement of globotriaosylceramide synthesis from S1P uptake and recycling
SPTLC1 KO HeLa cells, SGPP1, PLPP3 (PPAP2B), SPHK1, SPHK2, or SGPL1 KD HeLa cells were cultured with myriocin (2.5 μM, 0.1% dimethylsulfoxide (DMSO)) for 7 days to inhibit de novo sphingolipid synthesis. Cells were harvested after S1P (3 μM, 0.6% DMSO) treatment for 1, 3, 5 h. For controls, HeLa cells stably expressing Cas9 were used. In some experiments, S1P was bound to carrier proteins, HDL (Lee Biosolutions, Maryland Heights, MO) (15), and fatty acid–free BSA (Sigma, St. Louis, MO) (16) and exposed to SPTLC1 KO HeLa cells and control cells for 5h. Cell-surface globotriaosylceramide (Gb3) expression was measured by flow cytometry. Cells were stained with purified anti-CD77 (Gb3) antibody (clone 5B5, BioLegend, San Diego, CA) and labeled with secondary antibody phycoerythrin-conjugated goat anti-mouse IgM (heavy chain) (supplemental Table S1). Cell-surface CD77 expression was analyzed using a BD FACSAria™ III flow cytometer and FlowJo software (FlowJo LLC, Ashland, OR).
Subcellular localization of SGPP1, PLPP3, SPHK1, SPHK2, and SGPL1
HeLa cells were transfected either with pReceiver-M29 plasmids containing eGFP-SGPP1, eGFP-SPHK1, or eGFP-SPHK2, or with pReceiver-M03 plasmids containing PLPP3 (PPAP2B)-eGFP or SGPL1-eGFP (GeneCopoeia, Rockville, MD) using the Lipofectamine 3000 reagent. The mCherry2-C1 plasmid was simultaneously transfected to label cytosol. CellLight reagents were added according to the manufacturer’s instruction to label organelles 48 h after transfection. NucBlue Live Cell Stain Ready Probe Reagent was added per the manufacturer’s instruction prior to confocal microscopy for nuclear staining. The plasmids and organelle markers are listed in supplemental Table S1. The images were captured three days after transfection with a Zeiss confocal microscope (Jena, Germany, LSM710) and were analyzed for colocalization of fluorescent markers using ZEN 2012 SP5 software (Zeiss, Jena, Germany).
Statistical analyses
GraphPad Prism (v.9, GraphPad Software, San Diego, CA) was used for graphing and statistical analyses with one-way ANOVA and unpaired t-tests. All data are presented as mean ± SD. P < 0.05 was considered to be statistically significant and the P values are indicated by asterisks in the figures.
Results
Development of assay system for monitoring S1P cellular uptake and recycling into the sphingolipid synthesis pathway
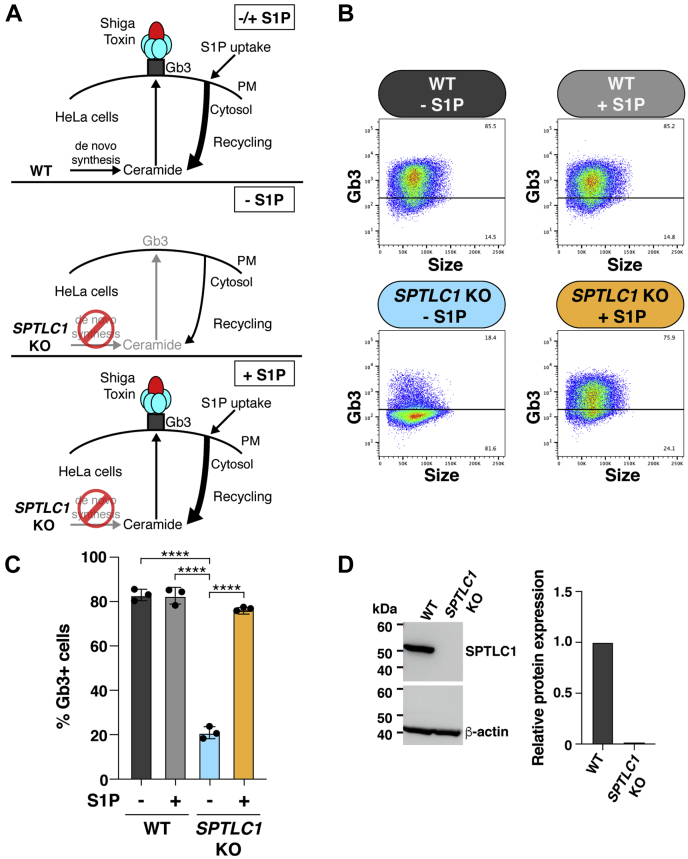
We devised a cellular assay based on the expression of a plasma membrane glycosphingolipid to monitor S1P cellular uptake and the salvage of its sphingoid base for recycling into the sphingolipid synthesis pathway. WT HeLa cells express Gb3, a glycosphingolipid that serves as a receptor for Shiga toxin, on their cell-surface (Fig. 1A [top], B and C) and are thus highly susceptible to Shiga toxin–induced cell death due to toxin uptake and subsequent protein-synthesis inhibition (17, 18, 19). We have previously shown that disabling de novo sphingolipid synthesis in HeLa cells through the KO of SPTLC1, a gene encoding an essential subunit for serine palmitoyltransferase, significantly reduced Gb3 expression (Fig. 1A [middle], B–D) and resulted in Shiga-toxin resistance (20). When the medium of SPTLC1 KO HeLa cells was supplemented with S1P, Gb3 cell-surface expression was restored to levels equivalent to those of WT HeLa cells (Fig. 1A [bottom], B and C), indicating that the sphingoid base of S1P was salvaged and recycled into the sphingolipid synthesis pathway under these conditions. The effectiveness of unbound S1P was similar to that of S1P bound to carrier proteins HDL and BSA for the stimulation of Gb3 cell-surface expression (supplemental Fig. S1).
Fig. 1.
Assay system for monitoring S1P cellular uptake and recycling into the sphingolipid synthesis pathway. A: Schematic of experimental design. Top, WT HeLa cells express Shiga toxin receptor, Gb3, on their cell surface regardless of exogenous supplementation with S1P. Middle, SPTLC1 KO HeLa cells express low levels of cell-surface Gb3. Bottom, supplementation with exogenous S1P restores Gb3 cell-surface expression in SPTLC1 KO HeLa cells to WT levels. B: Flow cytometry analysis of Gb3 cell-surface expression in WT HeLa and SPTLC1 KO HeLa cells. Top row, representative dot plots of Gb3 cell-surface expression in WT HeLa cells without S1P treatment and after 16 h of 2 μM S1P treatment. Bottom row, representative dot plots of Gb3 cell-surface expression in SPTLC1 KO HeLa cells without S1P treatment and after 16 h of 2 μM S1P treatment. C: Bar graph represents quantification of Gb3 cell-surface expression in WT HeLa and SPTLC1 KO HeLa cells determined by flow cytometry. Data represent the mean ± SD (n = 3). P values were determined by one-way ANOVA followed by Bonferroni’s multiple comparisons test; ∗∗∗∗P < 0.0001. D: Representative Western blot of SPTLC1 protein expression in WT HeLa and SPTLC1 KO HeLa cells. β-actin was used as a loading control. Bar graph presents quantification of relative SPTLC1 protein expression normalized to β-actin. SPTLC1 protein expression in WT HeLa cells is shown as 1.0. Gb3, globotriaosylceramide; PM, plasma membrane; S1P, sphingosine-1-phosphate.
Genome-wide CRISPR/Cas9 KO screening identifies regulators of S1P cellular uptake and recycling into the sphingolipid synthesis pathway
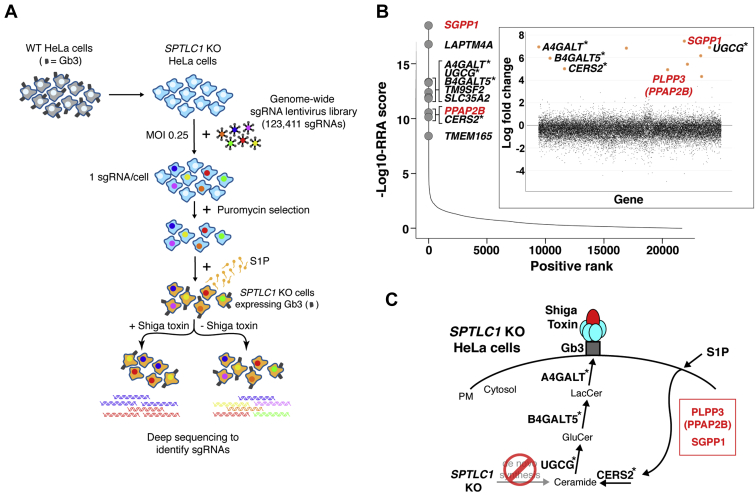
The S1P-mediated expression of Gb3 on the cell surface of SPTLC1 KO HeLa cells allowed us to undertake a genome-wide CRISPR/Cas9 KO screen to identify the genes required for S1P cellular uptake and incorporation into the sphingolipid synthesis pathway (Fig. 2A). The screen was based on the ability of Shiga toxin to kill SPTLC1 KO HeLa-Cas9 cells that express cell-surface Gb3 after exposure to S1P-supplemented medium (Fig. 1A [bottom]). We used a lentivirus-based genome-wide CRISPR/Cas9 KO (GeCKO v2) library containing 123,411 sgRNAs that target 19,050 human genes with generally six sgRNAs per gene (13). After transduction of SPTLC1 KO HeLa cells expressing Cas9 with the library at an MOI of 0.25, puromycin was used to select for cells that were successfully transduced with an sgRNA-containing lentivirus. The surviving cells were then grown in media containing S1P to induce Gb3 cell-surface expression and treated either with or without Shiga toxin. After a period of cell growth, the genomic DNA of cells in each group was subjected to deep sequencing to identify the sgRNAs.
Fig. 2.
Genome-wide CRISPR/Cas9 KO screen identifies regulators of S1P cellular uptake and recycling into the sphingolipid synthesis pathway. A: Schematic of the genome-wide CRISPR/Cas9 KO screening strategy undertaken to identify genes required for S1P uptake and recycling. WT Cas9-expressing HeLa cells (gray); SPTLC1 KO Cas9-expressing HeLa cells (light blue). Cells were transduced (MOI of 0.25) with a lentivirus-based genome-wide CRISPR/Cas9 KO library and selected in puromycin to yield cells that each contained approximately one viral genome. Cells were grown in media containing S1P (orange) to induce Gb3 cell-surface expression and treated either with or without Shiga toxin. The genomic DNA of cells from each group was subjected to deep sequencing to identify the sgRNAs. B: Scatterplot showing the ranking of genes from MAGeCK analysis. X-axis indicates positive ranking of individual genes, and y-axis indicates -Log10 values of corresponding robust ranking aggregation (RRA) score. The 10 top-ranking genes are highlighted and labeled. Inset panel, scatterplot showing log-fold change for all genes. The genes for enzymes directly involved in Gb3 synthesis are marked with asterisks. The genes shown in red are involved in the S1P uptake and recycling pathway. C: Schematic showing sphingolipid synthesis pathway leading to the production of Gb3 in SPTLC1 KO HeLa cells. Top-ranking genes from the screen that are directly involved in Gb3 synthesis are marked with asterisks. PLPP3 (PPAP2B) and SGPP1, which are not involved in de novo synthesis of Gb3 (20, 21, 22, 23), are involved in the S1P uptake and recycling pathway. CRISPR, clustered regularly interspersed short palindromic repeats; Gb3, globotriaosylceramide; MAGeCK, Model-based Analysis of Genome-wide CRISPR-Cas9 KO; MOI, multiplicity of infection; PM, plasma membrane; S1P, sphingosine-1-phosphate; sgRNA, single guide RNA.
Relative abundance of sgRNAs present in cells treated with or without Shiga toxin was compared using the MAGeCK algorithm (14). The genes represented by individual sgRNAs were ranked using the modified robust ranking aggregation score from MAGeCK analysis (Fig. 2B and supplemental Table S3). The 10 top-ranked genes included A4GALT (Gb3 synthase), B4GALT5 (lactosylceramide synthase), UGCG (glucosylceramide synthase), and CERS2 (ceramide synthase 2). Each of these genes is directly involved in the Gb3 synthesis pathway, starting with the formation of ceramide (Fig. 2C). The top 20 genes also included presumptive positive regulators of Gb3 expression, including LAPTM4A (lysosomal protein transmembrane 4 alpha), TM9SF2 (transmembrane 9 superfamily member 2), SLC35A2 (solute carrier family 35 member A2), TMEM165 (transmembrane protein 165), GOLPH3 (Golgi phosphoprotein 3), and AHR (Aryl hydrocarbon receptor) (Fig. 2B and supplemental Table S3). LAPTM4A interacts with Gb3 synthase and activates the enzyme’s activity (21, 22). TM9SF2 is a regulatory factor of endosomal trafficking and Golgi matrix assembly (21, 22). SLC35A2 transports UDP-galactose into the Golgi for glycosylation (24, 25). TMEM165, which is required for glycosylation in Golgi, is a transporter of Ca2+ and Mn2+ (26, 27). GOLPH3 mediates the retention of sphingolipid glycosyltransferases in the Golgi dictating their abundance (28). AHR is a transcription factor that elevates gene expression of the sphingolipid biosynthetic pathway (20).
The top 10 hits also included two lipid phosphatases, SGPP1 and PLPP3 (PPAP2B) (Fig. 2B, C and supplemental Table S3). SGPP1 is a S1P-specific phosphatase, and PLPP3 is a general phospholipid phosphatase that also acts on S1P. These two genes were not previously identified in Shiga toxin–based screens in cells with the de novo sphingolipid synthesis pathway intact, suggesting that they were specific for the S1P uptake and recycling pathway (20, 21, 22, 23).
PLPP3 or SGPP1 disruption reduces S1P-stimulated Gb3 cell-surface expression
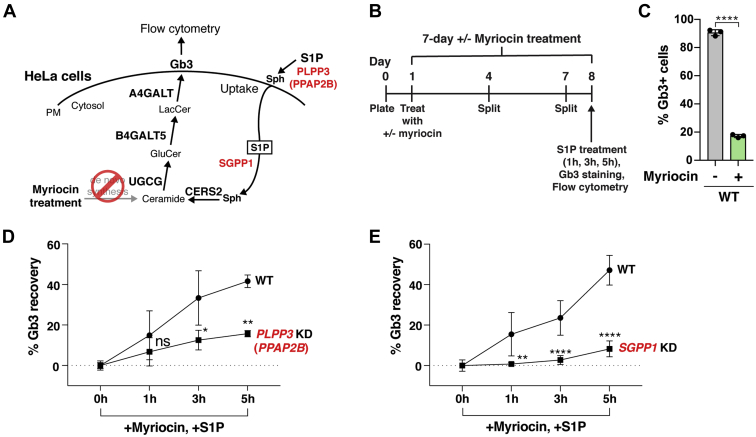
To verify that PLPP3 and SGPP1 regulate Gb3 cell-surface expression after S1P uptake, we produced PLPP3 and SGPP1 KD HeLa cell lines via Cas9-mediated gene disruption (supplemental Fig. S2A, B). Cells were treated with myriocin for 7 days to inhibit de novo sphingolipid synthesis (Fig. 3A, B), allowing Gb3 synthesis to be limited only to that which could be produced from S1P uptake and sphingoid base salvage. Without myriocin treatment, ∼90% of WT HeLa cells expressed Gb3 on their cell surface; myriocin treatment significantly reduced Gb3 cell-surface expression (Fig. 3C). When cells were then exposed to S1P for 1, 3, or 5 h and Gb3 cell-surface expression quantified, PLPP3 KD cells had significantly lower Gb3 cell-surface expression recovery after 3 h or 5 h S1P incubations than that of WT HeLa cells (Fig. 3D). SGPP1 KD cells similarly had significantly lower Gb3 cell-surface expression recovery after 1, 3, or 5 h S1P incubations than that of WT HeLa cells (Fig. 3E). These data validate the genome-wide CRISPR/Cas9 KO screening results that PLPP3 and SGPP1 are positive regulators of S1P uptake and recycling into the Gb3 sphingolipid synthesis pathway.
Fig. 3.
PLPP3 or SGPP1 disruption reduces S1P-stimulated Gb3 cell-surface expression. A: Schematic of the potential roles of PLPP3 (PPAP2B) and SGPP1 in the S1P uptake and recycling pathway. B: Timeline of experiment used to measure Gb3 recovery after 2.5 μM myriocin and 3 μM S1P treatment in KD HeLa cells and WT HeLa cells. C: Validation of myriocin inhibition of de novo Gb3 synthesis and subsequent cell-surface expression in WT HeLa cells. WT HeLa cells were cultured with 2.5 μM myriocin for 7 days and cell-surface Gb3 was measured by flow cytometry. Data represent the mean ± SD (n = 3). D: Gb3 cell-surface expression recovery rate after supplemental S1P treatment in myriocin-treated PLPP3 KD HeLa cells and WT HeLa cells, based on flow cytometry. Experiments were performed twice using one PLPP3 KD clone and WT HeLa cells (n = 6). E: Gb3 cell-surface expression recovery rate after supplemental S1P treatment in myriocin-treated SGPP1 KD HeLa cells and WT HeLa cells, based on flow cytometry. Experiments were performed three times using two SGPP1 KD clones and WT HeLa cells (n = 9 for WT, n = 12 for SGPP1). D and E: Gb3 cell-surface expression in the cells treated only with myriocin (0 h of S1P supplementation) was used as a baseline and subtracted from each data value. Experiments were conducted in triplicate wells for each condition. Data are presented as mean ± SD. P values were determined by unpaired t-tests; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001. ns, not significant. Gb3, globotriaosylceramide; KD, knockdown; PM, plasma membrane; Sph, sphingosine, S1P, sphingosine-1-phosphate.
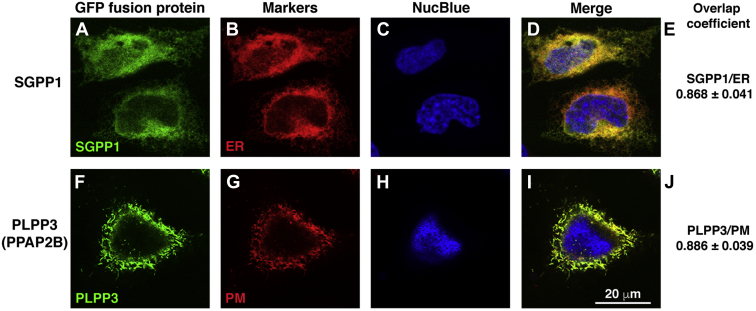
We next examined the subcellular localization of PLPP3 and SGPP1 in WT HeLa cells by coexpressing either SGPP1 or PLPP3, fused to eGFP, with red fluorescent protein–tagged organelle markers and analyzed the captured images for fluorescence colocalization using confocal fluorescence microscopy. SGPP1 appeared localized to the ER, as its fluorescent signal highly overlapped with the ER marker fluorescent signal (Fig. 4A–E). In contrast, PLPP3 highly colocalized with the plasma membrane marker fluorescent signal (Fig. 4F–J). These results show that PLPP3 and SGPP1 reside in distinct cellular compartments.
Fig. 4.
SGPP1 localizes to the ER and PLPP3 (PPAP2B) localizes to the plasma membrane. A: eGFP-SGPP1 and (F) PLPP3 (PPAP2B)-eGFP were transfected into HeLa cells along with RFP-tagged organelle markers (supplemental Table S1) for (B) ER and (G) plasma membrane (PM). C and H: Nuclei (blue) were stained by NucBlue Live Cell ReadyProbes Reagent (Hoechst 33,342). Cells were examined by confocal fluorescence microscopy. D and I: Merged images. E and J: Colocalization of fluorescent markers was determined by Manders overlap coefficient using ZEN Blue software (ZEN 2012 SP5). n=20–30 cells. RFP, red fluorescent protein.
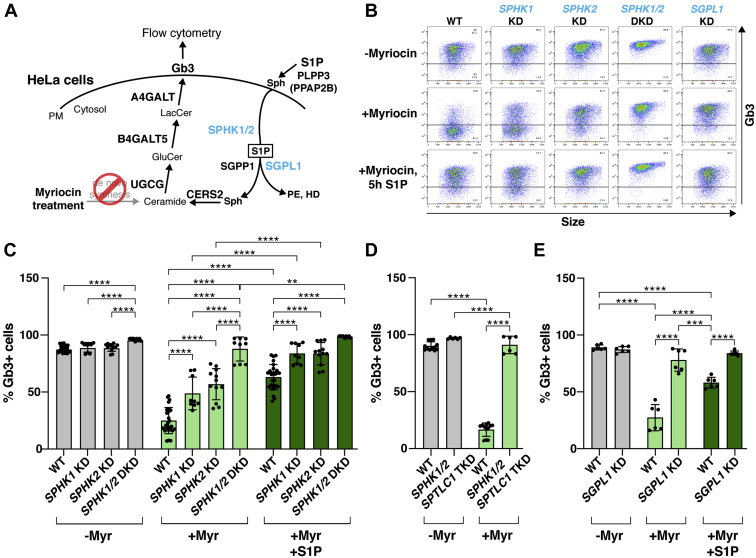
Sphingosine-kinase disruption enhances Gb3 cell-surface expression
Sphingosine kinases should connect PLPP3 and SGPP1 in the recycling pathway by rephosphorylating the sphingosine product of PLPP3 back to a S1P substrate available for SGPP1 dephosphorylation (Fig. 5A). However, the sphingosine kinases, SPHK1 and SPHK2, were not highly ranked in the genome-wide CRISPR/Cas9 KO screening we performed (i.e., they were not found in the top 100 positively ranked genes presented in supplemental Table S3). To investigate the role of sphingosine kinases in Gb3 cell-surface expression, we created individual SPHK1 KD and SPHK2 KD, as well as SPHK1/2 DKD, HeLa cells through Cas9-mediated gene disruption (supplemental Fig. S2C, D). In the absence of myriocin treatment, the SPHK1 KD, SPHK2 KD, and SPHK1/2 DKD HeLa cells all displayed Gb3 cell-surface expression comparable with that observed in WT HeLa cells (Fig. 5B, C). Extensive myriocin treatment to block de novo sphingolipid synthesis lowered Gb3 cell-surface expression in WT HeLa cells by approximately 75%. The remaining fraction of Gb3 cell-surface expression would presumably be due to salvage of sphingoid bases from residual sphingolipids in the cells and medium. Interestingly, the myriocin treatment was relatively ineffective in reducing Gb3 cell-surface expression in cells deficient in SPHKs (Fig. 5C). When SPHK1/2 DKD cells were treated with myriocin, Gb3 expression was reduced by only approximately 10%. Exposure of myriocin-treated SPHK1 KD, SPHK2 KD, and SPHK1/2 DKD HeLa cells to S1P significantly increased their Gb3 cell-surface expression compared with what was observed in myriocin-treated cells without S1P exposure (Fig. 5B, C).
Fig. 5.
SPHK1, SPHK2, or SGPL1 KD elevates Gb3 cell-surface expression. A: Schematic of the potential roles of SPHK1, SPHK2, and SGPL1 in the S1P recycling and degradation pathways. Myriocin treatment inhibits de novo Gb3 synthesis so that Gb3 synthesis via the recycling pathway can be analyzed. B: Representative dot plots of flow cytometry for WT, SPHK1 KD, SPHK2 KD, SPHK1/2 DKD, and SGPL1 KD HeLa cells under three different myriocin/S1P treatment conditions (no myriocin [7 days], 2.5 μM myriocin [7 days], 2.5 μM myriocin [7 days] followed by 3 μM S1P [5 h]). C, D, and E: Quantification of Gb3 cell-surface expression based on flow cytometry data for cells that were not treated with myriocin (gray), treated with myriocin (light green), or treated with myriocin and S1P (dark green). C: Experiments were performed using the SPHK1 KD cell pool, and two cell clones each for SPHK2 KD and SPHK1/2 DKD cells along with WT HeLa cells (WT, n = 24; SPHK1 KD, n = 6; SPHK2 KD, n = 12; SPHK1/2 DKD, n = 9). D: Experiments were performed with two cell clones of SPHK1/2;SPTLC1 TKD and WT HeLa cells (WT, n = 6; SPHK1/2;SPTLC1 TKD, n = 6). E: Experiments were performed using the SGPL1 KD cell pool and WT HeLa cells (WT, n = 6; SGPL1 KD, n = 6). C–E: All experiments for each KD cell line were conducted in triplicate wells for each condition and repeated at least twice. Data represent the mean ± SD. P values were determined by one-way ANOVA followed by Bonferroni’s multiple comparisons test; ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. DKD, double knockdown; Gb3, globotriaosylceramide; HD, hexadecenal; KD, knockdown; PE, phosphoethanolamine; PM, plasma membrane; Sph, sphingosine; S1P, sphingosine-1-phosphate; TKD, triple knockdown.
To ensure that the elevated Gb3 cell-surface expression in the SPHK KD lines was not the result of ineffective inhibition of de novo sphingolipid synthesis by myriocin, we introduced a genetic block in the de novo sphingolipid synthesis pathway in the SPHK1/2 DKD cells with the additional genetic disruption of SPTLC1 to produce TKD cells (supplemental Fig. S2C–E). Even though the disruption of SPTLC1 alone significantly reduced Gb3 cell-surface expression (Fig. 1B–D), when coupled to the disruption of both SPHK genes, significantly higher levels of Gb3 cell-surface expression were detected in myriocin-treated SPHK1/SPHK2/SPTLC1 TKD cells than in myriocin-treated WT HeLa cells (Fig. 5D). Collectively, the results with pharmacologic and genetic blocks of de novo sphingolipid synthesis indicate that sphingosine-kinase disruption enhances sphingoid base salvage for recycling into the sphingolipid synthesis pathway. This finding provides an explanation for the absence of SPHK1 and SPHK2 as top hits in the genome-wide CRISPR/Cas9 KO screen, which was geared toward detecting genes that, when knocked out, depress the incorporation of sphingoid bases into the sphingolipid synthesis pathway.
Sphingosine-kinase disruption may enhance sphingoid base salvage, and Gb3 cell-surface expression, by eliminating the S1P degradation pathway mediated by S1P lyase (SGPL1) and thereby shunting sphingoid base substrate into the sphingolipid synthesis pathway (6, 29, 30). If so, the Gb3 cell-surface expression pattern of SGPL1 KD HeLa cells should be similar to that of the SPHK1/2 DKD cells (Fig. 5C). We produced validated SGPL1 KD HeLa cells by Cas9-mediated gene disruption (supplemental Fig. S2F) and determined Gb3 cell-surface expression with and without treatment with myriocin to block de novo sphingolipid synthesis. In untreated SGPL1 KD cells, Gb3 cell-surface expression was comparable with that observed in untreated WT HeLa cells. Myriocin treatment was relatively ineffective in reducing Gb3 cell-surface expression in SGPL1 KD cells compared to myriocin-treated WT HeLa cells (Fig. 5B, E), similar to what was observed for SPHK1/2 DKD cells (Fig. 5C).
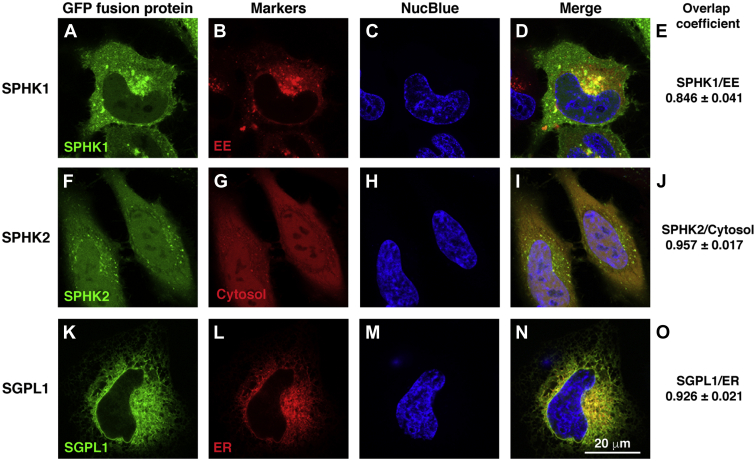
Lastly, we determined the subcellular localization of SPHK1, SPHK2, and SGPL1 by coexpressing eGFP-tagged SPHK1, SPHK2, or SGPL1 with organelle markers in WT HeLa cells and examining fluorescence colocalization. Similar to previous reports, SPHK1 was found to be largely expressed in early endosomes (31) (Fig. 6A–E). SPHK2 was expressed diffusely in the cytosol and nucleus (32, 33) (Fig. 6F–J). SGPL1 localized to the ER (Fig. 6K–O) as has been described (34). These results confirm that the sphingosine kinases are broadly expressed on vesicles and in cytoplasmic and nuclear compartments and indicate that they are poised to convert sphingosine to S1P throughout the cell. SGPL1, along with SGPP1, is confined to the ER where they compete for S1P substrate.
Fig. 6.
SPHK1 localizes to the early endosomes, SPHK2 localizes to the cytosol and nucleus, and SGPL1 localizes to the ER. A: eGFP-SPHK1, (F) eGFP-SPHK2, and (K) SGPL1-eGFP were transfected into HeLa cells along with RFP-tagged organelle markers (supplemental Table S1) for (B) early endosomes (EE) or (L) ER, or with mCherry2-C1 plasmid for (G) cytosol. C, H, and M: Nuclei (blue) were stained by NucBlue Live Cell Stain ReadyProbeReagent (Hoechst 33342). Cells were examined by confocal fluorescence microscopy. D, I, and N: Merged images. E, J, and O: Colocalization of fluorescent markers was determined by Manders overlap coefficient using ZEN Blue software (ZEN 2012 SP5). n=20–30 cells. RFP, red fluorescent protein.
Discussion
S1P is an extracellular-signaling sphingolipid whose metabolism regulates its signaling activity. It has two major intracellular metabolic fates (1, 2, 3): 1) degradation by S1P lyase and removal of sphingoid base substrate from the sphingolipid pathway or 2) salvage and recycling of its sphingoid base for sphingolipid synthesis. Recent studies have illuminated the pathway for the cellular uptake and S1P lyase–mediated degradation of extracellular S1P in hepatocytes (12). However, the pathway used for intracellular salvage of the sphingoid base from extracellular S1P and its recycling has not been clearly defined. We have used a genome-wide CRISPR/Cas9 KO screen to identify the genes responsible for this S1P cellular uptake and sphingoid base salvage for recycling into the sphingolipid synthesis pathway.
Among the genes we identified for uptake sphingoid base salvage and recycling were two distinct lipid phosphatases, PLPP3 and SGPP1. Also known as LPP3, PAP-2b, or PPAP2B, PLPP3 is an integral-membrane protein with an extracellular-facing active site that catalyzes the dephosphorylation of a variety of lipid phosphates, including S1P (35, 36, 37, 38). We found that PLPP3 was exclusively expressed at the plasma membrane in WT HeLa cells, which was in agreement with previous studies (39, 40, 41). At the hepatocyte surface, PLPP3 has been shown to dephosphorylate blood S1P, allowing sphingosine to enter hepatocytes for rephosphorylation by Sphk2 and subsequent degradation by S1P lyase (12). Similarly, in human myeloid-derived HAP1 cells, the three-member PLPP family, including PLPP3, was demonstrated to be important for the efficient handling of exogenous S1P, although some PLPP-independent uptake was also detected (42). In human lung endothelial cells, PLPP1 was found to stimulate uptake of the sphingoid base of S1P, which was then subjected to rephosphorylation by SPHK1 (43). Interestingly, adipocyte PLPP3 deficiency was found to regulate sphingolipid synthesis, resulting in reduced ceramide and sphingomyelin accumulation during adipose-tissue expansion (44). The other two PLPP family members, although expressed in HeLa cells (Human Protein Atlas proteinatlas.org), were not detected in the screen indicating that PLPP3 is dominant in initiating S1P uptake by dephosphorylation of exogenous S1P.
SGPP1 is a lipid phosphatase that specifically catalyzes dephosphorylation of S1P (45). We found that SGPP1 localized to the ER in HeLa cells, which is consistent with previous findings that SGPP1 and its homologs in yeast (46), mouse (47), and human (48) are ER integral-membrane proteins. Overexpression of SGPP1 has been shown to stimulate the incorporation of sphingosine into ceramide for the production of glycosphingolipids (49). SGPP2, while structurally and functionally similar to SGPP1 (50), is not well expressed in HeLa cells possibly explaining its absence among the hits in the screen (Human Protein Atlas proteinatlas.org).
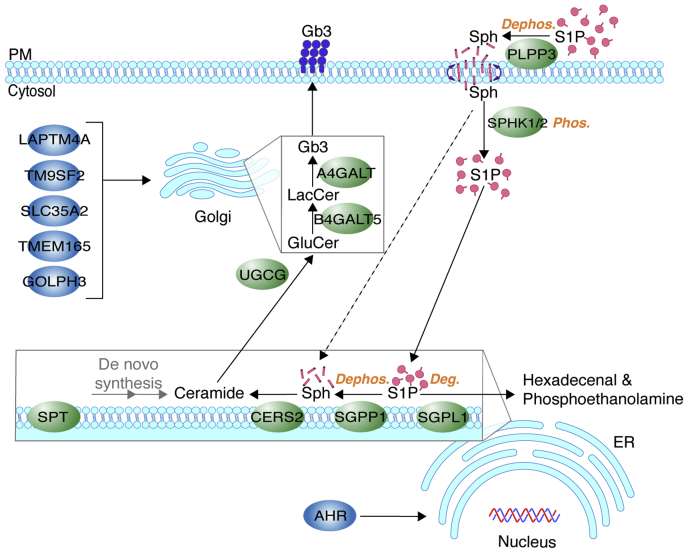
The different locations of PLPP3 and SGPP1 imply that extracellular S1P is dephosphorylated in two disparate subcellular compartments during a dephosphorylation-phosphorylation-dephosphorylation cycle prior to the incorporation of its sphingoid base into the sphingolipid synthesis pathway. In this scheme, the sphingosine product of PLPP3 generated at the plasma membrane would be rephosphorylated in the cytoplasm by the sphingosine kinases. The newly formed intracellular S1P would then be transferred to the ER surface, by a yet to-be-explained process, where it would be dephosphorylated by SGPP1 to generate sphingoid base substrate for ceramide production (Fig. 7). Other top hits in the screen included core biosynthetic enzymes responsible for Gb3 production (CERS2, UGCG, B4GALT5, A4GALT), proteins that support glycosphingolipid synthesis in the Golgi (LAPTM4A, TM9SF2, SLC35A2, TMEM165, GOLPH3), and a transcription factor (AHR) that regulates glycosphingolipid synthesis gene expression (Fig. 7).
Fig. 7.
Proposed pathway for S1P cellular uptake and incorporation into the sphingolipid synthesis pathway. Plasma membrane–resident PLPP3 dephosphorylates exogenous S1P to sphingosine, which enters cells by a flip-flop mechanism and is phosphorylated to S1P by SPHKs 1 and 2. Intracellular S1P serves as a branch-point substrate for ER-resident SGPP1 and SGPL1. SGPP1 dephosphorylates S1P to sphingosine, which is utilized for ceramide synthesis (CERS2) and sequential modifications for glycosphingolipid production. SGPL1 irreversibly degrades S1P to hexadecenal and phosphoethanolamine, allowing exit of the substrate from the sphingolipid synthesis pathway. These enzymes work in concert to drive an S1P dephosphorylation-phosphorylation-dephosphorylation/degradation cycle. In the absence of sphingosine kinases, sphingosine may bypass the cycle and directly serve as a CERS2 substrate (dashed line), thereby being excluded from the SGPL1-mediated degradation pathway. UGCG, B4GALT5, and A4GALT are Golgi core glycosphingolipid synthesis enzymes for synthesis of Gb3. LAPTM4A, TM9SF2, SLC35A2, TMEM165, and GOLPH3 are positive regulators of Gb3 synthesis in the Golgi. AHR is a transcriptional activator of genes involved in glycosphingolipid synthesis. Deg, degradation; Dephos, dephosphorylation; Gb3, globotriaosylceramide; Phos, phosphorylation; PM, plasma membrane; Sph, sphingosine; S1P, sphingosine-1-phosphate
In yeast, previous studies have indicated that exogenous sphingoid bases undergo a cycle of phosphorylation and dephosphorylation to be efficiently incorporated into ceramides and sphingolipids (51, 52, 53, 54). Although this cycle can be bypassed, sphingolipids are synthesized less efficiently if exogenous sphingoid bases cannot be phosphorylated (51, 52, 53, 54).
Our findings indicate that, in HeLa cells, the sphingosine kinases do not appear to be needed for Gb3 synthesis via the salvage of sphingoid base from extracellular S1P and its recycling into the sphingolipid synthesis pathway. First, our genome-wide CRISPR/Cas9 KO screen identified both phosphatases as positive regulators of sphingoid base recycling into the Gb3 synthesis pathway but did not pick up the sphingosine kinases as regulators of this process. Second, knocking down sphingosine-kinase expression in cells with the de novo sphingolipid synthesis pathway disabled led to elevated Gb3 cell-surface expression, apparently due to enhanced salvage of sphingoid bases for use in sphingolipid synthesis. The enhanced Gb3 cell-surface expression in cells deficient in sphingosine kinase may be due to the inability to produce a substrate for S1P lyase, effectively blocking the degradation pathway and thus salvaging sphingoid bases for incorporation into new sphingolipids (Fig. 7, dashed-line arrow). Indeed, we found a similar enhancement of Gb3 cell-surface expression when S1P lyase was knocked down in HeLa cells. Other studies have also confirmed that disruption of S1P lyase activity stimulates sphingoid base salvage and recycling (55, 56).
S1P, when in circulation, is predominately bound to carrier proteins, HDL, and serum albumin (4). In our screen, S1P was added to HeLa cells without carrier proteins, a form that was taken up as well as carrier-bound S1P. However, under these conditions, we may not have identified cell surface receptors involved in the uptake of carrier-bound S1P for subsequent entry into the recycling pathway.
The dephosphorylation-phosphorylation-dephosphorylation cycle for extracellular S1P provides several key functions. The dephosphorylation of extracellular S1P controls its extracellular levels by allowing lipid uptake into cells. The sphingoid base liberated by PLPP3 may pass though the plasma membrane by a flip-flop mechanism as proposed for hepatocytes (12) (Fig. 7). The subsequent sphingosine kinase–mediated rephosphorylation of sphingoid bases entering cells prevents severe disturbances in endocytic trafficking and autophagic function (31, 57, 58). This rephosphorylation step also serves to produce a critical branch-point substrate (59, 60), S1P, allowing either sphingoid base removal from the sphingolipid synthesis pathway via degradation by S1P lyase or its preservation in the pathway by the second S1P dephosphorylation step in the ER for sphingoid base salvage and subsequent recycling, a concerted process that is critical for the control of sphingolipid levels (49, 55).
Data availability
All data are included in the article and supporting information.
Supplemental data
This article includes supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We thank Linda Raab for editing and suggestions for improving the article. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, USA.
Author contributions
M. K., L. H.-H., S. M., R. S., C. B., H. Z., and R. L. P. conceptualization; M. K., L. H.-H., S. M., R. S., C. B., and H. Z. investigation; M. K., L. H.-H., S. M., R. S., C. B., H. Z., and R. L. P. formal analysis; M. K., L. H.-H., and R. L. P. writing–original draft; M. K., L. H.-H., S. M., R. S., C. B., H. Z., and R. L. P. Writing—review and editing.
Funding and additional information
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplemental data
References
- 1.Merrill A.H., Jr. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 2011;111:6387–6422. doi: 10.1021/cr2002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hannun Y.A., Obeid L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandhoff R., Schulze H., Sandhoff K. Ganglioside Metabolism in Health and Disease. Prog. Mol. Biol. Transl Sci. 2018;156:1–62. doi: 10.1016/bs.pmbts.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Proia R.L., Hla T. Emerging biology of sphingosine-1-phosphate: its role in pathogenesis and therapy. J. Clin. Invest. 2015;125:1379–1387. doi: 10.1172/JCI76369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green C.D., Maceyka M., Cowart L.A., Spiegel S. Sphingolipids in metabolic disease: the good, the bad, and the unknown. Cell Metab. 2021;33:1293–1306. doi: 10.1016/j.cmet.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saba J.D. Fifty years of lyase and a moment of truth: sphingosine phosphate lyase from discovery to disease. J. Lipid Res. 2019;60:456–463. doi: 10.1194/jlr.S091181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olivera A., Allende M.L., Proia R.L. Shaping the landscape: metabolic regulation of S1P gradients. Biochim. Biophys. Acta. 2013;1831:193–202. doi: 10.1016/j.bbalip.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baeyens A.A.L., Schwab S.R. Finding a way out: S1P signaling and immune cell migration. Annu. Rev. Immunol. 2020;38:759–784. doi: 10.1146/annurev-immunol-081519-083952. [DOI] [PubMed] [Google Scholar]
- 9.Peest U., Sensken S.C., Andreani P., Hanel P., Van Veldhoven P.P., Graler M.H. S1P-lyase independent clearance of extracellular sphingosine 1-phosphate after dephosphorylation and cellular uptake. J. Cell Biochem. 2008;104:756–772. doi: 10.1002/jcb.21665. [DOI] [PubMed] [Google Scholar]
- 10.Venkataraman K., Lee Y.M., Michaud J., Thangada S., Ai Y., Bonkovsky H.L., et al. Vascular endothelium as a contributor of plasma sphingosine 1-phosphate. Circ. Res. 2008;102:669–676. doi: 10.1161/CIRCRESAHA.107.165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salous A.K., Panchatcharam M., Sunkara M., Mueller P., Dong A., Wang Y., et al. Mechanism of rapid elimination of lysophosphatidic acid and related lipids from the circulation of mice. J. Lipid Res. 2013;54:2775–2784. doi: 10.1194/jlr.M039685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharel Y., Huang T., Salamon A., Harris T.E., Santos W.L., Lynch K.R. Mechanism of sphingosine 1-phosphate clearance from blood. Biochem. J. 2020;477:925–935. doi: 10.1042/BCJ20190730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Met. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sattler K., Graler M., Keul P., Weske S., Reimann C.M., Jindrova H., et al. Defects of high-density lipoproteins in coronary artery disease caused by low sphingosine-1-phosphate content: correction by sphingosine-1-phosphate-loading. J. Am. Coll. Cardiol. 2015;66:1470–1485. doi: 10.1016/j.jacc.2015.07.057. [DOI] [PubMed] [Google Scholar]
- 16.Lee M.J., Van Brocklyn J.R., Thangada S., Liu C.H., Hand A.R., Menzeleev R., et al. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 1998;279:1552–1555. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- 17.Jacewicz M., Clausen H., Nudelman E., Donohue-Rolfe A., Keusch G.T. Pathogenesis of shigella diarrhea. XI. Isolation of a shigella toxin-binding glycolipid from rabbit jejunum and HeLa cells and its identification as globotriaosylceramide. J. Exp. Med. 1986;163:1391–1404. doi: 10.1084/jem.163.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keusch G.T., Jacewicz M., Acheson D.W., Donohue-Rolfe A., Kane A.V., McCluer R.H. Globotriaosylceramide, Gb3, is an alternative functional receptor for Shiga-like toxin 2e. Infect. Immun. 1995;63:1138–1141. doi: 10.1128/iai.63.3.1138-1141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindberg A.A., Brown J.E., Stromberg N., Westling-Ryd M., Schultz J.E., Karlsson K.A. Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriae type 1. J. Biol. Chem. 1987;262:1779–1785. [PubMed] [Google Scholar]
- 20.Majumder S., Kono M., Lee Y.T., Byrnes C., Li C., Tuymetova G., et al. A genome-wide CRISPR/Cas9 screen reveals that the aryl hydrocarbon receptor stimulates sphingolipid levels. J. Biol. Chem. 2020;295:4341–4349. doi: 10.1074/jbc.AC119.011170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian S., Muneeruddin K., Choi M.Y., Tao L., Bhuiyan R.H., Ohmi Y., et al. Genome-wide CRISPR screens for Shiga toxins and ricin reveal Golgi proteins critical for glycosylation. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaji T., Sekizuka T., Tachida Y., Sakuma C., Morimoto K., Kuroda M., et al. A CRISPR screen identifies LAPTM4A and TM9SF proteins as glycolipid-regulating factors. iScience. 2019;11:409–424. doi: 10.1016/j.isci.2018.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pacheco A.R., Lazarus J.E., Sit B., Schmieder S., Lencer W.I., Blondel C.J., et al. CRISPR screen reveals that EHEC's T3SS and Shiga toxin rely on shared host factors for infection. mBio. 2018;9 doi: 10.1128/mBio.01003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witters P., Tahata S., Barone R., Ounap K., Salvarinova R., Gronborg S., et al. Clinical and biochemical improvement with galactose supplementation in SLC35A2-CDG. Genet. Med. 2020;22:1102–1107. doi: 10.1038/s41436-020-0767-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yates T.M., Suri M., Desurkar A., Lesca G., Wallgren-Pettersson C., Hammer T.B., et al. SLC35A2-related congenital disorder of glycosylation: defining the phenotype. Eur. J. Paediatr. Neurol. 2018;22:1095–1102. doi: 10.1016/j.ejpn.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Potelle S., Morelle W., Dulary E., Duvet S., Vicogne D., Spriet C., et al. Glycosylation abnormalities in Gdt1p/TMEM165 deficient cells result from a defect in Golgi manganese homeostasis. Hum. Mol. Genet. 2016;25:1489–1500. doi: 10.1093/hmg/ddw026. [DOI] [PubMed] [Google Scholar]
- 27.Stribny J., Thines L., Deschamps A., Goffin P., Morsomme P. The human Golgi protein TMEM165 transports calcium and manganese in yeast and bacterial cells. J. Biol. Chem. 2020;295:3865–3874. doi: 10.1074/jbc.RA119.012249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizzo R., Russo D., Kurokawa K., Sahu P., Lombardi B., Supino D., et al. Golgi maturation-dependent glycoenzyme recycling controls glycosphingolipid biosynthesis and cell growth via GOLPH3. EMBO J. 2021;40 doi: 10.15252/embj.2020107238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kihara A. Sphingosine 1-phosphate is a key metabolite linking sphingolipids to glycerophospholipids. Biochim. Biophys. Acta. 2014;1841:766–772. doi: 10.1016/j.bbalip.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Maceyka M., Harikumar K.B., Milstien S., Spiegel S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012;22:50–60. doi: 10.1016/j.tcb.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen H., Giordano F., Wu Y., Chan J., Zhu C., Milosevic I., et al. Coupling between endocytosis and sphingosine kinase 1 recruitment. Nat. Cell Biol. 2014;16:652–662. doi: 10.1038/ncb2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hait N.C., Allegood J., Maceyka M., Strub G.M., Harikumar K.B., Singh S.K., et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igarashi N., Okada T., Hayashi S., Fujita T., Jahangeer S., Nakamura S. Sphingosine kinase 2 is a nuclear protein and inhibits DNA synthesis. J. Biol. Chem. 2003;278:46832–46839. doi: 10.1074/jbc.M306577200. [DOI] [PubMed] [Google Scholar]
- 34.Ikeda M., Kihara A., Igarashi Y. Sphingosine-1-phosphate lyase SPL is an endoplasmic reticulum-resident, integral membrane protein with the pyridoxal 5'-phosphate binding domain exposed to the cytosol. Biochem. Biophys. Res. Commun. 2004;325:338–343. doi: 10.1016/j.bbrc.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 35.Hooks S.B., Ragan S.P., Lynch K.R. Identification of a novel human phosphatidic acid phosphatase type 2 isoform. FEBS Lett. 1998;427:188–192. doi: 10.1016/s0014-5793(98)00421-9. [DOI] [PubMed] [Google Scholar]
- 36.Jamal Z., Martin A., Gomez-Munoz A., Brindley D.N. Plasma membrane fractions from rat liver contain a phosphatidate phosphohydrolase distinct from that in the endoplasmic reticulum and cytosol. J. Biol. Chem. 1991;266:2988–2996. [PubMed] [Google Scholar]
- 37.Kai M., Wada I., Imai S., Sakane F., Kanoh H. Cloning and characterization of two human isozymes of Mg2+-independent phosphatidic acid phosphatase. J. Biol. Chem. 1997;272:24572–24578. doi: 10.1074/jbc.272.39.24572. [DOI] [PubMed] [Google Scholar]
- 38.Roberts R., Sciorra V.A., Morris A.J. Human type 2 phosphatidic acid phosphohydrolases. Substrate specificity of the type 2a, 2b, and 2c enzymes and cell surface activity of the 2a isoform. J. Biol. Chem. 1998;273:22059–22067. doi: 10.1074/jbc.273.34.22059. [DOI] [PubMed] [Google Scholar]
- 39.Ishikawa T., Kai M., Wada I., Kanoh H. Cell surface activities of the human type 2b phosphatidic acid phosphatase. J. Biochem. 2000;127:645–651. doi: 10.1093/oxfordjournals.jbchem.a022652. [DOI] [PubMed] [Google Scholar]
- 40.Kai M., Wada I., Imai S., Sakane F., Kanoh H. Identification and cDNA cloning of 35-kDa phosphatidic acid phosphatase (type 2) bound to plasma membranes. Polymerase chain reaction amplification of mouse H2O2-inducible hic53 clone yielded the cDNA encoding phosphatidic acid phosphatase. J. Biol. Chem. 1996;271:18931–18938. doi: 10.1074/jbc.271.31.18931. [DOI] [PubMed] [Google Scholar]
- 41.Sciorra V.A., Morris A.J. Sequential actions of phospholipase D and phosphatidic acid phosphohydrolase 2b generate diglyceride in mammalian cells. Mol. Biol. Cell. 1999;10:3863–3876. doi: 10.1091/mbc.10.11.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goto H., Miyamoto M., Kihara A. Direct uptake of sphingosine-1-phosphate independent of phospholipid phosphatases. J. Biol. Chem. 2021;296:100605. doi: 10.1016/j.jbc.2021.100605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y., Kalari S.K., Usatyuk P.V., Gorshkova I., He D., Watkins T., et al. Intracellular generation of sphingosine 1-phosphate in human lung endothelial cells: role of lipid phosphate phosphatase-1 and sphingosine kinase 1. J. Biol. Chem. 2007;282:14165–14177. doi: 10.1074/jbc.M701279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Federico L., Yang L., Brandon J., Panchatcharam M., Ren H., Mueller P., et al. Lipid phosphate phosphatase 3 regulates adipocyte sphingolipid synthesis, but not developmental adipogenesis or diet-induced obesity in mice. PLoS One. 2018;13 doi: 10.1371/journal.pone.0198063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandala S.M. Sphingosine-1-phosphate phosphatases. Prostaglandins Other Lipid Mediat. 2001;64:143–156. doi: 10.1016/s0090-6980(01)00111-3. [DOI] [PubMed] [Google Scholar]
- 46.Mao C., Saba J.D., Obeid L.M. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem. J. 1999;342:667–675. [PMC free article] [PubMed] [Google Scholar]
- 47.Le Stunff H., Peterson C., Thornton R., Milstien S., Mandala S.M., Spiegel S. Characterization of murine sphingosine-1-phosphate phosphohydrolase. J. Biol. Chem. 2002;277:8920–8927. doi: 10.1074/jbc.M109968200. [DOI] [PubMed] [Google Scholar]
- 48.Johnson K.R., Johnson K.Y., Becker K.P., Bielawski J., Mao C., Obeid L.M. Role of human sphingosine-1-phosphate phosphatase 1 in the regulation of intra- and extracellular sphingosine-1-phosphate levels and cell viability. J. Biol. Chem. 2003;278:34541–34547. doi: 10.1074/jbc.M301741200. [DOI] [PubMed] [Google Scholar]
- 49.Le Stunff H., Giussani P., Maceyka M., Lepine S., Milstien S., Spiegel S. Recycling of sphingosine is regulated by the concerted actions of sphingosine-1-phosphate phosphohydrolase 1 and sphingosine kinase 2. J. Biol. Chem. 2007;282:34372–34380. doi: 10.1074/jbc.M703329200. [DOI] [PubMed] [Google Scholar]
- 50.Ogawa C., Kihara A., Gokoh M., Igarashi Y. Identification and characterization of a novel human sphingosine-1-phosphate phosphohydrolase, hSPP2. J. Biol. Chem. 2003;278:1268–1272. doi: 10.1074/jbc.M209514200. [DOI] [PubMed] [Google Scholar]
- 51.Qie L., Nagiec M.M., Baltisberger J.A., Lester R.L., Dickson R.C. Identification of a Saccharomyces gene, LCB3, necessary for incorporation of exogenous long chain bases into sphingolipids. J. Biol. Chem. 1997;272:16110–16117. doi: 10.1074/jbc.272.26.16110. [DOI] [PubMed] [Google Scholar]
- 52.Mao C., Wadleigh M., Jenkins G.M., Hannun Y.A., Obeid L.M. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J. Biol. Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- 53.Mandala S.M., Thornton R., Tu Z., Kurtz M.B., Nickels J., Broach J., et al. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc. Natl. Acad. Sci. U. S. A. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Funato K., Lombardi R., Vallee B., Riezman H. Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:7325–7334. doi: 10.1074/jbc.M209925200. [DOI] [PubMed] [Google Scholar]
- 55.Bektas M., Allende M.L., Lee B.G., Chen W., Amar M.J., Remaley A.T., et al. Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver. J. Biol. Chem. 2010;285:10880–10889. doi: 10.1074/jbc.M109.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hagen-Euteneuer N., Lutjohann D., Park H., Merrill A.H., Jr., van Echten-Deckert G. Sphingosine 1-phosphate (S1P) lyase deficiency increases sphingolipid formation via recycling at the expense of de novo biosynthesis in neurons. J. Biol. Chem. 2012;287:9128–9136. doi: 10.1074/jbc.M111.302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young M.M., Takahashi Y., Fox T.E., Yun J.K., Kester M., Wang H.G. Sphingosine kinase 1 cooperates with autophagy to maintain endocytic membrane trafficking. Cell Rep. 2016;17:1532–1545. doi: 10.1016/j.celrep.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lima S., Milstien S., Spiegel S. Sphingosine and Sphingosine Kinase 1 Involvement in Endocytic Membrane Trafficking. J. Biol. Chem. 2017;292:3074–3088. doi: 10.1074/jbc.M116.762377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez Rafael S., Vallee-Belisle A., Fabregas E., Plaxco K., Palleschi G., Ricci F. Employing the metabolic "branch point effect" to generate an all-or-none, digital-like response in enzymatic outputs and enzyme-based sensors. Anal. Chem. 2012;84:1076–1082. doi: 10.1021/ac202701c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LaPorte D.C., Walsh K., Koshland D.E., Jr. The branch point effect. Ultrasensitivity and subsensitivity to metabolic control. J. Biol. Chem. 1984;259:14068–14075. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the article and supporting information.