Summary
EGFR cell surface density, stability, internalization, and recycling can be measured by cell surface ELISA (cs-ELISA). Performing this experiment on ice impedes receptor internalization; thus the physiological cell surface receptor levels can be measured by cs-ELISA. Cell surface EGFR levels are detected by measuring Amplex Red fluorescence intensity. Although cell surface receptor levels can be measured by flow cytometry, cs-ELISA does not include cell dissociation steps that might affect cell surface receptor levels.
For complete details on the use and execution of this protocol, please refer to Kazan et al. (2019).
Subject areas: Cell Biology, Cell Membrane, Cell-based Assays, Signal Transduction, Molecular/Chemical Probes
Graphical abstract

Highlights
-
•
Cs-ELISA assesses cell surface receptor trafficking in alive adherent cells
-
•
Cell dissociation is not required to assess receptor density by cs-ELISA
-
•
Cs-ELISA is a small-scale technique to assess single receptor at once
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
EGFR cell surface density, stability, internalization, and recycling can be measured by cell surface ELISA (cs-ELISA). Performing this experiment on ice impedes receptor internalization; thus the physiological cell surface receptor levels can be measured by cs-ELISA. Cell surface EGFR levels are detected by measuring Amplex Red fluorescence intensity. Although cell surface receptor levels can be measured by flow cytometry, cs-ELISA does not include cell dissociation steps that might affect cell surface receptor levels.
Before you begin
The protocol below describes the specific steps on how to perform a cell surface-ELISA (cs-ELISA) using HeLa cells. However, we have also used this protocol for MDA-MB-231 and 4T1 breast cancer cell lines.
Cell lines
Timing: 1 week
-
1.HeLa cells infected with a non-targeting shRNA (NT control cells) or with shRNA targeting an ESCRT component (e.g., HD-PTP-, Endofin-, Hrs-, Tsg101-, Ubap1- or STAM-depleted HeLa cells) are thawed.
-
a.Allow at least two passages before seeding for a cs-ELISA experiment. HeLa cells are passaged every 48 h at a 1:3 splitting ratio.
-
a.
Cell seeding
Timing: 48 h
-
2.Seed NT control and ESCRT-depleted HeLa cells in a 24-well plate.
-
a.Cells are seeded in quadruplets (40 × 103 cells/well in 0.5 mL DMEM media/well), including an IgG Ab control.
-
b.The experiment is performed 48 h post-seeding when cells reach 80%–90% confluency. The seeding density should be adjusted for other cell lines.
-
a.
Buffer preparation
Timing: On the day of the experiment
-
3.
All washes and antibody (Ab) dilutions should be done using ice cold 1× PBS buffer supplemented with 1 mM MgCl2 and 0.1 mM CaCl2 (PBS++). The PBS++ buffer decreases the risk of cell detachment during the washing steps.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| EGFR (ICR10) used for cs-ELISA and FRIA (1:1000 dilution) | Abcam | Cat# Ab231 https://www.abcam.com/egfr-antibody-icr10-ab231.html |
| anti-Rat HRP F(ab')2 (1:1500 dilution) |
Jackson ImmunoResearch | Cat# 712-036-153 https://www.jacksonimmuno.com/catalog/products/712-036-153 |
| mouse unconjugated anti-rat monovalent F(ab’)2 (1:1000 dilution) |
Jackson ImmunoResearch | Cat# 212-005-106 https://www.jacksonimmuno.com/catalog/products/212-005-106 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Human EGF (50 ng/mL final concentration) | Gibco | Cat# PHG0311 https://www.thermofisher.com/antibody/product/Human-EGF-Recombinant-Protein/PHG0311 |
| Amplex Red reagent | Invitrogen | Cat# A12222 https://www.thermofisher.com/order/catalog/product/A12222 |
| Experimental models: Cell lines | ||
| HeLa NT | Pause Lab | N/A |
| HeLa sh54Endofin | Pause Lab | N/A |
| HeLa sh75Endofin | Pause Lab | N/A |
| HeLa sh47HD-PTP | Pause Lab | N/A |
| HeLa sh51HD-PTP | Pause Lab | N/A |
| HeLa sh98Hrs | Pause Lab | N/A |
| Other | ||
| 96-Well Solid Black Polystyrene Microplates | Corning | Cat# 3650 https://ecatalog.corning.com/life-sciences/b2b/CA/en/Microplates/Assay-Microplates/96-Well-Microplates/Corning%C2%AE-96-well-Solid-Black-and-White-Polystyrene-Microplates/p/3650 |
Step-by-step method details
Measuring EGFR plasma membrane density by cs-ELISA
ESCRT downregulation disrupts cell surface receptor density, trafficking and homeostasis (Apaja and Lukacs, 2014; Apaja et al., 2010; Kazan et al., 2021; Kharitidi et al., 2015). Here, we discuss how to measure receptor plasma membrane density at steady states (i.e., in the absence of receptor activation or inhibition) by cs-ELISA.
EGFR labeling
Timing: 2 h
EGFR is labeled with a primary Ab followed with an HRP-conjugated F(ab’)2 secondary Ab. EGFR cell surface density is measured by the fluorescence generated by the addition of Amplex Red reagent.
48 h after cell seeding:
-
1.
Wash cells once with pre-warmed PBS++ at 37°C.
-
2.
Starve cells for 2 h in pre-warmed 0.5% FBS-DMEM media (0.5 mL/well) at 37°C. Serum-starvation induces cellular stress responses and accumulate receptors at the plasma membrane.
-
3.
After starvation, cells should be assessed by a light microscope if they are in good condition, then the plate is placed on ice for 5–10 min to impede receptor internalization. The next steps are all performed on ice.
-
4.Add 300 μL/well of EGFR primary Ab diluted 1:1000 (i.e., a final Ab concentration of 1 μg/mL) in ice cold 0.5% BSA/PBS++ and incubate for 1 h on ice.
-
a.As a background signal control, an IgG isotype control should be used (same dilution as the primary Ab is prepared).
-
a.
-
5.
Wash cells 3 times with ice cold PBS++ (500 μL/well). All washes should be done gently and quickly to avoid cell detachment and wells from getting dry. It is recommended to use an aspirator.
-
6.Add 300 μL/well of horseradish peroxidase (HRP)-conjugated F(ab’)2 secondary Ab diluted 1:1500 (i.e., a final Ab concentration of 1 μg/mL) in ice cold 0.5% BSA/PBS++ and incubate for 1 h on ice.
-
a.F(ab’)2 secondary Ab is used to avoid non-specific interactions of the Ab Fc portion with cell surface proteins.
-
a.
-
7.
Wash cells at least 4 times with ice cold PBS++ (500 μL/well).
Fluorescence measurement
Timing: 15 min/24-well plate
Amplex Red reagent is a highly sensitive probe for H2O2. In the presence of HRP, Amplex Red reacts with H2O2 to produce highly fluorescent resorufin. This facilitates the measurement of receptor plasma membrane density in live cells.
-
8.Prepare Amplex Red reagent dilution as follows (for 10 mL): 9.85 mL ice cold 1× PBS, 50 μL of 10 mM stock Amplex Red reagent and 100 μL of 20 mM H2O2.
-
a.Prepare 10 mM stock Amplex Red reagent according to manufacturer’s instructions, aliquot and store at −20°C protected from light.
-
b.To prepare 20 mM H2O2 working solution: dilute 3% H2O2 stock solution in the appropriate volume of dH2O (i.e., 1 mL 20 mM H2O2= 23 μL of 3% H2O2 + 977 μL of dH2O).
-
a.
-
9.
Add 300 μL/well of the prepared Amplex Red reagent dilution and incubate for 10 min on ice. The plate should be protected from light.
-
10.After incubation, transfer 200 μL of the added Amplex Red to a 96 well BLACK plate (Corning 96-Well Solid Black Polystyrene Microplates).
-
a.It is important to transfer all the quadruplets to the 96 well BLACK plate exactly after 10 min of incubation with Amplex Red reagent. For that, allow one minute difference between quadruplets once Amplex Red reagent is added in step 8 (Figure 1).
-
a.
-
11.Cell surface receptor density is measured by detecting Amplex Red fluorescence intensity (590 nm) using a fluorescence plate reader (Tecan).
-
a.In case of a high background signal (IgG control wells), increase washes to 6 times after secondary Ab incubation and add Amplex Red reagent again.
-
b.The average signal from IgG wells (i.e., the background signal) should be subtracted from the signal of each well labeled for EGFR (i.e., from NT and shESCRT wells) (Figure 1).
-
a.
Figure 1.
Schematic representation on how to add and remove Amplex Red reagent for equal incubation times among the different samples being tested and for an accurate signal quantification
Measuring EGFR plasma membrane stability by cs-ELISA
Receptor plasma membrane stability is the measure of cell surface receptor density after a stimulatory or inhibitory signal (e.g., activating EGFR with EGF or inhibiting it with Gefitinib). Receptor plasma membrane stability is regulated by several factors, such as ligand concentration, rate of receptor internalization and recycling. Here, we discuss how to measure EGFR plasma membrane stability following EGF stimulation.
Cell seeding
Timing: 48 h
-
12.
Same as seeding for cell surface receptor density measurement. However, to measure receptor plasma membrane stability, NT control and ESCRT-depleted HeLa cells should be seeded in two 24-well plates: one plate to be stimulated with EGF and the second is unstimulated.
EGF stimulation
Timing: 20 min
-
13.
48 h after cell seeding, starve cells for 2 h in 0.5% FBS-DMEM pre-warmed media at 37°C. After starvation, one plate is stimulated with EGF (50 ng/mL prepared in 0.5% FBS-DMEM media) for 20 min at 37°C, whereas the second plate is left unstimulated. The 20 min EGF stimulation is sufficient to deliver EGFR towards the endo-lysosomal compartment, however upon ESCRT depletion EGFR trafficking is disrupted and its plasma membrane stability is affected (Kazan et al., 2021).
-
14.
After EGF stimulation, the two plates are washed twice with PBS++ and placed on ice for 5–10 min to impede receptor internalization (keep PBS++ in wells).
Receptor labeling
Same as described for cell surface density measurement.
Fluorescence measurement
Same as described for cell surface density measurement.
Analysis of receptor plasma membrane stability
After subtracting the background signal from all wells labeled for EGFR, the average signal of each quadruplet in the EGF stimulated plate is normalized to the average of each quadruplet in the unstimulated plate. Results are plotted as the percentage of remaining EGFR at the plasma membrane (Figure 2).
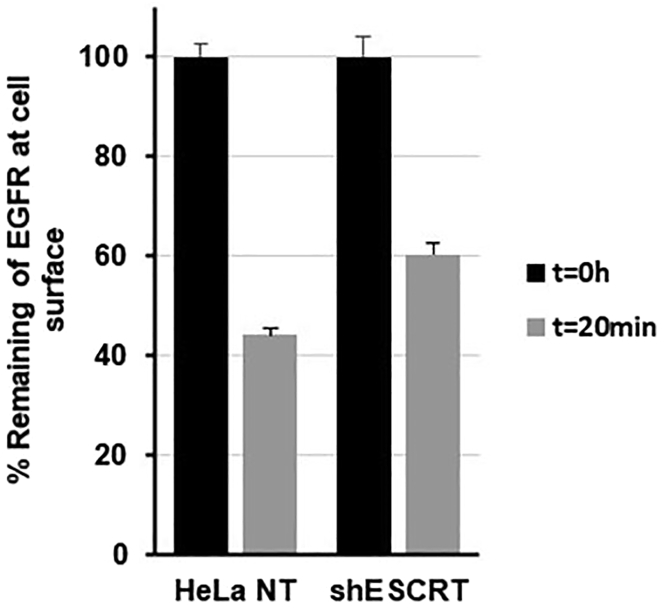
Figure 2.
Representative graph showing the plasma membrane stability of EGFR after 20 min of EGF stimulation (50 ng/mL, 37°C) in ESCRT-depleted HeLa cells (shESCRT) compared to control cells (HeLa NT)
EGFR levels after stimulation (t=20 min) are normalized to that in the unstimulated plate (t=0 h) and plotted as the percentage remaining at the cell surface. Values represent the average mean +-SEM.
Measuring EGFR internalization using cs-ELISA
Cell surface receptor is internalized via endocytosis which is catalyzed by several processes such as clathrin-dependent or -independent mechanisms (Scita and Di Fiore, 2010). EGFR internalization from the plasma membrane into the cytoplasm can be measured by cs-ELISA upon a short EGF stimulation.
Cell seeding
Same as described for cell surface stability measurement (i.e., cells should be seeded in two 24-well plates: one to be stimulated with EGF, the second is unstimulated).
EGF stimulation
Timing: 5 min
-
15.
48 h after cell seeding, starve cells for 2 h in 0.5% FBS-DMEM pre-warmed media at 37°C. After starvation, one plate is stimulated with EGF (50 ng/mL prepared in 0.5% FBS-DMEM media) for 5 min at 37°C, whereas the second plate is left unstimulated. The 5 min EGF stimulation is sufficient to induce the rapid internalization of EGFR (Kazan et al., 2021).
-
16.
After EGF stimulation, the two plates are washed twice with PBS++ and placed on ice for 5–10 min to impede receptor internalization (keep PBS++ in wells).
Receptor labeling
Same as described for cell surface density measurement.
Fluorescence measurement
Same as described for cell surface density measurement.
Analysis of cell surface receptor internalization
Same as described for cell surface stability measurement.
Measuring EGFR recycling using cs-ELISA
Upon ligand activation, several cell surface receptors, including EGFR, undergo ubiquitination and subsequent internalization, and then traverse the early endosomes (Kumari et al., 2010; Piper et al., 2014). Receptors in early endosomes can be sorted towards multivesicular bodies and then to lysosomes or recycled back to the plasma membrane via recycling endosomes (Kumari et al., 2010).
This experiment requires cell seeding in five different 24-well plates:
In plate one (P1): receptor internalization is not induced, hence recycling is not triggered. In plate two (P2): receptor internalization is induced, however, blocking is not performed, and recycling is not triggered. In plate three (P3): receptor internalization is induced, blocking is performed, but recycling is not triggered. In plate four (P4): receptor internalization is induced, blocking is performed and recycling is triggered. (P5): same as P1, however the Ab labeling is different which includes a blocking step. The blocking efficiency of the mouse anti-rat unconjugated F(ab’)2 secondary Ab will be assessed by comparing the signal of P5 to that of P1 (Table 1).
Table 1.
The different steps that should be performed for each plate used in a receptor recycling cs-ELISA assay
| Plate | Induction of receptor internalization | Blocking | Induction of receptor recycling |
|---|---|---|---|
| P1 | |||
| P2 | X | ||
| P3 | X | X | |
| P4 | X | X | X |
| P5 |
Cell seeding
Timing: 48 h
-
17.
Seed cells in quadruplets in five different 24-well plates (P1, P2, P3, P4 and P5). (Same as described for plasma membrane density measurement).
Receptor labeling
Timing: 2 h
-
18.
After starvation (as described for plasma membrane density measurement), plates are placed on ice for 5–10 min to impede receptor internalization.
-
19.Add 300 μL/well of rat anti-EGFR primary Ab diluted 1:1000 in ice cold 0.5% BSA/PBS++ and incubate for 1 h on ice.
-
a.As a background signal control, an IgG isotype control should be used in every plate (1:1000 dilution, i.e., a final Ab concentration of 1 μg/mL).
-
a.
-
20.
Wash cells 3 times with ice cold PBS++ (0.5 mL/well).
Induce receptor internalization
Timing: 10 min
-
21.
Add 1 mL of pre-warmed complete media (10% FBS-DMEM media) to P2, P3 and P4 and incubate for 10 min at 37°C to internalize receptors. Keep P1 and P5 on ice to avoid receptor internalization.
-
22.
Place P2, P3 and P4 on ice for 5–10 min to stop receptor internalization.
-
23.
Wash P2, P3 and P4 once with ice cold PBS++ (0.5 mL/well) on ice.
Blocking labeled/remaining cell surface EGFR
Timing: 1 h
-
24.
Add to P3, P4 and P5 300 μL/well of mouse anti-rat unconjugated monovalent F(ab’)2 secondary Ab 1:1000 in ice cold 0.5% BSA/PBS++, and incubate for 1 h on ice, to block remaining cell surface EGFR/Ab complexes. Keep PBS++ in the wells of P1 and P2 and keep the plates on ice.
-
25.
Wash P3, P4 and P5 five times with ice cold PBS++ (0.5 mL/well) on ice.
Induce receptor recycling
Timing: 10 min
-
26.
Add 1 mL of pre-warmed complete media (10% FBS-DMEM media) to P4 and incubate for 10 min at 37°C to trigger receptor recycling.
-
27.
Place P4 on ice for 5–10 min to stop receptor trafficking.
-
28.
Wash P4 once with ice cold PBS++ (0.5 mL/well) on ice.
Receptor labeling
Timing: 1 h
-
29.
Add 300 μL of anti-rat HRP-conjugated F(ab’)2 secondary Ab diluted 1:1500 in ice cold 0.5% BSA/PBS++ to P1, P2, P3 and P4, and incubate for 1 h on ice.
-
30.
Wash P1, P2, P3 and P4 four times with ice cold PBS++ (0.5 mL/well) on ice.
Fluorescence measurement
Same as described for plasma membrane density measurement.
Analysis of receptor plasma membrane stability
After background signal subtraction from all wells labeled for EGFR, the average signal of each quadruplet in each plate is normalized to the average signal of each quadruplet in P1 plate. Results are plotted as percentage of receptors remaining at the cell surface. To assess the blocking efficiency, the signal from P5 (with the minimum signal intensity) should be compared to P1 (with the maximum signal intensity) (Figure 3).
Figure 3.
Representative graph showing the percentage of EGFR remaining at the plasma membrane in the different plates used to detect EGFR recycling
Upon ESCRT depletion more EGFR are recycled back to the plasma membrane (P4). Values represent the average mean +-SEM.
Monitoring EGFR endocytic sorting by fluorescence ratiometric image analysis (FRIA)
This method allows tracking the post-endocytic fate of plasma membrane receptors based on pH measurement of internalized cargo-containing compartments in live cells. The detailed description of FRIA technique can be found in: (Barrière et al., 2011; Barriere and Lukacs, 2008; Kazan et al., 2019). Endocytic organelles (early endosomes, late endosomes and lysosomes) have characteristic pHs, which can be used to predict the endocytic compartment of the internalized/labeled EGFR. The recognition of EGFR is carried by a primary antibody which recognizes the extracellular domain of the receptor, followed by recognition with a pH sensitive FITC-conjugated secondary Ab. The determination of FITC fluorescence intensities at 490 nm excitation normalized to that at 450 nm excitation (i.e., ratio 490 nm/450 nm) allows to predict the pH value of the EGFR-containing vesicle in the presence of a multi-point pH calibration curve.
Expected outcomes
For plasma membrane stability measurement, it is expected to have more stable EGFR at the cell surface upon Gefitinib treatment (EGFR inhibitor) compared to control cells and upon EGF stimulation. Moreover, remaining cell surface EGFR levels in the EGF stimulated plate should be less than the levels detected in the unstimulated plate even with the Gefitinib control. The same is also expected for the internalization measurement (Kazan et al., 2021).
For receptor recycling measurement, signal in P1 should be the highest and it starts to drop as we move from P2 to P3, however if recycling occurs, a higher signal in P4 is expected compared to P3. The signal in P5 should be the lowest (Figure 3).
Limitations
Three possible limitations could render the protocol to be unreliable: 1. Cell seeding: unequal cell numbers among and between quadruplets, or variability in cell proliferation between cell lines might lead to inconsistent signal quantification. 2. Changes in temperature during Ab labeling might induce spontaneous receptor internalization. 3. Harsh washes and allowing the wells to dry during washes might lead to cell detachment and death. Plates should always be kept on ice throughout the whole experiment and buffers should always be ice cold. In addition, the user should be fast and delicate during the washing steps to avoid dryness of the wells and cell detachment.
The cs-ELISA is a small-scale application that allows the user to assess a single receptor at once. However, this technique could also be used to assess other receptors, other than EGFR, as long as the primary Ab recognizes the receptor’s extracellular domain and serves as a non-blocker to the receptor’s ligand binding site. Cs-ELISA might not be a suitable technique for cell surface proteins with a very short extracellular domain (e.g., ABC transporters).
Troubleshooting
Two main problems might arise when performing cs-ELISA:
Signal is either too high or too low.
High signal variability among quadruplets.
Cell detachment.
Cell death.
EGFR levels are too low to detect at the plasma membrane.
Problem 1
Signal is too high or too low after fluorescence measurement.
Potential solution
Antibody titration.
After purchasing the primary Ab, a titration cs-ELISA should be performed:
Same technique as described for plasma membrane density measurement; however only wild-type cells are seeded in a 24-well plate.
Different Ab dilutions should be prepared and tested (e.g., 1:250, 1:500, 1:1000, 1:2000 and 1:3000).
An IgG control should be used at the highest concentration (i.e., 1:250).
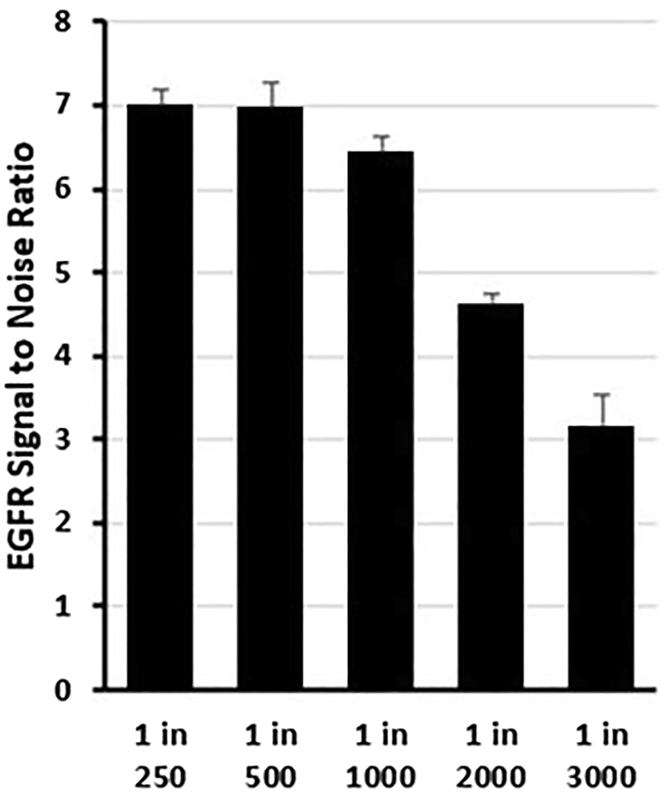
After reading Amplex Red signal, subtract the average IgG signal from all wells and plot a graph of the average signal of each Ab dilution (Figure 4). For instance, the best Ab dilution to be used in Figure 2 is 1:1000 or 1:2000. The 1:250 and 1:500 Ab dilutions show saturated signals, however 1:3000 Ab dilution shows a low signal compared to the other Ab dilutions.
Figure 4.
Primary EGFR Ab titration
EGFR signal to noise ratio is plotted, where the average Amplex Red signal from Ab dilution is normalized to the average signal of the IgG control. Values represent the average mean +-SEM.
Avoid Ab dilutions showing either a saturated signal or signal equal to that of the IgG control.
Problem 2
High signal variability among quadruplets.
Potential solution
Count cells after Amplex Red measurement.
Unequal cell seeding among wells or harsh washing steps may lead to signal variability among quadruplets. After reading Amplex Red signal, detach cells from wells by trypsin enzyme then count cells using trypan blue. Signal from each well could then be normalized to the number of viable cells.
Problem 3
Cell detachment.
Potential solution
Harsh washing steps may lead to cell detachment. To avoid cell detachment, washes should be performed using the PBS++ buffer. If cells with weak surface adhesion ability are used (e.g., 293T cells), wells could be treated with poly-L-Lysine before cell seeding.
Problem 4
Cell death.
Potential solution
Serum-starvation might decrease the viability of certain cells being used, even if the starvation is applied for 1 h only. This might induce cell detachment and negatively affects the accuracy and consistency of cs-ELISA. This could be solved by starving the cells either in 0.5% or 1% FBS supplemented media. The presence of FBS at very low concentrations maintains cell survival.
Problem 5
EGFR levels are too low to detect at the plasma membrane.
Potential solution
To assess cell surface EGFR in another cell line, we strongly recommend the inclusion of a positive control cell line (e.g., HeLa cells), which expresses high levels of EGFR at the plasma membrane.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Arnim Pause (Arnim.pause@mcgill.ca) and Gergely L. Lukacs (Gergely.lukacs@mail.mcgill).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work was supported by the Canadian Institutes for Health Research (CIHR) grant 245969 and the Canadian Cancer Society Research Institute (CCSRI) grant 245095.
Author contributions
J.M.K., A.P.B., G.L.L., and A.P. wrote and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Gergely L. Lukacs, Email: gergely.lukacs@mcgill.ca.
Arnim Pause, Email: arnim.pause@mcgill.ca.
Data and code availability
This paper does not report original code.
References
- Apaja P.M., Lukacs G.L. Protein homeostasis at the plasma membrane. Physiology. 2014;29:265–277. doi: 10.1152/physiol.00058.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apaja P.M., Xu H., Lukacs G.L. Quality control for unfolded proteins at the plasma membrane. J. Cell Biol. 2010;191:553–570. doi: 10.1083/jcb.201006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière H., Apaja P., Okiyoneda T., Lukacs G.L. Endocytic sorting of CFTR variants monitored by single-cell fluorescence ratiometric image analysis (FRIA) in living cells. Methods Mol. Biol. 2011;741:301–317. doi: 10.1007/978-1-61779-117-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere H., Lukacs G.L. Analysis of endocytic trafficking by single-cell fluorescence ratio imaging. Curr. Protoc. Cell Biol. 2008;40:15.13.1–15.13.21. doi: 10.1002/0471143030.cb1513s40. [DOI] [PubMed] [Google Scholar]
- Kazan J.M., Desrochers G., Martin C.E., Jeong H., Kharitidi D., Apaja P.M., Roldan A., Denis N.S., Gingras A.-C., Lukacs G.L. Endofin is required for HD-PTP and ESCRT-0 interdependent endosomal sorting of ubiquitinated transmembrane cargoes. iScience. 2021:103274. doi: 10.1016/j.isci.2021.103274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan J.M., Lukacs G.L., Apaja P.M., Pause A. Single cell fluorescence ratio image analysis for studying ESCRT function in receptor trafficking. Methods Mol. Biol. 2019;1998:93–103. doi: 10.1007/978-1-4939-9492-2_7. [DOI] [PubMed] [Google Scholar]
- Kharitidi D., Apaja P.M., Manteghi S., Suzuki K., Malitskaya E., Roldan A., Gingras M.-C., Takagi J., Lukacs G.L., Pause A. Interplay of endosomal pH and ligand occupancy in integrin α5β1 ubiquitination, endocytic sorting, and cell migration. Cell Rep. 2015;13:599–609. doi: 10.1016/j.celrep.2015.09.024. [DOI] [PubMed] [Google Scholar]
- Kumari S., Mg S., Mayor S. Endocytosis unplugged: multiple ways to enter the cell. Cell Res. 2010;20:256–275. doi: 10.1038/cr.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R.C., Dikic I., Lukacs G.L. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb. Perspect. Biol. 2014;6:a016808. doi: 10.1101/cshperspect.a016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scita G., Di Fiore P.P. The endocytic matrix. Nature. 2010;463:464–473. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This paper does not report original code.