Abstract
The emergence of new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants is of public health concern in case of vaccine escape. Described are 3 patients with advanced human immunodeficiency virus (HIV)-1 and chronic SARS-CoV-2 infection in whom there is evidence of selection and persistence of novel mutations that are associated with increased transmissibility and immune escape.
Keywords: SARS CoV-2, B.1.1.7, Alpha variant, immunocompromised, HIV-1 infection
Chronic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection persisting for several months has been described in immunocompromised patients [1, 2] but is less well characterized in patients with advanced human immunodeficiency virus (HIV)-1. In immunocompromised patients with prolonged virus replication, there is an optimal environment for virus evolution and the generation of novel variants [3]. This is a concern for the pandemic global response due to the possibility of vaccine escape: where novel mutations in the virus genome (in particular the spike protein [S]) lead to reduced susceptibility to neutralizing antibodies (nAbs). Novel variants with accumulated mutations in the S gene (Alpha, Beta, and Delta) have already been shown to have reduced susceptibility to nAbs in relation to wild-type (WT) Wuhan 1-Hu-1 [4, 5]. Neutralizing antibodies are predictive of immune protection from SARS-CoV-2 and correlate with virus clearance and survival [6].
Here we present 3 patients with advanced HIV-1 (CD4+ cells <200 × 106/L) and chronic SARS-CoV-2 infection in whom we have detected the emergence of novel mutations in vivo via next-generation sequencing (NGS). In 1 patient we describe the nAb response.
METHODS
Case Histories
Patient A, in their 40s, was admitted in early 2021 with shortness of breath, significant weight loss, fevers, and night sweats. SARS-CoV-2 infection was detected via reverse transcription–polymerase chain reaction (RT-PCR) of a combined nose and throat swab (CNTS). A new diagnosis of HIV-1 was confirmed during the admission. Chest radiograph changes were believed to be more consistent with miliary Mycobacterium tuberculosis than coronavirus disease 2019 (COVID-19) pneumonia. Pneumocystis jirovecii prophylaxis and empirical anti-tuberculous treatment was commenced. Antiretroviral therapy (ART) was started after discharge. This patient had persistent SARS-CoV-2 RNA detection from respiratory samples for 2 months.
Patient B is also in their 40s and has a history of diffuse large B-cell lymphoma and HIV-1. The patient was admitted in early 2021 with urosepsis managed with broad-spectrum antibiotics. The patient was diagnosed with HIV-1 20 years previously and had had poor adherence to ART. Four weeks into the admission, a CNTS was positive for SARS-CoV-2 RNA by RT-PCR. The patient required a short intensive care unit admission for high-flow oxygen. This patient had persistent detection of SARS-CoV-2 RNA for almost 3 months.
Patient C is in their 30s and was also admitted in early 2021 with a history of cough, fever, and odynophagia. HIV-1 had been diagnosed over 10 years previously and adherence to ART was intermittent since diagnosis. SARS-CoV-2 RT-PCR on a CNTS was positive on admission. The patient was swabbed weekly and SARS-CoV-2 RNA was detected from all subsequent respiratory samples for 8.5 months.
For detailed patient histories and materials and methods please refer to the Supplementary material.
RESULTS
Clinical and laboratory data (including CD4+ counts and HIV viral load) for patients A, B, and C were collected during their inpatient stay (Supplementary Table 1).
SARS-CoV-2 genome sequencing was performed on all positive samples. In all cases, sequencing confirmed B.1.1.7 (Alpha) lineage.
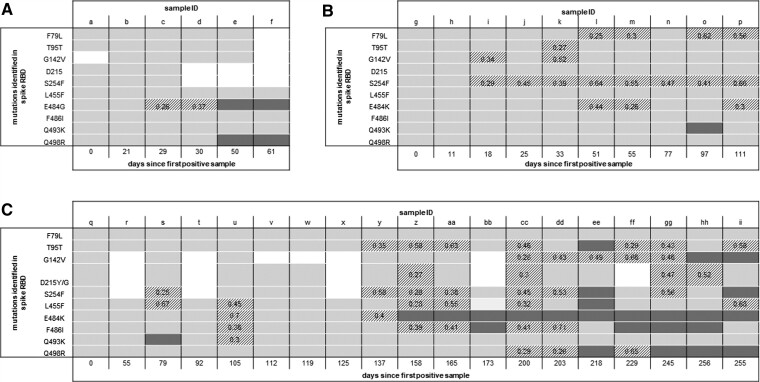
Figure 1 shows the amino acid (AA) mutations (located in the receptor binding domain [RBD] of the S protein) detected in patient samples over time. Supplementary Table 2 includes more detailed sequencing data including the AA loci, genome coverage, and read depth.
Figure 1.
Amino acid mutations in the RBD of the SARS-CoV-2 spike protein detected via NGS of respiratory samples from patient A (A), patient B (B), and patient C (C) over time since the first positive sample. Each column represents 1 sample, sample ID corresponds to sample ID in Supplementary Table 2. Light-gray boxes denote positions where >75% of reads are the wild-type allele, dark-gray boxes denote positions where >75% of reads are the mutant allele, and striped boxes denote mixed positions, with the number representing the proportion of reads with the mutant allele detected. White boxes are positions where there is no sequencing coverage. More details on genome coverage, read depth, and accession numbers are provided in Supplementary Table 2. Abbreviations: NGS, next-generation sequencing; RBD, receptor binding domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
We analyzed anti-S immunoglobulin G (IgG) using serum and ancestral strain virus neutralization for patient C. All serum samples tested were unable to neutralize Wuhan Hu-1 SARS-CoV-2 (half-maximal inhibitory concentration [IC50] <50; Supplementary Figure 1A) despite detection of anti-S IgG (>0.8; Supplementary Figure 1B).
DISCUSSION
Three patients, all with advanced HIV-1 infection, CD4+ lymphopenia, and chronic SARS-CoV-2 infection, are described. Persistent detection of SARS-CoV-2 RNA is common and does not always correspond with productive infection. All 3 patients had sufficient viral RNA detected in CNTSs sampled over months of follow-up for successful genome sequencing. This typically corresponds to a cycle threshold (Ct) value less than 30 in the majority of RT-PCR assays, and although these assays are frequently not quantitative, this normally equates to a moderate or high viral load [7]. In all 3 patients we observed the generation of novel mutations over time, some of which persisted either in a mixed population or as the majority variant (Figure 1). This persistence is evidence of active viral replication and is presumed to be due to a lack of CD4 T cells and nAbs. Virus culture was attempted and successful from 1 of patient C’s samples 245 days after the first positive sample (see Figure 1C sample “gg” for details of sequencing). This provides further evidence of replication-competent virus in this patient.
Cellular studies suggest that the mutations we observed in the recovered SARS-CoV-2 genomes from these patients will enhance transmissibility and may be associated with nAbs and vaccine escape. All patients were infected with the B.1.1.7 (Alpha) lineage that contains N501Y, a mutation associated with an approximately 10-fold higher binding affinity of the RBD for angiotensin-converting enzyme 2 (ACE2) [8]. All 3 patients developed a mutation at position E484, which is located in the receptor binding motif (Figure 1). Substitutions at E484, including E484K and E484G, enhance affinity of binding to ACE2 [9]. N501Y and E484K are together present in variant B.1.351 where a significant decrease in nAb activity from the sera of vaccinated individuals has been observed [10]. The summative effects of mutations N501Y, Q498R, and E484K/G observed in patients A and C (Figure 1A and 1C) are also predicted in vitro to increase ACE2 binding by 50-fold, compared with WT, which may translate into increased transmissibility [11]. Q493K (Figure 1B and 1C) located in the RBD has previously been detected in immunocompromised hosts and has been shown to lead to resistance to nAbs [12]. Q493K and Q498R are mutations identified in the Omicron SARS-CoV-2 variant.
There was no neutralization against WT-Wuhan SARS-CoV-2 detected from sera at different time points from patient C (Supplementary Figure 1). It is extrapolated that there will also be poor neutralization of SARS-CoV-2 B.1.1.7 lineage. The lack of neutralizing ability in patient C despite detectable anti-S antibodies emphasizes the importance of a functional assessment of the immune response in immunocompromised individuals.
Neutralizing antibody responses and CD8+ T-cell responses are CD4+ T-cell dependent [13, 14] and SARS-CoV-2–specific CD4+ cells have been associated with accelerated viral clearance [15], which may explain the observed chronic infections in our patients. Conversely, absent/delayed CD4+ cell responses have been associated with severe/fatal disease, whereas our patients had short-lived symptoms [15].
CONCLUSIONS
These 3 patients with advanced HIV-1 and chronic SARS-CoV-2 infection suggest a critical role for CD4+ T cells to nAb response and the clearance of SARS-CoV-2 infection. Our observations highlight the need for further research to understand the consequences of specific immune deficits in control of SARS-CoV-2 and other (viral) infections.
Early identification of patients with chronic SARS-CoV-2 infection is essential to prevent transmission of variants with potential for vaccine escape. Our patients highlight a novel use of viral genome-sequencing data to detect replication-competent virus in real time. We suggest that access to rapid viral sequencing in clinical diagnostic laboratories is essential in the management of this and future epidemics.
If or how patients with chronic infection influence the emergence of novel variants is unclear. Chronically infected patients pose a significant risk in resource-poor settings: their identification is challenging due to reduced access to healthcare and to RT-PCR and NGS. Furthermore, the majority of the population remain unvaccinated, and in sub-Saharan Africa there is a higher burden of people living with HIV (∼30% who are not receiving appropriate ART) [16]. This underlines the moral imperative to ensure equity of access to COVID-19 vaccines across the world.
Supplementary Material
Contributor Information
Anna C Riddell, Virology Department, Division of Infection, Barts Health NHS Trust, London, United Kingdom.
Beatrix Kele, Virology Department, Division of Infection, Barts Health NHS Trust, London, United Kingdom.
Kathryn Harris, Virology Department, Division of Infection, Barts Health NHS Trust, London, United Kingdom.
Jon Bible, Virology Department, Division of Infection, Barts Health NHS Trust, London, United Kingdom.
Maurice Murphy, Department of Infection and Immunity, Barts Health NHS Trust, London, United Kingdom.
Subathira Dakshina, Department of Infection and Immunity, Barts Health NHS Trust, London, United Kingdom.
Nathaniel Storey, Microbiology, Virology, and Infection Prevention and Control, Great Ormond Street Hospital for Children NHS Foundation Trust, London, United Kingdom.
Dola Owoyemi, Virology Department, Division of Infection, Barts Health NHS Trust, London, United Kingdom.
Corinna Pade, Blizard Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, United Kingdom.
Joseph M Gibbons, Blizard Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, United Kingdom.
David Harrington, Virology Department, Division of Infection, Barts Health NHS Trust, London, United Kingdom.
Eliza Alexander, Virology Department, Division of Infection, Barts Health NHS Trust, London, United Kingdom.
Áine McKnight, Blizard Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, United Kingdom.
Teresa Cutino-Moguel, Virology Department, Division of Infection, Barts Health NHS Trust, London, United Kingdom.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the Rare and Imported Pathogens Laboratory, Salisbury, United Kingdom, for virus culture.
Financial support. This work was supported by the Rosetrees Trust (CF1\10003 to J. B. and A. M.).
Potential conflicts of interest. M. M. reports an honorarium to self for a presentation for ViiV, February 2020, and for an HIV workshop for Janssen in November 2018 and support for attendance at the 17th EACS Conference, November 2019, from Gilead; registration for the EACS Conference, October 2021, from ViiV; and registration for the International AIDS Conference, July 2021, from ViiV. S. D. reports receiving support for attending the EACS Conference from ViiV (virtual conference) in October 2021. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Baang JH, Smith C, Miarbelli C, et al. prolonged severe acute respiraotry syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2020; 223:23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avanzato VA, Matson MJ, Seifert SN, et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immuncompromised individual with cancer. Cell 2020; 183:1901–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cele S, Karim F, Lustig G, et al. SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe 2022; 30:154–62. doi:10.1016/j.chom.2022.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Milcochova P, Kemp SA, Dhar MS, et al. SARS COV 2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021; 599:114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reynolds CJ, Pade C, Gibbons JM, et al. Prior SARS-CoV-2 infection recuses B and T cell responses to variants after first vaccine dose. Science 2021; 372:1418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dispinseri S, Secchi M, Pirillo MF, et al. Neutralizing antibody responses to SARS-COV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun 2021; 12:2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans D, Cowen S, Kammel M, et al. The dangers of using Cq to quantify nucleic acid in biological samples: a lesson from COVID-19. Clin Chem 2022; 68:153–62. [DOI] [PubMed] [Google Scholar]
- 8. Liu H, Zhang Q, Wei P, et al. The basis of a more contagious 501Y.V1 variant of SARS-CoV-2. Cell Res 2021; 31:720–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yi H, Wang J, Wang J, et al. The emergence and spread of novel SARS-CoV-2 variants. Front Public Health 2021; 9:696664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Planas D, Bruel T, Grzelak L, et al. Sensitivity of infectious SARS-CoV-2, B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 2021; 27:917–24. [DOI] [PubMed] [Google Scholar]
- 11. Zahradnik J, Marciano S, Shemesh M, et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol 2021; 6:1188–98. [DOI] [PubMed] [Google Scholar]
- 12. Clark S, Clark LE, Pan J, et al. SARS-COV-2 evolution in immuncompromised host reveals shared neutralization escape mechanisms. Cell 2021; 184:2605–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity 2019; 50:1132–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grifoni A, Weiskopf D, Ramirez S, et al. Targets of T cell responses to SARS CoV 2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020; 181:1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tan AT, Linster M, Tan CW, et al. Early induction of functional SARS CoV 2 specific T cells associated with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep 2021; 34:108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Msomi N, Lessells R, Mlisana K, de Oliveira T. Africa: tackle HIV and COVID-19 together. Nature 2021; 600:33–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.