Abstract
Background
Comparison of humoral responses in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccinees, those with SARS-CoV-2 infection, or combinations of vaccine/ infection (“hybrid immunity”) may clarify predictors of vaccine immunogenicity.
Methods
We studied 2660 US Military Health System beneficiaries with a history of SARS-CoV-2 infection-alone (n = 705), vaccination-alone (n = 932), vaccine-after-infection (n = 869), and vaccine-breakthrough-infection (n = 154). Peak anti-spike–immunoglobulin G (IgG) responses through 183 days were compared, with adjustment for vaccine product, demography, and comorbidities. We excluded those with evidence of clinical or subclinical SARS-CoV-2 reinfection from all groups.
Results
Multivariable regression results indicated that vaccine-after-infection anti-spike–IgG responses were higher than infection-alone (P < .01), regardless of prior infection severity. An increased time between infection and vaccination was associated with greater post-vaccination IgG response (P < .01). Vaccination-alone elicited a greater IgG response but more rapid waning of IgG (P < .01) compared with infection-alone (P < .01). BNT162b2 and mRNA-1273 vaccine-receipt was associated with greater IgG responses compared with JNJ-78436735 vaccine-receipt (P < .01), regardless of infection history. Those with vaccine-after-infection or vaccine-breakthrough-infection had a more durable anti-spike–IgG response compared to infection-alone (P < .01).
Conclusions
Vaccine-receipt elicited higher anti-spike–IgG responses than infection-alone, although IgG levels waned faster in those vaccinated (compared to infection-alone). Vaccine-after-infection elicits a greater humoral response compared with vaccine or infection alone; and the timing, but not disease severity, of prior infection predicted these post-vaccination IgG responses. While differences between groups were small in magnitude, these results offer insights into vaccine immunogenicity variations that may help inform vaccination timing strategies.
Keywords: SARS-CoV-2, IgG, antibody response, vaccine, vaccine breakthrough
Prior severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection followed by vaccination, or vice versa, provides a greater and more durable immunoglobulin G response than SARS-CoV-2 infection- or vaccine-induced immunity alone. The timing between SARS-CoV-2 infection and vaccination shapes the magnitude of post vaccine responses.
Coronavirus disease 2019 (COVID-19) messenger RNA (mRNA) vaccines induce severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein expression in vaccine recipients, resulting in robust humoral and T-cell responses [1]. In the United States, two mRNA vaccines were authorized in December 2020 [2–4]. Since their widespread use in the United States and elsewhere, these mRNA vaccines proved highly effective in prevention and reduced hospitalization, morbidity, and death [5–7]. The US Food and Drug Administration (FDA) also authorized use of a viral vectored vaccine using an adenovirus-26 backbone in February 2021 [8].
While there is strong evidence of COVID-19 vaccines’ effectiveness and safety [2, 3, 6, 9, 10], full characterization of the durability of vaccine-induced humoral immunity and predictors of a durable vaccine immune response remains important to investigate. Comparison of humoral immune responses of SARS-CoV-2 vaccine recipients with and without a history of SARS-CoV-2 infection to immunity induced by SARS-CoV-2 infection alone helps to identify such predictors of vaccine immune response and potentially inform vaccination timing strategies.
Prior studies noted that infection-induced and vaccine-induced immunity were durable for at least 6 months, but those vaccinated with prior infection (ie, vaccine after infection) mounted a greater and more durable level of humoral response [11–13]. While these studies emphasized the value of vaccination following a prior infection, additional data that replicate these findings add to consensus for vaccination regardless of infection history. However, several questions surround SARS-CoV-2 “hybrid immunity” that is induced by infection and subsequent vaccination. It is unclear whether initial illness severity or time between infection and subsequent vaccination modifies this post-infection vaccine immunoglobulin G (IgG) “boost.” The emergence of the Omicron variant underscores the importance of understanding predictors of durable and robust vaccine immunity, particularly as a rapidly growing proportion of the population with a history of vaccination before or after infection develops hybrid immunity.
Here, we sought to compare the magnitude and durability of vaccine-induced humoral immunity against humoral immune response elicited by SARS-CoV-2 infection, adjusting for vaccine product and vaccine recipient baseline health and age. We then confirmed whether vaccine-induced immunity following prior infection (hybrid immunity) offered more robust humoral response than that afforded by prior infection/vaccination alone and whether this was affected by host characteristics (age and comorbidities), severity or timing of the prior infection, specific product vaccine received, or number of vaccine doses received.
METHODS
Study Population and General Study Design
The Epidemiology, Immunology, and Clinical Characteristics of Emerging Infectious Diseases with Pandemic Potential (EPICC) study is a longitudinal cohort study of US Military Health System (MHS) beneficiaries with a history of SARS-CoV-2 infection and/or vaccination (see Supplementary Materials for extended details). Briefly, eligibility criteria for enrollment include those MHS beneficiaries who present to a military treatment facility (MTF) for SARS-CoV-2 polymerase chain reaction (PCR) testing. In early 2021, eligibility for EPICC was expanded to enroll those who receive an FDA-authorized SARS-CoV-2 vaccine, that is, BNT162b2 (Pfizer/BioNTech), mRNA-1273 (Moderna), or JNJ-78436735 (Janssen) within the MHS, with enrollment on the day of vaccination.
EPICC enrollment occurs at 10 MTFs: Brooke Army Medical Center, Fort Belvoir Community Hospital, Madigan Army Medical Center, Naval Medical Center Portsmouth, Naval Medical Center San Diego, Tripler Army Medical Center, Walter Reed National Military Medical Center, Carl R. Darnall Army Medical Center, William Beaumont Army Medical Center, and Womack Army Medical Center. To permit scalability of enrollment across other geographic areas (including non-US locations) and to enhance convenience to participants, beginning on 19 October 2020, MHS beneficiaries who were either tested for SARS-CoV-2 and/or vaccinated were also eligible for enrollment and followed remotely via an online pathway (Supplementary Table 1).
The EPICC study therefore enrolls and follows participants who are SARS-CoV-2 test-positive, SARS-CoV-2 test-negative, and those vaccinated with or without a history of SARS-CoV-2 infection. MHS beneficiaries enrolled in EPICC who were diagnosed with SARS-CoV-2 and/or vaccinated against SARS-CoV-2 from 20 March 2020 through 15 November 2021 were included in this analysis.
We excluded those participants who tested SARS-CoV-2–negative and had not received any vaccination. We also excluded those who received a vaccine as part of a clinical trial. In addition, children (aged <18 years) were excluded from the analysis due to later implementation of vaccines in pediatric populations (Supplementary Figure 1).
Diagnosis of SARS-CoV-2 Infection and Case Definition
SARS-CoV-2 infection was determined using a positive PCR clinical laboratory test performed at the enrolling MTF or follow-up positive upper respiratory swab collected as part of the EPICC study by day 17 post-enrollment and tested by PCR [2] and/or supplemented with self-reported testing history in online-enrolled participants (see Supplementary Materials).
Ascertainment and Definitions of COVID-19 Vaccine History
Vaccination status was established, including the date and product of vaccine receipt, through the Military Health System Data Repository (MDR) in addition to a case report form and supplemented by questionnaire self-report (see Supplementary Materials). We defined “completed primary vaccine series” by 14 days after a 2-dose series of mRNA vaccine administered 21 days (ie, BNT162b2) or 28 days (ie, mRNA-1273) apart or a single dose of viral vector vaccine (ie, JNJ-78436735).
Measurement of SARS-CoV-2 Anti-Spike–IgG Response
Anti-spike–IgG responses were measured using a multiplex microsphere-based immunoassay, described in detail previously [14, 15], which has been used in concurrent testing with the Mt. Sinai emergency use authorization enzyme-linked immunosorbent assay and performed with 99% concordance [16] (see Supplementary Materials). Antigen-specific SARS-CoV-2 spike and nucleocapsid protein (NP) reactive IgG levels were reported as a median fluorescence intensity (MFI).
Comparisons of Anti-Spike–IgG Responses to SARS-CoV-2 Infection and Vaccines
The primary outcome of interest in this analysis was peak observed SARS-CoV-2 anti-spike–IgG MFI assessed out to 183 days post-vaccine/infection exposure (ie, final vaccine date or symptom onset date, respectively). This period was constrained by the duration of post-vaccine follow-up time available. Log10 transformations were applied to anti-spike–IgG MFI values to normalize the data, and exponentiated coefficients were used to interpret the results. We compared 4 groups for anti-spike–IgG responses to SARS-CoV-2 infection and/or vaccines: infection alone (those who tested SARS-CoV-2–positive and did not receive any subsequent vaccination), vaccination alone (completed primary vaccination series without a known history of SARS-CoV-2 infection), vaccination after infection (those who tested SARS-CoV-2–positive and then received complete primary vaccination series), and vaccine breakthrough infection (those who were infected by SARS-CoV-2 two or more weeks after complete primary vaccination series). Given the small number of participants who received booster doses and limited follow-up time, these participants were excluded from analysis. We retained a subset of participants classified as vaccine after infection who received only 1 dose of mRNA vaccine (compared with 2 doses) to examine humoral immune response (see Supplementary Materials for extended details).
Adjusted Comparisons of Anti-Spike–IgG Responses to SARS-CoV-2 Infection and Vaccines and Identification of Predictors of Humoral Immune Response
We performed unadjusted and adjusted comparisons of peak observed SARS-CoV-2 anti-spike–IgG MFI between 4 groups using univariable and multivariable linear regression. These models considered other potential predictors of vaccine and/or infection humoral response, including serum specimen sampling time, that is, time between date of serum specimen collection of peak anti-spike–IgG and date of final dose of vaccine receipt or SARS-CoV-2 infection symptom onset date (whichever was latest); comorbidities; sex; age; and vaccine product received. We examined post-vaccination anti-spike–IgG response in participants with a prior infection history with a multivariable regression model to examine how severity of initial infection changed peak post-vaccination anti-spike–IgG. We conducted a Spearman correlation (ρ) to determine whether increasing time from infection to vaccination correlated with increased peak post-vaccination IgG.
We used the Charlson comorbidity index (CCI) [17] to quantify comorbidities measured in MDR or self-reported by participant. Age, sex, race and ethnicity were reported by participants. We categorized infection severity by the need for hospitalization for COVID-19. Descriptive statistics were calculated for demographic characteristics, comorbidities, and illness severity by 4 groups, with P values computed using the Fisher exact test. The nonparametric Mann–Whitney U test and Kruskal–Wallis variance analysis were used to evaluate the data. Figures were generated and statistical analyses were performed using RStudio version 4.0.2 software [18].
Repeated Measures Sensitivity Analysis
We extended the above analyses (which used a single time point anti-spike–IgG value with adjustment for sampling time after latest antigenic exposure) and confirmed findings with a repeated measures model that incorporated within-participant changes in anti-spike–IgG over time and restricted to those with two or more sera specimens collected. For this mixed linear model, we used anti-spike–IgG as the dependent variable while controlling for random effects of participants and the fixed effect of time since infection to vaccination. The coefficients derived from this estimate indicated whether vaccination alone, vaccine after infection, and vaccine breakthrough infection were associated with larger peak IgG response compared with SARS-CoV-2 infection alone. We fit an interaction term of sampling time by group to estimate whether IgG quantity waned faster by group.
Ethics
The Uniformed Services University of the Health Sciences Institutional Review Board (under protocol IDCRP-085) approved this study. All participants or their legally authorized representative provided informed consent to participate.
RESULTS
Demographic, Clinical, and Antigenic Exposure Characteristics of EPICC Participants
Among 3475 SARS-CoV-2–tested and/or vaccinated participants who were enrolled in EPICC, 2660 (84%) had sera or dried blood spot collected and were included in this analysis: SARS-CoV-2 infection alone (n = 705), SARS-CoV-2 vaccinated alone (n = 932), vaccine after infection (n = 869), and vaccine breakthrough infection (n = 154; Supplementary Figure 1). This total number of participants included 189 and 217 participants who received 1 dose of vaccine in the vaccination-alone and vaccine-after-infection groups, respectively, at the time of data cutoff for analysis. These participants were not used for 4-group comparison but were used for supplemental analyses. This analysis set excluded (n = 209) clinically apparent infections and (n = 502) subclinical reinfections across all 4 groups (Supplementary Figure 1). Most (79.9%) vaccinees received the BNT162b2 vaccine, 18.9% received the mRNA-1273 vaccine, and 1.2% received the JNJ-78436735 vaccine.
More than half of these participants were male (59.4%), 66.2% were aged 18–44 years, and 81% had no comorbidities at enrollment (Table 1). The median age of these enrolled participants was 38 years (interquartile range, 20.7). The median sampling time for each group was evaluated based on the latest antigenic exposure (SARS-CoV-2 infection/vaccination) and is presented in Table 1.
Table 1.
Clinical and Demographic Characteristics of 2660 Military Health System Beneficiaries by SARS-CoV-2 Infection and/or Vaccination History
| Characteristic | SARS-CoV-2 Infection Alone (N = 705) | Vaccination Alone (N = 932) | Vaccine After Infection (N = 869) | Vaccine Breakthrough Infection (N = 154) | Total (N = 2660) | P Valuea |
|---|---|---|---|---|---|---|
| Vaccine | <.01 | |||||
| Pfizer/BioNTech-BNT162b2 | 727 (78.0%) | 701 (80.7%) | 134 (87.0%) | 1562 (79.9%) | ||
| Moderna/mRNA-1273 | 200 (21.5%) | 150 (17.3%) | 19 (12.3%) | 369 (18.9%) | ||
| Janssen/JNJ-78436735 | 5 (0.5%) | 18 (2.1%) | 1 (0.6%) | 24 (1.2%) | ||
| Sex | .81 | |||||
| Female | 284 (40.3%) | 390 (41.8%) | 347 (39.9%) | 60 (39.0%) | 1081 (40.6%) | |
| Male | 421 (59.7%) | 542 (58.2%) | 522 (60.1%) | 94 (61.0%) | 1579 (59.4%) | |
| Age group, years | < .01 | |||||
| 18–44 | 448 (63.5%) | 726 (77.9%) | 492 (56.6%) | 94 (61.0%) | 1760 (66.2%) | |
| 45–64 | 203 (28.8%) | 169 (18.1%) | 283 (32.6%) | 41 (26.6%) | 696 (26.2%) | |
| ≥65 | 54 (7.7%) | 37 (4.0%) | 94 (10.8%) | 19 (12.3%) | 204 (7.7%) | |
| Race/ethnicity | < .01 | |||||
| White | 314 (44.5%) | 593 (63.6%) | 468 (53.9%) | 90 (58.4%) | 1465 (55.1%) | |
| Hispanic or Latino | 200 (28.4%) | 125 (13.4%) | 192 (22.1%) | 30 (19.5%) | 547 (20.6%) | |
| Black | 102 (14.5%) | 64 (6.9%) | 109 (12.5%) | 19 (12.3%) | 294 (11.1%) | |
| Asian | 21 (3.0%) | 49 (5.3%) | 28 (3.2%) | 3 (1.9%) | 101 (3.8%) | |
| Others | 68 (9.6%) | 101 (10.8%) | 72 (8.3%) | 12 (7.8%) | 253 (9.5%) | |
| Charlson comorbidity index | < .01 | |||||
| ≥5 | 23 (3.3%) | 3 (0.3%) | 34 (3.9%) | 4 (2.6%) | 64 (2.4%) | |
| 3–4 | 28 (4.0%) | 11 (1.2%) | 44 (5.1%) | 5 (3.2%) | 88 (3.3%) | |
| 1–2 | 124 (17.6%) | 37 (4.0%) | 180 (20.7%) | 17 (11.0%) | 358 (13.5%) | |
| 0 | 530 (75.2%) | 881 (94.5%) | 611 (70.3%) | 128 (83.1%) | 2150 (80.8%) | |
| Severity | < .01 | |||||
| Inpatient | 168 (23.8%) | 142 (16.3%) | 9 (5.8%) | 319 (18.5%) | ||
| Outpatient | 537 (76.2%) | 727 (83.7%) | 145 (94.2%) | 1409 (81.5%) | ||
| Sampling timeb | ||||||
| Time since infection | 32 (20) | |||||
| Time since latest antigenic exposurec | 76 (79) | 78 (70) | 89 (63) | |||
Vaccination status determined through Military Health System Data Repository record, case report form, and/or self-report questionnaire.
n × k Fisher exact test.
Median (interquartile range).
Time since final dose of vaccine or infection, whatever is latest.
Unadjusted and Adjusted Comparison of Anti-Spike–IgG Response to SARS-CoV-2 Infection and/or Vaccination
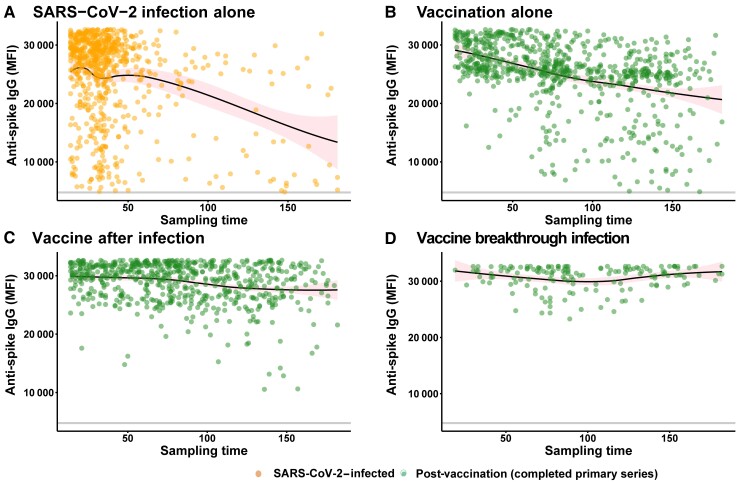
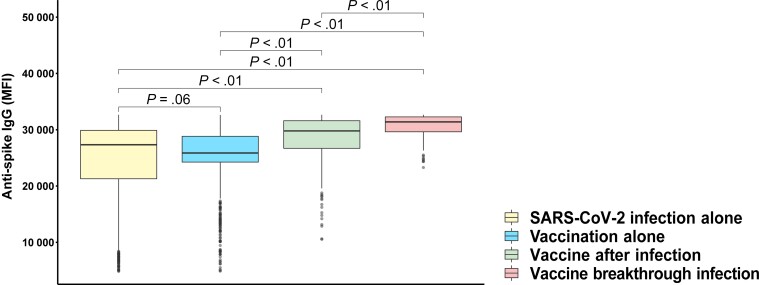
The observed maximum (peak) SARS-CoV-2 anti-spike–IgG levels by sampling time are presented in Figure 1. The mean unadjusted peak anti-spike–IgG level was higher in vaccine-after-infection participants compared with infection-alone or vaccination-alone participants (P < .01) and highest in those with vaccine breakthrough infection (Figure 2). Age group stratification revealed similar findings related to peak anti-spike–IgG responses (Supplementary Figure 2A). Even after a single dose of mRNA vaccine following prior infection, peak anti-spike–IgG response was greater than peak anti-spike–IgG response after infection alone (P < .01; Supplementary Figure 2B).
Figure 1.
Peak anti-spike–IgG MFI by sampling time (sampling time defined as time since vaccination or infection, whatever is latest). The y-axis depicts anti-spike–IgG MFI values, and the x-axis depicts sampling time (days). Each data point represents a single participant with a single peak humoral response. The locally estimated scatterplot smoothing (LOESS) curves were fit to SARS-CoV-2 infection alone (those who tested SARS-CoV-2–positive and did not receive any subsequent vaccination) (A), vaccination alone without a known history of SARS-CoV-2 infection (B), vaccine after infection (those who tested SARS-CoV-2–positive and then received a complete series of vaccination) (C), and vaccine breakthrough infection (those who were infected by SARS-CoV-2 after complete doses of vaccination) (D). These curves report moving averages but are not adjusted rates of decay; 95% confidence intervals are shaded in pink. Orange dots depict pre-vaccination samples, and green data points depict sampling greater than 2 weeks after complete vaccination. Abbreviations: IgG, immunoglobulin G; MFI, median fluorescence intensity; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Figure 2.
Unadjusted comparison of peak observed anti-spike–IgG MFI by category of infection and/or vaccination. P value determined using the Mann-Whitney U test. These comparisons do not adjust for sampling time, which varies by group. Box plots denote median, first quartile (25th percentile), and third quartile (75th percentile) of anti-spike–IgG MFI levels (y-axis) and each group (x-axis) representing SARS-CoV-2 infection alone (yellow), vaccination alone (blue), vaccine after infection (light green), and vaccine breakthrough infection (coral). SARS-CoV-2 infection alone and vaccination alone did not portray any statistically significant difference, but vaccination after vaccine-breakthrough SARS-CoV-2 infection shows greater humoral response compared with infection alone or vaccination alone. Abbreviations: IgG, immunoglobulin G; MFI, median fluorescence intensity; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
To account for differences in post-infection/post-vaccine sera sampling time and host characteristics between groups (Table 1), we examined the differences in peak anti-spike–IgG between groups and demonstrated that vaccination-alone status was associated with a 1.07 MFI increase in peak anti-spike–IgG compared with those with SARS-CoV-2 infection alone, after adjusting for host characteristics (ie, age, sex, CCI, and sampling time; P < .01; Table 2). We confirmed that SARS-CoV-2 vaccination after infection is associated with a larger peak anti-spike–IgG response (increase of 1.13 MFI compared with SARS-CoV-2 infection alone, adjusting for group and host characteristics; P < .01; Table 2) and that vaccine breakthrough infection is associated with the highest peak anti-spike–IgG (1.2 MFI; P < .01; Table 2).
Table 2.
Univariable and Multivariable Models to Compare Adjusted Anti-Spike Log10 Median Fluorescence Intensity Response in Different Categories of Infection and Vaccination
| Covariates | Coefficient | P Value | Adjusted Coefficienta | P Value |
|---|---|---|---|---|
| (95% CI) | (95% CI) | |||
| Age group, years | ||||
| 45–64 | 0.03 (.02 to .05) | <0.01 | 0.02 (.01 to .04) | <.01 |
| ≥65 | 0.06 (.04 to .08) | <.01 | 0.03 (.01 to .05) | .01 |
| Sex, male | <0.01 (−.01 to .01) | .81 | <0.01 (−.01 to .01) | .5 |
| Charlson comorbidity index | ||||
| ≥5 | 0.04 (<0.01 to .08) | .04 | <0.01 (−.04 to .03) | .91 |
| 3–4 | 0.05 (.01 to .08) | .01 | 0.02 (−.01 to .05) | .25 |
| 1–2 | 0.05 (.03 to .07) | <.01 | 0.03 (.01 to .04) | <.01 |
| Group | ||||
| Vaccination after infectionb | 0.1 (.08 to .11) | <.01 | 0.13 (.12 to .15) | <.01 |
| Vaccine breakthrough infectionb | 0.13 (.1 to .15) | <.01 | 0.18 (.16 to .21) | <.01 |
| Vaccination aloneb | 0.03 (.01 to .04) | <.01 | 0.07 (.06 to .09) | <.01 |
| Sampling time | −<0.01 (−<.01 to −<.01) | < .01 | −<0.01 (−<.01 to −<.01) | < .01 |
Number of observations, 2254. Sampling time refers to last antigenic exposure, that is, time since final dose of vaccine or infection, whatever is latest. Log10 median fluorescence intensity coefficients are exponentiated in the text for interpretability.
Abbreviation: CI, confidence interval.
Adjusted for age group, sex, CCI, group, and sampling time.
Ref: SARS-CoV-2 infection alone.
Comparison of Vaccine Product Responses, Accommodating Infection History, and Host Characteristics
When stratified by each vaccine product, we consistently noted the highest median anti-spike–IgG response in the vaccine-breakthrough-infection group (Supplementary Figure 2C). The vaccination-after-infection group had a higher peak anti-spike–IgG response compared with the vaccination-alone group (Supplementary Figure 2C).
When directly compared between vaccine types (unadjusted), a receipt of an mRNA vaccine (BNT162b2, mRNA-1273) induced a higher peak anti-spike–IgG compared with JNJ-78436735 (P < .01; Supplementary Figure 2D). Even after adjusting for differences in host characteristics and prior infection history between vaccine recipient groups, JNJ-78436735 receipt produced lower peak response compared with BNT162b2 and mRNA-1273 (Table 3). These adjusted comparisons also noted a statistically significant difference in the magnitude of peak responses between BNT162b2 and mRNA-1273, but the magnitude of this difference was small (Table 3, Supplementary Table 2).
Table 3.
Univariable and Multivariable Models to Identify Correlates of Peak Post-Vaccine Anti-Spike Log10 IgG Among Those Vaccinated With and Without a History of Infection
| Covariates | Coefficient (95% CI) | P Value | Adjusted Coefficienta (95% CI) | P Value |
|---|---|---|---|---|
| Age group, years | ||||
| 45–64 | <0.01 (−.01 to .02) | .65 | −0.01 (−.02 to 0) | .09 |
| ≥65 | 0.03 (.01 to .05) | .01 | −<0.01 (−.02 to .02) | .89 |
| Sex, male | <0.01 (−.01 to .01) | .8 | −<0.01 (−.01 to .01) | .49 |
| Charlson comorbidity index | ||||
| ≥5 | 0.04 (0 to .08) | .04 | −0.01 (−.05 to .03) | .66 |
| 3–4 | 0.06 (.02 to .09) | < .01 | 0.03 (0 to .06) | .04 |
| 1–2 | 0.04 (.02 to .06) | < .01 | 0.01 (−.01 to .03) | .26 |
| Sampling time | −<0.01 (−<.01 to −<.01) | < .01 | −<0.01 (−<.01 to <.01) | < .01 |
| Vaccine | ||||
| Pfizer/BioNTech BNT162b2b | 0.06 (.01 to .11) | .01 | 0.09 (.05 to .13) | < .01 |
| Moderna/mRNA-1273b | 0.08 (.03 to .12) | < .01 | 0.11 (.07 to .16) | < .01 |
| History of SARS-CoV-2 infectionc | 0.07 (.06 to .08) | < .01 | 0.08 (.06 to .09) | < .01 |
Number of observations 1395. Sampling time refers to last antigenic exposure, that is, time since final dose of vaccine or infection, whatever is latest.
Abbreviation: CI, confidence interval.
Adjusted for age group, sex, CCI, sampling time, vaccine product, and prior SARS-CoV-2 infection.
Ref: Janssen/JNJ-78436735.
Ref: No history of SARS-CoV-2 infection.
Correlation of Severity and Timing of Initial Infection With Post-Vaccine Anti-Spike–IgG Response
We examined predictors of vaccine-after-infection anti-spike–IgG responses, specifically whether this was dependent on severity of initial infection. We fit a multivariable regression model restricted to vaccine-after-infection participants (Table 4). The result indicated that being hospitalized for COVID-19 (compared with outpatient COVID-19 infection history) was not associated with higher observed post-vaccine peak anti-spike–IgG log10 (P = .88). However, we noted an increasing post-vaccine IgG peak with increasing time since infection to vaccination (Supplementary Figure 3).
Table 4.
Univariable and Multivariable Models to Identify Correlates of Peak Anti-Spike log10 Median Fluorescence Intensity Response in Those Vaccinated With Prior Infection History
| Covariates | Coefficient (95% CI) | P Value | Adjusted Coefficienta (95% CI) | P Value |
|---|---|---|---|---|
| Age group, years | ||||
| 45–64 | <0.01 (−.01 to .01) | .66 | <0.01 (−.01 to .01) | .72 |
| 65+ | 0.01 (−<.01 to .03) | .18 | 0.01 (−.01 to .02) | .52 |
| Sex, male | 0.01 (−<.01 to .01) | .3 | <0.01 (−.01 to .01) | .41 |
| Charlson comorbidity index | ||||
| ≥5 | 0.01 (−.01 to .04) | .33 | <0.01 (−.02 to .03) | .86 |
| 3–4 | 0.02 (<.01 to .04) | .03 | 0.02 (0 to .04) | .06 |
| 1–2 | 0.01 (−.01 to .02) | .38 | <0.01 (−.01 to .02) | .67 |
| Severity of prior infection Inpatientb | <0.01 (−.01 to .02) | .46 | <0.01 (−.01 to .01) | .88 |
| Sampling timec | −<0.01 (−<.01 to −<.01) | <.01 | −<0.01 (−<.01 to −<.01) | <.01 |
Number of observations, 653. Vaccination refers to messenger RNA vaccine.
Abbreviation: CI, confidence interval.
Adjusted for age, sex, CCI, severity, sampling time.
Ref: Outpatient.
Sampling time refers to time since final dose of vaccination.
Estimation of Anti-Spike–IgG Waning Rates by Vaccination and/or Infection
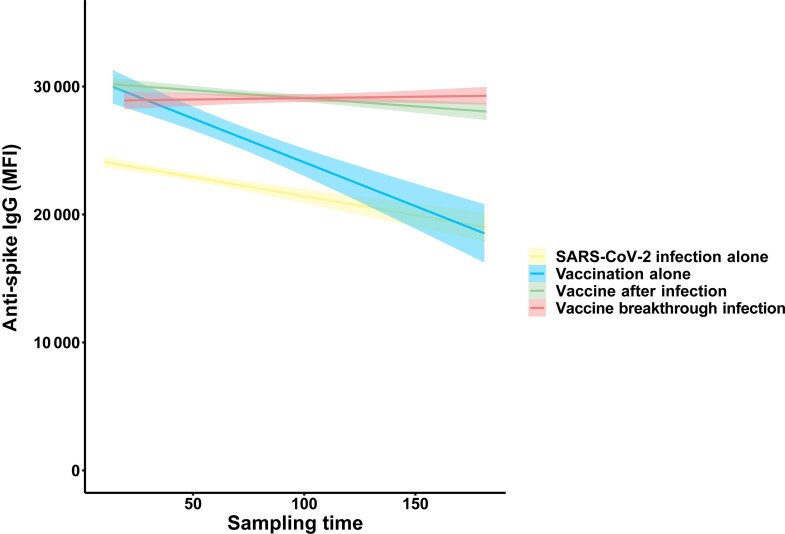
A mixed effects model derived from longitudinally collected sera compared within-participant changes of anti-spike–IgG levels among groups. The findings confirmed prior single time point analyses, that is, that vaccination alone was associated with a higher peak IgG than infection alone, that vaccine breakthrough infection was associated with the highest peak spike IgG response, and that vaccination after infection was associated with a higher peak anti-spike–IgG response than vaccine alone or infection alone (Supplementary Table 3).
To better understand anti-spike–IgG durability, we included a regression model interaction term of vaccine/infection history and sampling time to specifically estimate the rate of anti-spike–IgG waning. These longitudinal time-varying interaction coefficients permitted valid estimation of waning rates that weighted moving averages of cross-sectional data (Figure 1) could not provide. The coefficient estimates indicated that anti-spike–IgG waned faster in those who only received a vaccine compared with SARS-CoV-2 infection (Table 5, Figure 3). Post-vaccination anti-spike–IgG waned slowest in those with vaccine breakthrough infection and those who received vaccine after infection (Table 5, Figure 3). We performed 2 sensitivity analyses: removing potentially influential data points and compared standardized differences between regression coefficients (using the dfbeta command) and restricting to nonsevere (ie, outpatients) SARS-CoV-2 infections. The findings from both sensitivity analyses were similar to the findings from the overall analysis (Supplementary Tables 4, 5).
Table 5.
Longitudinal Linear Mixed Modeling to Estimate Decay Kinetics of Log10 Anti-Spike IgG Response to Antigenic Exposure
| Covariates | Coefficient | P Value |
|---|---|---|
| Sampling time | −<0.01 (−<0.01 to −<0.01) | <.01 |
| Vaccination after infectiona | 0.13 (0.09 to 0.16) | <.01 |
| Vaccine breakthrough infectiona | 0.1 (0.06 to 0.14) | <.01 |
| Vaccination alonea | 0.15 (0.09 to 0.21) | <.01 |
| Sampling time * Vaccine after infection | <0.01 (<0.01 to <0.01) | .03 |
| Sampling time * Vaccine breakthrough infection | <0.01 (<0.01 to <0.01) | <.01 |
| Sampling * Vaccination alone | −<0.01 (−<0.01 to −<0.01) | .02 |
Number of observations, 1160. Sampling time refers to last antigenic exposure, that is, time since final dose of vaccine or infection, whatever is latest. Model fit to all longitudinal data and including group, sampling time, and an interaction term of sampling time and group. The asterisk (*) is used to indicate interactions among the variables that it joins.
Ref: SARS-CoV-2 infection alone.
Figure 3.
Anti-spike–IgG MFI by sampling time (time since vaccination or infection, whatever is latest), restricted to longitudinal analysis using at least 2 sera samples per participant, stratified by SARS-CoV-2 infection alone (yellow), vaccination alone (blue), vaccine after infection (light green), and vaccine breakthrough infection (coral). The y-axis depicts anti-spike–IgG MFI values, and the x-axis depicts sampling time. Each data point represents a sera sample. The solid line is the estimated slope derived from mixed-effects regression models in Table 5; shaded area depicts confidence interval. Abbreviations: IgG, immunoglobulin G; MFI, median fluorescence intensity; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
DISCUSSION
Our findings confirm that hybrid immunity arising from COVID-19 vaccine-induced immunity after prior infection offers greater humoral immunogenicity than either vaccination or prior infection alone. Furthermore, humoral immune response to prior infection is substantially augmented by even a single dose of vaccine, corroborating the findings from other studies [19–24]. These findings are particularly relevant as the Delta and Omicron variants have led to a large proportion of the population having hybrid immunity. Our findings expand on those from prior studies on this topic and show that increasing infection to vaccine time, but not initial infection severity, correlates with a greater post-vaccination response. This finding may help guide strategies on timing of mRNA vaccine dosing after initial SARS-CoV-2 infection, although this requires further study. In addition, we noted that vaccine breakthrough infections offer the highest peak IgG response (even as breakthrough infections were noted to be milder than pre-vaccine infections), emphasizing that repeated exposures to SARS-CoV-2 antigens leads to higher IgG responses. This latter finding is consistent with recent data that show that mRNA vaccine booster doses lead to significant increases in immunogenicity [25]. Importantly, these analyses only refer to antibody kinetics and do not measure cellular immune response or vaccine effectiveness directly.
Our results indicate that receipt of an mRNA vaccine (in those without a history of infection) offers higher short- to -medium-term peak humoral responses compared with SARS-CoV-2 infection alone, after adjustment for other predictors (Table 2). However, we noted that the vaccine-alone group had more rapid waning of IgG compared with those with SARS-CoV-2 infection alone and those who received vaccine after infection. The difference in waning rates between groups was small, and their clinical significance is unclear [26]. These data contribute to the discussion about the significance of recent comparisons of infection frequency and clinical outcomes among vaccinees when compared with infection-alone participants and those with a history of both infection and vaccination [27–32]. These findings also contribute to our understanding of how vaccine-induced immunity compares with SARS-CoV-2 infection and optimal timing of vaccine boosting.
Our study also permitted a head-to-head comparison of COVID-19 vaccines. As with other studies, we noted a greater peak anti-spike–IgG response in those who received an mRNA vaccine (mRNA-1273, BNT162b2) compared with the JNJ-78436735 vaccine when adjusting for host characteristics and infection history. Among mRNA vaccinees, we also noted that mRNA-1273 vaccine showed a higher peak IgG compared with BNT162b2 (Table 3, Supplementary Table 2) [33–35]. This is consistent with published findings that show a greater humoral immune response from mRNA-1273 and, in some studies, apparent increased effectiveness of the mRNA-1273 vaccine.
This analysis has several limitations. First, these results refer only to binding antibody assays due to prohibitive logistics for performing neutralizing antibody assays on these many specimens. However, this binding assay (and other binding anti-spike–IgG assays) has moderate to high correlation with neutralizing antibody titers previously performed on a small number of EPICC participants [15]. Second, quantitative differences of anti-spike–IgG binding responses, measured as MFI at a single blood specimen dilution (1:400), between groups were small overall, which may reflect upper limits of assay quantification and anti-spike–IgG MFI saturation in blood samples collected after multiple immunogen stimulations. Third, follow-up time did not exceed 183 days, yet the study spanned a period of sequential dominant variants in the United States. Finally, our understanding of how antibody responses serve as a correlate of clinical protection is still developing [36]. While a landmark meta-analysis estimated a neutralizing antibody titer that could protect against infection [37], definition of a protective titer remains challenging and is subject to assay-to-assay comparability and the particular clinical end point of interest. Further, there is increasing evidence on the protective role of T-cell immune responses against infection and severity, further complicating estimations of a protective antibody titer [38, 39]. These limitations are important to acknowledge before inference is undertaken of relative protection between those with different antigenic exposure histories. They also underscore the need for further study in this and other cohorts to include integration of clinical outcomes with a range of immune readout data from B-cell, T-cell, and innate arms of the immune system, as well as in pediatric age groups and booster recipients, which were not part of this analysis.
The strengths of this study include a large sample size, measurement of a broad range of participant and illness characteristics to account for confounding between 4 groups, and careful ascertainment of subclinical repeat infections that could have biased comparisons. Taken together, these findings provide additional data on the importance of antiviral immunogenicity by vaccination beyond that afforded by prior SARS-CoV-2 infection and offer further insights into host responses to sequential SARS-CoV-2 antigenic exposure that may inform future vaccination strategies and their development, including evaluation on timing of post-infection vaccine administration.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments . We sincerely thank the members of the Epidemiology, Immunology, and Clinical Characteristics of Emerging Infectious Diseases with Pandemic Potential (EPICC) COVID-19 Cohort Study Group for their many contributions in conducting the study and ensuring effective protocol operations. The authors acknowledge the following who have contributed to the EPICC COVID-19 study: Brooke Army Medical Center, Fort Sam Houston, TX: COL J. Cowden, LTC M. Darling, S. DeLeon, Maj D. Lindholm, LTC A. Markelz, K. Mende, S. Merritt, T. Merritt, LTC N. Turner, CPT T. Wellington. Carl R. Darnall Army Medical Center, Fort Hood, TX: LTC S. Bazan, P. K. Love. Fort Belvoir Community Hospital, Fort Belvoir, VA: N. Dimascio-Johnson, MAJ E. Ewers, LCDR K. Gallagher, LCDR D. Larson, A. Rutt. Henry M. Jackson Foundation, Inc, Bethesda, MD: P. Blair, J. Chenoweth, D. Clark. Madigan Army Medical Center, Joint Base Lewis McChord, WA: S. Chambers, LTC C. Colombo, R. Colombo, CAPT C. Conlon, CAPT K. Everson, COL P. Faestel, COL T. Ferguson, MAJ L. Gordon, LTC S. Grogan, CAPT S. Lis, COL C. Mount, LTC D. Musfeldt, CPT D. Odineal, LTC M. Perreault, W. Robb-McGrath, MAJ R. Sainato, C. Schofield, COL C. Skinner, M. Stein, MAJ M. Switzer, MAJ M. Timlin, MAJ S. Wood. Naval Medical Center Portsmouth, Portsmouth, VA: S. Banks, R. Carpenter, L. Kim, CAPT K. Kronmann, T. Lalani, LCDR T. Lee, LCDR A. Smith, R. Smith, R. Tant, T. Warkentien. Naval Medical Center San Diego, San Diego, CA:CDR C. Berjohn, S. Cammarata, N. Kirkland, D. Libraty, CAPT (Ret) R. Maves, CAPT (Ret) G. Utz. Tripler Army Medical Center, Honolulu, HI: S. Chi, LTC R. Flanagan, MAJ M. Jones, C. Lucas, LTC (Ret) C. Madar, K. Miyasato, C. Uyehara. Uniformed Services University of the Health Sciences (USUHS), Bethesda, MD: B. Agan, L. Andronescu, A. Austin, C. Broder, CAPT T. Burgess, C. Byrne, COL (Ret) K. Chung, J. Davies, C. English, N. Epsi, C. Fox, M. Fritschlanski, M. Grother, A. Hadley, COL P. Hickey, E. Laing, LTC C. Lanteri, LTC J. Livezey, A. Malloy, R. Mohammed, C. Morales, P. Nwachukwu, C. Olsen, E. Parmelee, S. Pollett, S. Richard, J. Rozman, J. Rusiecki, E. Samuels, P. Nwachukwu, M. Tso, M. Sanchez, A. Scher, CDR M. Simons, A. Snow, K. Telu, D. Tribble, L. Ulomi. US Air Force School of Aerospace Medicine, Dayton, OH: TSgt T. Chao, R. Chapleau, M. Christian, A. Fries, C. Harrington, V. Hogan, S. Huntsberger, K. Lanter, E. Macias, J. Meyer, S. Purves, K. Reynolds, J. Rodriguez, C. Starr. US Coast Guard, Washington, DC: CAPT J. Iskander, CDR I. Kamara. Womack Army Medical Center, Fort Bragg, NC: B. Barton, LTC D. Hostler, LTC J. Hostler, MAJ K. Lago, C. Maldonado, J. Mehrer. William Beaumont Army Medical Center, El Paso, TX: MAJ T. Hunter, J. Mejia, J. Montes, R. Mody, R. Resendez, P. Sandoval, M. Wayman. Walter Reed National Military Medical Center, Bethesda, MD: I. Barahona, A. Baya, A. Ganesan, MAJ N. Huprikar, B. Johnson. Walter Reed Army Institute of Research, Silver Spring, MD: S. Peel.
Group authors . We thank the members of the EPICC COVID-19 Cohort Study Group for their many contributions in conducting the study and ensuring effective protocol operations. The following members were all closely involved with the design, implementation, and oversight of the study and have met group authorship criteria for this article: Brooke Army Medical Center, Fort Sam Houston, TX: LTC M. Darling, T. Merritt, CPT T. Wellington. Fort Belvoir Community Hospital, Fort Belvoir, VA: N. Dimascio-Johnson. Austere environments Consortium for Enhanced Sepsis Outcomes (ACESO), Henry M. Jackson Foundation, Inc, Bethesda, MD: J. Chenoweth, D. Clark, P. Blair. Madigan Army Medical Center, Joint Base Lewis McChord, WA: S. Chambers, R. Colombo, COL P. Faestel, CPT S. Lis, CPT D. Odineal, LTC M. Perreault, C. Schofield, M. Stein. Naval Medical Center San Diego, San Diego, CA: CDR C. Berjohn, N. Kirkland. Tripler Army Medical Center, Honolulu, HI: LTC (Ret) C. Madar, C. Uyehara. USUHS, Bethesda, MD: COL (Ret) K. Chung, COL P. Hickey, LTC J. Livezey, A. Malloy, E. Parmelee. US Air Force School of Aerospace Medicine, Dayton, OH: TSgt T. Chao, R. Chapleau, C. Harrington, S. Huntsberger, E. Macias, J. Meyer, C. Starr. US Coast Guard, Washington, DC: CAPT J. Iskander. Womack Army Medical Center, Fort Bragg, NC: B. Barton, LTC D. Hostler, MAJ K. Lago, C. Maldonado. William Beaumont Army Medical Center, El Paso, TX: M. Wayman.
Disclaimer. Some of the authors are service members or employees of the US government. This work was prepared as part of their official duties. The contents of this publication are the sole responsibility of the author(s) and do not necessarily reflect the views, opinions, or policies of the USUHS; the Department of Defense; the Defense Health Agency; the departments of the Army, Navy, or Air Force; Brooke Army Medical Center; Walter Reed National Military Medical Center; Naval Medical Center San Diego; Madigan Army Medical Center; US Air Force School of Aerospace Medicine; Naval Medical Center Portsmouth; Tripler Army Medical Center; Fort Belvoir Community Hospital; or the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc Mention of trade names, commercial products, or organizations does not imply endorsement by the US government. The investigators have adhered to the policies for protection of human participants as prescribed in 45 CFR 46.
Financial support. This work was supported by awards from the Defense Health Program (HU00012020067) and the National Institute of Allergy and Infectious Diseases (NIAID) (HU00011920111). The protocol was executed by the Infectious Disease Clinical Research Program (IDCRP), a Department of Defense program executed by the USUHS through a cooperative agreement by the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. This project has been funded in part by the NIAID at the National Institutes of Health under an interagency agreement (Y1-AI-5072).
Conflicts of interest. S. D. P., T. H. B., D. T., and M. P. S. report that the Uniformed Services University (USU) IDCRP, a US Department of Defense institution, and the Henry M. Jackson Foundation were funded under a cooperative research and development agreement to conduct an unrelated phase 3 COVID-19 monoclonal antibody immunoprophylaxis trial sponsored by AstraZeneca. The Henry M. Jackson Foundation, in support of the USU IDCRP, was funded by the Department of Defense Joint Program Executive Office for Chemical, Biological, Radiological, and Nuclear Defense to augment the conduct of an unrelated phase 3 vaccine trial sponsored by AstraZeneca. Both of these trials were part of the US government COVID-19 response. Neither is related to the work presented here. C. M. B. reports a leadership or fiduciary role on the Infectious Diseases Society of America Clinical Affairs Committee. R. C. M. reports grants or contracts to his institution and unrelated to this work from AiCuris, Sound Pharmaceutical, and AstraZeneca; consulting fees and honorarium for advisory panel membership from the Society of Critical Care Medicine; honorarium for a lecture from the California Thoracic Society; travel support from the American Thoracic Society, American College of Chest Physicians, and Society of Critical Care Medicine; a US patent for investigational dengue vaccine candidate (no payments made or current commercial development planned); data and safety monitoring board membership (funds to author) for Trauma Insights, LLC; member of The Society of Critical Care Medicine (SCCM) Congress Program Committee (travel support for official meetings [pre-March 2020]), chair of the American College of Chest Physicians (CHEST) COVID-19 Task Force and Disaster/Global Health Section (travel support for official meetings), and member of the CHEST Scientific Program Committee (travel support for official meetings). All remaining authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Supplementary Material
Contributor Information
Nusrat J Epsi, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA.
Stephanie A Richard, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA.
David A Lindholm, Brooke Army Medical Center, Fort Sam Houston, Texas, USA; Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Katrin Mende, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA; Brooke Army Medical Center, Fort Sam Houston, Texas, USA.
Anuradha Ganesan, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA; Walter Reed National Military Medical Center, Bethesda, Maryland, USA.
Nikhil Huprikar, Walter Reed National Military Medical Center, Bethesda, Maryland, USA.
Tahaniyat Lalani, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA; Naval Medical Center Portsmouth, Portsmouth, Virginia, USA.
Anthony C Fries, US Air Force School of Aerospace Medicine, Dayton, Ohio, USA.
Ryan C Maves, Wake Forest School of Medicine, Winston-Salem, North Carolina, USA.
Rhonda E Colombo, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA; Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Madigan Army Medical Center, Joint Base Lewis McChord, Washington, USA.
Derek T Larson, Fort Belvoir Community Hospital, Fort Belvoir, Virginia, USA; Naval Medical Center San Diego, San Diego, California, USA.
Alfred Smith, Naval Medical Center Portsmouth, Portsmouth, Virginia, USA.
Sharon W Chi, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA; Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Carlos J Maldonado, Womack Army Medical Center, Fort Bragg, North Carolina, USA.
Evan C Ewers, Fort Belvoir Community Hospital, Fort Belvoir, Virginia, USA.
Milissa U Jones, Tripler Army Medical Center, Honolulu, Hawaii, USA.
Catherine M Berjohn, Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Naval Medical Center San Diego, San Diego, California, USA.
Daniel H Libraty, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA; Naval Medical Center San Diego, San Diego, California, USA.
Margaret Sanchez Edwards, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA.
Caroline English, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA.
Julia S Rozman, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA.
Rupal M Mody, William Beaumont Army Medical Center, El Paso, Texas, USA.
Christopher J Colombo, Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Madigan Army Medical Center, Joint Base Lewis McChord, Washington, USA.
Emily C Samuels, Department of Microbiology and Immunology, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Princess Nwachukwu, Department of Microbiology and Immunology, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Marana S Tso, Department of Microbiology and Immunology, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Ann I Scher, Department of Preventive Medicine & Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA.
Celia Byrne, Department of Preventive Medicine & Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA.
Jennifer Rusiecki, Department of Preventive Medicine & Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA.
Mark P Simons, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
David Tribble, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Christopher C Broder, Department of Microbiology and Immunology, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Brian K Agan, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA.
Timothy H Burgess, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Eric D Laing, Department of Microbiology and Immunology, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA.
Simon D Pollett, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, Maryland, USA.
for the Epidemiology, Immunology, and Clinical Characteristics of Emerging Infectious Diseases with Pandemic Potential COVID-19 Cohort Study Group:
J Cowden, M Darling, S DeLeon, D Lindholm, A Markelz, K Mende, S Merritt, T Merritt, N Turner, T Wellington, S Bazan, P K Love, N Dimascio-Johnson, E Ewers, K Gallagher, D Larson, A Rutt, P Blair, J Chenoweth, D Clark, S Chambers, C Colombo, R Colombo, C Conlon, K Everson, P Faestel, T Ferguson, L Gordon, S Grogan, S Lis, C Mount, D Musfeldt, D Odineal, M Perreault, W Robb-McGrath, R Sainato, C Schofield, C Skinner, M Stein, M Switzer, M Timlin, S Wood, S Banks, R Carpenter, L Kim, K Kronmann, T Lalani, T Lee, A Smith, R Smith, R Tant, T Warkentien, C Berjohn, S Cammarata, N Kirkland, D Libraty, R Maves, G Utz, S Chi, R Flanagan, M Jones, C Lucas, C Madar, K Miyasato, C Uyehara, B Agan, L Andronescu, A Austin, C Broder, T Burgess, C Byrne, K Chung, J Davies, C English, N Epsi, C Fox, M Fritschlanski, M Grother, A Hadley, P Hickey, E Laing, C Lanteri, J Livezey, A Malloy, R Mohammed, C Morales, P Nwachukwu, C Olsen, E Parmelee, S Pollett, S Richard, J Rozman, J Rusiecki, E Samuels, P Nwachukwu, M Tso, M Sanchez, A Scher, M Simons, A Snow, K Telu, D Tribble, L Ulomi, T Chao, R Chapleau, M Christian, A Fries, C Harrington, V Hogan, S Huntsberger, K Lanter, E Macias, J Meyer, S Purves, K Reynolds, J Rodriguez, C Starr, J Iskander, I Kamara, B Barton, D Hostler, J Hostler, K Lago, C Maldonado, J Mehrer, T Hunter, J Mejia, J Montes, R Mody, R Resendez, P Sandoval, M Wayman, I Barahona, A Baya, A Ganesan, N Huprikar, B Johnson, and S Peel
References
- 1. Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med 2020; 383:1920–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020; 383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 2021; 384:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sadoff J, Gray G, Vandebosch A, et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med 2021; 384:2187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amit S, Regev-Yochay G, Afek A, Kreiss Y, Leshem E. Early rate reductions of SARS-CoV-2 infection and COVID-19 in BNT162b2 vaccine recipients. Lancet 2021; 397:875–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haas EJ, Angulo FJ, McLaughlin JM, et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet 2021; 397:1819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bubar KM, Reinholt K, Kissler SM, et al. Model-informed COVID-19 vaccine prioritization strategies by age and serostatus. Science 2021; 371:916–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Livingston EH, Malani PN, Creech CB. The Johnson & Johnson vaccine for COVID-19. JAMA 2021; 325:1575. [DOI] [PubMed] [Google Scholar]
- 9. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med 2021; 385:585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nanduri S, Pilishvili T, Derado G, et al. Effectiveness of Pfizer-BioNTech and Moderna vaccines in preventing SARS-CoV-2 infection among nursing home residents before and during widespread circulation of the SARS-CoV-2 B.1.617.2 (Delta) variant—National Healthcare Safety Network, March 1–August 1, 2021. MMWR Morb Mortal Wkly Rep 2021; 70:1163–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anichini G, Terrosi C, Gandolfo C, et al. SARS-CoV-2 antibody response in persons with past natural infection. N Engl J Med 2021; 385:90–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Centers for Disease Control and Prevention . Science Brief: SARS-CoV-2 infection-induced and vaccine-induced immunity. Atlanta, GA: Centers for Disease Control and Prevention, 2020. [PubMed] [Google Scholar]
- 13. Gilbert PB, Montefiori DC, McDermott AB, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022; 375:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Epsi NJ, Richard SA, Laing ED, et al. Clinical, immunological and virological SARS-CoV-2 phenotypes in obese and non-obese military health system beneficiaries. J Infect Dis 2021; 224:1462–72. doi: 10.1093/infdis/jiab396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laing ED, Sterling SL, Richard SA, et al. Antigen-based multiplex strategies to discriminate SARS-CoV-2 natural and vaccine induced immunity from seasonal human coronavirus humoral responses. medRxiv 2021. doi: 10.1101/2021.02.10.21251518. [DOI] [Google Scholar]
- 16. Clifton GT, Pati R, Krammer F, et al. SARS-CoV-2 infection risk among active duty military members deployed to a field hospital—New York City, April 2020. MMWR Morb Mortal Wkly Rep 2021; 70:308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373–83. [DOI] [PubMed] [Google Scholar]
- 18. Team TRDC . R: a language environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2020. [Google Scholar]
- 19. Schmidt F, Weisblum Y, Rutkowska M, et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature 2021; 600:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gobbi F, Buonfrate D, Moro L, et al. Antibody response to the BNT162b2 mRNA COVID-19 vaccine in subjects with prior SARS-CoV-2 infection. Viruses 2021; 13:422. doi: 10.3390/v13030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021; 592:616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saadat S, Rikhtegaran Tehrani Z, Logue J, et al. Binding and neutralization antibody titers after a single vaccine dose in health care workers previously infected with SARS-CoV-2. JAMA 2021; 325:1467–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krammer F, Srivastava K, Alshammary H, et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med 2021; 384:1372–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bradley T, Grundberg E, Selvarangan R, et al. Antibody responses after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med 2021; 384:1959–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krammer F. SARS-CoV-2 vaccines in development. Nature 2020; 586:516–27. [DOI] [PubMed] [Google Scholar]
- 26. Siggins MK, Thwaites RS, Openshaw PJM. Durability of immunity to SARS-CoV-2 and other respiratory viruses. Trends Microbiol 2021; 29:648–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gazit S, Shlezinger R, Perez G, et al. SARS-CoV-2 Naturally Acquired Immunity vs. Vaccine-induced Immunity, Reinfections versus Breakthrough Infections: a Retrospective Cohort Study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, ciac262. 2022. doi: 10.1093/cid/ciac262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cavanaugh AM, Spicer KB, Thoroughman D, Glick C, Winter K. Reduced risk of reinfection with SARS-CoV-2 after COVID-19 vaccination—Kentucky, May-June 2021. MMWR Morb Mortal Wkly Rep 2021; 70:1081–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cerqueira-Silva T, Andrews JR, Boaventura VS, et al. Effectiveness of CoronaVac, ChAdOx1 nCoV-19, BNT162b2, and Ad26.COV2.S among individuals with previous SARS-CoV-2 infection in Brazil: a test-negative, case-control study. Lancet Infect Dis 2022; 22:P791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nordström P, Ballin M, Nordström A. Risk of SARS-CoV-2 reinfection and COVID-19 hospitalisation in individuals with natural and hybrid immunity: a retrospective, total population cohort study in Sweden. Lancet Infect Dis 2022; 22:781-790. doi: 10.1016/S1473-3099(22)00143-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hall V, Foulkes S, Insalata F, et al. Protection against SARS-CoV-2 after Covid-19 vaccination and previous infection. 2022; 386:1207–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sidik SM. COVID vaccine plus infection can lead to months of immunity. Nature 2022. 10.1038/d41586-022-00961-3 [DOI] [PubMed] [Google Scholar]
- 33. Steensels D, Pierlet N, Penders J, Mesotten D, Heylen L. Comparison of SARS-CoV-2 antibody response following vaccination with BNT162b2 and mRNA-1273. JAMA 2021; 326:1533–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Self WH, Tenforde MW, Rhoads JP, et al. Comparative effectiveness of Moderna, Pfizer-BioNTech, and Janssen (Johnson & Johnson) vaccines in preventing COVID-19 hospitalizations among adults without immunocompromising conditions—United States, March–August 2021. MMWR Morb Mortal Wkly Rep 2021; 70:1337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chung H, He S, Nasreen S, et al. Effectiveness of BNT162b2 and mRNA-1273 covid-19 vaccines against symptomatic SARS-CoV-2 infection and severe covid-19 outcomes in Ontario, Canada: test negative design study. BMJ 2021; 374:n1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krause PR, Fleming TR, Peto R, et al. Considerations in boosting COVID-19 vaccine immune responses. Lancet 2021; 398(10308). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021; 27:1205–11. [DOI] [PubMed] [Google Scholar]
- 38. Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020; 183:996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Naranbhai V, Nathan A, Kaseke C, et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell 2022; 185:1041–51.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.