Abstract
The effects of dexamethasone (DXM) treatment on pulmonary immunity in COVID-19–associated acute respiratory distress syndrome (CARDS) remain insufficiently understood. We performed transcriptomic RNA-seq analysis of bronchoalveolar lavage fluid from 20 mechanically ventilated patients: 12 with CARDS (with or without DXM) and 8 non–COVID-19 critically ill controls. CARDS with DXM was characterized by upregulation of genes related to B-cell and complement pathway activation, antigen presentation, phagocytosis, and FC-γ receptor signaling. Most interferon-stimulated genes were upregulated in CARDS, particularly in CARDS without DXM. In conclusion, DXM treatment was not associated with regulation of proinflammatory pathways in CARDS but with regulation of other local immune responses.
Clinical Trials Registration. NCT04354584.
Keywords: acute respiratory distress syndrome, bronchoalveolar lavage fluid, coronavirus disease 2019, inflammation, interferon stimulated genes, transcriptome profiling
A distinct dexamethasone-dependent transcriptomic signature was detected in the lungs of mechanically ventilated COVID-19 patients.
Coronavirus disease 2019 (COVID-19) acute respiratory distress syndrome (CARDS) is associated with a mortality rate of 40%–50%, which may be even higher in patients requiring invasive mechanical ventilation [1]. Understanding of CARDS immunopathology was improved by a few studies in particular of bronchoalveolar lavage fluid (BALF) and showing prominent pulmonary immune cell invasion, most notably inflammatory myeloid cells [2–4], and expression of several proinflammatory mediators. However, a more detailed understanding of the specific mechanisms of COVID-19–mediated lung injury is pertinent to uncover how various immunomodulatory therapies, including dexamethasone (DXM), affect lung immunity.
We investigated the transcriptomic profiles of the local pulmonary host responses during early-stage severe CARDS by per protocol BALF sampling early after intubation. Specifically, we compared the immune transcriptomic profile of CARDS patients, with and without DXM treatment, and non–COVID-19 critically ill patients (controls) to elucidate how this specific immune-targeted treatment modifies the pulmonary host defense, including the gene expression profiles of proinflammatory mediators and interferon-stimulated genes (ISGs).
METHODS
Study Population and Ethics
Inclusion criteria of CARDS patients were age >18 years, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection confirmed by reverse transcription polymerase chain reaction (RT-PCR), presence of ARDS (Berlin definition 2012), and less than 72 hours of invasive mechanical ventilation. All patients were recruited during admission to the intensive care unit (ICU) at Hvidovre Hospital, Denmark. All patients were recruited between 6 April 2020 and 12 April 2021. This period included the first and second COVID-19 wave in Denmark, which corresponded to a time prior to and after DXM treatment was introduced as standard of care. Thus, patients recruited during the first wave did not receive DXM treatment. Supportive ICU treatment was not changed during this period. We have previously presented flow cytometric results of 4 CARDS patients, who were also included in the present study [3]. None of the previous published data was reused in the present study.
Patients with ARDS from bacterial respiratory pathogens as well as mechanically ventilated patients with sepsis but without ARDS were included as controls. Except for SARS-CoV-2 infection, inclusion criteria were identical for CARDS and controls. All controls had ARDS and/or sepsis according to consensus criteria (Berlin definition 2012; Sepsis-3 2016).
All patients were sedated and unable to provide oral and written informed consent, which was obtained from next of kin [3]. The study was approved by the Regional Ethics Committee of Copenhagen (H-20023159/H-22011021/H-22009131) and the Knowledge Center for Data Review of Copenhagen (P-2020-399) and registered at ClinicalTrials.gov (NCT04354584).
Bronchoalveolar Lavage Procedure and Sample Processing
The BAL procedure was performed per protocol in a standardized fashion as described in the Supplementary material.
RNA Extraction and Sequencing
Information about RNA extraction and library preparation [5] has been included in the Supplementary material. Differential gene analysis was performed in R, as described previously [6]. Briefly, we used the Limma-voom and DeSeq2 package for preprocessing and principal component (PC) analysis, respectively. The following 4 contrasts were compared: CARDS versus controls, CARDS with DXM treatment versus controls, CARDS without DXM treatment versus controls, and CARDS with DXM treatment versus CARDS without DXM treatment. Gene ontology analysis was performed using Goseq on differentially expressed genes separately for each contrast. Analysis of ISGs was performed using ROAST for each contrast using a curated list of 399 ISGs [7]. Finally, we analyzed selected inflammatory, including interleukin (IL)-1A, IL-1B, IL-2, IL-6, IL-8, IL-11, IL-17A, IL-17B, IL-18, tumor necrosis factor-α (TNF-α), interferon-λ (IFN-λ), granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte colony-stimulating factor (GCSF), monocyte chemoattractant protein-1 (MCP-1), interferon-λ–induced protein (IP-10), and macrophage inflammatory protein-1 (MIP-1-α), which were selected based on previous BALF-based studies.
RESULTS
Patient Characteristics and Bronchoalveolar Lavage Procedure
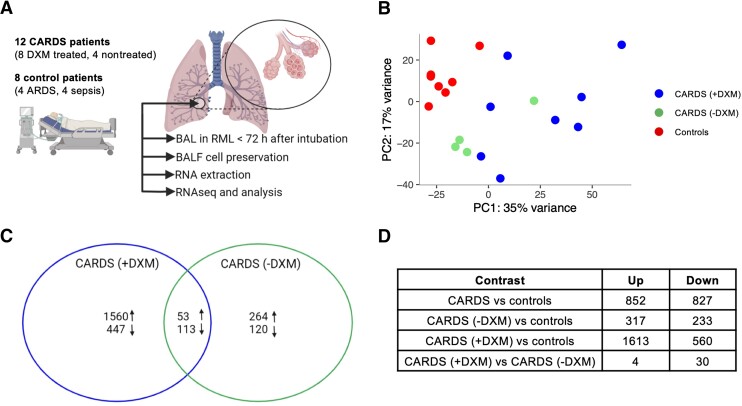
The BAL procedures in CARDS and critically ill control patients were carried out after a mean of 28.8 hours (range, 6–72 hours) after intubation by the same physician (author, R. R. P.). All samples were obtained from the right middle lobe. A total of 13 CARDS patients and 8 controls were included (Supplementary Table 1). One DXM-treated CARDS patient was excluded from analysis due to insufficient RNA extraction. All CARDS patients had moderate-to-severe impairment of oxygenation judged by Pao2/Fio2 ratio; 8/12 were treated with vasopressors at the time of the BAL procedure and median duration of COVID-19 symptoms was 11 days (range, 8–20 days). A total of 4 CARDS patients were not treated with DXM whereas 9 CARDS patients received DXM (2–6 days prior to the procedure). Controls encompassed 4/8 with ARDS and 4/8 with sepsis but without ARDS (Figure 1A). None of the controls were treated with corticosteroids.
Figure 1.
BALF transcriptome profile for CARDS patients: (A) experimental setup; (B) PC analysis depicting the 3 patient groups; and (C) Venn diagram representing the DE gene relationship for CARDS without DXM treatment and CARDS with DXM treatment compared to controls. Arrows pointing up depict upregulated DE genes, and arrows pointing down depict downregulated DE genes. D, Table of number of differentially expressed genes. The number of up- or downregulated genes for CARDS patients is not a sum of up- or downregulated genes of the 2 subgroups (with [+] and without [−] DXM treatment) as the differential expression analyses are independent. B and C, The color coding represents patient groups. Abbreviations: ARDS, acute respiratory distress syndrome; BAL, bronchoalveolar lavage; CARDS, COVID-19–associated acute respiratory distress syndrome; DE, differentially expressed; DXM, dexamethasone; PC, principal component; RML, right middle lobe.
Transcriptional Features in the Lungs During DXM Treatment
The levels of viral RNA in samples from CARDS patients varied from 0.001% to 1.378% (Supplementary Figure 1). The 7 samples with high fractions (above 0.1%) of viral reads showed differential coverage distribution skewed towards higher depth at the 3′ end of the viral genome compared to coverage at the 5′ end. This was concordant with the presence of subgenomic RNA from replicating genomes indicating active infection. Furthermore, we found traces of negative-strand RNA in those samples indicating active viral replication. PC analysis did not reveal clustering of human gene expression related to high versus low viral load and was not accounted for in subsequent analysis.
PC analysis showed controls clustering closely together, thus justifying grouping of these patients, while CARDS clustered more broadly but distinctly separate from the controls (Figure 1B). Furthermore, CARDS without DXM treatment clustered more closely than CARDS with DXM treatment. In the CARDS patients compared to the controls, 852 genes were differentially upregulated and 827 downregulated (false discovery rate, < 0.05). Similar analysis performed with only CARDS without DXM treatment revealed 317 up- and 233 downregulated genes, while CARDS with DXM treatment had 1613 up- and 560 downregulated genes (Figure 1C and 1D, and Supplementary Figure 2). This indicated DXM as a primary driver of transcriptome changes in CARDS.
Functional Signatures in the Lungs During DXM Treatment
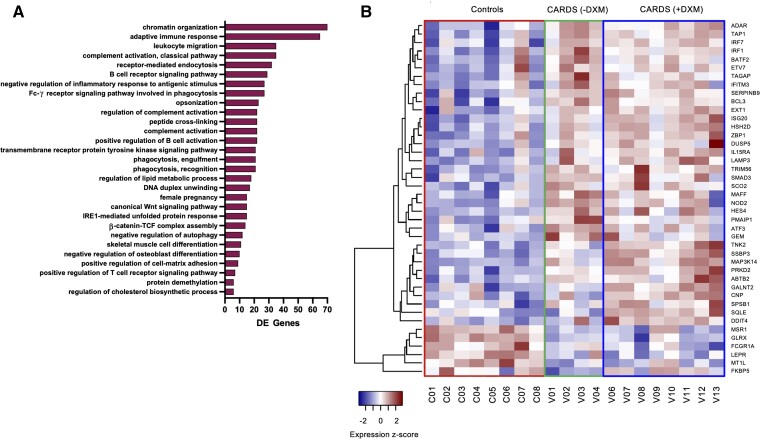
CARDS with DXM treatment gene signatures were enriched for several B-cell activation and antigen-presenting pathways, leukocyte migration, and activation of complement. Interestingly, phagocytosis was also upregulated, especially FC-γ receptor signaling (Figure 2A). Only few downregulated gene categories were found, including lysozyme function, MHC class II responses, and neutrophil degranulation.
Figure 2.
Gene ontology analysis and ISG regulation in CARDS patients. A, Gene ontology (biological process) analysis of upregulated DE genes for CARDS with DXM treatment. Adjusted P value set to P < .05 and only categories where DE genes accounts for more than 20% of total with 10 or more genes in the category displayed. X-axis shows the number of DE genes upregulated in each category. B, Heatmap of 42 differentially expressed ISGs showing expression z-scores. The 42 ISGs were identified from a list of 399 known ISGs [7]. Rows represent individual ISGs and columns individual patients with patient IDs shown below. Abbreviations: CARDS, COVID-19–associated acute respiratory distress syndrome; DE, differentially expressed; DXM, with (+) and without (−) dexamethasone; ISG, interferon-stimulated gene.
IFN-Stimulated Genes in CARDS
ISGs were enriched among upregulated genes in CARDS, but specifically for CARDS patients without DXM treatment (Supplementary Figure 3). Among genes differentially expressed across any condition (CARDS with DXM treatment, CARDS without DXM treatment, controls), 42 ISGs were identified (Figure 2B). Most of these (36 genes) were elevated for CARDS (both with and without DXM treatment) and included typical responders to acute viral infection like ADAR, ATF3, IFITM3, IRF1, IRF7, ISG20, and TRIM56. A subset primarily enriched for CARDS with DXM treatment included ABTB2, CNP, DDIT4, GALNT2, MAP3K14, PRKD2, SPSB1, SQLE, SSBP3, and TNK2. Six genes, FCGR1A, FKBP5, GLRX, LEPR, MSR1, and MT1L, were specifically upregulated for controls and may represent responses to bacterial infection. One control patient (C07) had an ISG expression signature similar to CARDS patients, suggesting a putative undiagnosed viral infection in that patient.
Inflammatory Signatures During DXM Treatment
When comparing cytokine/chemokine gene expression of IL-1A, IL-1B, IL-2, IL-6, IL-8, IL-11, IL-17A, IL-17B, IL-18, TNF, IFN-λ, GM-CF, GCSF, MCP-1, IP-10, and MIP-1-α, only the IL-18 gene was differentially expressed (downregulated) in CARDS compared to controls. None of the genes were differentially regulated in CARDS with DXM treatment versus CARDS without DXM treatment.
DISCUSSION
We identified a distinct RNA expression profile of DXM-treated CARDS patients. Functional analysis of CARDS with DXM treatment revealed upregulation of cellular metabolism and rearrangement functions, enriched pathways for B-cell activation, antigen presentation, and complement responses. ISGs were particularly upregulated in nontreated CARDS patients and contrary to our expectations, key proinflammatory genes did not differ according to DXM treatment.
ARDS involves complex immunopathogenesis, which include influx of inflammatory cells (most notably neutrophils and macrophages), proinflammatory cytokines, proteases, and procoagulant factors in the lungs. A local hyperactive immune response has also been noted in CARDS and the term “cytokine storm” has been widely used to describe its pathophysiology. Although we and others have previously shown a high expression of inflammatory cytokines and chemokines in CARDS patients compared to healthy controls [3, 8], we did not find a differential expression when compared to critically ill controls. However, here we did find a total of 1613 and 317 other differentially expressed genes in CARDS patients with and without DXM treatment, respectively, of which only 53 were upregulated in both groups compared to controls. Although glucocorticoid treatment is known to suppress the production of at least some inflammatory cytokines in other diseases, and has indeed been posited to improve outcome in CARDS by suppressing the “cytokine storm”, we did not observe a differential gene expression according to DXM treatment of proinflammatory mediators in the lungs.
Clinical trials have provided evidence that treatment with DXM reduces mortality in patients hospitalized with COVID-19. Glucocorticoid treatment has, on the other hand, also been associated with increased mortality in other infectious diseases, including influenza. This discrepancy suggests a COVID-19–specific response profile that renders DXM effective, and it has been hypothesized that COVID-19 may induce glucocorticoid insensitivity in the process of modulating host cell activities. In our gene ontology analysis, complement responses, particularly genes related to the classical pathway initiated by antigen-antibody complexes, were upregulated in CARDS with DXM treatment. Complement activation is an important component of the innate immune response to viral infections. However, the clinical role of local complement activation in CARDS remains insufficiently understood. One study found increased levels of soluble C5a correlating with severity of COVID-19 with high expression levels of C5aR1 receptors in blood and pulmonary myeloid cells [9]. In contrast to our finding, studies unrelated to COVID-19 have shown reduced local complement activation following DXM treatment [10]. In addition, we found enriched B-cell signatures in gene ontology analysis, suggesting that DXM preferentially stimulates B-cell immunity to SARS-CoV-2. In line with this, early studies have revealed a direct effect of glucocorticoids on human B-cell survival and development [11]. We also found upregulation of the phagocytic pathway especially FC-γ receptor signaling, which is associated with antibody-bound viruses and concomitant phagocytosis.
Type I IFNs (eg, IFN-α and IFN-β) provide protective immunity against SARS-CoV-2 as highlighted by studies showing that inborn errors of TLR3- and TLR7-dependent antiviral innate immune pathways may underlie severe COVID-19 [12]. Previous in vitro studies of other viral infections have shown that glucocorticoids may suppress type-I IFN-mediated responses by inhibiting intracellular signaling pathways and subsequent expression of ISGs [13]. This correlated well with our observation of enrichment of ISGs specifically in the BALF of CARDS patients without DXM treatment. In addition, we found one distinct cluster of ISGs to be upregulated in CARDS patients with DXM treatment (TNK2, SSBP3, MAP3K14, PPKD2, ABTB2, GALNT2, CNP, SPSB1, SQLE, and DDIT4). Whether differential expression of these ISGs can be exclusively attributed to DXM, or whether enrichment of ISGs in these patients is beneficial, is unknown. Another study performing single-cell RNA sequencing revealed higher levels of ISGs in peripheral blood monocyte-macrophages and T cells from mild versus severe COVID-19 [14], which could suggest that an increased IFN response may be beneficial. Moreover, lung expression profiles in autopsy material from COVID-19 patients found a significantly longer hospitalization time for patients with low ISG expression compared to patients with high ISG expression [15]. Taken together, these results could point toward a dual and time-dependent role of the interferon response.
In contrast to prior COVID-19 studies assessing the local pulmonary immune response, in which BALF was primarily collected on clinical indication at a later stage and at the clinician’s discretion regarding site and volume of lavage, a strength of our study is the per protocol BAL sampling in the early course of severe disease progression using a uniform cross-sectional approach. Although we were able to identify specific DXM treatment effects, our sample size is small, which prevents us from adjusting for certain clinical factors such as symptom or treatment duration, and results should be confirmed in larger studies with several time points. Our results provide further insight into the pulmonary host response of CARDS and potential effects of DXM treatment, but they also raise several important questions and specifically challenge the idea that DXM dampens a local “cytokine storm”.
Supplementary Material
Contributor Information
Ulrik Fahnøe, Copenhagen Hepatitis C Program, Department of Infectious Diseases, Hvidovre Hospital and Department of Immunology and Microbiology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; Department of Infectious Diseases, Copenhagen University Hospital-Amager and Hvidovre Hospitals, Hvidovre, Denmark.
Andreas Ronit, Department of Infectious Diseases, Copenhagen University Hospital-Amager and Hvidovre Hospitals, Hvidovre, Denmark.
Ronan M G Berg, Department of Biomedical Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; Department of Clinical Physiology and Nuclear Medicine, Copenhagen University Hospital-Rigshospitalet, Copenhagen, Denmark; Centre for Physical Activity Research, Copenhagen University Hospital-Rigshospitalet, Copenhagen, Denmark; Neurovascular Research Laboratory, Faculty of Life Sciences and Education, University of South Wales, Pontypridd, United Kingdom.
Sofie E Jørgensen, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark.
Trine H Mogensen, Department of Infectious Diseases, Aarhus University Hospital, Aarhus, Denmark; Department of Biomedicine, Aarhus Research Center for Innate Immunology, Aarhus University, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Alexander P Underwood, Copenhagen Hepatitis C Program, Department of Infectious Diseases, Hvidovre Hospital and Department of Immunology and Microbiology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; Department of Infectious Diseases, Copenhagen University Hospital-Amager and Hvidovre Hospitals, Hvidovre, Denmark.
Troels K H Scheel, Copenhagen Hepatitis C Program, Department of Infectious Diseases, Hvidovre Hospital and Department of Immunology and Microbiology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; Department of Infectious Diseases, Copenhagen University Hospital-Amager and Hvidovre Hospitals, Hvidovre, Denmark; Laboratory of Virology and Infectious Disease, The Rockefeller University, New York, New York, USA.
Jens Bukh, Copenhagen Hepatitis C Program, Department of Infectious Diseases, Hvidovre Hospital and Department of Immunology and Microbiology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; Department of Infectious Diseases, Copenhagen University Hospital-Amager and Hvidovre Hospitals, Hvidovre, Denmark.
Ronni R Plovsing, Department of Anesthesiology and Intensive Care, Copenhagen University Hospital-Amager and Hvidovre Hospitals, Hvidovre, Denmark; Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank all the patients and their families for their participation. We thank Anna Louise Sørensen for her outstanding technical laboratory assistance. We thank the clinical staff and nurses at the Department of Anesthesiology and Intensive Care for their dedicated contribution.
Author contributions. U. F., A. R., R. M. G. B., J. B., and R. R. P. contributed to the initial analysis protocol and design of the study. R. R. P. was responsible for patient recruitment. A. R., A. U., and R. R. P. were responsible for bronchoalveolar lavage fluid collection and processing. U. F. was responsible for transcriptomic experiments and U. F., A. R., and T. K. H. S. were responsible for transcriptomic analyses and data processing. U. F. and A. R. prepared the first draft of the manuscript and completed all revisions. R. M. G. B., J. B., and R. R. P. handled funding. All authors provided critical input at all stages, interpreted the data, and were involved in drafting and editing of the manuscript.
Financial support. The study was supported by the Lundbeck Foundation (grant number R349-2020-540 to A. R., R. P., and R. M. G. B.); the Independent Research Fund Denmark (grant number 8020-00391B to J. B.); the Danish Cancer Society (grant number R204-A12639 to J. B.); and the Novo Nordisk Foundation (grant numbers NNF19OC0054518 and NNF19OC0055462 to J. B., NNF21OC0067157 to T. H. M.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data availability. All data produced in the present study are available upon reasonable request to the authors.
References
- 1. Tzotzos SJ, Fischer B, Fischer H, Zeitlinger M. Incidence of ARDS and outcomes in hospitalized patients with COVID-19: a global literature survey. Crit Care 2020; 24:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liao M, Liu Y, Yuan J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 2020; 26:842–84. [DOI] [PubMed] [Google Scholar]
- 3. Ronit A, Berg RMG, Bay JT, et al. Compartmental immunophenotyping in COVID-19 ARDS: a case series. J Allergy Clin Immunol 2021; 147:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saris A, Reijnders TDY, Nossent EJ, et al. Distinct cellular immune profiles in the airways and blood of critically ill patients with COVID-19. Thorax 2021; 76:1010–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ramirez S, Fernandez-Antunez C, Galli A, et al. Overcoming culture restriction for SARS-CoV-2 in human cells facilitates the screening of compounds inhibiting viral replication. Antimicrob Agents Chemother 2021; 65:e0009721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tomlinson JE, Wolfisberg R, Fahnøe U, et al. Pathogenesis, microrna-122 gene-regulation, and protective immune responses after acute equine hepacivirus infection. Hepatology 2021; 74:1148–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dittmann M, Hoffmann HH, Scull MA, et al. A serpin shapes the extracellular environment to prevent influenza a virus maturation. Cell 2015; 160:631–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou Z, Ren L, Zhang L, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 2020; 27:883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carvelli J, Demaria O, Vély F, et al. Association of COVID-19 inflammation with activation of the C5a–C5aR1 axis. Nature 2020; 588:146–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Surace L, Lysenko V, Fontana AO, et al. Complement is a central mediator of radiotherapy-induced tumor-specific immunity and clinical response. Immunity 2015; 42:767–77. [DOI] [PubMed] [Google Scholar]
- 11. Kovacs WJ. To B or not to B? Glucocorticoid impact on B lymphocyte fate and function. Endocrinology 2014; 155:339–41. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Q, Liu Z, Moncada-Velez M, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020; 370:eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marcellini A, Swieboda D, Guedán A, et al. Glucocorticoids impair type I IFN signalling and enhance rhinovirus replication. Eur J Pharmacol 2021; 893:173839. [DOI] [PubMed] [Google Scholar]
- 14. Xu G, Qi F, Li H, et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov 2020; 6:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nienhold R, Ciani Y, Koelzer VH, et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun 2020; 11:5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.