Abstract
Human germ cell development is a highly regulated process beginning soon after embryo implantation with the specification of primordial germ cells (PGCs) and ending in adulthood with the differentiation of gametes. Here, we show that fibroblast growth factor receptor 3 (FGFR3) is expressed by human PGCs during the first and second trimester, becoming repressed as PGCs differentiate into primordial oocytes. Using fluorescence-activated cell sorting (FACS) with antibodies that recognize FGFR3 followed by single-cell RNA sequencing, we show that isolating FGFR3-positive cells enriches for human PGCs. Taken together, FGFR3 could be used in future studies as a strategy to identify maturing PGCs in vitro.
Keywords: germ cells, FACS, FGFR3, PGCLCs, embryonic ovaries, embryonic testes
Highlights
-
•
Human primordial germ cells (PGCs) express FGFR3
-
•
FGFR3 RNA and protein are repressed as PGCs enter prophase I of meiosis I
-
•
FACS using FGFR3 antibodies can be used to enrich for PGCs from prenatal ovaries
-
•
PGC-like cells (PGCLCs) express FGFR3 following differentiation from hESCs
Stem cell biology benefits from in vivo reference maps of human cell types to enable comparisons with human cells generated in vitro using stem cell differentiation. In this study, Chitiashvili and colleagues use single cell RNA-seq to show that FGFR3 is expressed by human primordial germ cells (PGCs) in vivo and, using fluorescence-activated cell sorting with antibodies that recognize FGFR3, that PGCs can be enriched from prenatal ovarian tissue.
Introduction
Human reproduction depends on the correct establishment and differentiation of germ cells. Each generation, germ cell formation begins with the specification of primordial germ cells (PGCs) in the post-implantation embryo at the end of week 2 post-fertilization (pf) (Chen et al., 2019). At the end of week 3 pf, a cluster of PGCs can be identified in the yolk-sac endoderm, corresponding to specified PGCs that ultimately differentiate into eggs and sperm in the adult ovary or testis, respectively. Following specification, PGCs migrate from the yolk-sac niche through the dorsal mesentery of the hindgut where, starting at around week 5 pf, the migrating PGCs colonize a new niche, the genital-ridge epithelium (the future gonad). Once colonized in the developing fetal gonads, PGCs are variably referred to as gonocytes, late PGCs, fetal germ cells (FGCs), or oogonia when in fetal ovaries (Chen et al., 2017; Tang et al., 2016). Sex determination of the genital-ridge epithelial niche cells initiates at around week 6, with testicular PGCs beginning the process of differentiating into fetal spermatogonia also called prospermatogonia or fetal 0 (F0) at around week 14–16 (Guo et al., 2021). In the embryonic ovary, PGCs initiate the process of meiotic differentiation at week 9 and will arrest in prophase I of meiosis I as primordial oocytes starting from week 13–16. Creation of primordial follicles composed of a primordial oocyte and its surrounding layer of squamous granulosa cells begins at approximately 20 weeks pf (Konishi et al., 1986). Given the importance of PGC formation to egg and sperm formation in the adult, the cell and molecular basis of human PGC development is of significant importance to our basic understanding of human fertility and reproduction.
Isolation of PGCs from embryonic and fetal tissue using fluorescence-activated cell sorting (FACS) has transformed our ability to study PGC development in vivo and to enrich for PGC-like cells (PGCLCs) differentiated from human pluripotent stem cells in vitro. In addition, molecular analysis of FACS-isolated in vivo PGCs has led to the creation of important benchmarks for comparing and staging in vivo PGCs with in-vitro-differentiated PGCLCs (Gkountela et al., 2012, 2015; Irie et al., 2015; Guo et al., 2015; Chen et al., 2017; Chen et al., 2018).
So far, a small number of proteins expressed at the cell membrane of human PGCs have been used to enrich for PGCs from a single-cell suspension of human embryonic and fetal tissue in vivo, as well as PGCLCs in vitro. These include cKIT, TNAP, PDPN, CD38, ITGA6, and EPCAM (Gkountela et al., 2012; MacGregor et al. 1995; Sasaki et al., 2015; Irie et al., 2015; Fernandes et al., 2018; Mishra et al., 2021). In most cases, combinations of antibodies that recognize two or more of these cell-surface proteins are used in FACS strategies to isolate highly enriched PGC populations for further analysis. Therefore, the identification of additional surface molecules that could facilitate isolation of PGCs from somatic cells is warranted. A candidate new cell-surface receptor identified previously as being expressed by human testicular FGCs is fibroblast growth factor receptor 3 (FGFR3) (Ewen et al., 2013). However, it is not known whether FGFR3 protein is expressed by PGCs in the human fetal ovary or whether anti-FGFR3 antibodies conjugated to fluorescent tags could be used to isolate in vivo PGCs from prenatal ovarian tissue consented to research or PGCLCs generated in vitro.
To dissect the expression of FGFR3 in embryonic and fetal ovaries, we utilized previously published single-cell RNA (scRNA) sequencing datasets (Chitiashvili et al., 2020; Li et al., 2017) as well as immunofluorescence. We show that the FGFR3 gene is transcribed, translated, and present at the surface of embryonic PGCs from at least 5 weeks pf and that it becomes repressed as ovarian meiotic germ cells upregulate SCP3 and SPO11 in prophase I of meiosis I. In addition, we show that FACS can be used to enrich for FGFR3-positive PGCs from the human embryonic and fetal ovary but not PGCLCs differentiated from human pluripotent stem cells. Taken together, our data identify FGFR3 as a new surface marker that can enrich for PGCs from a single-cell suspension of embryonic ovarian cells and this approach could be used in future studies to identify PGCLCs differentiated from human pluripotent stem cells that have progressed toward gonadal-stage PGCs.
Results
FGFR3 is expressed by germ cells in the fetal testis and ovary
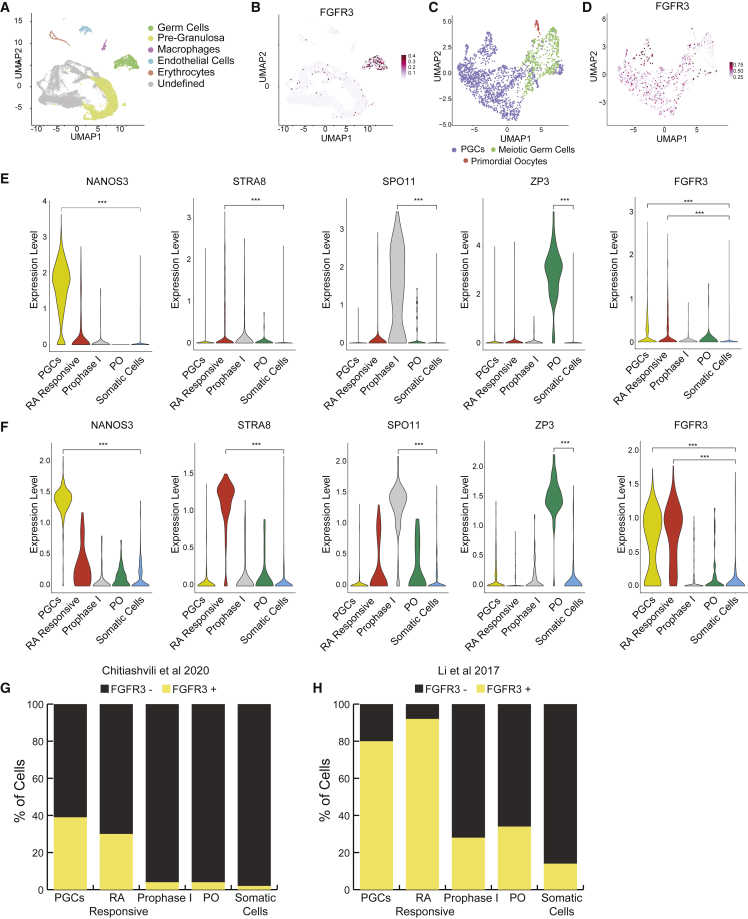
Previous studies have shown that FGFR3 protein is dynamically expressed during fetal testicular germ cell development (Ewen et al., 2013); however, the expression of FGFR3 by embryonic and fetal ovarian germ cells has not been reported. Utilizing a previously published scRNA sequencing (scRNA-seq) 10x Genomics dataset of human embryonic and fetal ovaries (Figure 1A) and testes (Figure S1A) from 6–16 weeks pf (Chitiashvili et al., 2020), we identify FGRF3 mRNA as being enriched in the germ cell population of each sex, with some expression of FGFR3 mRNA by rare somatic cells (Figures 1A–1D, S1A, and S1B).
Figure 1.
FGFR3 mRNA is expressed by PGCs in the prenatal human ovary
(A) Annotation of ovary cell types based on expression of cell-type-specific markers.
(B and C) Expression of (B) FGFR3 in ovarian cells data from (C) stage-specific annotation of ovarian germ cells.
(D) FGFR3 expression of female germ cells.
(E) Expression of marker genes including FGFR3 in PGCs, RA-responsive and prophase I meiotic germ cells primordial oocytes (PO) and ovarian somatic cells . Data from Chitiashvili et al. (2020).
(F) Similar to (E). Data from Li et al. (2017).
(G and H) FGFR3+ and FGFR-cell counts in PGCs, RA-responsive and prophase I meiosis I meiotic germ cells, primordial oocytes (POs), and somatic cells from Chitiashvili et al. (2020) and Li et al. (2017) datasets. ∗∗∗p < 0.001, Statistical significance was assessed by Wilcoxon test.
Given that FGFR3 is more enriched in germ cells of the prenatal ovary relative to the somatic cells, we focused our analysis on the germ cells. To identify the stage-specific expression of FGFR3 mRNA in embryonic and fetal ovarian germ cells, we defined the PGC population (positive for NANOG, POU5F1, BLIMP1, TFAP2C, and NANOS3), the meiotic population (positive for STRA8, ZGLP1, SPO11, and SYCP1), and primordial oocytes (positive for ZP3) (Figures S2A and S2B). Using these annotations, we discovered that FGFR3 mRNA is predominantly expressed in PGCs and retinoic acid (RA)-responsive meiotic germ cells (Figure 1E). Given that FGFR3 mRNA appears to be detectable in more cells within the PGC cluster relative to the meiotic cluster (Figures 1C and 1D), we hypothesized that FGFR3 may be repressed as the ovarian germ cells progress through meiosis. To address this, we separated the meiotic germ cell cluster into the initial RA-responsive stage defined by expression of ZGLP1 and STRA8 (Li et al., 2017), and prophase I of meiosis I, defined by expression of SPO11 and SYCP1 (Li et al., 2017; Vértesy et al., 2018; Chitiashvili et al., 2020). This analysis shows that as PGCs respond to RA to enter meiosis, FGFR3 mRNA is still expressed and becomes repressed as the meiotic germ cells express SPO11. To confirm these results in a second dataset, we examined FGFR3 expression using the scSMART-seq dataset published by Li et al., which covers 5–26 weeks pf (Li et al., 2017). This analysis corroborated that FGFR3 mRNA is predominantly expressed by PGCs and RA-responsive meiotic germ cells (Figure 1F). However, due to methodological differences between SMART-seq2 and 10x Genomics platforms, higher dropout rate of FGFR3 detection was observed in 10x datasets (Figures 1G and 1H) (Stegle et al. 2015; Wang et al., 2021). While FGFR3 was below the detection limit in the vast majority of somatic cells, rare somatic cells were detected in both the 10x Genomics and SMART-seq2 datasets that expressed FGFR3 mRNA (Figures 1G and 1H). In summary, utilizing scRNA-seq data from either the 10x Genomics (Chitiashvili et al., 2020) or the SMART-seq (Li et al., 2017) platform, we show that FGFR3 mRNA is expressed by PGCs in the prenatal ovary and becomes repressed as the PGCs enter prophase I of meiosis I toward the formation of primordial oocytes.
FGFR3 protein is expressed by ovarian PGCs and small VASA+ germ cells in ovarian cortical cords
Since RNA and protein may have different expression dynamics (Cheng et al., 2016), we next evaluated expression of FGFR3 protein in embryonic and fetal ovaries using immunofluorescence from weeks 7–14 pf (Figure 2). This time window of prenatal life was chosen because it corresponds to the window of human development when PGCs initiate the process of meiotic entry within the cortical cords of the developing ovary (Figure 2A) (Gkountela et al., 2012). Human PGCs are detected by immunostaining for TFAP2C+/VASAlow (Gkountela et al., 2012), whereas meiotic germ cells in the cortical cords are defined as TFAP2C-/VASA+ (Figure 2A). Using these criteria at weeks 7, 13, and 14 pf, we show that >90% of TFAP2C+/VASAlow PGCs express FGFR3 (Figure 2B). In contrast, when focusing on the TFAP2C-/VASA+ germ cells in the cortical cords, ∼60% of VASA+ germ cells are positive for FGFR3 (Figure 2C). These results indicate that FGFR3 protein is expressed by the majority of PGCs in the embryonic and fetal ovary between weeks 7–14 pf before becoming repressed in the VASA + meiotic germ cells located in the cortical cords.
Figure 2.
FGFR3 protein is expressed by PGCs in the prenatal human ovary
(A) Schematic representation of germ cell differentiation in prenatal ovaries. TFAP2C+/VASAlow PGCs mostly reside at the cortex of the ovary. With time, PGCs differentiate and advance toward meiosis by upregulating expression of VASA and silencing TFAP2C. Right-hand side: immunostaining of ovarian tissues at weeks 7, 10, and 14 with TFAP2C (magenta), FGFR3 (yellow), VASA (cyan), and DAPI (blue).
(B) Quantification of the percentage of FGFR3+ and FGFR- cells that express TFAP2C and low levels of VASA (50 cells were counted for each sample).
(C) Quantification of FGFR3+ and FGFR3- cells that are negative for TFAP2C and positive for VASA (n = 3 samples for each category; 50 cells were counted from each sample).
(D) Immunostaining of a 14 week ovary with TFAP2C (magenta), FGFR3 (yellow), VASA (cyan), and DAPI (blue).
(E) Nuclei size of VASA+/FGFR3+ and VASA+/FGFR3- ovarian germ cells (25 cells were measured for each category from a 14 week ovary). ∗∗∗p < 0.001. Statistical significance was assessed by Wilcoxon test.
Upon closer inspection of VASA+ germ cells in the cortical cords, it appeared that the FGFR3+ germ cells were smaller than VASA+/FGFR3- germ cells (Figure 2D). This is important, as the transition of germ cells into meiosis is associated with an increase in cell size (Gondos et al. 1971). To quantify this, we measured the size of VASA+/FGFR3- and VASA+/FGFR3+ germ cell nuclei in the cords. This approach revealed that the FGFR3+ germ cells are significantly smaller than the FGFR3- germ cells (Figure 2E). Taken together, these results show that FGFR3 protein is expressed by the majority of PGCs in the prenatal ovary from weeks 7–14 pf. Moreover, during cortical-cord formation, the smaller VASA+ germ cells continue to express FGFR3+, whereas the larger VASA+ germ cells are FGFR3-.
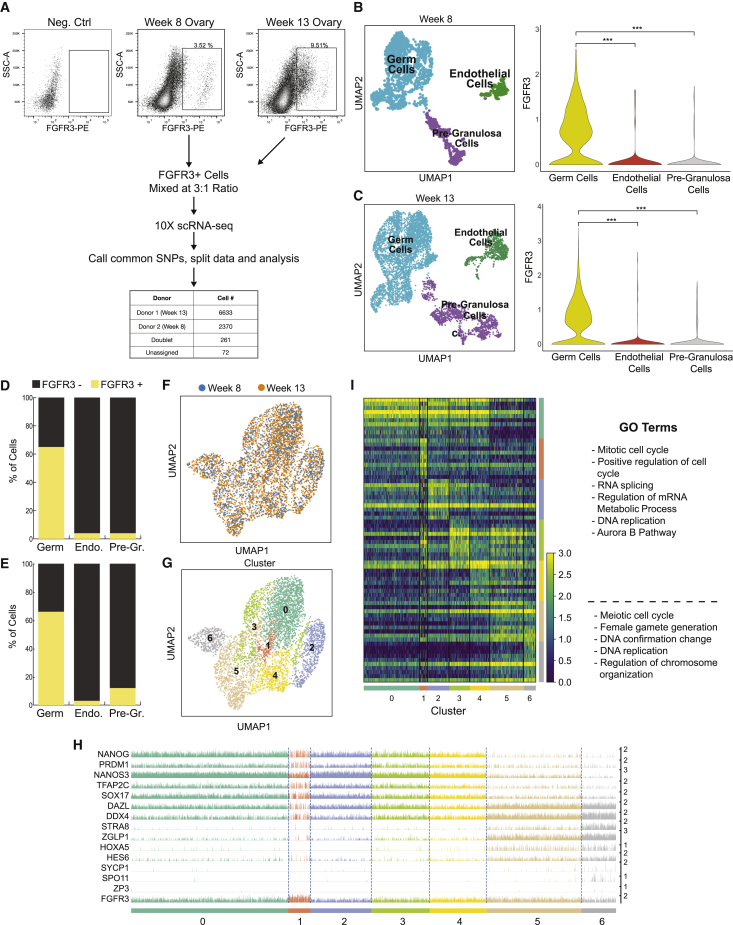
Flow cytometry for FGFR3 enriches for ovarian PGCs and meiotic germ cells
Given that FGFR3 defines PGCs in the prenatal ovary between 7 and 14 weeks, we next evaluated whether FGFR3 could be used to enrich for germ cells from single-cell suspensions of embryonic and fetal ovarian cells using FACS. For this purpose, we dissociated ovaries from two different developmental time points, one at week 8 pf and the second at week 13 pf, and stained each single-cell suspension with antibodies that recognize FGFR3 and are conjugated to phycoerythrin (PE) (Figure 3A). Since the absolute number of female germ cells in a single ovary at week 8 or 13 pf is below the limit of cells required for a single 10x Genomics scRNA-seq run (Gkountela et al., 2012), we collected FGFR3+ cells from each sample and combined them after FACS. We reasoned that the samples could be demultiplexed during subsequent data analysis by discriminating between single-nucleotide polymorphisms (SNPs) present in the different donor samples. Since we did not have access to parental reference genomes, we combined the samples using a discordant ratio of 3 (week 13 pf) to 1 (week 8 pf) (Figure 3A). Following common SNP analysis, the assignment of cells to the samples was in the predicted 3:1 ratio. Therefore, we assigned 6,633 FGFR3+ cells to the week 13 pf ovary and 2,371 cells to the week 8 pf ovary (Figure 3A). 261 and 72 cells were designated as doublets or could not be assigned, respectively, and were therefore excluded from further analysis.
Figure 3.
FACS for FGFR3 enriches for PGCs from single-cell suspensions of prenatal ovaries
(A) Fluorescence activated cell sorting (FACS) strategy to isolate female germ cells based on FGFR3 antibody labeling. Cells from week 8 and 13 ovaries were mixed at 1:3 ratio accordingly to create one 10x Genomics scRNA-seq library. After sequencing, the library was split into two by identifying common single nucleotide polymorphisms (SNPs).
(B) Annotation of FGFR3-sorted cells from the week 8 ovary and expression of FGFR3 on the right-hand side.
(C) Similar to (B) for cells from the week 13 ovary.
(D and E) FGFR3+ and FGFR3- cell counts in germ cells (Germ), endothelial cells (Endo), and pre-granulosa cells (Pre-Gr) from FGFR3-sorted week 8 and 13 ovaries, respectively.
(F) UMAP clustering of germ cells from week 8 and 13 ovaries.
(G) Sorted germ cells displayed on UMAP plot by their cluster numbers.
(H) Ordering of sorted germ cells along the developmental trajectory from cluster 0 to 6, with classification as PGCs (clusters 0–4) and meiotic germ cells (clusters 5–6), based on diagnostic germ cell marker expression. Each cluster contains individual cells (columns) for which the expression of the indicated marker is given (rows).
(I) Heatmap representation of top 10 expressed genes per cluster of germ cells from (F). Gene Ontology analysis highlighting terms per category. ∗∗∗p < 0.001. Statistical significance was assessed by Wilcoxon test.
Analysis of cell identity within the FACS isolated cells of the embryonic ovaries at week 8 pf and fetal ovaries at week 13 pf revealed that ∼70% of the cells isolated by FACS corresponded to germ cells (5,213 cells). These germ cells were of equivalent identity regardless of whether the cells were isolated at week 8 or 13 (Figures 3F–3G, S3A, and S3B). Surprisingly, we also identified non-germ cells (2,447 cells) at each time point, and these are predicted to correspond to pre-granulosa (FOXL2) and endothelial (PECAM1) cells (Figures 3B, 3C, S3A, and S3B). The mRNA expression levels of FGFR3 in these somatic cells were on average lower than germ cells at both week 8 and 13 pf samples (Figures 2B and 2C, right panels). However, when selecting only the FGFR3+ pre-granulosa (164 cells), endothelial (33 cells) and germ (3,416 cells) cells at 8 and 13 pf for comparison of FGFR3 levels, the expression of FGFR3 in these cells is similar to or, in the case of some endothelial cells, even higher than PGCs (Figures 3D, 3E, S3C, and S3D). These data support the notion that FGFR3 protein is detectable on rare sub-populations of pre-granulosa cells and on even rarer sub-populations of endothelial cells, and this can be detected by FACS. Next, we asked whether FGFR3 protein can be detected on pre-granulosa cells of the fetal ovary using immunofluorescence. For this, we performed immunofluorescence for the pre-granulosa cell marker FOXL2 together with FGFR3. After counting 70 FOXL2+ cells from two independent experiments, we could not detect FGFR3 protein expression on FOXL2+ cells, suggesting that FGFR3+/FOXL2+ cells are very rare in the fetal ovary and are difficult to detect by immunofluorescence (Figures S3E and S3F). Taken together, FACS for FGFR3+ enriches for embryonic and fetal ovarian germ cells; however, our data indicate that performing FACS for FGFR3 does not lead to a pure population of PGCs, as ∼30% of analyzed cells after scRNA-seq correspond to granulosa and endothelial cells.
To evaluate whether the ovarian prenatal germ cells enriched by FACS correspond to both PGCs and RA-responsive meiotic germ cells (which would be predicted based on the unsorted 10x Genomics data of Figure 1), we re-clustered the FGFR3+ germ cells, resulting in 7 different groups (Figures 3F and 3G). Gene expression analysis revealed that clusters 0–4 correspond to PGCs that express genes such as NANOG, PRDM1, TFAP2C, SOX17, DAZL, and VASA, with Gene Ontology (GO) analysis indicating an enrichment in terms associated with mitotic cell-cycle genes (Figures 3H and 3I). In contrast, cells in clusters 5 and 6 correspond to the meiotic germ cells, with GO analysis indicating an enrichment in terms associated with gamete generation (Figures 3H and 3I). Rare populations of cells expressing the meiotic prophase I genes including SPO11 were also identified (Figures 3H and S3B). Whereas, primordial oocytes characterized by expression of zona pellucida 3 (ZP3), were absent in FGFR3-sorted germ cells (Figure 3H). Taken together, these results demonstrate that separation of prenatal ovarian cells using FACS from week 8 and 13 with antibodies that recognize FGFR3 results in the enrichment of female PGCs and RA-responsive meiotic germ cells but not ZP3+ primordial oocytes.
FGFR3 RNA and protein is expressed by early PGCs in vivo
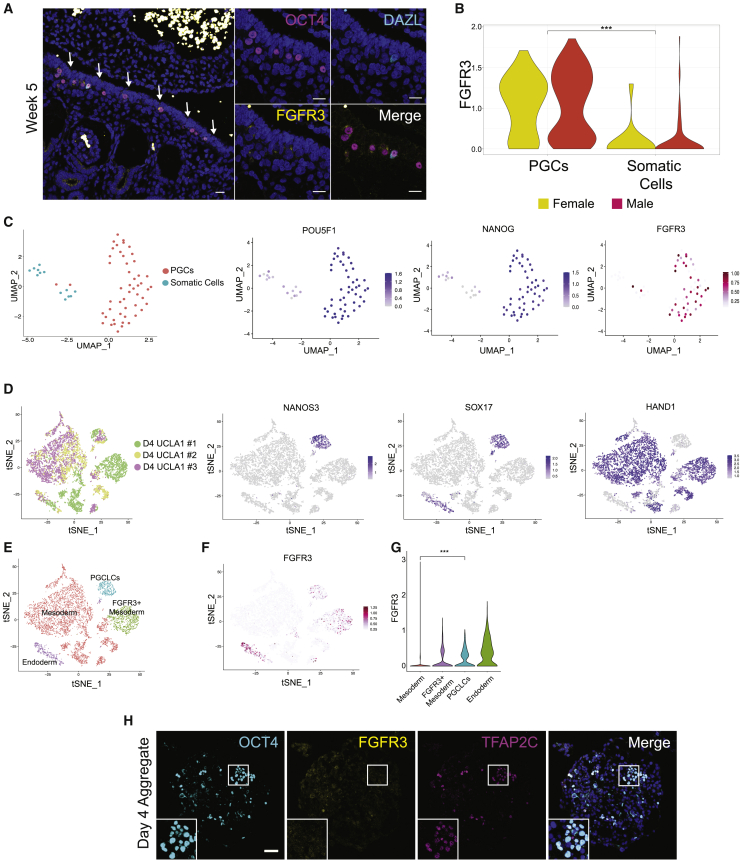
The specification of PGCs begins at the end of week 2 pf (Chen et al., 2019). This is approximately 3–4 weeks before the somatic cells of the ovary begin to form from the mesoderm of the genital ridge between week 5–6 pf. In order to evaluate FGFR3 expression in PGCs at the time of genital-ridge colonization, we performed immunofluorescence of a single human embryo at week 5, where both early PGCs, defined as OCT4+/DAZL-, and late-PGCs, defined as OCT4+/DAZL+, are identified (Figure 4A). Evaluation of FGFR3 in this specimen revealed rather weak but detectable punctate expression of FGFR3 protein signal on all OCT4+/DAZL- PGCs (11 cells) as well as the single OCT4+/DAZL+ PGC (Figure 4A). This result indicates that FGFR3 is expressed by early PGCs at the time of gonadal colonization. To confirm this result, we examined the week 4 and 5 pf SMART-seq data of Li et al., (2017) (Figures 4B and 4C) and found FGFR3 expression in the majority of POU5F1 (OCT4+) PGCs at this early gestational time point.
Figure 4.
FGFR3 mRNA and protein are expressed by PGCs before gonadal colonization
(A) Immunostaining of a week 5 embryo showing the genital ridge. OCT4 (magenta), DAZL (cyan), FGFR3 (yellow) and DAPI (blue). n = 1 sample.
(B) Expression of FGFR3 in female and male PGCs at weeks 4 and 5 from Li et al. (2017) dataset.
(C) Week 4 and 5 germ cells and somatic cells depicted on a UMAP displaying expression of POU5F1 (OCT4), NANOG, and FGFR3.
(D) Day 4 PGCLCs in three replicates (Chen et al., 2019) differentiated from UCLA1 hESCs and expression of diagnostic markers: NANOS3 for PGCLCs, SOX17 for endoderm cells, HAND1 for mesoderm cells and SOX17/NANOS3 double positive cells for PGCLCs.
(E) Annotation of D4 aggregate cells based on marker expression from (D).
(F) Expression of FGFR3 depicted at t-SNE maps and violin plots (G).
(H) UCLA1 female hESCs differentiated for 4 days to create aggregates and immunostained for OCT4 (cyan), FGFR3 (yellow), TFAP2C (magenta), and DAPI (blue) (n = 2 differentiation experiments). ∗∗∗p < 0.001, Statistical significance was assessed by Wilcoxon test.
To evaluate the expression of FGFR3 in PGCLCs in vitro, we differentiated human embryonic stem cells (hESCs) using a two-step differentiation protocol involving an incipient mesoderm-like cell (iMELC) intermediate, as previously described (Sasaki et al., 2015). In this model, PGCLC induction begins 24–48 h after BMP4 exposure, with fully specified PGCLCs identified at day 4 (Chen et al., 2019). Examining three independent differentiation experiments of the hESC line UCLA1 at D4 using the 10x Genomics RNA-seq pipeline reveals a distinct cluster of PGCLCs defined as co-expressing NANOS3, NANOG, and SOX17 (Figure 4D) (Chen et al., 2017b). Displaying FGFR3 RNA expression on this dataset shows that FGFR3 mRNA is detected in the PGCLC population and the SOX17+ endodermal cells as well as a sub-population of HAND1+ mesoderm cells (Figures 4E–4G). To evaluate protein expression, we performed immunofluorescence at day 4 following BMP4 exposure. Using immunofluorescence, our results suggest that FGFR3 protein is below the limit of detection in the clusters of OCT4+/TFAP2C+ PGCLCs at day 4 (Figures 4H and S4). Taken together, FGFR3 protein is expressed by PGCs in vivo from as early as week 5 pf and, at the RNA level is expressed by hESC-derived hPGCLCs at day 4 in vitro.
Discussion
In this work, we show that FGFR3 is expressed by PGCs in prenatal ovaries and testis. Additional characterization of FGFR3 expression by scRNA-seq and immunofluorescence revealed that female PGCs maintain FGFR3 expression into the initial stages of meiotic progression, including the RA-responsive stage of meiosis and the beginning of prophase I of meiosis I. However, ZP3+ primordial oocyte formation is associated with the repression of FGFR3.
In adult mouse ovaries, FGFR3 is expressed on the membrane of granulosa cells in the growing follicle but not by the oocyte or the theca cells (Amsterdam et al., 2001). Consistent with this expression pattern, FGFR3 activating mutations in anchondroplastic female mice causes loss of granulosa cells and ultimately infertility (Amsterdam et al., 2001). In adult bovine ovaries, FGFR3 is also detected on granulosa cells, with FGFR3 expression levels positively associated with increasing exposure to follicle-stimulating hormones (Buratini et al., 2005). In the current study, we show that the expression pattern of FGFR3 in the embryonic and fetal ovary is different from the adult, with FGFR3 predominantly expressed by the PGCs and meiotic germ cells (Figure 1). Our data suggest that a small sub-population of FOXL2+ pre-granulosa cells begin to express FGFR3 and can be isolated by FACS; however, these cells are very rare and difficult to detect using immunofluorescence (Figures 1G, 1H, 3D, 3E, S3C, and S3D).
In the current study, we show that FGFR3 is expressed by male PGCs and fetal prospermatogonia (Figure S1). Although the expression of FGFR3 has not been evaluated in the adult human testis, in the adult mouse testis, FGFR3 is expressed by spermatogonia in vivo (von Kopylow et al., 2012; Mayerhofer et al., 1991; Steger et al., 1998) and also in vitro (Kubota et al. 2004). As adult spermatogonia are recruited into meiosis, FGFR3 is repressed (Guo et al., 2017). This suggests that FGFR3 might have a similar role in the survival and proliferation of diploid germline cells prior to entering meiosis (Guo et al., 2017). Indeed, rare gain-of-function mutations in FGFR3 gene have been associated with abnormal sperm formation and testicular cancer (Goriely and Wilkie 2012; Goriely et al., 2009).
In this work, we also demonstrate that sorting for FGFR3+ cells is a viable strategy that will enrich for human PGCs and meiotic germ cells from the embryonic and fetal ovary. However, FACS for FGFR3 also leads to the isolation of a smaller population of somatic cells that we found difficult to detect using immunofluorescence, but do express appreciable levels of FGFR3 mRNA when compared with PGCs (Figures S3). Therefore, moving forward, using anti-FGFR3 antibodies could be a powerful marker in combination with other markers such as cKIT (Gkountela et al., 2012) to isolate PGCs and meiotic germ cells from prenatal ovaries.
In summary, although FGFR3 is expressed by some somatic cells in the prenatal ovary, it is predominantly expressed by PGCs and RA-responsive meiotic germ cells. Therefore, we propose that FGFR3 can be used to enrich for those germ cell populations and potentially be used as a diagnostic surface marker for further improvements of PGCLC differentiation protocols.
Experimental procedures
Human fetal tissues
University of Washington Birth Defects Research Laboratory (BDRL) provided all prenatal gonads (5–15 weeks pf) for this study. At the BDRL, prenatal gonads were obtained with regulatory oversight from the University of Washington IRB-approved Human Participants protocol, combined with a Certificate of Confidentiality from the federal government. BDRL collected the fetal ovaries and shipped them overnight in HBSS with an ice pack for immediate processing at UCLA. All human fetal tissues used here were obtained following informed consent. No personal identifiers were carried about the tissues sent to UCLA. Donors have not received any payments as they knowingly and willingly consented to provide research materials without restrictions for research and for use without identifiers. Developmental age was documented by the BDRL.
scRNA-seq data analysis
For Figures 1, 4, S1, and S2, previously published scRNA-seq datasets were downloaded and re-analyzed for this paper. All scRNA-seq datasets, from prenatal ovarian and testicular cells (Chitiashvili et al., 2020), FGCs (Li et al., 2017), to day 4 aggregates (Chen et al., 2019) generated for this paper were aligned to the human hg38 genome assembly using 10x Genomics Cell Ranger v.3.4. Expression matrixes generated by Cell Ranger were imported into Seurat (Hao et al., 2021) or Scanpy (Wolf et al. 2018) for downstream analysis. First, all libraries were merged, and cells were filtered in the same manner. All the genes that were expressed in less than five cells were discarded and cells with less than 250 detected genes were filtered out. The unique molecular identifier (UMI) counts were then normalized for each cell by the total expression, multiplied by 10,000, and log transformed. Using Scanpy’s default method, highly variable genes were identified, and data were scaled to regress out variation from UMI counts and mitochondrial genes. Cells were clustered using the Louvain algorithm, and the uniform manifold approximation and projection (UMAP) and t-distributed stochastic neighbor embedding (t-SNE) packages were used to visualize cells in a two-dimensional plot. Germ cell clusters were identified by expression of germ-cell-specific markers, such as NANOS3, DAZL, DDX4, and SYCP1. Gonadal somatic cells were annotated by previously published literature (Li et al., 2017; Chitiashvili et al., 2020; Guo et al., 2021).
Demultiplexing samples using SNPs
To demultiplex cells from scRNA-seq library and assign them to a week 8 or 13 sample, we have first aligned the data to human hg38 genome assembly using 10x Genomics Cellranger. Afterward, a bam file generated by Cell Ranger was used in a cellSNP-lite v.0.3 tool that can detect expressed alleles in scRNA-seq datasets (Huang and Huang 2021). cellSNP-lite uses a list of candidate SNPs from 1000 Genome Project. After creating the list of expressed SNPs, an output file was passed into Vireo v.0.4.1 to deconvolve the donors (Huang et al., 2019). Following deconvolution of the donors, we got 6,633 cells from donor 1 and 2,370 cells from donor 2, while 261 cells were assigned as doublets and 72 cells were not assigned to any donor. Doubles and unassigned cells were filtered out from downstream analysis. Since we mixed the cells at a 1:3 ratio at the beginning of the experiment, we could determine that 6,633 cells were from the week 13 ovary and 2,370 were from week 8.
Data and code availability
The scRNA-seq data of prenatal tissues reported in this paper are available under the following accession numbers: Database: GSM5808297 (FGFR3-sorted cells) and Database: GSM5808298 (UCLA1 day 4 aggregate). Previously published scRNA-seq data from prenatal ovarian and testicular cells (Chitiashvili et al., 2020), FGCs (Li et al., 2017), and day 4 aggregates (Chen et al., 2019) are under following accession numbers: Database: GSE143380 (ovarian cells), Database: GSE143356 (testicular cells) (Chitiashvili et al., 2020), Database: GSE86146 (Li et al., 2017), and Database: GSE140021(Chen et al., 2019).
Custom scripts used for aligning scRNA-seq, data processing, and plotting are available upon request.
Author contributions
T.C., K.P., and A.C. designed the experiments. T.C. conducted immunofluorescent experiments on tissues and PGCLC sections. F.-m.H. contributed to hPGCLC differentiation experiments. T.C. performed scRNA-seq experiments, and the resulting data were analyzed by T.C and I.D. T.C., K.P., and A.C. interpreted all data and wrote the manuscript.
Conflicts of interests
A.C. and K.P. are members of Stem Cell Reports editorial board. A.C. is a member of the executive committee of ISSCR.
Acknowledgments
We would like to thank the Flow Cytometry, Next Generation Sequencing, and Microscopy cores at the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Center (BSCRC) for help with cell sorting, sequencing, and imaging. Also, we would like to thank the Technology Center for Genomics and Bioinformatics at the UCLA Johnson Comprehensive Cancer Center (JCCC) and the Translational Pathology Core Laboratory for help with histology. T.C. was supported by a Boehringer Ingelheim PhD Fellowship. This work (excluding the research with human fetal tissue) was supported by funds from the NIH to A.C. (R01HD079546). K.P. was supported by the BSCRC at UCLA, the David Geffen School of Medicine at UCLA, and the UCLA JCCC, the NIH (R01HD098387, P01GM099134), and a Faculty Scholar grant from the Howard Hughes Medical Institute. Human fetal tissue research is supported by a grant to Ian Glass at the University of Washington Birth Defects laboratory (5R24HD000836-53). Human conceptus tissue requests can be made to bdrl@u.washington.edu.
Published: May 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.04.015.
Supplemental information
References
- Amsterdam A., Kannan K., Givol D., Yoshida Y., Tajima K., Ada D. Apoptosis of granulosa cells and female infertility in achondroplastic mice expressing mutant fibroblast growth factor receptor 3G374R. Mol. Endocrinol. 2001;15:1610–1623. doi: 10.1210/mend.15.9.0700. [DOI] [PubMed] [Google Scholar]
- Buratini J., Teixeira A.B., Costa I.B., Glapinski V.F., Pinto M.G.L., Giometti I.C., Barros C.M., Cao M., Nicola E.S., Price C.A. Expression of fibroblast growth factor-8 and regulation of cognate receptors, fibroblast growth factor receptor-3c and -4, in bovine antral follicles. Reproduction. 2005;130:343–350. doi: 10.1530/rep.1.00642. [DOI] [PubMed] [Google Scholar]
- Chen D., Liu W., Lukianchikov A., Hancock G.V., Zimmerman J., Lowe M.G., Kim R., Galic Z., Irie N., Surani M.A., et al. Germline competency of human embryonic stem cells depends on eomesodermin. Biol. Reprod. 2017;97:850–861. doi: 10.1093/biolre/iox138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Liu W., Zimmerman J., Pastor W.A., Kim R., Hosohama L., Ho J., Aslanyan M., Gell J.J., Jacobsen S.E., Clark A.T. The TFAP2C-regulated OCT4 naive enhancer is involved in human germline formation. Cell Rep. 2018;25:3591–3602.e5. doi: 10.1016/j.celrep.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Sun N., Hou L., Kim R., Faith J., Aslanyan M., Yu T., Zheng Y., Fu J., Liu W., et al. Human primordial germ cells are specified from lineage-primed progenitors. Cell Rep. 2019;29:4568–4582.e5. doi: 10.1016/j.celrep.2019.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Teo G., Krueger S., Rock T.M., Koh H.W.L., Choi H., Vogel C. Differential dynamics of the mammalian MRNA and protein expression response to misfolding stress. Mol. Syst. Biol. 2016;12:855. doi: 10.15252/msb.20156423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitiashvili T., Dror I., Kim R., Hsu F.M., Chaudhari R., Pandolfi E., Chen D., Liebscher S., Schenke-Layland K., Plath K., Clark A. Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation. Nat. Cell Biol. 2020;22:1436–1446. doi: 10.1038/s41556-020-00607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen K.A., Olesen I.A., Winge S.B., Nielsen A.R., Nielsen J.E., Graem N., Juul A., Rajpert-De Meyts E. Expression of FGFR3 during human testis development and in germ cell-derived tumours of young adults. Int. J. Dev. Biol. 2013;57:141–151. doi: 10.1387/ijdb.130022er. [DOI] [PubMed] [Google Scholar]
- Gkountela S., Li Z., Vincent J.J., Zhang K.X., Chen A., Pellegrini M., Clark A.T. The ontogeny of CKIT+ human primordial germ cells proves to Be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat. Cell Biol. 2012;15:113–122. doi: 10.1038/ncb2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S., Zhang K.X., Shafiq T.A., Liao W.W., Hargan-Calvopiña J., Chen P.Y., Clark A.T. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M.G., Bialecka M., Salvatori D.C.F., Chuva de Sousa Lopes S.M. Characterization of migratory primordial germ cells in the aorta-gonad-mesonephros of a 4.5-week-old human embryo: a toolbox to evaluate in vitro early gametogenesis. Mol. Hum. Reprod. 2018;24:233–243. doi: 10.1093/molehr/gay011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondos B., Bhiraleus P., Hobel C.J. Ultrastructural observations on germ cells in human fetal ovaries. Am. J. Obstet. Gynecol. 1971;110:644–652. doi: 10.1016/0002-9378(71)90245-6. [DOI] [PubMed] [Google Scholar]
- Goriely A., Hansen R.M.S., Taylor I.B., Olesen I.A., Jacobsen G.K., McGowan S.J., Pfeifer S.P., McVean G.A.T., Rajpert-De Meyts E., et al. Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat. Genet. 2009;41:1247–1252. doi: 10.1038/ng.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriely A., Wilkie A.O.M. Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am. J. Hum. Genet. 2012;90:175–200. doi: 10.1016/j.ajhg.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Yan L., Guo H., Lin L., Hu B., Zhao Y., Yong J., Wang X., Wei Y., Wang W., et al. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Guo J., Grow E.J., Yi C., Mlcochova H., Maher G.J., Lindskog C., Patrick J., Carrell D.T., Goriely A., Hotaling J.M., et al. Chromatin and single-cell RNA-seq profiling reveal dynamic signaling and metabolic transitions during human spermatogonial stem cell development. Cell Stem Cell. 2017;21:533–546.e6. doi: 10.1016/j.stem.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Sosa E., Chitiashvili T., Nie X., Javier Rojas E., Oliver E., DonorConnect, Plath K., Hotaling J.M., Stukenborg J.B., et al. Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell. 2021;28:764–778.e4. doi: 10.1016/j.stem.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Stephanie H., Andersen-Nissen E., Mauck W.M., III, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., McCarthy D.J., Oliver S. Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol. 2019;20:273. doi: 10.1186/s13059-019-1865-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Huang Y. Cellsnp-lite: an efficient tool for genotyping single cells. Bioinformatics. 2021;37:4569–4571. doi: 10.1093/bioinformatics/btab358. [DOI] [PubMed] [Google Scholar]
- Irie N., Weinberger L., Tang W.W.C., Kobayashi T., Viukov S., Manor Y.S., Dietmann S., Hanna J.H., Azim Surani M. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi I., Fujii S., Okamura H., Parmley T., Mori T. Development of interstitial cells and ovigerous cords in the human fetal ovary: an ultrastructural study. J. Anat. 1986;148:121–135. [PMC free article] [PubMed] [Google Scholar]
- Kubota H., Avarbock M.R., Brinster R.L. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. U S A. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X., et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20:891–892. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- MacGregor G.R., Zambrowicz B.P., Soriano P. Tissue non-specific alkaline phosphatase is expressed in both embryonic and extraembryonic lineages during mouse embryogenesis but is not required for migration of primordial germ cells. Development. 1995;121:1487–1496. doi: 10.1242/dev.121.5.1487. [DOI] [PubMed] [Google Scholar]
- Mayerhofer A., Russell L.D., Grothe C., Rudolf M., Gratzl M. Presence and localization of a 30-KDa basic fibroblast growth factor-like protein in rodent testes. Endocrinology. 1991;129:921–924. doi: 10.1210/endo-129-2-921. [DOI] [PubMed] [Google Scholar]
- Mishra S., Taelman J., Chang Y.W., Boel A., de Sutter P., Heindryckx B., De Sousa Lopes S.M.C. Sex-specific isolation and propagation of human premeiotic fetal germ cells and germ cell-like cells. Cells. 2021;10:1214. doi: 10.3390/cells10051214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Yokobayashi S., Nakamura T., Okamoto I., Yabuta Y., Kurimoto K., Ohta H., Moritoki Y., Iwatani C., Tsuchiya H., et al. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Steger K., Frank T., Seitz J., Seitz J., Grothe C., Bergmann M. Localization of fibroblast growth factor 2 (FGF-2) protein and the receptors FGFR 1–4 in normal human seminiferous epithelium. Histochem. Cell Biol. 1998;110:57–62. doi: 10.1007/s004180050265. [DOI] [PubMed] [Google Scholar]
- Stegle O., Teichmann S.A., Marioni J.C. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- Tang W.W.C., Kobayashi T., Irie N., Dietmann S., Azim Surani M. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- Vértesy Á., Arindrarto W., Roost M.S., Reinius B., Torrens-Juaneda V., Bialecka M., Moustakas I., Ariyurek Y., Kuijk E., Mei H., et al. Parental haplotype-specific single-cell transcriptomics reveal incomplete epigenetic reprogramming in human female germ cells. Nat. Commun. 2018;9:1873. doi: 10.1038/s41467-018-04215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kopylow K., Hannah S., Schulze W., Will H., Kirchhoff C. Fibroblast growth factor receptor 3 is highly expressed in rarely dividing human type A Spermatogonia. Histochem. Cell Biol. 2012;138:759–772. doi: 10.1007/s00418-012-0991-7. [DOI] [PubMed] [Google Scholar]
- Wang X., He Y., Zhang Q., Ren X., Zhang Z. Direct comparative analyses of 10X genomics chromium and smart-seq2. Genomics Proteomics Bioinformatics. 2021;19:253–266. doi: 10.1016/j.gpb.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data of prenatal tissues reported in this paper are available under the following accession numbers: Database: GSM5808297 (FGFR3-sorted cells) and Database: GSM5808298 (UCLA1 day 4 aggregate). Previously published scRNA-seq data from prenatal ovarian and testicular cells (Chitiashvili et al., 2020), FGCs (Li et al., 2017), and day 4 aggregates (Chen et al., 2019) are under following accession numbers: Database: GSE143380 (ovarian cells), Database: GSE143356 (testicular cells) (Chitiashvili et al., 2020), Database: GSE86146 (Li et al., 2017), and Database: GSE140021(Chen et al., 2019).
Custom scripts used for aligning scRNA-seq, data processing, and plotting are available upon request.