Summary
Mouse embryonic stem cells (mESCs), a model for differentiation into primed epiblast-like cells (EpiLCs), have revealed transcriptional and epigenetic control of early embryonic development. The control and significance of morphological changes, however, remain less defined. We show marked changes in morphology and actin architectures during differentiation that depend on Arp2/3 complex but not formin activity. Inhibiting Arp2/3 complex activity pharmacologically or genetically does not block exit from naive pluripotency, but attenuates increases in EpiLC markers. We find that inhibiting Arp2/3 complex activity delays formative pluripotency and causes globally defective lineage specification as indicated by RNA sequencing, with significant effects on TBX3-depedendent transcriptional programs. We also identify two previously unreported indicators of mESC differentiation, namely, MRTF and FHL2, which have inverse Arp2/3 complex-dependent nuclear translocation. Our findings on Arp2/3 complex activity in differentiation and the established role of formins in EMT indicate that these two actin nucleators regulate distinct modes of epithelial plasticity.
Keywords: mouse embryonic stem cells, actin cytoskeleton, morphology, pluripotency, differentiation, lineage specification, Arp2/3 complex, TBX3, MRTF, FHL2
Highlights
-
•
Arp2/3 complex activity is necessary for morphology changes during differentiation
-
•
Arp2/3 complex activity regulates transcriptional markers of differentiation
-
•
Inhibiting Arp2/3 complex activity delays entry into formative pluripotency
-
•
Arp2/3 complex activity-dependent shuttling of FHL2 and MRTF occurs in mESCs
Aloisio and Barber report a role for Arp2/3 complex activity in morphological and transcriptional changes with mESC differentiation. The authors reveal that inhibiting Arp2/3 complex activity delays entry into formative pluripotency, offering further mechanistic insight on this recently identified transition. Global examination of lineage specification using RNA sequencing suggests a role for Arp2/3 complex activity in TBX3-dependent transcriptional programs during differentiation.
Introduction
As an in vitro model, mouse embryonic stem cells (mESCs) have provided insights on the regulated transition to primed pluripotent epiblast-like cells (EpiLCs) of the post-implantation blastocyst (Martin, 1981; Evans and Kaufman, 1981), which is one of the earliest known transitions in embryonic differentiation (Nichols and Smith, 2009, 2012; Weinberger et al., 2016). While studies with mESCs have revealed how biochemical cues, transcriptional programs, and epigenetics drive differentiation, less is known about morphological changes, how they are controlled, and their importance for differentiation or lineage specification (Gilmour et al., 2017; Villeneuve and Wickström, 2021).
Actin remodeling is a major driver of morphological changes that facilitates diverse cell behaviors. Actin filament architectures are predominantly generated by two classes of actin nucleators: the Arp2/3 complex comprising seven subunits that nucleates branched actin filaments and formins that nucleate unbranched actin filaments. While neither actin nucleator has been reported for roles in pluripotency transition or lineage specification, the processes they regulate, including cellular stiffness (Bongiorno et al., 2018), the formation of ventral cortex F-actin asters (Xia et al., 2019), apparent membrane tension (De Belly et al., 2021), and membrane-to-cortex attachment (Bergert et al., 2021), have roles in mESC differentiation. Here we show that with mESC differentiation, morphological changes and actin filament remodeling depend on activity of the Arp2/3 complex but not formins, and that Arp2/3 complex activity is necessary for transition from naive mESCs to EpiLCs, including timing entry into intermediate formative pluripotency with global effects on lineage specification.
Formative pluripotency, a recently identified intermediate stage during the differentiation of naive mESCs to EpiLCs (Kalkan and Smith, 2014), is considered an executive phase when naive signaling networks are dismantled and cells acquire competence for lineage specification. A major limitation for a mechanistic understanding of formative pluripotency timing and regulation is the ambiguity of experimentally isolating and continuously propagating formative pluripotent cells. Our findings reveal a previously unrecognized role for the Arp2/3 complex in timing entry into formative pluripotency and subsequent lineage specification, which identifies new approaches for studying pluripotency transition states that could be applicable in regenerative medicine. Additionally, our work identifies opposing nuclear localization of myocardin-related transcription factor (MRTF) and FHL2, which are competing co-factors for serum response factor (SRF) transcriptional activity, as previously unreported markers of mESC differentiation that are regulated by Arp2/3 complex activity.
Results
Inhibiting Arp2/3 complex but not formin activity blocks morphological changes and actin remodeling during mESC differentiation
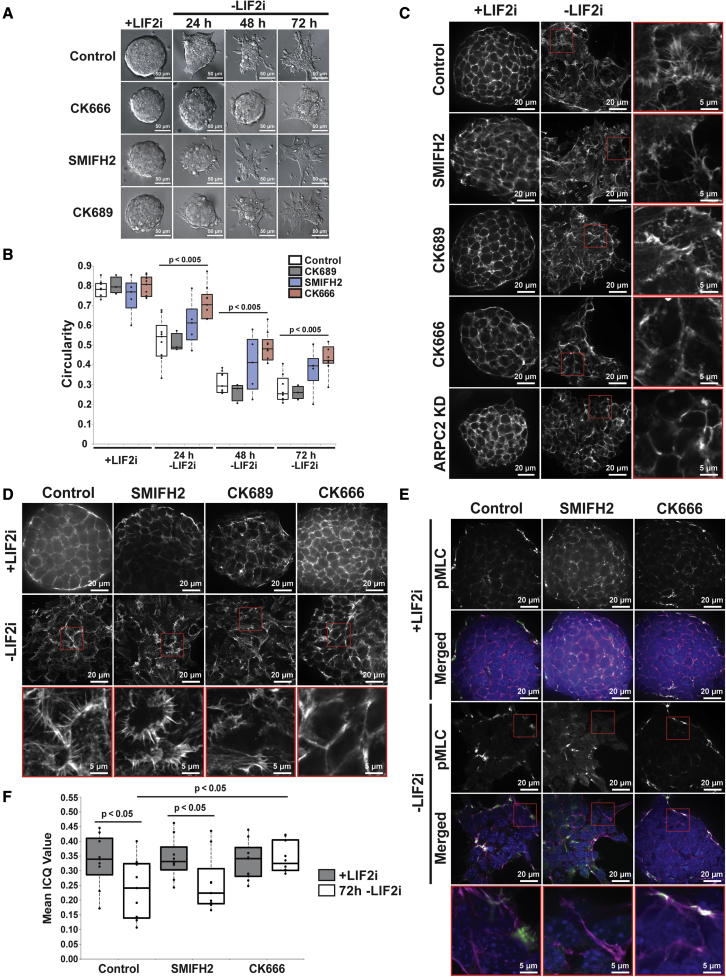
A regulatory mechanism for actin-dependent changes in mESC morphology in the context of differentiation and lineage specification remains incompletely understood. Recent advances propose roles for apparent cell membrane tension and polarity as requisite regulators of both mESC differentiation (Xia et al., 2019; Bergert et al., 2021) and in vivo embryonic development (Molé et al., 2021). To our knowledge, however, a direct link between actin-dependent changes in morphology and the transcriptional programming of lineage specification has not been reported. To determine changes in colony morphology in real time, we quantified differential interference contrast (DIC) images of E14 mESCs maintained in the presence of LIF2i and spontaneously differentiated for 72h after removal of LIF2i (Ying et al., 2008). Naive colonies in LIF2i have a static circular morphology as quantified using circularity = 4 pi (area/perimeter2) with a value of 1.0 indicating a perfect circle and values approaching 0.0 indicating an elongated polygon shape (Figures 1A and 1B, Video S1). In control cells, colony circularity progressively decreases after removing LIF2i (Figures 1B and Video S2), in agreement with previous reports (Bongiorno et al., 2018; Bergert et al., 2021). In determining how these morphological changes are regulated, we find that decreases in colony circularity are significantly attenuated by CK666, a selective pharmacological inhibitor of Arp2/3 complex activity (Nolen et al., 2009; Yang et al., 2012), but not by SMIFH2, a broad-spectrum inhibitor of formin activity, or CK689, an inactive analog of CK666 (Rizvi et al., 2009; Ganguly et al., 2015; Nolen et al., 2009) (Figures 1A and 1B). In contrast, we recently showed that SMIFH2 but not CK666 blocks morphological changes during epithelial to mesenchymal transition (EMT) (Rana et al., 2018).
Figure 1.
Inhibiting Arp2/3 complex but not formin activity blocks morphological changes and actin remodeling during mESC differentiation
(A) DIC images of E14 mESCs over 72h –LIF2i.
(B) Circularity quantified from (A) with data points representing means obtained from 15 to 20 colonies per condition (n = 3–6).
(C and D) Confocal images of E14 mESCs (C) and V6.5 mESCs (D) +LIF2i and at 72h –LIF2i stained for F-actin with phalloidin.
(E) Confocal images of E14 mESCs +LIF2i and at 72h –LIF2i immunolabeled for pMLC (green) and stained for F-actin with phalloidin (magenta) or nuclei with Hoechst (blue).
(F) Colocalization of pMLC with phalloidin from (E), mean ICQ values obtained from nine colonies per condition (n = 3). Box plots in (B) and (F) show median, first and third quartile, with whiskers extending to observations within 1.5 times the interquartile range. See also Figure S1.
Images taken every 10 min for 20h on gelatin-coated glass, 40x.
Images taken every 10 min for 20h on gelatin-coated glass, 40x.
We next analyzed actin architectures during differentiation. Using high-resolution spinning disc confocal imaging, we find that naive E14 mESCs in LIF2i have a compact polygonal cell shape with a cortical ring of actin filaments that are remodeled to an elongated cell shape with prominent membrane protrusions containing ribbed, fan-like actin filaments after 72h –LIF2i (Figure 1C). In the presence of CK666 but not SMIFH2 or CK689, actin filaments retain a cortical ring after 72h –LIF2i and fan-like filament networks are rarely seen (Figure 1C). We also find that effects with CK666 are phenocopied with CRISPR/Cas9 knockdown of ARPC2, an Arp2/3 complex subunit. We confirmed CRISPR/Cas9 editing of the Arpc2 locus by immunoblotting showing decreased ARPC2 in E14 mESCs (Figures S1A and S1B) and by sequencing (Figures S1E and S1F). Consistent with the stability of Arp2/3 complex subunits being dependent on their assembly (Di Nardo et al., 2005), Arpc2 silencing in mESCs significantly decreases the abundance of the Arp2/3 complex subunit ARP2 (Figures S1C and S1D). Inhibiting Arp2/3 complex activity in E14 mESCs by either CK666 or ARPC2 knockdown has no effect on the morphology or cortical actin organization of naive cells, but blocks pronounced fan-like actin filaments after 72h -LIF2i (Figure 1C). Similar to our findings with E14 mESCs, we confirmed that genetically distinct V6.5 mESCs show a similar remodeling of actin filament architectures with spontaneous differentiation that is blocked by CK666 but not SMIFH2 or CK689 (Figure 1D). Taken together, these findings indicate that changes in filament architectures occur during mESC differentiation in two mESC lines, which are blocked by inhibiting Arp2/3 complex but not formin activity.
Changes in cell morphology are often not driven by actin filament remodeling alone, but also in combination with actomyosin contractility (Murrell et al., 2015). Consistent with our finding that CK666 attenuates changes in morphology, immunolabeling reveals that phosphorylated myosin light chain (pMLC), an indicator of actomyosin contractility, decorates the cortical actin ring around cells and the peripheral ring around free margins of colonies in control naive mESCs but is diffuse in the cytoplasm after 72h –LIF2i (Figure 1E). In contrast, CK666 but not SMIFH2 retains a cortical pMLC localization after 72h –LIF2i (Figure 1E). We used an intensity correlation analysis to quantify the colocalized distribution of F-actin and pMLC as previously described (Li et al., 2004; Brown et al., 2010), wherein a mean intensity correlation quotient (ICQ) value ranges between −0.5 (to indicate complete segregation) and 0.5 (to indicate complete overlap). In control cells, the removal of LIF2i induces a significantly decreased mean ICQ value between pMLC and phalloidin-stained F-actin after differentiation (Figure 1F). In contrast, the presence of CK666 but not SMIFH2 blocks the loss of colocalized pMLC and F-actin (Figure 1F). There is no change in pMLC abundance during differentiation in controls or with CK666 or SMIFH2, however, as determined by immunoblotting of E14 mESC lysates (Figures S1G and S1H). Taken together, these data indicate that activity of the Arp2/3 complex but not formins regulates changes in colony morphology, actin architectures, and pMLC localization during mESC differentiation.
Inhibiting Arp2/3 complex but not formin activity impairs differentiation to EpiLCs
We used several approaches to show that Arp2/3 complex activity is also necessary for transcriptional changes during mESC differentiation. We first used a V6.5 dual-reporter (DR) mESC line engineered to express distinct fluorophores as cells transition from naive to primed pluripotency. In brief, Parchem et al. (2014) found that V6.5 mESCs in LIF2i express a naive-specific miR-290 cluster and with –LIF2i spontaneous differentiation miR-290 expression decreases while expression of the primed-specific miR-302 cluster increases. They generated cells that express mCherry driven by the miR-290 promoter and GFP driven by the miR-302 promoter. Using flow cytometry, DR mESCs can be used to score for decreased mCherry expression and increased GFP expression as an index of differentiation on the cell population level, while intermediate cells are double positive for both markers (Figure 2A). Our analysis indicates that control DR mESCs in LIF2i are more than 90% mCherry positive (Figure S2A), which is decreased to 16.3% after 72h –LIF2i in controls but is significantly greater at 44.0% with CK666 (Figure 2B). In contrast, the percent of mCherry single-positive cells is not different with CK689, SMIFH2, or DMSO compared with controls (Figure 2B). Cell death and proliferation in the presence of CK666, CK689, SMIFH2, or DMSO are not significantly different from control cells at any time point during differentiation (Figures S2B, S2C, and S2E). These data indicate that Arp2/3 complex but not formin activity is necessary for changes in stage-specific microRNA (miRNA) expression during naive to primed pluripotency, suggesting a broader role for Arp2/3 complex beyond changes in mESC morphology.
Figure 2.
Inhibiting Arp2/3 complex but not formin activity impairs differentiation to EpiLCs
(A) Schematic of DR mESCs.
(B) FACS data of DR mESCs after 72h –LIF2i show a mean (n = 6).
(C–E) RT-qPCR for Fgf5 (C) and Brachyury (D) in DR mESCs and for Fgf5 in E14 mESCs (E) +LIF2i and at 72h –LIF2i, means ± standard error of the mean normalized to Tbp (n = 3).
(F) Confocal images of E14 mESCs +LIF2i and at 72h –LIF2i immunolabeled for TFE3 (green) and stained for F-actin with phalloidin (magenta) or nuclei with Hoechst (blue).
(G) Quantified nuclear to cytoplasmic ratio of TFE3 immunolabeling in (F), means ± standard error of the mean (n = 3). See also Figure S2.
As a second approach, we confirmed that CK666 and ARPC2 silencing attenuates the expression of established primed EpiLC markers. Reverse transcriptase quantitative PCR for Fgf5 (Figure 2C) and Brachyury (Figure 2D) in V6.5 DR mESCs indicates significantly increased expression in controls and with CK689 after 72h –LIF2i, but not with CK666. We used a similar approach to show that expression of Fgf5 (Figure 2E) and Brachyury (Figure S2D) in E14 mESCs significantly increases after 72h –LIF2i in controls, but is attenuated with ARPC2 silencing. These data support a role for Arp2/3 complex activity in the transcriptional changes associated with the differentiation of naive to primed EpiLCs, as indicated by pharmacologically or genetically inhibiting Arp2/3 complex activity in V6.5 and E14 mESCs.
Our third approach scored for the cytosolic and nuclear localization TFE3, a bHLH transcription factor that is predominantly nuclear in naive mESCs but mostly cytoplasmic in primed EpiLCs (Betschinger et al., 2013; Kalkan et al., 2017). By quantifying immunolabeling of E14 mESCs, we find that the nuclear to cytoplasmic ratio of endogenous TFE3 significantly decreases after 72h –LIF2i in controls but not in the presence of CK666 (Figures 2F and 2G). Taken together, these data reveal a role for Arp2/3 complex activity beyond morphology to include transcriptional indicators, such as miRNA expression, primed marker gene expression, and transcription factor localization during differentiation.
Inhibiting Arp2/3 complex activity has no effect on exit from naive self-renewal, but delays entry into formative pluripotency
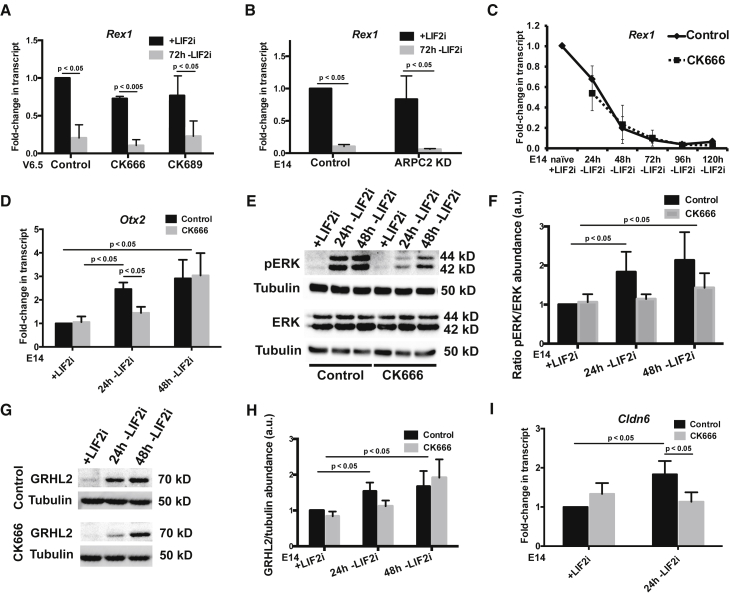
To further understand how Arp2/3 complex activity enables mESC differentiation, we tested whether it is necessary for exit from naive pluripotency. While DR mESCs can be used as a general index of differentiation at the population level, expression (or lack thereof) of these miRNAs does not reveal robust temporal regulation as other naive markers, such as Rex1 or Stra8, at the individual cell level. At 72h –LIF2i, we observe that the naive marker Rex1 (also called Zfp42) significantly decreases in V6.5 and E14 cells in the absence and presence of CK666 and CK689 (Figures 3A and 3B), as well as in E14 cells with ARPC2 silencing (Figure 3B). Moreover, the time-dependent decrease in the expression of Rex1 as well as Stra8 over 120h –LIF2i in E14 cells is similar in the absence or presence of CK666 (Figures 3C and S3). These data suggest that Arp2/3 complex activity is not necessary for maintaining naive markers or for exit from naive pluripotency.
Figure 3.
Inhibiting Arp2/3 complex activity has no effect on exit from naive self-renewal, but delays entry into formative pluripotency
(A and B) Reverse transcriptase quantitative PCR for Rex1 in DR mESCs (A) and E14 mESCs (B) +LIF2i and at 72h –LiF2i, means ± standard error of the mean normalized to Tbp (n = 3).
(C) Reverse transcriptase quantitative PCR for Rex1 in E14 mESCs during 120h time-course –LIF2i, means ± standard error of the mean normalized to Tbp (n = 4).
(D) Reverse transcriptase quantitative PCR for Otx2 in E14 mESCs during 48h time-course –LIF2i, means ± standard error of the mean normalized to Tbp (n = 4).
(E) Representative immunoblot of lysates from E14 mESCs during 48h time-course –LIF2i probed for pERK, total ERK, or tubulin as a loading control.
(F) Semiquantitative densitometry of (E), means ± standard error of the mean (n = 3).
(G) Representative immunoblot of lysates from E14 mESCs during 48h time-course –LIF2i probed for GRHL2 or tubulin as a loading control.
(H) Semiquantitative densitometry of (G), means ± standard error of the mean (n = 7).
(I) Reverse transcriptase quantitative PCR for Cldn6 in E14 mESCs after 24h –LIF2i, means ± standard error of the mean normalized to Tbp (n = 5). Given directional a priori predictions in panels D-H, data were analyzed by one-tailed unpaired Student’s t test with a significance level of p < 0.05. See also Figure S3.
An intermediate state between naive and primed pluripotency, termed formative pluripotency, was recently identified. Formative pluripotency is considered an executive state, when cells are most responsive to differentiation cues and receptive for lineage commitment (Smith, 2017; Kalkan et al., 2017, 2019). The formative pluripotent state is currently defined by decreased expression of Rex1, which we confirmed is not impaired when Arp2/3 activity is inhibited (Figures 3A–3C), increased Otx2 (Kalkan et al., 2017; Mulas et al., 2017), increased phosphorylated ERK (pERK) (Kalkan et al., 2019), and increased Grhl2 (Chen et al., 2018). We confirmed that Otx2 significantly increases in control E14 cells within 24h –LIF2i (Figure 3D). In contrast, with CK666 Otx2 expression at 24h –LIF2i is significantly less compared with control cells and not different than in naive cells (Figure 3D). After 48h –LIF2i, however, CK666-treated cells have a delayed increase in Otx2 (Figure 3D). These results are consistent with previous reports for delayed Otx2 expression in the presence of a pharmacological inhibitor of NODAL signaling, which is suggested to function as a timing mechanism for pluripotency transition (Mulas et al., 2017).
Increased pERK, another marker of formative pluripotency, is required for activating the downstream formative pluripotent gene regulatory networks (Kalkan et al., 2019; Azami et al., 2019). We find increased pERK in control E14 cells at 24 and 48h –LIF2i compared with total ERK, which does not change during differentiation, as determined by immunoblotting cell lysates (Figures 3E and 3F). In contrast, increased pERK at 24 and 48h after the removal of LIF2i is attenuated in the presence of CK666 (Figures 3E and 3F). These findings agree with other studies indicating a strong link between pERK and actin polymerization, cell motility, and myosin contractility, as previously reviewed (Tanimura and Takeda, 2017).
Finally, we used immunoblotting of E14 cell lysates to confirm increased abundance of GRHL2 in control E14 cells at 24 and 48h –LIF2i (Figures 3G and 3H), which is similar to reported findings using V6.5 cells (Chen et al., 2018). In contrast, with CK666 increased GRHL2 is delayed with a significant increase at 48h but not at 24h in –LIF2i in cells (Figures 3G and 3H). Further, expression of Cldn6, a downstream target gene of GRHL2 in mESCs, significantly increases in control E14 cells at 24h –LIF2i, but not with CK666 (Figure 3I). Hence, inhibiting Arp2/3 complex activity in two different mESC lines has no effect on maintenance of naive self-renewal or exit from naive pluripotency, but delays entry into the intermediate formative pluripotency.
Inhibiting Arp2/3 complex activity disrupts lineage commitment with pronounced effects on TBX3 target genes across all three germ layers
Our findings that Arp2/3 complex activity is necessary for actin remodeling, attenuated expression of primed marker expression, and timing for formative pluripotency during mESC differentiation suggest a role in promoting lineage specification. To investigate the global effects of inhibiting Arp2/3 complex activity on lineage specification, we performed RNA sequencing (RNA-seq) on E14 cells differentiated in the absence and presence of CK666 for 72h –LIF2i and control naive cells maintained in LIF2i (Figures 4A and 4B). We find that control naive +LIF2i and –LIF2i cells have a total of 6,576 differentially expressed genes (DEGs) with an adjusted qval of less than 0.05 after batch correction (Figure 4C). Of these DEGs, 1,662 are unique to control –LIF2i cells compared with naive +LIF2i cells and are not differentially expressed in CK666 –LIF2i compared with naive +LIF2i cells (Figure 4C). CK666 –LIF2i cells compared with naive +LIF2i have 4,796 DEGs, with 457 unique DEGs (Figure 4C). In CK666, –LIF2i cells compared with both control +/–LIF2i cells, there are 972 unique DEGs (Figure 4C). A gene ontology enrichment analysis of this latter subset suggests that unique CK666-specific DEGs are associated with biological processes related to extracellular matrix organization, endothelial cell migration, sprouting angiogenesis, and the MAPK/ERK cascade (Figure 4D). As a general summary, these data indicate global transcriptomic differences in CK666-treated cells.
Figure 4.
Inhibiting Arp2/3 complex activity disrupts lineage commitment with pronounced effects on TBX3 target genes across all three germ layers.
(A and B) Volcano plot showing the transcriptome fold-changes (beta values) in control –LIF2i compared with +LIF2i E14 mESCs (A) or in CK666 –LIF2i compared with control –LIF2i E14 mESCs (B) after 72h. Each dot represents one gene, with significantly changed genes (q-value < 0.05) indicated in blue and significantly changed genes-of-interest indicated in red.
(C) Venn diagram showing the number of shared and distinct DEGs, (D) gene ontology biological process (2019) enrichment analysis of 972 DEGs uniquely indicated in CK666 –LIF2i compared with control –LIF2i after 72h.
(E–H) Clustermap showing naive mESC marker (E), mesoderm marker (F), endoderm marker (G), and ectoderm marker (H) expression indicated by beta values from RNA-seq analysis.
(I) Enriched bar graph with beta value fold-changes indicated from RNA-seq analysis of E14 mESCs for TBX3 target genes having significantly different expression (qval < 0.05) in CK666 –LIF2i compared with control –LIF2i. See also Figure S4.
Consistent with our data indicating that CK666 has no effect on exit from naive self-renewal (Figures 3A–3C), RNA-seq data also show down-regulated naive markers in the presence of CK666 compared with control cells with the exception of Tbx3, a master regulator of pluripotency and lineage specification (Figure 4E) (Lu et al., 2011). Further, our findings include dysregulated Fgf5, Fn1, Zeb1, Kctd12b, and Mat2a (Figure 4B) and targets specific to mesoderm such as Pdlim3, Adm, and Fhl2 (Figure 4F), endoderm such as Eomes and Kit (Figure 4G), and ectoderm such as Mycn, Prickle1, and Nes (Figure 4H) in the presence of CK666. Taken together, these data indicate a role for Arp2/3 complex activity in timing lineage specification related to extinction of Tbx3, which has been reported to counteract initiation of formative pluripotency (Kalkan et al., 2019).
TBX3 is an established regulator of early development with dynamic context-dependent roles in embryonic organogenesis across germ layers (Chapman et al., 1996). With binding to a number of transcription factors such as KLF4, OCT4, SOX2, and NANOG, TBX3 plays a complex role at the center of pluripotency circuitry (Han et al., 2010; Russell et al., 2015) with the potential to act as either an activator or inhibitor of gene expression dependent on cofactor binding (Carlson et al., 2001). To determine the extent to which inhibiting Arp2/3 complex activity globally affects TBX3 target gene expression, we compared our DEGs with three publicly available mESC datasets related to target genes that change expression relative to a TBX3 reporter (Russell et al., 2015), change expression with short hairpin RNA (shRNA) knockdown of Tbx3 (Nishiyama et al., 2013), and bind TBX3 as indicated by chromatin immunoprecipitation sequencing (Han et al., 2010). Comparing datasets indicates that CK666 treatment generally shows a contrasting transcriptional profile of TBX3 target genes compared with that of a control differentiation (Figures S4A–S4C). Enriching for TBX3 target genes common to all three datasets that are significantly dysregulated with CK666 compared with control (Figure 4I) suggests effects on a number of key regulators. Other noteworthy gene expression changes include Tfe3 (Figures 2F, 2G, and S4A), Cldn6 (Figures 3I and S4A), and Fhl2, a mesodermal gene encoding a recently identified tension-dependent actin-binding protein (Sun et al., 2020) (Figures 4F and S4A). Collectively, these data suggest that Arp2/3 complex activity times entry into formative pluripotency, possibly by delayed loss of Tbx3 expression, resulting in defective downstream global and distinct lineage specification programs.
Inhibiting Arp2/3 complex activity blocks cytoplasmic and nuclear shuttling of MRTF and FHL2
Our findings on TBX3 target genes regulated by Arp2/3 complex activity led us to identify two previously unreported markers of mESC differentiation—the cytoplasmic and nuclear localization of FHL2 and MRTF, which are SRF co-transcriptional activators. MRTF is an actin polymerization-responsive transcriptional co-activator that translocates to the nucleus with increased actin polymerization (Miralles et al., 2003; Posern and Treisman, 2006). FHL2 is a TBX3 target gene and a transcriptional co-activator that is predominantly nuclear in response to decreased F-actin tension (Philippar et al., 2004; Nakazawa et al., 2016). Although formin-dependent nuclear translocation of MRTF is well described for adult mesenchymal stem cell differentiation, neither MRTF nor FHL2 translocation has been reported during mESC differentiation.
We scored for changes in MRTF localization during mESC differentiation and found that in control and SMIFH2-treated naive E14 cells MRTF is diffuse in the cytoplasm but after 72h –LIF2i is predominantly nuclear (Figures 5A and 5B). In contrast, with CK666 or ARPC2 knockdown nuclear translocation of MRTF is inhibited at 72h –LIF2i (Figures 5A and 5B) and 120h –LIF2i (Figures S5A and S5B). These data indicate that MRTF nuclear translocation occurs during mESC differentiation and depends on activity of the Arp2/3 complex but not formins. Further, our RNA-seq data confirm that MRTF target genes (Esnault et al., 2014) have a contrasting transcriptional profile with CK666 compared with controls (Figure 5C).
Figure 5.
Inhibiting Arp2/3 complex activity blocks cytoplasmic and nuclear shuttling of MRTF and FHL2
(A) Confocal images of E14 mESCs +LIF2i and at 72h –LIF2i immunolabeled for MRTF (green) and stained for F-actin with phalloidin (magenta) and for nuclei with Hoechst (blue).
(B) Quantified nuclear to cytoplasmic ratio of MRTF immunolabeling shown in (A), means ± standard error of the mean (n = 3).
(C) Clustermap showing expression of MRTF target genes identified by Esnault et al. (2014) with beta values indicated from RNA-seq analysis of E14 mESCs in a control differentiation (control –LIF2i vs +LIF2i) and how they are affected in the presence of CK666 (CK666 –LIF2i vs control –LIF2i).
(D) Confocal images of E14 mESCs +LIF2i and at 72h –LIF2i immunolabeled for FHL2 (green) and stained for F-actin with phalloidin (magenta) and for nuclei with Hoechst (blue).
(E) Quantified nuclear to cytoplasmic ratio of FHL2 immunolabeling shown in (D), means ± standard error of the mean (n = 4).
(F) Proposed model of competing inverse actin-dependent MRTF/FHL2 nuclear translocation for mESCs in the presence and absence of LIF2i. Created with BioRender.com. See also Figure S5.
Similar to MRTF, nuclear FHL2 binds to SRF to promote target gene expression (Esnault et al., 2014; Philippar et al., 2004; Russell et al., 2015). We scored for changes in FHL2 localization during mESC differentiation and found that in control and SMIFH2-treated naive E14 cells FHL2 is nuclear, but after 72h –LIF2i undergoes translocation to the cytoplasm, quantified by a significant decrease in the nuclear to cytoplasmic ratio (Figures 5D and 5E). In contrast, with CK666 or ARPC2 knockdown cytoplasmic translocation of FHL2 is attenuated with no significant decrease in nuclear abundance at 72h –LIF2i (Figures 5D and 5E). Taken together with data showing F-actin binding of FHL2 (Sun et al., 2020), our findings reveal three previously unrecognized events during mESC differentiation; first is changes in the localization of MRTF and FHL2, second is the opposing nuclear and cytoplasmic translocation of these SRF transcriptional co-activators, and third is that their translocation depends on Arp2/3 complex activity (Figure 5E).
Discussion
We report a previously unrecognized function of Arp2/3 complex activity in enabling mESC differentiation. We find that changes in mESC colony morphology and actin architectures depend on the activity of the Arp2/3 complex but not formins. We also show that Arp2/3 complex activity is necessary for transition to distinct pluripotency states; although not necessary for exit from the naive state, the loss of Arp2/3 complex activity delays entry into the formative pluripotent state, contributing further mechanistic insight on how this intermediate state is controlled. Further, our data include a global examination of actin-dependent lineage specification, which aligns with evolutionarily conserved roles for force-sensitive differentiation and development in other organisms, both in vitro (Chowdhury et al., 2010; Lee et al., 2013) and in vivo (Keller et al., 2003; Krieg et al., 2008). Last, we show for the first time that MRTF and FHL2, both actin-responsive transcriptional co-activators to SRF, undergo inverse and Arp2/3 complex activity-dependent translocation during mESC differentiation.
We show marked Arp2/3 complex-dependent changes in colony morphology and actin architectures during differentiation. These findings are consistent with reports on morphology and dynamic cellular stiffness during differentiation (Bongiorno et al., 2018) and an acute role for the Arp2/3 complex in mESC actin remodeling (Xia et al., 2019). We observed Arp2/3 complex-dependent mESC actin architectures, which are established to generate protrusive forces for membrane dynamics (Bailly et al., 2001) and are of particular interest with regard to recent reports (Bergert et al., 2021; De Belly et al., 2021). An important question to resolve is how Arp2/3 complex activity regulates mESC transcriptional changes. Previous studies indicated force-sensitive lineage specification (Keller et al., 2003; Krieg et al., 2008; Gilmour et al., 2017; Villeneuve and Wickström, 2021) as well as mechanosensing and contractility in fate specification for mESCs (Janmey et al., 2013; Happe and Engler, 2016; Tatapudy et al., 2017). The role of the Arp2/3 complex as a central node between biochemical cues and biophysical responses (Iskratsch et al., 2014; Charras and Yap, 2018) suggests a mechanosensitive mechanism whereby Arp2/3 complex activity enables mESC differentiation.
Our data also suggest that Arp2/3 complex activity times entry to intermediate formative pluripotency, an executive state when cells are most receptive for lineage specification cues (Smith, 2017; Kalkan et al., 2017, 2019). Inhibiting Arp2/3 complex activity delays entry into formative pluripotency, as indicated by a delayed increase in Otx2 and Cldn6 expression, and GRHL2 abundance as well as no change in pERK with –LIF2i compared with controls. In related findings, inhibiting NODAL signaling has no effect on exit from naive pluripotency but delays formative pluripotent marker expression from 24h to 48h –LIF2i (Mulas et al., 2017). Taken together, our data indicate that Arp2/3 complex activity is a previously unrecognized node in the signaling network of formative pluripotent regulators.
We also reveal that inhibiting Arp2/3 complex activity disrupts lineage commitment across all three germ layers with pronounced effects in TBX3 target genes. TBX3 is a context-dependent master regulator of both naive self-renewal (Niwa et al., 2009; Han et al., 2010; Russell et al., 2015) and lineage specification (Costello et al., 2011; Lu et al., 2011; Weidgang et al., 2013; Kartikasari et al., 2013) with continued expression during mESC differentiation reported to destabilize entry into formative pluripotency (Kalkan et al., 2019). Dysregulated Tbx3 expression is associated with atypical cell and colony morphology in mESCs (Han et al., 2010; Russell et al., 2015), which we also see with loss of Arp2/3 complex activity. Smith et al. (see Kalkan et al., 2019) proposed a relationship between the timing of formative pluripotency and RBPJ, a regulator of mESC morphology, whereby RBPJ inhibits TBX3 expression to block formative cells from returning to self-renewal. Further, ERK signaling is reported to inhibit Tbx3 expression and mESCs null for ERK pathway components have sustained Tbx3 expression (Niwa et al., 2009; Hamilton et al., 2013; Chen et al., 2015). Our data indicating attenuated pERK and persistent Tbx3 abundance with CK666 suggest a relationship between Arp2/3 complex activity, mESC morphology, formative pluripotency, and TBX3-dependent lineage specification. Future studies on the link between Arp2/3 complex activity and formative pluripotency timing will be important to resolve the interface between morphology and lineage specification.
We also show that MRTF and FHL2, actin-responsive transcriptional co-activators for SRF, undergo previously unreported translocation events during mESC differentiation. MRTF, a transcriptional co-activator of SRF (Posern and Treisman, 2006; Sun et al., 2006; Vartiainen et al., 2007), is retained in the cytoplasm by binding to monomeric actin and with increased actin polymerization translocates to the nucleus where it binds to SRF to promote differentiation programs in adult mesenchymal stem cells (Miralles et al., 2003; Nobusue et al., 2014; McDonald et al., 2015; Bian et al., 2016) and was recently shown to block the induction of pluripotency during iPSC reprogramming (Hu et al., 2019). Our data show two previously unreported findings on MRTF nuclear translocation; first, that it occurs with mESC differentiation and, second, that it depends on Arp2/3 complex activity. Although MRTF has not been reported for direct roles in embryonic differentiation, SRF is confirmed to regulate embryonic mesoderm formation (Weinhold et al., 2000) and Srf−/− ESCs have altered cell morphology and decreased cortical actin (Schratt et al., 2002). FHL2, dysregulated in many cancers and developmental disorders, is another SRF-binding transcriptional co-activator with a direct transcriptional response to the actin cytoskeleton (Sun et al., 2020) and is a shared TBX3 target gene (Russell et al., 2015). In the cytosol, FHL2 contains LIM domains that mechanoaccumulate on strain sites of tensed actin filaments (Sun et al., 2020). FHL2 is released from F-actin upon loss of filament strain, causing it to translocate to the nucleus where it competes with MRTF for SRF binding (Philippar et al., 2004). In tandem, MRTF and FHL2 are both direct actin-responsive transcriptional co-activators with opposing roles both in the cytosol and in the nucleus. Taken together, our data suggest that inversely mechanosensitive translocation events of MRTF and FHL2 could serve as a novel marker for pluripotency status and provide motivation for understanding how basic cell biology can provide a framework for elucidating mechanisms of mESC differentiation and lineage specification.
As recently indicated (Gilmour et al., 2017; Villeneuve and Wickström, 2021), a current challenge is to identify the connection between the cellular machines that generate shape and the genes that control cell fate decisions. Our observations, compared with previous findings that activity of formins but not Arp2/3 complex is necessary for EMT and the assembly of unbranched contractile actin filaments (Li et al., 2010; Jurmeister et al., 2012; Rana et al., 2018), indicate that these different classes of actin nucleators and the architectures they generate have selective roles in distinct types of epithelial plasticity. With known functions in migration (Suraneni et al., 2012) and adherens junction tension (Verma et al., 2012; Fierro-Gonzales et al., 2013), there are abundant potential mechanisms whereby Arp2/3 complex activity might regulate mESC pluripotency transition (Rotty et al., 2013; Pieters and van Roy, 2014; Wagh et al., 2021; Molé et al., 2021). Our study provides a step toward closing the gap between phenotype and genotype, opening new directions and advancing new approaches to understand how morphological changes and actin filament dynamics promote pluripotency transition.
Experimental procedures
Cell culture
Wild-type and DR V6.5 ESCs, obtained from R. Blelloch (University of California San Francisco), and E14 ESCs, provided by A. Smith (University of Cambridge) were maintained in tissue culture dishes coated with 0.2% gelatin (G1393; Sigma) at 37°C and 5% CO2 in DMEM (10569; Gibco) supplemented with 15% fetal bovine serum (FB-11, Omega Scientific, Inc.), glutamine (2 mM), non-essential amino acids (0.1 mM), penicillin-streptomycin (100 U/mL Penicillium and 100 μg/mL Streptomycin), and 2-mercaptoethanol (55 μM). Cells received fresh medium every 24 h and were passaged every 3 days after dissociating with 0.25% Trypsin-EDTA (25200-056; Gibco). For self-renewal, cells were maintained in medium containing LIF (ESGRO Cat#ESG1106; EMD Millipore) and inhibitors for MEK (1 μM; PD0325901, Cat#S1036; Selleck Chemicals) and glycogen synthase kinase-3β (1 μM; CHIR99021, Cat#S2924; Selleck Chemicals), collectively termed LIF2i. To induce spontaneous differentiation, cells were washed in PBS and then incubated in medium without LIF2i as described in Parchem et al. (2014). CK666 (80 μM final; 182515; EMD Millipore), CK689 (80 μM final; 182517; EMD Millipore), and SMIFH2 (25 μM final; S4826; Sigma) were added at 1:000 from stock solutions prepared in DMSO and included in medium replacements every 24 h. See the supplemental experimental procedures for CRISPR/Cas9 gene editing of E14 ESCs and cell line authentication.
DIC image acquisition
Naive E14 ESCs were plated for 24 h on gelatin-coated glass bottom microwell dishes (P35G-1.5-14-C; MatTek) in medium containing LIF2i, washed with PBS, and then maintained for the indicated times in medium without LIF2i. CK666 and SMIFH2 were added at the time of LIF2i removal and replaced every 24 h until the completion of imaging. Live cells were imaged using a Plan Apo 40 0.95 NA objective on an inverted spinning disc microscope system (Nikon Eclipse TE2000 Perfect Focus System; Nikon Instruments) equipped with D-C DIC Slider 40× I (MBH76240; Technical Instruments), a multipoint stage (MS-2000; Applied Scientific Instruments), a CoolSnap HQ2 cooled charge-coupled camera (Photometrics), and camera-triggered electronic shutters controlled with NIS-Elements Imaging Software (Nikon). See the supplemental experimental procedures for circularity analysis.
Immunolabeling and staining
Cells were plated on gelatin-coated coverslips prepared in an ultrasonic cleaning bath. In brief, coverslips were sonicated for 20 min in the presence of double distillation H2O (ddH2O) and Versa detergent, washed in ddH2O, sonicated again for 20 min, and stored in 70% ethanol (EtOH). Cells were maintained for the indicated times, washed with PBS, and fixed with 4% formaldehyde for 15 min at room temperature (RT). Cells were then permeabilized with 0.1% Triton X-100 for 5 min, incubated with blocking buffer of 5% horse serum and 1% BSA in PBS for 1 h, and then incubated with primary antibodies overnight at 4°C. Primary antibodies included in Table S1. The cells were then washed with PBS, incubated for 1 h at RT with secondary antibodies conjugated with fluorophores, and washed with PBS. One wash included Hoechst 33342 (1:10,000; H-3570; Molecular Probes) to stain nuclei. Actin filaments were labeled with rhodamine-phalloidin (1:400; Invitrogen) added during secondary antibody incubations. See the Supplemental experimental procedures for image acquisition and analysis.
RNA extraction, cDNA synthesis, and quantitative PCR
Total RNA was isolated from ESCs by using TRIzol Reagent (15596026; Ambion) according to the manufacturer’s protocol with the following modifications: after washing cells with PBS, 800 μL TRIzol were added to cells in a six-well plate and the pellet was rinsed in 75% EtOH. RNA purity was assessed on a Nanodrop spectrometer. cDNA was synthesized using the iScript cDNA Synthesis Kit according to manufacturer’s protocol (170–8891; Bio-Rad Laboratories). Quantitative PCR was performed with iQ SYBR Green Supermix (170–8880; Bio-Rad Laboratories) according to the manufacturer’s protocol on a QuantStudio six Flex Real-Time PCR System (Applied Biosystems), with data analyzed using GraphPad Prism 6 software. Sequences for primers used in this study are included in Table S2.
Immunoblotting
Cells were lysed for 10 min in RIPA buffer (2.5 mM HEPES, pH 7.5, 150 mM NaCl, 3 mM KCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 1 mM vanadate, and 5 mM NaF supplemented with protease and phosphatase inhibitors). Lysates were centrifuged at 13,000 rpm for 15 min to obtain a post-nuclear supernatant. Proteins were separated by SDS-PAGE and transferred onto Immobilon-P PVDF transfer membranes (IPVH00010; EMD Millipore) as previously described (Haynes et al., 2011; Rana et al., 2015). Membranes were blocked with 5% non-fat milk in TBS containing 0.1% Tween (TBST) and incubated with primary antibodies overnight at 4°C. Primary antibodies included in Table S1. After washing, membranes were incubated in TBST with 5% non-fat milk and horseradish peroxidase-conjugated secondary antibodies (1:10,000; 170-6516 and 172-1019; Bio Rad Laboratories) for 1 h at room temperature. After washing, immunoreactivity was developed with enhanced femto chemiluminescence (1859022 and 1859023; ThermoFisher Scientific) and imaged using a BioRad Chemidoc XRS. ImageJ software was used for semi-quantitative densitometry analysis. Data presentation and statistical analysis were preformed using Excel Analyze-it and GraphPad Prism 6 software.
Dataset acquisition
CHIP-seq data of TBX3 binding in mESCs was available from NCBI (GEO: GSE19219) (Han et al., 2010). Microarray data from shRNA Tbx3 knockdown mESCs was available from NCBI (GEO: GSE26520) (Nishiyama et al., 2013). RNA-seq data from TBX3-HI and TBX3-LO mESCs was available from NCBI (GEO: GSE73862) (Russell et al., 2015). CHIP-seq data of MRTF binding in NIH3T3 fibroblasts was available from the NCBI (GEO: GSE45888) (Esnault et al., 2014).
Statistics
For all statistical analysis, PRISM 6 (GraphPad Software, Inc) or Excel Analyse-It (Microsoft) was used. All data were analyzed by two-tailed unpaired Student’s t test with a significance level of p < 0.05, unless otherwise noted. Shapiro-Wilk test was used to test for normality. N represents number of independent biological replicates. Pooled independent experiments are used in dot plots.
Author contributions
D.L.B. conceived the hypothesis, which was developed by F.M.A. D.L.B. and F.M.A. obtained and analyzed data as well as assembled figures and wrote the manuscript.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
We thank A. Smith (University of Cambridge) for providing the E14 cell line and R. Blelloch (UCSF) for providing V6.5 wild-type and DR cells. We also thank T. Nystul (UCSF), M. Welch (UC Berkeley), and the Barber Lab for helpful discussions. This work was supported by NIH grant CA197855 and NSF grant P0538109 to D.L.B. F.M.A. was supported by an HHMI Gilliam Fellowship and an NIGMS T32GM008568 training grant.
Published: June 2, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.05.002.
Supplemental information
Data and code availability
RNA-seq data generated during this study have been deposited in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). The accession number for the data reported in this paper is GEO: GSE175391. Software/packages used to analyze the dataset are freely available. See the supplemental experimental procedures for RNA-seq library preparation and analysis.
References
- Azami T., Bassalert C., Allègre N., Valverde Estrella L., Pouchin P., Ema M., Chazaud C. Regulation of the ERK signalling pathway in the developing mouse blastocyst. Development. 2019;146:dev177139. doi: 10.1242/dev.177139. [DOI] [PubMed] [Google Scholar]
- Bailly M., Ichetovkin I., Grant W., Zebda N., Machesky L.M., Segall J.E., Condeelis J. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr. Biol. 2001;11:620–625. doi: 10.1016/s0960-9822(01)00152-x. [DOI] [PubMed] [Google Scholar]
- Bergert M., Lembo S., Sharma S., Russo L., Milovanović D., Gretarsson K.H., Börmel M., Neveu P.A., Hackett J.A., Petsalaki E., Diz-Muñoz A. Cell surface mechanics gate embryonic stem cell differentiation. Cell Stem Cell. 2021;28:209–216.e4. doi: 10.1016/j.stem.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Nichols J., Dietmann S., Corrin P.D., Paddison P.J., Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian H., Lin J.Z., Li C., Farmer S.R. Myocardin-related transcription factor A (MRTFA) regulates the fate of bone marrow mesenchymal stem cells and its absence in mice leads to osteopenia. Mol. Metab. 2016;5:970–979. doi: 10.1016/j.molmet.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiorno T., Gura J., Talwar P., Chambers D., Young K.M., Arafat D., Wang G., Jackson-Holmes E.L., Qiu P., McDevitt T.C., Sulchek T. Biophysical subsets of embryonic stem cells display distinct phenotypic and morphological signatures. PLoS One. 2018;13 doi: 10.1371/journal.pone.0192631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M., Adyshev D., Bindokas V., Moitra J., Garcia J.G.N., Dudek S.M. Quantitative distribution and colocalization of non-muscle myosin light chain kinase isoforms and cortactin in human lung endothelium. Microvasc. Res. 2010;80:75–88. doi: 10.1016/j.mvr.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson H., Ota S., Campbell C.E., Hurlin P.J. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum. Mol. Genet. 2001;10:2403–2413. doi: 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- Chapman D.L., Garvey N., Hancock S., Alexiou M., Agulnik S.I., Gibson-Brown J.J., Cebra-Thomas J., Bollag R.J., Silver L.M., Papaioannou V.E. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev. Dyn. 1996;206:379–390. doi: 10.1002/(sici)1097-0177(199608)206:4<379::aid-aja4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Charras G., Yap A.S. Tensile forces and mechanotransduction at cell-cell junctions. Curr. Biol. 2018;28:R445–R457. doi: 10.1016/j.cub.2018.02.003. [DOI] [PubMed] [Google Scholar]
- Chen A.F., Liu A.J., Krishnakumar R., Freimer J.W., DeVeale B., Blelloch R. GRHL2-Dependent enhancer switching maintains a pluripotent stem cell transcriptional subnetwork after exit from naive pluripotency. Cell Stem Cell. 2018;23:226–238.e4. doi: 10.1016/j.stem.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Guo R., Zhang Q., Guo H., Yang M., Wu Z., Gao S., Liu L., Chen L. ERK signaling is indispensable for genomic stability and self-renewal of mouse embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2015;112:E5936–E5943. doi: 10.1073/pnas.1516319112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury F., Li Y., Poh Y.C., Yokohama-Tamaki T., Wang N., Tanaka T.S. Soft substrates promote homogeneous self-renewal of embryonic stem cells via downregulating cell-matrix tractions. PLoS One. 2010;5:e15655. doi: 10.1371/journal.pone.0015655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I., Pimeisl I.M., Dräger S., Bikoff E.K., Robertson E.J., Arnold S.J. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 2011;13:1084–1091. doi: 10.1038/ncb2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Belly H., Stubb A., Yanagida A., Labouesse C., Jones P.H., Paluch E.K., Chalut K.J. Membrane tension gates ERK-mediated regulation of pluripotent cell fate. Cell Stem Cell. 2021;28:273–284.e6. doi: 10.1016/j.stem.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A., Cicchetti G., Falet H., Hartwig J.H., Stossel T.P., Kwiatkowski D.J. Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc. Natl. Acad. Sci. U S A. 2005;102:16263–16268. doi: 10.1073/pnas.0508228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C., Stewart A., Gualdrini F., East P., Horswell S., Matthews N., Treisman R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014;28:943–958. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fierro-González J.C., White M.D., Silva J.C., Plachta N. Cadherin-dependent filopodia control preimplantation embryo compaction. Nat Cell Biol. 2013;15:1424–1433. doi: 10.1038/ncb2875. [DOI] [PubMed] [Google Scholar]
- Ganguly A., Tang Y., Wang L., Ladt K., Loi J., Dargent B., Leterrier C., Roy S. A dynamic formin-dependent deep F-actin network in axons. J. Cell Biol. 2015;210:401–417. doi: 10.1083/jcb.201506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D., Rembold M., Leptin M. From morphogen to morphogenesis and back. Nature. 2017;541:311–320. doi: 10.1038/nature21348. [DOI] [PubMed] [Google Scholar]
- Hamilton W.B., Kaji K., Kunath T. ERK2 suppresses self-renewal capacity of embryonic stem cells, but is not required for multi-lineage commitment. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Yuan P., Yang H., Zhang J., Soh B.S., Li P., Lim S.L., Cao S., Tay J., Orlov Y.L., et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happe C.L., Engler A.J. Mechanical forces reshape differentiation cues that guide cardiomyogenesis. Circ. Res. 2016;118:296–310. doi: 10.1161/circresaha.115.305139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Liu Z.Z., Chen X., Schulz V.P., Kumar A., Hartman A.A., Weinstein J., Johnston J.F., Rodriguez E.C., Eastman A.E., et al. MKL1-actin pathway restricts chromatin accessibility and prevents mature pluripotency activation. Nat. Commun. 2019;10:1695. doi: 10.1038/s41467-019-09636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes J., Srivastava J., Madson N., Wittmann T., Barber D.L. Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol Biol Cell. 2011;22:4750–4764. doi: 10.1091/mbc.E11-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T., Wolfenson H., Sheetz M.P. Appreciating force and shape – the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey P.A., Wells R.G., Assoian R.K., McCulloch C.A. From tissue mechanics to transcription factors. Differentiation. 2013;86:112–120. doi: 10.1016/j.diff.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurmeister S., Baumann M., Balwierz A., Keklikoglou I., Ward A., Uhlmann S., Zhang J.D., Wiemann S., Sahin O. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol. Cell Biol. 2012;32:633–651. doi: 10.1128/mcb.06212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T., Smith A. Mapping the route from naive pluripotency to lineage specification. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130540. doi: 10.1098/rstb.2013.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T., Bornelöv S., Mulas C., Diamanti E., Lohoff T., Ralser M., Middelkamp S., Lombard P., Nichols J., Smith A. Complementary activity of ETV5, RBPJ, and TCF3 drives formative transition from naïve pluripotency. Cell Stem Cell. 2019;24:785–801.e7. doi: 10.1016/j.stem.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T., Olova N., Roode M., Mulas C., Lee H.J., Nett I., Marks H., Walker R., Stunnenberg H.G., Lilley K.S., et al. Tracking the embryonic stem cell transition from ground state pluripotency. Development. 2017;144:1221–1234. doi: 10.1242/dev.142711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartikasari A.E.R., Zhou J.X., Kanji M.S., Chan D.N., Sinha A., Grapin-Botton A., Magnuson M.A., Lowry W.E., Bhushan A. The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 2013;32:1393–1408. doi: 10.1038/emboj.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R., Davidson L.A., Shook D.R. How we are shaped: the biomechanics of gastrulation. Differentiation. 2003;71:171–205. doi: 10.1046/j.1432-0436.2003.710301.x. [DOI] [PubMed] [Google Scholar]
- Krieg M., Arboleda-Estudillo Y., Puech P.H., Käfer J., Graner F., Müller D.J., Heisenberg C.P. Tensile forces govern germ-layer organization in zebrafish. Nat. Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- Lee J., Abdeen A.A., Zhang D., Kilian K.A. Directing stem cell fate on hydrogel substrates by controlling cell geometry, matrix mechanics and adhesion ligand composition. Biomaterials. 2013;34:8140–8148. doi: 10.1016/j.biomaterials.2013.07.074. [DOI] [PubMed] [Google Scholar]
- Li Q., Lau A., Morris T.J., Guo L., Fordyce C.B., Stanley E. A syntaxin 1, gao, and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 2004;24:4070–4081. doi: 10.1523/jneurosci.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhu X., Zeng Y., Wang J., Zhang X., Ding Y.Q., Liang L. FMNL2 enhances invasion of colorectal carcinoma by inducing epithelial-mesenchymal transition. Mol. Cancer Res. 2010;8:1579–1590. doi: 10.1158/1541-7786.mcr-10-0081. [DOI] [PubMed] [Google Scholar]
- Lu R., Yang A., Jin Y. Dual functions of T-box 3 (Tbx3) in the control of self-renewal and extraembryonic endoderm differentiation in mouse embryonic stem cells. J. Biol. Chem. 2011;286:8425–8436. doi: 10.1074/jbc.m110.202150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M.E., Li C., Bian H., Smith B.D., Layne M.D., Farmer S.R. Myocardin-related transcription factor A regulates conversion of progenitors to beige adipocytes. Cell. 2015;160:105–118. doi: 10.1016/j.cell.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A.I., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Molé M.A., Weberling A., Fässler R., Campbell A., Fishel S., Zernicka-Goetz M. Integrin β1 coordinates survival and morphogenesis of the embryonic lineage upon implantation and pluripotency transition. Cell Rep. 2021;34:108834. doi: 10.1016/j.celrep.2021.108834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulas C., Kalkan T., Smith A. NODAL secures pluripotency upon embryonic stem cell progression from the ground state. Stem Cell Rep. 2017;9:77–91. doi: 10.1016/j.stemcr.2017.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell M., Oakes P.W., Lenz M., Gardel M.L. Forcing cells into shape: the mechanics of actomyosin contractility. Nat. Rev. Mol. Cell Biol. 2015;16:486–498. doi: 10.1038/nrm4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa N., Sathe A.R., Shivashankar G.V., Sheetz M.P. Matrix mechanics controls FHL2 movement to the nucleus to activate p21 expression. Proc. Natl. Acad. Sci. U S A. 2016;113:E6813–E6822. doi: 10.1073/pnas.1608210113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Pluripotency in the embryo and in culture. Cold Spring Harb. Perspect. Biol. 2012;4:a008128. doi: 10.1101/cshperspect.a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A., Sharov A.A., Piao Y., Amano M., Amano T., Hoang H.G., Binder B.Y., Tapnio R., Bassey U., Malinou J.N., et al. Systematic repression of transcription factors reveals limited patterns of gene expression changes in ES cells. Sci. Rep. 2013;3 doi: 10.1038/srep01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Nobusue H., Onishi N., Shimizu T., Sugihara E., Oki Y., Sumikawa Y., Chiyoda T., Akashi K., Saya H., Kano K. Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nat. Commun. 2014;5:3368. doi: 10.1038/ncomms4368. [DOI] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., Pollard T.D. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem R.J., Ye J., Judson R.L., LaRussa M.F., Krishnakumar R., Blelloch A., Oldham M.C., Blelloch R. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014;14:617–631. doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar U., Schratt G., Dieterich C., Müller J.M., Galgóczy P., Engel F.B., Keating M.T., Gertler F., Schüle R., Vingron M., Nordheim A. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell. 2004;16:867–880. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- Pieters T., van Roy F. Role of cell-cell adhesion complexes in embryonic stem cell biology. J. Cell Sci. 2014;127:2603–2613. doi: 10.1242/jcs.146720. [DOI] [PubMed] [Google Scholar]
- Posern G., Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Rana M.K., Aloisio F.M., Choi C., Barber D.L. Formin-dependent TGF-β signaling for epithelial to mesenchymal transition. Mol. Biol. Cell. 2018;29:1465–1475. doi: 10.1091/mbc.e17-05-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana M.K., Srivastava J., Yang M., Chen C.S., Barber D.L. Hypoxia increases the abundance but not the assembly of extracellular fibronectin during epithelial cell transdifferentiation. J Cell Sci. 2015;128:1083–1089. doi: 10.1242/jcs.155036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., Kovar D.R. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty J.D., Wu C., Bear J.E. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Russell R., Ilg M., Lin Q., Wu G., Lechel A., Bergmann W., Eiseler T., Linta L., Kumar P., Klingenstein M., et al. A dynamic role of Tbx3 in the pluripotency circuitry. Stem Cell Rep. 2015;5:1155–1170. doi: 10.1016/j.stemcr.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G., Philippar U., Berger J., Schwarz H., Heidenreich O., Nordheim A. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. Formative pluripotency: the executive phase in a developmental continuum. Development. 2017;144:365–373. doi: 10.1242/dev.142679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Chen G., Streb J.W., Long X., Yang Y., Stoeckert C.J., Miano J.M. Defining the mammalian CArGome. Genome Res. 2006;16:197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Phua D.Y.Z., Axiotakis L., Jr., Smith M.A., Blankman E., Gong R., Cail R.C., Espinosa de los Reyes S., Beckerle M.C., Waterman C.M., Alushin G.M. Mechanosensing through direct binding of tensed F-actin by LIM domains. Dev. Cell. 2020;55:468–482.e7. doi: 10.1016/j.devcel.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P., Rubinstein B., Unruh J.R., Durnin M., Hanein D., Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura S., Takeda K. ERK signalling as a regulator of cell motility. J. Biochem. 2017;162:145–154. doi: 10.1093/jb/mvx048. [DOI] [PubMed] [Google Scholar]
- Tatapudy S., Aloisio F., Barber D., Nystul T. Cell fate decisions: emerging roles for metabolic signals and cell morphology. EMBO Rep. 2017;18:2105–2118. doi: 10.15252/embr.201744816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen M.K., Guettler S., Larijani B., Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Verma S., Han S.P., Michael M., Gomez G.A., Yang Z., Teasdale R.D., Ratheesh A., Kovacs E.M., Ali R.G., Yap A.S. A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol. Biol. Cell. 2012;23:4601–4610. doi: 10.1091/mbc.e12-08-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve C., Wickström S.A. Shaping the stem cell field. Nat. Rev. Mol. Cell Biol. 2021;22:305. doi: 10.1038/s41580-021-00347-9. [DOI] [PubMed] [Google Scholar]
- Wagh K., Ishikawa M., Garcia D.A., Stavreva D.A., Upadhyaya A., Hager G.L. Mechanical regulation of transcription: recent advances. Trends Cell Biol. 2021;31:457–472. doi: 10.1016/j.tcb.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidgang C.E., Russell R., Tata P.R., Kühl S.J., Illing A., Müller M., Lin Q., Brunner C., Boeckers T.M., Bauer K., et al. TBX3 directs cell-fate decision toward mesendoderm. Stem Cell Rep. 2013;1:248–265. doi: 10.1016/j.stemcr.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N., Hanna J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Weinhold B., Schratt G., Arsenian S., Berger J., Kamino K., Schwarz H., Rüther U., Nordeim A. Srf-/- ES cells display non-cell-autonomous impairment in mesodermal differentiation. EMBO J. 2000;19:5835–5844. doi: 10.1093/emboj/19.21.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Lim Y.B., Zhang Z., Wang Y., Zhang S., Lim C.T., Yim E.K.F., Kanchanawong P. Nanoscale architecture of the cortical actin cytoskeleton in embryonic stem cells. Cell Rep. 2019;28:1251–1267.e7. doi: 10.1016/j.celrep.2019.06.089. [DOI] [PubMed] [Google Scholar]
- Yang Q., Zhang X.F., Pollard T.D., Forscher P. Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J. Cell Biol. 2012;197:939–956. doi: 10.1083/jcb.201111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Images taken every 10 min for 20h on gelatin-coated glass, 40x.
Images taken every 10 min for 20h on gelatin-coated glass, 40x.
Data Availability Statement
RNA-seq data generated during this study have been deposited in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). The accession number for the data reported in this paper is GEO: GSE175391. Software/packages used to analyze the dataset are freely available. See the supplemental experimental procedures for RNA-seq library preparation and analysis.