Summary
Cell cycle and differentiation decisions are linked; however, the underlying principles that drive these decisions are unclear. Here, we combined cell-cycle reporter system and single-cell RNA sequencing (scRNA-seq) profiling to study the transcriptomes of embryonic stem cells (ESCs) in the context of cell-cycle states and differentiation. By applying retinoic acid, to G1 and G2/M ESCs, we show that, while both populations can differentiate toward epiblast stem cells (EpiSCs), only G2/M ESCs could differentiate into extraembryonic endoderm cells. We identified Esrrb, a pluripotency factor that is upregulated during G2/M, as a driver of extraembryonic endoderm stem cell (XEN) differentiation. Furthermore, enhancer chromatin states based on wild-type (WT) and ESRRB knockout (KO) ESCs show association of ESRRB with XEN poised enhancers. G1 cells overexpressing Esrrb allow ESCs to produce XENs, while ESRRB-KO ESCs lost their potential to differentiate into XEN. Overall, this study reveals a vital link between Esrrb and cell-cycle states during the exit from pluripotency.
Keywords: Exit from pluripotency, Esrrb transcription factor, cell cycle, embryonic stem cells, cellular differentiation and lineage specification, extraembryonic endoderm stem cells(XEN), epiblast stem cells (EpiSC), ChIP-seq and single-cell RNA-seq (scRNA-seq)
Highlights
-
•
The ESC G2/M cell-cycle stage promotes XEN differentiation
-
•
ESRRB accumulation during G2/M is an inducer of XEN
-
•
ESRRB associates with ESC XEN poised enhancers
-
•
Loss of ESRRB impairs XEN differentiation potential
In this article, Ram and colleagues show that Esrrb, a pluripotency factor that is upregulated during G2/M, is a driver of XEN differentiation. Furthermore, enhancer chromatin states based on WT and ESRRB-KO ESCs show association of ESRRB with XEN poised enhancers. G1 ESCs overexpressing Esrrb allow XEN differentiation, while ESRRB-KO ESCs lost their potential to differentiate into XEN.
Introduction
Embryonic stem cells (ESCs) are pluripotent cells derived from the inner cell mass of the preimplantation blastocyst (Martin, 1981). These cells hold unique properties of self-renewal and the ability to give rise to all definitive structures of the fetus. The first cellular decision distinguishes between the epiblast, which produces the embryo body, and the hypoblast, which contributes to the extraembryonic endoderm cells (XENs) (O’Shea, 2004; Tam and Behringer, 1997).
The transition from an uncommitted to a differentiated state requires rapid and global execution of specific gene programs, including the gradual silencing of pluripotency genes and the activation of lineage-specific genes. External signals sensed by each cell drive its fate decisions. ESCs must coordinately alter their transcriptomes, chromatin architectures, and epigenetic landscapes throughout the differentiation process (Dixon et al., 2015; Graf and Enver, 2009; Kurimoto et al., 2015).
The cell cycle is a critical process in the development of an organism, and it is closely linked to cell-fate decisions (Lu et al., 2018; Pauklin and Vallier, 2014). Lineage decision making is essentially a single cell process and the response to lineage-specifying signals relies on the state of each individual cell. Thus, each cell exhibits lineage biases related to, among other factors, cell-cycle phase. Cell cycle consists of four distinct phases dedicated to the replication and transmission of genetic material to daughter cells; in S-phase and M-phase cells, chromosome replication and chromosome transmission occur, respectively. These key events are separated by gap phases, G1 and G2, that serve as regulatory windows to ensure that cell-cycle events occur at the correct time and order (mac Auley et al., 1993; Nurse et al., 1998). The cell-cycle structure of ESCs is characterized by a short G1 phase and a high proportion of cells in S phase (Coronado et al., 2013). This associates with pluripotency factors that influence cyclin-dependent protein kinases (CDKs) (Stead et al., 2002).
Previous studies showed that the cell-cycle stage is a major determinant of cell-fate decisions (Arai et al., 2011; Boward et al., 2016; Chen et al., 2015; Dalton, 2013; Lu et al., 2018; Salomoni and Calegari, 2010; Soufi and Dalton, 2016). Evidence suggests that G1/S is the cellular stage at which differentiation decisions are made, while the G2 phase is mostly dedicated to mitosis control (Hunter et al., 2016). Specifically, previous work showed that human ESCs (hESCs) commit toward specific differentiation path in early G1 but fully adopt a specific differentiation program only in G2 (Gonzales et al., 2015). The intersection between cell-cycle regulation and cell-fate decision mechanisms involves developmental signals and CDK activities, which mediate cell-cycle-dependent changes in the epigenetic landscape and chromosome architecture of developmental genes (Singh et al., 2015). CDKs are also responsible for recruitment of transcription factors (TFs) to cell-fate-related genes (Singh et al., 2015; Pauklin et al., 2016). TF activities can counter CDK activities and drive the cells to exit the pluripotent state. Thus, the balance between CDK and TF determines cell fate.
Retinoic acid (RA) is crucial in early embryonic development and in maintenance of many organ systems in the adult organism (Niederreither and Dollé, 2008). It has been shown that RA represses pluripotency-associated genes and activates lineage-specific markers in ESCs (Zhang et al., 2015). RA promotes a variety of lineage outcomes, such as ectodermal (Janesick et al., 2015; Okada et al., 2004; Takahashi et al., 1999), endodermal, and XEN (Borowiak et al., 2009; Cho et al., 2012; McDonald et al., 2014; Semrau et al., 2017; Simandi et al., 2010). The ability of RA to promote various differentiation phenotypes implies that RA is involved in the switch between proliferation and differentiation (Coronado et al., 2013; Janesick et al., 2015). Another study (Semrau et al., 2017) showed that the exit from pluripotency can be traced by single-cell RNA sequencing (scRNA-seq) after 24 h with RA. After 96 h, ectodermal and XEN subpopulations arose. However, the link between the cell-cycle states of ESCs, exit from pluripotency, and the effect on differentiation outcomes has not been explored.
Pluripotent cell identity is sustained by the activity of a highly interconnected network of TFs such as Pou5f1, Nanog, and Sox2 (Young, 2011) and a large group of ancillary factors such as estrogen-related receptor beta (Esrrb) (Okamura et al., 2019). Esrrb is an orphan nuclear receptor that is required for self-renewal and pluripotency of ESCs (Percharde et al., 2012). In ESCs, Esrrb function is controlled by extrinsic cues mediated by kinases such as GSK3i (Adachi and Niwa, 2013) and intrinsic regulators such as Nanog (Pfeuty et al., 2018). This confers flexibility to the pluripotency network, as changes in the activity of these factors modulate the balance between maintenance and loss of pluripotency (Festuccia et al., 2018). In the early post-implantation mouse embryo, Esrrb is specifically expressed in the extraembryonic ectoderm and plays a crucial role in trophoblast development (Okamura et al., 2019). Moreover, it was demonstrated that induced extraembryonic endoderm stem cells require high levels of Esrrb (Benchetrit et al., 2019), pointing out the possible role of Esrrb in regulating XEN lineage differentiation.
Here, by combining a sensitive cell-cycle reporter system with scRNA-seq, we studied the crosstalk between cell-cycle state and cell-fate decisions during the exit from pluripotency. We revealed that, following RA, ESCs in G1 exclusively differentiate to epiblast stem cells (EpiSCs) and mesodermal cells. Strikingly, if cells are exposed to RA during G2/M phase, they have the unique capacity to differentiate into primitive endoderm and, more specifically, into XENs. We identified that this capacity is driven by upregulation of Esrrb during G2/M state. Overexpressing Esrrb in G1 ESCs cells enabled XEN differentiation. ESRRB knockout (KO) ESCs lost their potential to produce XEN cells. We showed that this loss involves changes in the ESC enhancer landscape and, more specifically, the loss of H3K4me2 on endodermal enhancers. In addition, ESRRB chromatin immunoprecipitation sequencing (ChIP-seq) further supported the direct association of ESRRB with XEN enhancers (Festuccia et al., 2016). Overall, our results demonstrate that exit from pluripotency happens earlier during pluripotency, suggesting that cell-cycle-dependent pluripotency factors regulate cell-fate decision outcomes.
Results
ESC differentiation is coupled with decrease proliferation and cell-cycle state composition
To visualize cell-cycle states without complications of cell-to-cell variation, we produced a FUCCI (fluorescent ubiquitination-based cell cycle indicator) ESC line (Sakaue-Sawano et al., 2008). The FUCCI system is based on cycled translation and ubiquitination-based degradation of Cdt1 (mKO2 conjugated, red) and Geminin (mAG1 conjugated, green) (Figures 1A and 1B). To induce differentiation, we used RA, driving ESCs to differentiate toward intra-embryo progenitor cells (e.g., EpiSCs) but also toward extra-embryo cells such as XEN (Janesick et al., 2015).
Figure 1.
Temporal ESC FUCCI-based system
ESCs were maintained in LIF/2i ESC medium and then switched to differentiation medium (1 μM RA without LIF and 2i) and analyzed after 2 and 4 days (2d Diff. and 4d Diff., respectively).
(A) Representative ×20 bright-field images; scale bar, 50 μm.
(B) Representative ×20 confocal images; scale bar, 50 μm.
(C) Cell-cycle phases in pluripotent state and during differentiation calculated by a combination of FUCCI and Hoechst DNA staining.
(D) Growth rates of ESCs and RA treated cells (Diff.). Shown are mean values of three independent experiments (∗p < 0.05, Wilcoxon test).
(E) Differential expression of transcriptomes using RNA-seq. Transcriptomes of ESCs (WT) versus cells treated with RA for 4 days were plotted. The x axis represents expression fold changes in logarithmic scales; The y axis represents p values based on three replicates. Genes upregulated in ESCs are indicated in purple and genes upregulated in the differentiated cells are indicated in blue. See also Figures S1 and S5 and Table S1.
Upon RA treatment, cell morphology typically changed (Chen et al., 2013) (Figures 1A and 1B). Confocal imaging and fluorescence-activated cell sorting (FACS) analysis revealed that, prior to RA addition, around 8% of the cells were in G1 phase (Figure 1C), which increased to 32% after 4 days of differentiation. G1 increasement is accompanied by replication rate reduction (Figure 1D). CellTrace proliferation analysis verified that cell division rate was significantly slower in differentiating cells than in pluripotent cells (Figure S1B). Over differentiation, we observed transcriptional activation of genes that regulate EpiSC- and XEN-like states in the embryo (such as Sox4, Sox9, Gata4, and Gata6), while there was downregulation of pluripotency factors (such as Pou5f1, Nanog, and Esrrb) (Figure 1E). Additional genes associated with differentiation or pluripotent state with altered expression over time are shown in Table S1.
Cell-cycle phase of ESCs affects XEN differentiation potential
To map the potential links between cell-cycle states during pluripotency and differentiation trajectories, ESCs were sorted into G1 and G2/M states and immediately initiated differentiation (Figure 2A). To analyze the heterogeneous population of cells resulting from RA treatment, we used the inDrop scRNA-seq system (Zhang et al., 2019a) and Seurat pipeline for analysis (Stuart et al., 2019). Based on two independent replicates and analysis of transcriptomes of ∼2,500 cells, we detected four main subpopulations (Figure 2B): (1) XEN cells (cluster 0), (2) EpiSCs (clusters 1 and 3), and (3) mesodermal progenitor cells (cluster 2). A complete list of differentially expressed gene contributions for each cluster formation is given in Table S2. We further eliminated the possibility that cluster separation could be explained by single-cell coverage (Figure 2C). Cluster 3 presented slightly higher coverage compared with the other clusters; however, it shows overall similar markers as cluster 1, both supporting EpiSC differentiation states and thus not compromising our biological interpretations. Next, we revealed that G1 cells predominantly exhibit epiblast differentiation capacities, with expression profiles characteristic of EpiSCs and mesodermal progenitor cells (Figures 2D and 2E). In Figure 2F, we marked EpiSC marker genes highly expressed in clusters 1 and 3 (Sox4, Sox9, Sox11, Nrp1, Vimentin, Smad6, and Gata2). Gata2, Sin3b, Rhox6, and Rhox9 specifically mark the mesodermal subpopulation in cluster 2 (Lugus et al., 2007). As with G1 ESCs, G2/M ESCs also enabled differentiation toward EpiSCs; however, most of the G2/M ESCs differentiated into XEN cells that expressed primitive endoderm markers such as Gata4, Foxq1, Foxa2, Dab2, Lama1, and Lamc1 (Figure 2F). Finally, to rule out the possibility that, upon LIF/2i withdrawal, high levels of BMPs (bone morphogenetic proteins) in the serum may have an impact on the differentiation propensities, we repeated the experiment described in Figure 2 with ESCs growing in a fully chemically defined medium and obtained similar results (Figure S2). To validate our results, we used real-time PCR, which suggests that ESC culturing method does not affect differentiation potential and outcomes. Primer sequences for real-time PCR are given in Table S4.
Figure 2.
scRNA-seq analysis of differentiated cells based on G1 and G2/M ESC populations
(A) Schematic illustration of the sorting and scRNA-seq experiments.
(B) UMAP visualization of ∼2,000 cells clustered into four groups using the Seurat pipeline (Stuart et al., 2019), EpiSC (marked as 1 and 3), XEN (marked as 0), and mesodermal-like cluster (marked as 2) are also represented by different colors.
(C) Violin plots representing number of genes and UMIs for each cluster.
(D) UMAP visualization of the same data colored based on the initial sorting of ESCs; G1 are in red, and G2/M are in green.
(E) Heatmap describing proportion of cells in each cluster (x axis) based on cell-cycle-initiated ESC populations (y axis). Gray to purple scale indicates normalized cell number in each cluster.
(F) Dot plot of differentially expressed genes explaining the four different clusters. Dot size indicates percentage of cells that express the gene, and gray to purple scale indicates average expression within the cluster. See also Figure S3 and Tables S2 and S3.
Next, we captured the dynamics of RA-based differentiation using diffusion map analysis to examine unsorted ESCs together with G1 versus G2/M sorted ESCs followed by 2 and 4 days of RA differentiation (Figure S3). Interestingly, following 4 days of differentiation, endoderm and XEN states are spatially adjacent on diffusion map space, but they could still be distinguished, supporting the observation that only G2/M ESCs hold the capacity to produce XEN cells (Figures S3D and S3G).
Overall, our results show that the differentiation capacity of ESCs is strongly influenced by the state of the cell cycle and suggests that, during exit from pluripotency, ESCs had already acquired a propensity toward epiblast versus hypoblast.
ESRRB is strongly associated with RA-based XEN differentiation
To identify potential XEN-promoting genes expressed during pluripotency in the context of cell cycle, we profiled bulk mRNA of ESCs in the G1 and G2/M states. Based on three independent replicates of G1 and G2/M ESCs, we detected ∼2,000 genes expressed at higher levels in G2/M and ∼400 genes in G1 (Figure 3A; Table S1). The majority of these genes were cell-cycle regulators, and, thus, only cell-cycle Gene Ontology (GO) terms were significantly enriched. However, small number of the identified genes are involved in regulation of pluripotency and early differentiation. For example, genes expressed at higher levels in G1 ESCs included Nanog, Klf4, Otx2, Leafty1, and Pax3, which are central regulators of EpiSCs initiation (Mitsui et al., 2003). Genes with higher expression in G2/M cells did not show any known direct drivers of the XEN lineage. However, we identified two potential candidates: Sall4 and Esrrb (Figure 3A). Sall4 is expressed in both ESCs and XEN cells and encodes a TF that regulates expression of key XEN lineage-associated genes(Lim et al., 2008). Esrrb encodes a pluripotent TF that is involved in regulation of the trophectodermal lineage (Jaber et al., 2017) and, together with Oct4, Sox2, and Klf4, in maintaining ESCs (Betschinger et al., 2013). To test if the difference in Esrrb expression levels observed between G1 and G2/M ESCs is kept also at the protein level, we performed western blot analysis. Based on five independent replicates, we found small but significantly higher levels of ESRRB in G2/M ESCs (Figure S4). We concluded that this small difference is apparently sufficient to push G2/M cells toward XENs (Figures 2D–2F).
Figure 3.
ESRRB is an inducer of XEN cells
(A) Bulk RNA-seq data of ESCs in the G1 state (left) versus the G2/M state (right). The fold change in G2/M versus G1 ESCs is plotted versus t test p values (n = 3). Esrrb and Sall4 genes show G2/M upregulation. Nanog and Klf4 genes show G1 upregulation and Pou5f1 is insensitive to ESC cycle state.
(B) (1) FUCCI cells non-infected (WT) versus infected with Esrrb-YFP lentiviral construct. The y axis represents expression levels based on RNA-seq read counts of lentiviral transcripts (FUCCI also contributes to the overall count, hence WT counts). (2) Confocal imaging of cells following 4 days with RA showing an overlap between Esrrb-YFP and G1 cells (marked by RFP). Upper-: representative ×20 bright-field image; scale bar, 50 μm. Lower: representative ×20 confocal images; scale bar, 50 μm.
(C) Western blot of (left to right) WT ESCs, WT 4 days Diff. with RA, ESCs infected with Esrrb-YFP, 4-day RA differentiated cells infected with Esrrb-YFP.
(D) UMAP of 3025 G1 ESCs following 4-day differentiation process clustered into three groups using the Seurat pipeline (Stuart et al., 2019). The clusters correspond to EpiSC in red, XENs in green, and mesoderm-like in blue.
(E) UMAP plots highlighting expression levels of indicated marker genes. Gray (low) to purple (high) scale indicates average expression signal. See also Figures S2 and S4 and Tables S1, S2, and S3.
To further validate Esrrb involvement in G2/M-specific XEN induction, we produced a FUCCI ESC line that overexpresses Esrrb-YFP. We used FACS to select the Esrrb-YFP-expressing cells, verified nuclear YFP expression of the selected cells by confocal microscopy (Figure 3B), and validated ESRRB exogenous translation using western blot (Figure 3C). Then, we sorted G1-YFP ESCs and immediately initiated differentiation for 4 days, which resulted in the analysis of 3,024 cells from three independent replicates. In support of our hypothesis, the subpopulations observed for differentiated G1 Esrrb-YFP ESCs included EpiSC (Figure 3D) and a small subpopulation of Gata4- and Dab2-expressing XEN cells (Figure 3E; Table S2). Rescue of XEN differentiation potential derived from G1 ESCs suggests direct involvement of ESRRB upregulation. Furthermore, because our exogenous ESRRB expression is lower than native endogenous levels, we hypothesize that even a small increase in ESRRB levels is sufficient to restore XEN differentiation potential. Another possible explanation is that ESCs with exogenous ESRRB keep ESRRB levels higher during exit from pluripotency and differentiation. This further points out the involvement of Esrrb as a key factor in the induction of XEN and that XEN differentiation is regulated in a cell-cycle-dependent manner.
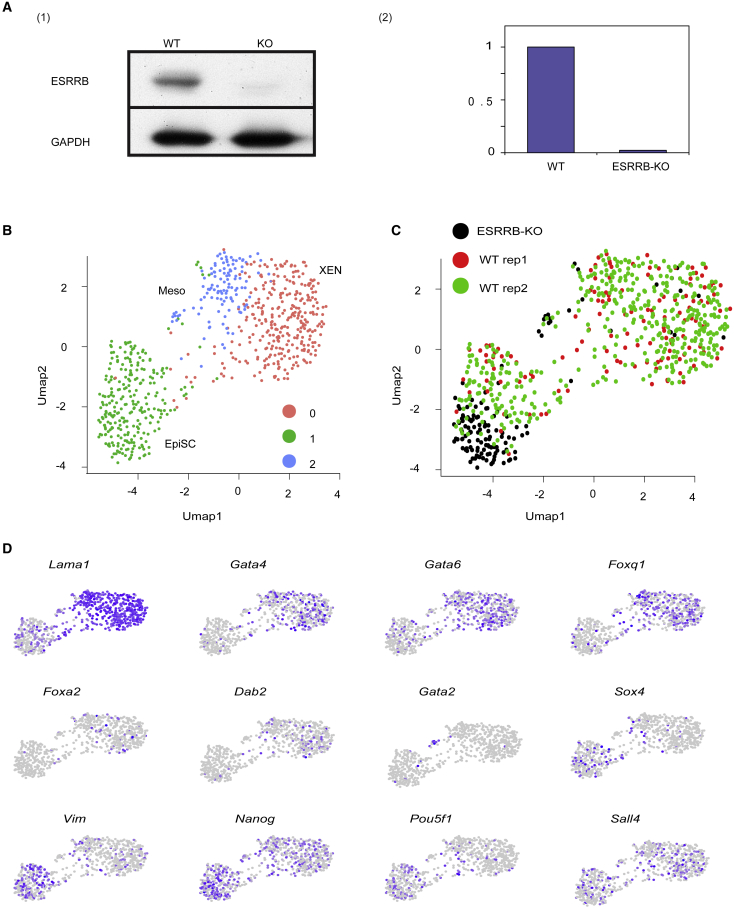
Finally, to directly test the function of ESRRB during the exit from pluripotency, we created ESRRB-deficient ESCs using CRISPR/CAS9, validated by WB (Figure 4A), and then differentiated them for 4 days. Supporting our previous observation, ESRRB-KO ESCs contributed only to the formation of EpiSC (Figures 4B–4D; Table S2). Additionally, in line with our previous results (Figure 2), a small population of Gata2 cells (mesodermal-like progenitor cells) could be detected even though unbiased clustering did not highlight it as a standalone cluster.
Figure 4.
ESRRB-KO ESCs fail to differentiate into XEN cells
(A) (1) Western blot of WT and ESRRB-KO ESCs. (2) ImageJ pixel-based interpretation (y axis: WT-based normalization).
(B) UMAP of 700 cells of 4-day Diff. clustered into three groups using the Seurat pipeline (Stuart et al., 2019). EpiSC in green, XEN in red and blue. Gata2 mesodermal-like cells are a small subpopulation within the blue cluster.
(C) The same UMAP as in Figure 4B colored for WT (red and green) and ESRRB-KO (black).
(D) UMAP plots highlighting expression levels of selected marker genes. Gray (low) to purple (high) scale indicates average expression signal. See also Tables S2 and S3.
During cellular differentiation, lineage outcomes are not affected by cell-cycle states
During early differentiation, once progenitor cells emerge, the cells replicate constantly. Therefore, we aimed to explore whether cell-cycle states also influence fate decisions in cells that have already exited from their pluripotent state. To this end, we treated ESCs with RA for 2 days, when the main pluripotency factors are significantly downregulated (Figure S4), and replication time becomes much slower (Figure 1D). Then, we sorted the cells based on the cell-cycle phase as described above. Sorted cells were subjected to scRNA-seq directly or after another 2 days of differentiation (Figure S5A). To validate the precision of cell-cycle-based sorting, we focused our analysis on known cell-cycle genes (Waisman et al., 2019), calculated the principal component matrix, performed clustering (Turkmen et al., 2019), and visualized the results using Uniform Manifold Approximation and Projection (UMAP). The cells clustered based on our FUCCI/Hoechst sorting gates and GO annotations showed a very clear separation based on G1 and G2/M states (Figure S5B). In EpiSC (cluster 0 in Figure S5C), a separation between G1 and G2/M cells could be detected (Figure S5D). G1-derived cells expressed higher levels of Vimentin, Ecm1, and Acta2 (Chung et al., 2019; Zhang et al., 2019b), which are associated with later stages of differentiation, whereas G2/M expressed higher levels of Nanog, which is associated with premature differentiation status (Kim et al., 2008).
Overall, these results suggest that, at 2 days after differentiation signal, the differentiation potential is no longer influenced by the cell-cycle state. Interestingly, single-cell profiling of 4-day differentiated cells, which were sorted at day 2 based on cell cycle (Figure S5A), shows two major clusters of EpiSC- and XEN-like cells (Figure S5B) with similar cell-cycle-derived proportion of EpiSC and XEN differentiation (Figures S5C and S5D). Taken together, our study suggests that ESC fate is strongly influenced by the cell-cycle state at the moment of exposure to the differentiation signal. However, once cells start to differentiate and cellular commitments are made, the cell cycle becomes irrelevant to fate decisions.
ESRRB associates with poised enhancers of XEN genes
Esrrb expression is quickly downregulated during the exit from pluripotency. Therefore, to promote XEN differentiation, we hypothesized that, in addition to the association of ESRRB with poised enhancers in general, it should specifically mark XEN enhancers. To test this hypothesis, we mapped H3K4me1 and H3K4me2 enhancers of WT and ESRRB-KO ESCs. We first extracted H3K4me1 and H3K4me2 signal over TSSs. Surprisingly, we found no significant changes between WT and KO ESCs over promoters and proximal enhancers (Figure S6).
Next, we performed an unbiased comparison of peaks over distal enhancers. We extracted differential peaks, hence candidate enhancers, and linked them with their target genes using enhancer atlas (Shen et al., 2012). We found a significant enrichment for WT distal enhancers involved in endodermal differentiation. On the other hand, ESRRB-KO ESCs were enriched with enhancers involved in metabolic and apoptotic pathways rather than specific differentiation pathways (Figure 5A). Having said that, many differentiation genes have several enhancers with additive regulatory effects (Figures 5B–5D). In most cases, ESRRB-KO reduced H3K4me1/2 from only one or a small number of enhancers (Figure 5B) and in many other enhancers, mainly involved in EpiSC and pluripotency, no difference between WT and KO ESCs could be captured (Figures 5C–5D). This might explain why ESRRB-KO ESCs can still differentiate to embryonic tissues, including endoderm.
Figure 5.
ESRRB associates with poised enhancers of XEN genes
(A) H3K4me2 ChIP-seq analysis of distal enhancers comparing WT and ESRRB-KO ESCs. MsigDB enriched annotations corresponding to WT versus ESRRB-KO are shown in each side of the Venn diagram.
(B–D) ChIP-seq IGV tracks for representative (B) XEN marker genes (Gata4 and Foxa2), (C) EpiSC marker genes (Smad6 and Sox4), and (D) pluripotent marker genes (Nanog and Pou5f1) mapped using H3K4me1 and H3K4me2 antibodies for WT and ESRRB-KO ESCs. Distal enhancer regions depleted in ESRRB-KO ESCs marked in gray boxes.
(E) ChIP-seq IGV tracks of H3K4me1, H3K27ac, and ESRRB (Festuccia et al., 2016) are shown for ESCs Gata6, Gata4, and Foxa2 XEN marker genes. H3K4me1 and H3K27ac are also shown for 4-day RA differentiated cells. Distal enhancers marked with gray boxes. See also Figure S6.
Later, we mapped H3K4me1 and H3K27ac enhancers in ESCs after 4 days of differentiation. We also obtained and re-analyzed ESRRB ChIP-seq maps of interphase ESCs (Festuccia et al., 2016) enriched for S/G2 ESCs (Coronado et al., 2013; ter Huurne et al., 2017). Both histone marks and ESRRB chip seq data obtained from ESCs grown in LIF-serum medium condition without 2i. Next, in examination of XEN marker genes (e.g., Gata6, Gata4, Foxa2, Dab2, and Foxq1), we found that ESRRB associates with their enhancers. Specifically, these candidate enhancers were poised (H3K4me1 positive and H3K27ac negative) during pluripotency but were activated upon differentiation, as shown by the gaining of H3K27ac (Figure 5E).
ESRRB is a cell-cycle-associated inducer of XEN
To assess if ESRRB is a specific inducer of XEN, we tested two differentiation protocols. We first produced embryoid bodies (EBs) (Kim et al., 2020), which contain cells of the three germ layers, ectoderm, mesoderm, and endoderm. We observed that EB size and structure is similar between WT and ESRRB-KO cells (Figure 6A left panel) and that replication rate is quite comparable (Figure 6B). Moreover, pluripotent transcription signature of ESRRB-KO ESCs is significantly lower compared with WT ESCs (Figure 6D1), and the overall differentiation scores of EB ESRRB-KO cells were slightly higher for all differentiation lineages (Figure S7). We concluded that ESRRB-KO ESCs retain the potential to produce healthy EBs.
Figure 6.
ESRRB is a XEN-specific inducer
(A) WT and ESRRB-KO ESCs followed by 10-day EBs and 9-day XEN direct differentiation protocols. Representative ×10 images scale bar, 50 μm.
(B and C) Live cell count of WT and ESRRB-KO cells for (B) EBs and (C) XEN direct differentiations. Statistics based on three biological replicates (∗∗p < 0.05, ∗p < 0.1 Wilcoxon test).
(D) Bulk RNA-seq data of WT ESCs, ESRRB-KO ESCs, and WT direct XEN differentiated cells (∗∗p < 0.05, ∗p < 0.1 Wilcoxon test). (1) Expression levels of selected pluripotent marker genes. (2) Expression levels of selected XEN marker genes (Brown et al., 2010; Lim et al., 2008; Lin et al., 2016). See also Figure S7.
Next, we tested 9 days of a direct XEN differentiation protocol (Ngondo et al., 2020). Reassuringly, the majority of ESRRB-KO ESCs could not differentiate into XEN but preferred to activate apoptotic pathways (Figure 6C). Conversely, WT ESCs could efficiently differentiate into cells expressing XEN markers (Brown et al., 2010; Lim et al., 2008; Lin et al., 2016) (Figure 6D2), supporting our initial hypothesis that ESRRB is a specific inducer of XEN.
Discussion
The molecular mechanisms that specify embryo and extraembryonic germ layer identities are only partially understood. Different TFs such as Gata, Fox, and Sox protein families, act to regulate extensive networks in ESCs (Boyer et al., 2005; Chen et al., 2013; Kim et al., 2008) and are associated with early differentiated states (Gökbuget and Blelloch, 2019). However, most of these factors are not expressed during pluripotency, hence they will not play a key role during exit from pluripotency. It has been suggested that the cell cycle of ESCs plays a key role in specifying differentiation outcomes during exit from pluripotency. To study ESC cycle and the extent to which it dictates differentiation outcomes, we used the FUCCI system (Sakaue-Sawano et al., 2008), which allowed us to monitor cell-cycle progression in living cells, and combined it for the first time with scRNA-seq measurements, which enable us to detect the underlying heterogeneity of differentiation.
While bulk transcriptome analysis suggested that early differentiation is mainly dictated from G1 ESCs, scRNA-seq in the context of cell cycle identifies heterogeneity in G2/M ESC differentiation capabilities. These cells included subpopulations of differentiated and less differentiated cell states. This suggests pre-existing lineage biases in G2/M ESCs result in heterogeneous populations of cells states. Therefore, we concluded that population-based measurements lead to a misleading assumption that only G1 cells are prone to differentiation, while averaging G2/M cells hides their underlying heterogeneity.
Our results demonstrate that cell-cycle states of ESCs, at the moment they get exposed to RA differentiation signals, dictate the decision to differentiate into XENs or EpiSCs/mesodermal cells. We focused on RA-driven mouse ESC (mESC) differentiation, which is widely used in differentiation assays (Semrau et al., 2017) and has significant functions in embryonic development.
We further demonstrated that Esrrb is a key factor upregulated during G2/M state of ESCs, which is also associated with the XEN differentiation pathway. In agreement with our finding, a previous study showed that Esrrb, in conjunction with Gata3, Eomes, Tfap2c, and Myc, can induce pluripotency by the activation of a unique XEN-like state (Benchetrit et al., 2019). In addition, Esrrb regulates expression of many TFs that are critical for maintaining pluripotency and self-renewal. Previously, a significant overlap between chromatin binding of Esrrb and Tfe3, which is a key regulator of the hypoblast/PrE circuitry, was also revealed (Betschinger et al., 2013). We functionally validated that Esrrb is an inducer of the XEN lineage as follows: (1) following exogenous expression of Esrrb in G1 ESCs, the cells restored the capacity to differentiate into XEN in addition to their non-compromised ability to differentiate to EpiSC and mesoderm. Thus, the integration of Esrrb into the core transcriptional network of G1 cells stimulated XEN initiation and overcame cell-cycle dependency. (2) We showed that ESRRB-KO ESCs lack the potential to form XEN-like cells regardless of their cell-cycle state at the moment of differentiation activation. This emphasizes that Esrrb is a key regulator of XEN differentiation, and cell-cycle-dependent expression of Esrrb allows G2/M cells to become XEN-like cells. (3) We next validated the importance of Esrrb for XEN formation by applying a direct XEN differentiation protocol. Indeed, the majority of ESRRB-KO ESCs could not produce XEN cells. Reassuringly, EB formation, a simple differentiation protocol that direct ESCs toward epiblast cells, was not affected by ESRRB-KO. This aligns with our observation for XEN RA-based differentiation.
Other known members of the XEN differentiation circuitry are Tfap2c, Sox17, Eomes, and Cdx2 (Kojima et al., 2017). However, although we detected some of these marker genes in bulk RNA sequencing (RNA-seq), we could not detect differential expression of mRNAs encoding these proteins in our scRNA-seq experiments, most probably due to the sensitivity limitation of the single-cell assay. In addition, as these genes are not expressed in ESCs, they are not likely to be involved in the exit from the pluripotency step.
Sall4 is critical for XEN differentiation through regulation of expression of Gata4, Gata6, Sox7, and Sox17, and due to interconnections in the pluripotent regulatory circuitry with Oct4, Sox2, and Nanog (Lim et al., 2008). We observed that Sall4 expression has a cell-cycle dependency, and its expression was upregulated during G2/M phase in ESCs. SALL4 chromatin-binding profile is correlated with that of ESRRB, suggesting that SALL4 and ESRRB may be co-regulated during XEN differentiation. TF recruitment can be modulated by epigenetic modifications to the chromatin (Lupien et al., 2008; Niwa et al., 2000) and cell-cycle-specific ESRRB enhancer occupancy could be regulated by methylation of DNA and covalent modification of histone proteins. Further experiments are needed to identify additional epigenetic players that may be involved in the exit from pluripotency specifically to the XEN state.
Overall, the presented results support the tight association between cell-cycle stage and cell-fate determination. We demonstrated that the cell-cycle state affects linage specification at the exit from pluripotency, and, although the complete cellular signaling is yet to be comprehensively revealed, we demonstrated that Esrrb plays a key role in this regulatory pathway. We therefore suggest a model for which Esrrb upregulated expression and protein accumulation during S-G2 expands its enhancer-binding capacity toward poised differentiation enhancers in general and XEN-specific enhancers in particular. Thus, in addition to the differentiation potential toward epiblast cells, G2/M cells also support differentiation toward XEN lineage (Figure 7).
Figure 7.
A model for ESRRB-dependent XEN induction during exit from pluripotency
ESRRB accumulates in ESCs during interphase with low expression level at G1 phase and high expression during G2/M phase. When cells are at G1, ESRRB’s limited abundance is mainly dedicated for pluripotency maintenance through co-binding with pluripotent factors such as Pou5f1, Sox2, and Nanog. When cells are at G2, accumulation of ESRRB allows excess ESRRB to bind XEN-specific distal enhancers, thus promoting XEN differentiation.
Further validation experiments and expanding our cell-cycle-based scRNA-seq protocol on other differentiation pathways will allow us to further increase our understanding of the link between cell cycle and differentiation outcomes, exposing other cell-cycle or cycling genes that are central for the elusive exit from pluripotency stage and early differentiation decisions.
Experimental procedures
Cell culture
Mouse R1 ESCs were a gift from A. Nagy (Mount Sinai Hospital, Toronto, Ontario, Canada). ESRRB-KO ZHBTc4 ESCs were produced by the Smith laboratory (Niwa et al., 2000). ESCs were cultured as described in Bavli et al. (2021). See also in supplemental experimental procedures. RA differentiation was induced using RA (Sigma-Aldrich, R2625) in dimethyl sulfoxide (ENCO, 0219605580) at 1 μM with 10% FBS, on 0.1% gelatin-coated plates. EBs were induced using LIF-free medium with 10% FBS, on a Petri dish.
Establishment of a stably transfected FUCCI cell line
Plasmids expressing mKO2-hCdt1 or mAG-hGem were a kind gift from Prof. Itamar Simon (The Hebrew University, Jerusalem, Israel). 293T cells were kindly gifted from Prof. Meshorer (The Hebrew University, Jerusalem, Israel). Plasmids were transfected to 293T cells using TransIT-LT1 Transfection Reagent (Mirusbio, MC-MIR-2300). After 24 h, the samples were filtered through a 0.45-mm filter, and ESCs were resuspended in polybrene-supplemented medium (8 μg/mL PB [Sigma-Aldrich, 107689] in ES medium). The procedure was repeated after 24 h. The cells were then two-step FACS sorted: (1) S, G2, and M cells were sorted by green fluorescence (488 filter) using a FACSAria II cell sorter (Becton Dickinson); (2) the sorted cells were then reseeded and, after a week, were sorted for red fluorescence (461 filter). For clonal selection, cells were seeded in 96-well plates at a density of one cell per well.
Establishment of Esrrb stable cell line
Esrrb-YFP plasmid was a kind gift from Prof. Yosef Buganim (The Hebrew University, Jerusalem, Israel). Plasmids were transfected into 293T cells using TransIT-LT1 Transfection Reagent (Mirusbio, MC-MIR-2300). After 24 h, samples were filtered through a 0.45-mm filter, and cells were resuspended in PB-supplemented medium. This step was repeated after 24 and 72 h.
FACS
For cell-cycle analysis, cells were trypsinized, washed twice in PBS, stained with Hoechst (Sigma-Aldrich, B2261), and resuspended in 2% FBS. The G1 and M/G2 populations were sorted using a FACSAria II cell sorter (Becton Dickinson). Sorted samples were processed for scRNA-seq or cDNA sequencing.
mRNA extraction
mRNA extraction was performed using Invitrogen Dynabeads mRNA DIRECT Purification Kit (61011) according to the manufacturer’s protocol.
Library preparation
The Drop-seq libraries were prepared according to Macosko et al. (2015). All the primers used in the library preparation are listed in Table S3. The inDrop libraries were prepared according to Bavli et al. (2021).
ChIP
Chromatin immunoprecipitation (ChIP) was performed as described in Sailaja et al. (2012). Antibodies used: H3K4me1 abcam ab8895, H3K27ac abcam ab4729, H3K36me abcam ab9050, 2 μg of antibody per 2 × 107 cells.
ChIP-seq data analysis
Sequencing data were aligned using Bismark and Bowtie (https://www.bioinformatics.babraham.ac.uk/projects/bismark/Bismark_User_Guide.pdf) using a paired-ended approach. TDF genomic browser files were produced using IGV count. We applied HOMER to find peaks using ChIP-seq criteria and used BEDTools to intersect bins with genomic intervals such as promoters, genes, and predicted enhancers.
Western blot and antibodies
Proteins were separated by SDS-PAGE 4%–20% polyacrylamide gradient gels and transferred to 0.45-μm nitrocellulose membranes (iBlot2, PVDF, mini-Transfer Stacks, Thermo Scientific; IB24002v). The membranes were incubated with the appropriate primary and secondary antibodies and washed in PBS-Tween 20. Horseradish-peroxidase-conjugated secondary antibodies were detected by Super-Signal West Pico Chemiluminescent Substrate (Thermo Scientific; PI-34080). Antibodies used: anti-human ESRRB/NR3B2 (R&D systems; PP-H6705-00), anti-GAPDH (Abcam; AB-ab8245), and goat anti-mouse (Jackson ImmunoResearch; 115-035-062).
Deep sequencing
Deep sequencing was performed with Illumina NextSeq using commercially available kits from Illumina (Danyel Biotech FC-404- 2005) following the manufacturer’s protocols.
Data analysis
The Illumina output was analyzed using an in-house Perl script that produced a read matrix that was aligned using RSEM (Li and Dewey, 2011) with Bowtie (Langmead et al., 2009). The resulting matrix was analyzed in R. For bulk data analysis, transcript per million (TPM) values were used to compare between libraries. Differential gene expression was visualized using volcano plots. Statistical analysis was performed for two independent replicates using a two-sided t test, and p values of <0.05 were deemed significant. scRNA-seq data was analyzed using the Seurat v2.4 pipeline (Stuart et al., 2019). Cells with more than 10,000 unique molecular identifiers were retained for further analysis. Global-scaling normalization was performed on the filtered dataset using LogNormalize with a scale factor of 10,000. Identification of highly variable genes was performed with the following parameters: x.low.cutoff = 0.2, x.high.cutoff = 5, y.cutoff = 0.5, and y.high.cutoff = 10. Cell-to-cell variation in gene expression driven by batch, cell alignment rate, and number of detected molecules was regressed out and a linear transformation was applied. Principal component analysis (PCA) was performed on the scaled data with 12 principal components. Clustering was done with resolution of 0.6, and t-distributed stochastic neighbor embedding (tSNE) or UMAP was used for visualization.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was done using GSEA software (Ray et al., 2003; Subramanian et al., 2005) with a false-negative discovery q value <0.01.
Statistical analyses
| G1 phase | G2 phase | |
|---|---|---|
| Specific cluster: X | no. of G1 cells in X | no. of G2 cells in X |
| All other clusters | total −o. of G1 cells − no. of G1 cells in X | total no. of G2 cells − no. of G2 cells in X |
Chi-square goodness-of-fit test was performed using chisq.test function in R. For small numbers, Fisher’s exact test was performed instead.
Author contributions
S.H.L., S.F., and O.R. conceived the study and prepared the figures. S.H.L., L.A., S.F., Y.B., and O.R. designed the experiments. S.H.L., L.A., S.F., and M.A. performed tissue culture, FACS sorting, and library preparation for bulk and scRNA-seq. D.B. and X.S. prepared the microfluidics system and performed scRNA-seq experiments. O.R. and S.L. performed the computational analysis. S.H.L., L.A., S.F., M.A., and O.R. wrote the manuscript.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
O.R. is supported by research grants from the European Research Council (ERC, # 715260 SC-EpiCode), the Israeli Center of Research Excellence (I-CORE) program, the Israel Science Foundation (ISF, #1618/16), and Azriely Foundation Scholar Program for Distinguished Junior Faculty. This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 765966 (EpiSyStem). We offer special thanks to Prof. Eran Meshorer, Dr. Yonatan Tzur, and Prof. Itamar Simon from The Hebrew University for helpful discussions and critical reading of the manuscript.
Published: May 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.04.016.
Supplemental information
Data and code availability
The accession number for the data reported in this paper is GEO: GSE178390. Correspondence and requests for materials should be addressed to O.R. (oren.ram@mail.huji.ac.il).
References
- Adachi K., Niwa H. A liaison between intrinsic and extrinsic regulators of pluripotency. EMBO J. 2013;32:2531–2532. doi: 10.1038/EMBOJ.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai Y., Pulvers J.N., Haffner C., Schilling B., Nüsslein I., Calegari F., Huttner W.B. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat. Commun. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavli D., Sun X., Kozulin C., Ennis D., Motzik A., Biran A., Brielle S., Alajem A., Meshorer E., Buxboim A., Ram O. CloneSeq: a highly sensitive analysis platform for the characterization of 3D-cultured single-cell-derived clones. Dev. Cell. 2021;56:1804–1817.e7. doi: 10.1016/J.DEVCEL.2021.04.026. [DOI] [PubMed] [Google Scholar]
- Benchetrit H., Jaber M., Zayat V., Sebban S., Pushett A., Makedonski K., Zakheim Z., Radwan A., Maoz N., Lasry R., et al. Direct induction of the three pre-implantation blastocyst cell types from fibroblasts. Cell Stem Cell. 2019;24:983–994.e7. doi: 10.1016/j.stem.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Nichols J., Dietmann S., Corrin P.D., Paddison P.J., Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M., Maehr R., Chen S., Chen A.E., Tang W., Fox J.L., Schreiber S.L., Melton D.A. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boward B., Wu T., Dalton S. Concise review: control of cell fate through cell cycle and pluripotency networks. Stem Cells. 2016;34:1427–1436. doi: 10.1002/stem.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K., Legros S., Artus J., Doss M.X., Khanin R., Hadjantonakis A.K., Foley A. A comparative analysis of extra-embryonic endoderm cell lines. PLoS One. 2010;5:e12016. doi: 10.1371/journal.pone.0012016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Gu Q., Wang X., Ma Q., Tang H., Yan X., Guo X., Yan H., Hao J., Zeng F. Directed neuronal differentiation of mouse embryonic and induced pluripotent stem cells and their gene expression profiles. Int. J. Mol. Med. 2013;32:25–34. doi: 10.3892/ijmm.2013.1372. [DOI] [PubMed] [Google Scholar]
- Chen X., Hartman A., Guo S. Choosing cell fate through a dynamic cell cycle. Curr. Stem Cell Rep. 2015;1:129–138. doi: 10.1007/s40778-015-0018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho L.T.Y., Wamaitha S.E., Tsai I.J., Artus J., Sherwood R.I., Pedersen R.A., Hadjantonakis A.K., Niakan K.K. Conversion from mouse embryonic to extra-embryonic endoderm stem cells reveals distinct differentiation capacities of pluripotent stem cell states. Development. 2012;139:2866–2877. doi: 10.1242/dev.078519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.H., Shen W., Davidson K.C., Pébay A., Wong R.C.B., Yau B., Gillies M. Differentiation of retinal glial cells from human embryonic stem cells by promoting the notch signaling pathway. Front. Cell Neurosci. 2019;13:527. doi: 10.3389/fncel.2019.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado D., Godet M., Bourillot P.Y., Tapponnier Y., Bernat A., Petit M., Afanassieff M., Markossian S., Malashicheva A., Iacone R., et al. A short G1 phase is an intrinsic determinant of naïve embryonic stem cell pluripotency. Stem Cell Res. 2013;10:118–131. doi: 10.1016/j.scr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Dalton S. G1 compartmentalization and cell fate coordination. Cell. 2013;155:13–14. doi: 10.1016/j.cell.2013.09.015. [DOI] [PubMed] [Google Scholar]
- Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W., et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N., Dubois A., Vandormael-Pournin S., Gallego Tejeda E., Mouren A., Bessonnard S., Mueller F., Proux C., Cohen-Tannoudji M., Navarro P. Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat. Cell Biol. 2016;18:1139–1148. doi: 10.1038/ncb3418. [DOI] [PubMed] [Google Scholar]
- Festuccia N., Owens N., Navarro P. Esrrb, an estrogen-related receptor involved in early development, pluripotency, and reprogramming. FEBS Lett. 2018;592:852–877. doi: 10.1002/1873-3468.12826. [DOI] [PubMed] [Google Scholar]
- Gökbuget D., Blelloch R. Epigenetic control of transcriptional regulation in pluripotency and early differentiation. Development. 2019;146:dev164772. doi: 10.1242/dev.164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales K.A.U., Liang H., Lim Y.-S., Chan Y.-S., Yeo J.-C., Tan C.-P., Gao B., Le B., Tan Z.-Y., Low K.-Y., et al. Deterministic restriction on pluripotent state dissolution by cell-cycle pathways. Cell. 2015;162:564–579. doi: 10.1016/J.CELL.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Graf T., Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Hunter G.L., Hadjivasiliou Z., Bonin H., He L., Perrimon N., Charras G., Baum B. Coordinated control of Notch-Delta signalling and cell cycle progression drives lateral inhibition mediated tissue patterning. Development. 2016;143:2305–2310. doi: 10.1242/dev.134213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber M., Sebban S., Buganim Y. Acquisition of the pluripotent and trophectoderm states in the embryo and during somatic nuclear reprogramming. Curr. Opin. Genet. Dev. 2017;46:37–43. doi: 10.1016/j.gde.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Janesick A., Wu S.C., Blumberg B. Retinoic acid signaling and neuronal differentiation. Cell Mol. Life Sci. 2015;72:1559–1576. doi: 10.1007/s00018-014-1815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I.S., Wu J., Rahme G.J., Battaglia S., Dixit A., Gaskell E., Chen H., Pinello L., Bernstein B.E. Parallel single-cell RNA-seq and genetic recording reveals lineage decisions in developing embryoid bodies. Cell Rep. 2020;33:108222. doi: 10.1016/j.celrep.2020.108222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., Orkin S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y., Sasaki K., Yokobayashi S., Sakai Y., Nakamura T., Yabuta Y., Nakaki F., Nagaoka S., Woltjen K., Hotta A., et al. Evolutionarily distinctive transcriptional and signaling programs drive human germ cell lineage specification from pluripotent stem cells. Cell Stem Cell. 2017;21:517–532.e5. doi: 10.1016/j.stem.2017.09.005. [DOI] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Hayashi K., Ohta H., Kiyonari H., Mitani T., Moritoki Y., Kohri K., Kimura H., Yamamoto T., et al. Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell. 2015;16:517–532. doi: 10.1016/J.STEM.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323–416. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.Y., Tam W.L., Zhang J., Ang H.S., Jia H., Lipovich L., Ng H.H., Wei C.L., Sung W.K., Robson P., et al. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell. 2008;3:543–554. doi: 10.1016/j.stem.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Lin J., Khan M., Zapiec B., Mombaerts P. Efficient derivation of extraembryonic endoderm stem cell lines from mouse postimplantation embryos. Scientific Rep. 2016;6:39457–39516. doi: 10.1038/srep39457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.C., Sanada C., Wang L., Zhang P.X., Grimes H.L., Venkatasubramanian M., Chetal K., Aronow B., Salomonis N., Krause D.S. The molecular signature of megakaryocyte-erythroid progenitors reveals a role for the cell cycle in fate specification. Cell Rep. 2018;25:2083–2093.e4. doi: 10.1016/j.celrep.2018.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugus J.J., Chung Y.S., Mills J.C., Kim S.I.L., Grass J.A., Grass J., Kyba M., Doherty J.M., Bresnick E.H., Choi K. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134:393–405. doi: 10.1242/dev.02731. [DOI] [PubMed] [Google Scholar]
- Lupien M., Eeckhoute J., Meyer C.A., Wang Q., Zhang Y., Li W., Carroll J.S., Liu X.S., Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- mac Auley A., Werb Z., Mirkes P.E. Characterization of the unusually rapid cell cycles during rat gastrulation. Development. 1993;117:873–883. doi: 10.1242/dev.117.3.873. [DOI] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald A.C.H., Biechele S., Rossant J., Stanford W.L. Sox17-mediated XEN cell conversion identifies dynamic networks controlling cell-fate decisions in embryo-derived stem cells. Cell Rep. 2014;9:780–793. doi: 10.1016/j.celrep.2014.09.026. [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Ngondo R.P., Cohen-Tannoudji M., Ciaudo C. Fast in vitro procedure to identify extraembryonic differentiation defect of mouse embryonic stem cells. STAR Protoc. 2020;1:100127. doi: 10.1016/j.xpro.2020.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K., Dollé P. Retinoic acid in development: towards an integrated view. Nat. Rev. Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J.I., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Nurse P., Masui Y., Hartwell L. Understanding the cell cycle. Nat. Med. 1998;4:1103–1106. doi: 10.1038/2594. [DOI] [PubMed] [Google Scholar]
- O’Shea K.S. Self-renewal vs. Differentiation of mouse embryonic stem Cells1. Biol. Reprod. 2004;71:1755–1765. doi: 10.1095/biolreprod.104.028100. [DOI] [PubMed] [Google Scholar]
- Okada Y., Shimazaki T., Sobue G., Okano H. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev. Biol. 2004;275:124–142. doi: 10.1016/j.ydbio.2004.07.038. [DOI] [PubMed] [Google Scholar]
- Okamura E., Tam O.H., Posfai E., Li L., Cockburn K., Lee C.Q.E., Garner J., Rossant J. Esrrb function is required for proper primordial germ cell development in presomite stage mouse embryos. Dev. Biol. 2019;455:382–392. doi: 10.1016/j.ydbio.2019.07.008. [DOI] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2014;156:1338. doi: 10.1016/J.CELL.2014.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S., Madrigal P., Bertero A., Vallier L. Initiation of stem cell differentiation involves cell cycle-dependent regulation of developmental genes by Cyclin D. Genes Dev. 2016;30:421–433. doi: 10.1101/GAD.271452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M., Lavial F., Ng J.H., Kumar V., Tomaz R.A., Martin N., Yeo J.C., Gil J., Prabhakar S., Ng H.H., et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 2012;26:2286–2298. doi: 10.1101/gad.195545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeuty B., Kress C., Pain B. Network features and dynamical landscape of naive and primed pluripotency. Biophys. J. 2018;114:237–248. doi: 10.1016/j.bpj.2017.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray H.N., Mootha V.K., Boxwala A.A. Building an application framework for integrative genomics. AMIA Annu. Symp. Proc. 2003;2003:981. [PMC free article] [PubMed] [Google Scholar]
- Sailaja B.S., Takizawa T., Meshorer E. Chromatin immunoprecipitation in mouse hippocampal cells and tissues. Methods Mol. Biol. 2012;809:353–364. doi: 10.1007/978-1-61779-376-9_24. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/J.CELL.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Salomoni P., Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20:233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Semrau S., Goldmann J.E., Soumillon M., Mikkelsen T.S., Jaenisch R., van Oudenaarden A. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. Nat. Commun. 2017;8:1096. doi: 10.1038/s41467-017-01076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Yue F., Mc Cleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V., et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simandi Z., Balint B.L., Poliska S., Ruhl R., Nagy L. Activation of retinoic acid receptor signaling coordinates lineage commitment of spontaneously differentiating mouse embryonic stem cells in embryoid bodies. FEBS Lett. 2010;584:3123–3130. doi: 10.1016/j.febslet.2010.05.052. [DOI] [PubMed] [Google Scholar]
- Singh A.M., Sun Y., Li L., Zhang W., Wu T., Zhao S., Qin Z., Dalton S. Cell-cycle control of bivalent epigenetic domains regulates the exit from pluripotency. Stem Cell Rep. 2015;5:323–336. doi: 10.1016/j.stemcr.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Dalton S. Cycling through developmental decisions: how cell cycle dynamics control pluripotency, differentiation and reprogramming. Development. 2016;143:4301–4311. doi: 10.1242/dev.142075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead E., White J., Faast R., Conn S., Goldstone S., Rathjen J., Dhingra U., Rathjen P., Walker D., Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 2002;21:8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi J., Palmer T.D., Gage F.H. Retinoic acid and neurotrophins collaborate to regulate neurogenesis in adult-derived neural stem cell cultures. J. Neurobiol. 1999;38:65–81. doi: 10.1002/(SICI)1097-4695(199901)38:1<65::AID-NEU5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Tam P.P.L., Behringer R.R. Mouse gastrulation: the formation of a mammalian body plan. Mech. Dev. 1997;68:3–25. doi: 10.1016/S0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- ter Huurne M., Chappell J., Dalton S., Stunnenberg H.G. Distinct cell-cycle control in two different states of mouse pluripotency. Cell Stem Cell. 2017;21:449–455.e4. doi: 10.1016/j.stem.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkmen A.S., Yuan Y., Billor N. Evaluation of methods for adjusting population stratification in genome wide association studies: standard versus categorical principal component analysis. Ann. Hum. Genet. 2019;83:454–464. doi: 10.1111/ahg.12339. [DOI] [PubMed] [Google Scholar]
- Waisman A., Sevlever F., Elías Costa M., Cosentino M.S., Miriuka S.G., Ventura A.C., Guberman A.S. Cell cycle dynamics of mouse embryonic stem cells in the ground state and during transition to formative pluripotency. Sc. Rep. 2019;9:8051. doi: 10.1038/s41598-019-44537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/J.CELL.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Gao Y., Yu M., Wu H., Ai Z., Wu Y., Liu H., Du J., Guo Z., Zhang Y. Retinoic acid induces embryonic stem cell differentiation by altering both encoding RNA and microRNA expression. PLoS One. 2015;10:e0132566. doi: 10.1371/journal.pone.0132566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li T., Liu F., Chen Y., Yao J., Li Z., Huang Y., Wang J. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol. Cell. 2019;73:130–142.e5. doi: 10.1016/j.molcel.2018.10.020. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang J., Chen Y., Suo L., Chen H., Zhu L., Wan G., Han X. Activin a promotes myofibroblast differentiation of endometrial mesenchymal stem cells via STAT3-dependent Smad/CTGF pathway. Cell Commun. Signal. 2019;17:45. doi: 10.1186/s12964-019-0361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the data reported in this paper is GEO: GSE178390. Correspondence and requests for materials should be addressed to O.R. (oren.ram@mail.huji.ac.il).