Abstract
We enrolled arriving international air travelers in a severe acute respiratory syndrome coronavirus 2 genomic surveillance program. We used molecular testing of pooled nasal swabs and sequenced positive samples for sublineage. Traveler-based surveillance provided early-warning variant detection, reporting the first US Omicron BA.2 and BA.3 in North America.
Keywords: SARS-CoV-2, genomic surveillance, international travelers
Despite layered mitigation measures, international travel during the coronavirus disease 2019 (COVID-19) pandemic continues to facilitate global spread of severe acute respiratory syndrome coronavirus (SARS-CoV-2), including novel variants of concern (VOCs). On 26 November 2021, B.1.1.529 (Omicron) was designated a VOC by the World Health Organization [1]. On 6 December 2021, as part of measures to reduce Omicron introduction and spread, the requirement for a negative SARS-CoV-2 test taken before air travel to the United States was shortened from 3 days to 1 day [1]. Although SARS-CoV-2 genomic sequencing has increased significantly during the pandemic [2], gaps remain in early detection of emerging variants among arriving travelers.
In September 2021, the Centers for Disease Control and Prevention (CDC), in collaboration with private partners, implemented a voluntary SARS-CoV-2 genomic surveillance pilot program. Initially, we enrolled travelers on certain flights from India during the Delta surge. On 28 November, we expanded the program to include travelers arriving from countries with high travel volumes, including those where Omicron was first detected.
METHODS
Design, Setting, and Participants
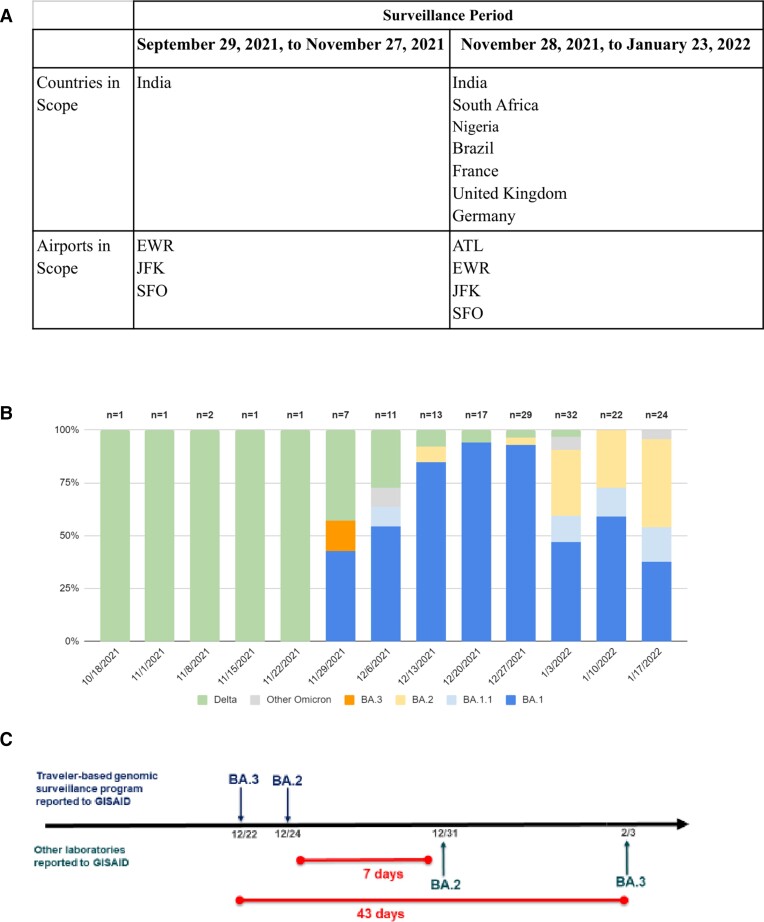
During 29 September 2021–27 November 2021, the surveillance program included travelers arriving on 7 direct flights from India at 3 international airports: John F. Kennedy, New York (September 29); Newark Liberty, New Jersey (October 4); and San Francisco, California (October 12). Hartsfield-Jackson Atlanta International Airport, Georgia, was added on 28 November 2021. During 28 November 2021–23 January 2022, travelers on flights from India, South Africa, Nigeria, the United Kingdom, France, Germany, and Brazil on approximately 50 flights per day were enrolled (Figure 1A). Participants were aged ≥18 years; provided informed consent; and answered demographic, clinical, and travel history questions.
Figure 1.
A, Program scope for countries of flight origin and airports for traveler-based severe acute respiratory syndrome coronavirus genomic surveillance program during 29 September 2021–23 January 2022. B, Proportions of variants detected, by collection week, pooled testing. C, Comparison of timeliness of Omicron sublineage detection in the United States (US) based on GISAID reporting for this program compared to other US laboratories 22 December 2021–3 February 2022. Abbreviations: ATL, Hartsfield-Jackson Atlanta International Airport, Georgia; EWR, Newark Liberty International Airport, GISAID, Global Initiative on Sharing All Influenza Data; JFK, John F. Kennedy Airport, New York, New York; SFO, San Francisco Airport, California.
Sample Collection
Participants could opt in for in-airport pooled nasal swab self-collection, at-home saliva sample collection 3–5 days after arrival, or both (Supplementary Figure 1). For in-airport pooled sampling, travelers self-collected a dry lower nasal swab sample. Samples were placed in collection tubes with 5–25 other samples and shipped to the Concentric Laboratory Network. During September 29–November 27, samples were pooled based on the flight number. During November 28–January 23, samples were pooled based by country of flight origination. For at-home kits, travelers were asked to collect a saliva sample on day 3–5 after arrival and send it to the laboratory.
Laboratory Testing
All samples underwent SARS-CoV-2 reverse-transcription polymerase chain reaction testing. After November 27, samples were tested for S-gene target failure (SGTF) using the TaqPath COVID-19 assay [3]. All positive samples underwent whole-genome sequencing and variant sublineage determination. Reverse-transcribed RNA was amplified using the ARTICv3 protocol [4]. Amplicons were pooled and prepared using standard protocols. For Illumina sequencing, samples underwent tagmentation and were sequenced on NovaSeq 6000 (2 × 50 bp; Illumina). For rapid sublineage identification, a ligation-based library was prepared and sequenced on GridION (Oxford Nanopore) as described in the Supplementary Methods.
Reporting
All travelers who participated in pooled testing were advised to submit their at-home kit for individual testing. Individual results were reported to participants via a secure digital portal and to public health authorities per CDC reporting guidelines; pooled results were not reported to participants [5]. Sequence data from positive samples were uploaded to the Global Initiative on Sharing All Influenza Data, and select samples were provided to the CDC for viral culture and further characterization.
Statistical Analysis
For this analysis, we focused on pooled testing for variant detection and thus included pooled results only. Using χ2 tests conducted in R 4.0.3, we assessed differences in pooled positivity rates by flight country of origin. This activity was reviewed by the CDC and conducted consistent with applicable federal law and CDC policy (see, eg, 45 C.F.R. § 46.102(l)(2); 21 C.F.R. part 56; 42 U.S.C. §241(d); 5 U.S.C. §552a; 44 U.S.C. §3501 et seq).
RESULTS
During 29 September 2021–23 January 2022, we enrolled 16 149 (approximately 10%) of an estimated 161 000 eligible travelers, yielding 1454 sample pools. Overall, 221 (16%) of 1367 pooled samples (average pool size, 11 swabs) tested were SARS-CoV-2–positive. The median turnaround time from sample collection to sequencing was 11 business days (range, 5–20). For select samples, we performed expedited sequencing within 48 hours to confirm the validity of SGTF as an early indicator for Omicron. Positivity among pooled samples was 1.8% (6 of 338) during September 29–November 27. After 27 November 2021, it was 20.9% (215 of 1029), and it varied by country of flight origin: 43.5% (40 of 92) in South Africa, 32.6% in Brazil (15 of 46), 25% in France (30 of 120), 18.4% in the United Kingdom (30 of 163), 17.8% in Germany (38 of 123), and 15.7% (62 of 395) in India (P < .001; Supplementary Table 1).
Before November 28, all sublineages were the Delta variant (B.1.617-like), except for 1 undetermined sublineage. During November 28–January 23, 67% (145 of 215) of positive pooled samples collected were the Omicron variant (B.1.1.529-like), 5% (11 of 215) were the Delta variant (B.1.617-like), and the remaining 27% (59 of 215) sublineages could not be determined due to low sample sequencing coverage (Figure 1B, Supplementary Table 2). Of 145 Omicron sequences, 112 exhibited complete or partial SGTF sublineage. Omicron sublineages included BA.1 (100), BA1.1 (12), BA.2 (26), BA.3 (1), BA.2 + Orf1a:M85 (1), and BA.2 + S:R346K (1). Four samples were identified as Omicron, but the sublineage could not be determined due to low sequencing coverage. A sample collected on 14 December was the first reported as BA.2 in the United States, 7 days earlier than any other US report (Figure 1C, Supplementary Table 3). Similarly, a sample collected on December 3 was the first reported BA.3 in North America, 43 days before the next report [6].
DISCUSSION
Through this traveler-based SARS-CoV-2 genomic surveillance program, we were able to identify early importation of variants, including Omicron sublineages BA.2 and BA.3, before they were reported elsewhere in the United States and North America, respectively. Overall, 16% of pooled tests were positive, with 21% positivity following Omicron emergence. We detected a large proportion of positive post-arrival pooled samples even though passengers were required to have a negative sample collected within 1 day pre-departure
Possible reasons for high pooled test positivity on arrival despite negative pre-departure testing include timing of infection and testing (ie, before infection was detectable), use of testing modalities with lower sensitivity [7], or infection soon after pre-departure testing [8, 9]. If passengers had infections that were undetected in pre-departure testing, longer flight times may have allowed for passengers in their incubation period to convert to a positive result after arrival [9]. Finally, it is possible that fraudulent test results were used to meet pre-departure testing requirements [10].
Pooled testing in this program is advantageous as it enables efficient, large-volume sampling and increases testing throughput while conserving resources. This can be valuable for continued detection when prevalence of SARS-CoV-2 infection is low. The pooled testing design minimizes dilution and reduces loss of sensitivity by pooling during collection. Each Concentric Network laboratory is validated to ensure molecular assay sensitivity of 1500 viral copies/mL. The disadvantage of pooled testing is an inability to directly link test results with individual-level data. Follow-up individual testing, such as the at-home test kits collected in our program (data not presented), provide an additional opportunity to capture linkable metadata.
With an approximately 10% participation rate, we detected sublineage BA.2 and BA.3 weeks before they were reported by other US and North American sequencing efforts. The country-level proportions of variants that we identified were consistent with those reported by national and global sequencing programs [2]. Our study suggests that when COVID-19 rates are high, as during the Omicron surge, a 10% participation rate would be sufficient to detect relatively rare sublineages. Sample size calculations for variant detection require a more complicated approach that will include models and simulations to maximize variant detection at different global prevalence rates while also reducing resource allocation. As the pandemic evolves, the program may include additional modalities, such as wastewater sample collection or air sampling from aircrafts, that enable SARS-CoV-2 monitoring in low-prevalence settings and are not dependent on individual passenger participation.
Detection of imported emerging infectious diseases has traditionally focused on travelers who present to health clinics after symptom onset [11]. COVID-19 presents unique challenges since transmission often occurs before symptom onset or in asymptomatic persons [7]. By the time of variant detection, there is often widespread community transmission. Many countries have required testing for arriving travelers to limit introduction and spread of SARS-CoV-2 [12], yet few use traveler-based viral genomic surveillance to detect novel variants and provide detailed epidemiological data. Earlier detection of novel SARS-CoV-2 variants allows researchers and public health officials the time needed to gather information about transmissibility, virulence, and vaccine effectiveness, enabling adjustments to treatment and prevention strategies [2].
This traveler-based genomic surveillance program underscores the importance of public–private partnerships in achieving public health priorities in an ever-changing pandemic and of the utility of surveillance tools beyond traditional individual testing. The program’s scalability and adaptability, including the ability to rapidly add locations and expedite sequencing, were key factors for success. Traveler-based SARS-CoV-2 genomic surveillance provides a model of pathogen detection that can be used as an early-warning sentinel system for future outbreaks.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Renee D Wegrzyn, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Grace D Appiah, Division of Global Migration and Quarantine, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Robert Morfino, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Scott R Milford, XpresCheck™, XpresSpa™, New York, NY, USA.
Allison Taylor Walker, Division of Global Migration and Quarantine, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Ezra T Ernst, XpresCheck™, XpresSpa™, New York, NY, USA.
William W Darrow, XpresCheck™, XpresSpa™, New York, NY, USA.
Siyao Lisa Li, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Keith Robison, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Duncan MacCannell, Office of Advanced Molecular Detection, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Dongjuan Dai, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Brintha P Girinathan, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Allison L Hicks, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Bryan Cosca, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Gabrielle Woronoff, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Alex M Plocik, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Birgitte B Simen, Ginkgo Bioworks, Inc, Boston, Massachusetts, USA.
Leah Moriarty, Division of Global Migration and Quarantine, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Sarah Anne J Guagliardo, Division of Global Migration and Quarantine, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Martin S Cetron, Division of Global Migration and Quarantine, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Cindy R Friedman, Division of Global Migration and Quarantine, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Notes
Acknowledgments. The authors thank the XpresCheck (Erica Mares, Lesley Shirley, Rob Stein, Henry Streich, Miguel Yapor), Concentric by Ginkgo (Thomas Aichele, Dan Bayley, Juskarun Cheema, Tyler Clarkson, Aakash Desai, Joseph Fridman, Alix Hamilton, Corey Hoehn, Erica Jackson, Hannah Knoll, Frank Langston, John Mcbride, Justin Montgomery, Jason Ng, Rich Nordin, Ben Rome, Andrew Rothstein, Sarah Rush, Zach Smith, Sativa Turner, Erika Gute, Maria Pis-Lopez, Cherish Weiler), the Centers for Disease Control and Prevention (CDC) Division of Global Migration and Quarantine Travelers’ Health (Teresa Smith, Jessica Allen, Laura Leidel, Igor Ristic, Robin Rinker), CDC Quarantine and Border Health Services Branches (Clive Brown, Tai-Ho Chen, Alida Gertz, Matthew Palo), CDC COVID-19 Response Health Department liaisons, US Customs and Border Protection, and state and local public health authority staff who supported the operational implementation of the program on the ground in airports.
Disclaimer. The findings and conclusions presented here are those of the authors and do not necessarily represent the official position of the CDC. This activity was conducted in partnership with commercial partners XpresCheck and Concentric by Ginkgo. The use of product or service names is for identification purposes and does not mean endorsement by the CDC.
Financial support. This work was supported by the CDC (contract award 75D30121C12036).
References
- 1. Centers for Disease Control and Prevention COVID-19 Response Team . SARS-CoV-2 B. 1.1. 529 (Omicron) variant—United States, 1–8 December 2021. MMWR Morb Mortal Wkly Rep 2021; 70:1731–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambrou AS, Shirk P, Steele MK, et al. Genomic surveillance for SARS-CoV-2 variants: predominance of the Delta (B. 1.617. 2) and Omicron (B. 1.1. 529) variants—United States, June 2021–January 2022. MMWR Morb Mortal Wkly Rep 2022; 71:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. TaqPath COVID-19 Tests Support Detection of SARS-CoV-2 in Samples Containing the Omicron Variant. Available at: https://www.thermofisher.com/blog/clinical-conversations/taqpath-covid-19-tests-support-detection-of-sars-cov-2-in-samples-containing-the-omicron-variant-3/. Accessed 21 December 2021.
- 4. Tyson JR, James P, Stoddart D, et al. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv 2020. [Google Scholar]
- 5. Centers for Disease Control and Prevention . How to report COVID-19 laboratory data. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/reporting-lab-data.html. Accessed 28 December 2021.
- 6. Concentric by Ginkgo and XpresCheck™ confirm first North American detections of novel BA.3 subsublineage of Omicron variant through CDC COVID-19 air travel biosecurity program. Available at: https://www.prnewswire.com/news-releases/concentric-by-ginkgo-and-xprescheck-confirm-first-north-american-detections-of-novel-ba3-sublineage-of-omicron-variant-through-cdc-covid-19-air-travel-biosecurity-program-301447761.html. Accessed 4 March 2022.
- 7. Johansson MA, Wolford H, Paul P, et al. Reducing travel-related SARS-CoV-2 transmission with layered mitigation measures: symptom monitoring, quarantine, and testing. BMC Med 2021; 19:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swadi T, Geoghegan JL, Devine T, et al. Genomic evidence of in-flight transmission of SARS-CoV-2 despite predeparture testing. Emerg Infect Dis 2021; 27:687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khanh NC, Thai PQ, Quach H-L, et al. Transmission of SARS-CoV 2 during long-haul flight. Emerg Infect Dis 2020; 26:2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mac V, Taylor M, Guendel I, et al. Falsification of travel-required COVID-19 laboratory reports—United States Virgin Islands, March–April 2021. Paper presented at the 71st Annual Epidemic Intelligence Service Conference; 3 May 2022; Atlanta, GA.
- 11. Hamer DH, Rizwan A, Freedman DO, Kozarsky P, Libman M. Geosentinel: past, present and future. J Travel Med 2020; 27:taaa219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abdalhamid B, Bilder CR, McCutchen EL, Hinrichs SH, Koepsell SA, Iwen PC. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am J Clin Pathol 2020; 153:715–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.