Summary
Indication extension or repositioning of drugs can, if done well, provide a faster, cheaper, and derisked route to the approval of new therapies, creating new options to address pockets of unmet medical need for patients and offering the potential for significant commercial and clinical benefits. We look at the promises and challenges of different repositioning strategies and the disease insights and scalability that new high-resolution patient stratification methodologies can bring. This is exemplified by a systematic analysis of all development candidates and on-market drugs, which identified 477 indication extension opportunities across 30 chronic disease areas, each supported by patient stratification biomarkers. This illustrates the potential that new artificial intelligence (AI) and combinatorial analytics methods have to enhance the rate and cost of innovation across the drug discovery industry.
Keywords: indication extension, drug repositioning, patient stratification, biomarkers, disease signatures, combinatorial analytics
The bigger picture
The cost of healthcare is at an all-time high in the United States, consuming 19.7% of gross domestic product (GDP). Chronic diseases, such as dementia, diabetes, and cardiovascular disease, are especially expensive and also complex, involving multiple genetic, clinical, and epidemiological factors, making finding druggable targets particularly difficult. This leaves many patients with unmet medical needs but also creates opportunities to reposition active molecules that were originally designed to treat another disease but which act on mechanisms relevant to poorly treated patient subgroups.
To do this systematically, we first need to accurately determine which targets are driving disease within clinically relevant patient subgroups in the new indication. High-resolution patient stratification based on combinatorial analytics offers an accurate and scalable way to map safe and effective drugs to patient subgroups across multiple new disease indications, providing a faster, cheaper, and derisked route to their approval.
Indication extension or drug repurposing holds the potential to deliver significant potential benefit both from a commercial perspective as well as for patients. In this perspective, the authors discuss the challenges of repositioning and the opportunities provided by applying AI methods, including high-resolution patient stratification using combinatorial analytics, to large-scale patient datasets. The authors demonstrate the scalability of this approach by performing a systematic search for repurposing opportunities across over 30 diseases and highlighting examples with supporting clinical evidence.
Introduction
Despite huge investment in pharmaceutical research and development (R&D) in recent years, success in translating this into novel therapeutic treatments with better patient outcomes has been lower than expected. We are starting to see signs of a recovery in R&D productivity,1 but still in the last decade, fewer than 10% of the targets investigated in discovery programs have led to approved drugs.2 Unfortunately, this has too often manifested as expensive late-stage phase III failures, mainly due to inability to demonstrate clinical efficacy in patients. In large part, this is due to poor understanding of the complexities and differences in disease biology across heterogeneous patient populations in many of the chronic disorders that are so expensive for health systems to treat.
In this period, there have been major technological advances in the generation and analysis of biological and patient datasets. However, our approach to disease characterization and the study of the underlying pathogenesis of complex diseases has remained relatively basic and rooted around single targets. While effective in relatively monogenic diseases, traditional drug discovery approaches have been concentrated on a small fraction of well-studied genes and pathways, leading to pools of unmet medical need, annotation bias, a lack of innovation, and even to dozens of repeated expensive failures within a single mechanism.3,4
Too often, drug discovery projects become focused on targets and disease mechanisms very early, implicitly assuming that patients who share a clinical diagnosis have a single common disease cause and that those mechanisms remain relevant and/or druggable through different stages of their disease.5 This is too simplistic to capture the biological complexity of chronic disease processes and the varied disease etiologies and influences in different subgroups of patients.6
It is clear that diagnoses such as schizophrenia,7 asthma,8 and type 2 diabetes9 are umbrella terms for various distinct disease subgroups (or endotypes) that have different underlying mechanisms, even though patients may ultimately exhibit similar symptoms. This heterogeneity can result in significant variations in prognosis and therapy response across the patient population. Consequently, a “one size fits all” clinical pathway or “blockbuster” discovery strategy does not work well for complex, chronic diseases, leading to late-stage clinical trial failures and patients often enduring a largely trial-and-error process before they get access to the right treatment.10,11
New precision medicine approaches, driven by patient stratification insights, can identify subgroups of patients who have similar disease etiologies and who are therefore likely to exhibit similar treatment responses. This provides both a route to innovation and also generates the patient stratification biomarker tools required to accelerate and derisk clinical development of novel targets.
At the same time, it is clear that many existing development and on-market compounds have potentially useful effects on pathways and mechanisms that may be shared between multiple disease indications. Exploiting such secondary uses within patients whose disease etiology involves these mechanisms can offer a faster, cheaper, and derisked opportunity to bring to market new medicines that address significant pockets of unmet medical need, to the benefit of both the inventor of the drug and patients. New artificial intelligence (AI)-based combinatorial analytics methods are enabling identification of such opportunities at unprecedented scale.
Drug indication extension
Identification of new indication extension (also known as repositioning) opportunities for approved or investigational drugs has long been recognized as a potentially interesting commercial strategy for pharmaceutical companies, especially if the compound has good remaining composition of matter patent life and well-established safety profiles, and the dosage and route of administration are similar in the new indication.
Until recently, many repositioning examples have been discovered serendipitously,12 but with the availability of larger biological datasets,13 computational approaches are now being used to do this systematically. Few validated repositioning candidates have, however, yet been identified, and success has often been limited by the quality of the data used in the analysis.14,15 Even some high-profile repositioned drugs with good mechanistic hypotheses, such as the potent anti-inflammatory activity of tocilizumab used in severe coronavirus disease 2019 (COVID-19) patients, have sometimes failed to show clear benefit once in the clinic.16,17
While potentially quicker, cheaper, and less risky, repositioning still faces many of the same challenges as novel drug discovery—hypothesis generation, understanding of the mechanism(s) of action, identification of the patient subgroups in the new indication area who would benefit from the drug, and establishing a robust patent position. In addition, for repositioning candidates, drug safety data from adverse event report databases18, 19, 20 and toxicity assessment21 or prediction data22 should be used, along with an evaluation of the dosage and route of administration needed, to identify any potential safety concerns associated with the drug and new indication(s) in question.
The most widely used drug-repositioning methods involve comparison of drug characteristics, such as transcriptomic or adverse event profiles, with a disease or clinical phenotype (phenotypic drug discovery [PDD]). These methods utilize data from resources such as Library of Integrated Network-based Cellular Signatures (LINCS),23,24 which holds gene and protein expression profiles from cell lines perturbed with a wide range of chemical compounds, and NCATS OpenData Portal,25 which contains phenotype data from high-throughput drug screening assays. Despite the richness and quantity of these data sources, concerns have been raised about the quality and reproducibility of the cellular phenotype data and challenges remain for deconvoluting druggable targets. These issues have affected the accuracy, clinical relevance, and scalability of such drug repositioning and PDD studies.26,27
Other drug-repositioning methods utilize knowledge-driven approaches that make use of graph- or network-based data mining methods13,28 that integrate data from genome-wide association studies (GWASs), gene expression, biological pathways, and molecular interactions to search for new indication opportunities for drugs. The biggest limitation of such annotation-driven methods is that the sparsity and biases of our knowledge of systems biology make it challenging to identify opportunities beyond the relatively obvious “low-hanging fruits.” This is because functional annotation of even well-researched genes cannot be considered as complete; experimental assays are limited in the type of information they can discover and, most notably, experimental designs are often guided by what is already known or expected.
Neither of these PDD- or network-based approaches addresses the major challenge that almost 60% of late-stage clinical trials failures are due to an inability to demonstrate clinical efficacy.11 The key factor here is a poor understanding of the mapping between the proposed mechanism of action of the target(s) involved and the subgroups of patients who will benefit from this approach and should therefore be recruited into the clinical trial. This detailed patient stratification is an essential component both of novel drug discovery and effective indication extension, but it cannot be delivered by existing approaches, such as GWASs.
High-resolution patient stratification using combinatorial analytics
Targets with strong genetic evidence are known to be more likely to succeed in clinical trials.29 However, although GWASs have revealed several disease-associated genes, their translation into the clinic has been far from successful, especially in complex, chronic diseases.30 This is largely because GWAS is inherently a low-resolution technique designed to identify single variants that exhibit a relatively large effect size across a whole study population, characteristics that are much more relevant to relatively monogenic diseases in homogeneous populations.
With non-linear contributions of multiple interacting genetic variants playing such an important role in chronic disease populations, and their inherent heterogeneity, GWAS results account for only a small proportion of genetic variation in these diseases and fail to delineate different patient subgroups.31 This fundamentally constrains its potential to identify targets relevant to specific patient subgroups and thus guide precision medicine outside of monogenic diseases.32
The key to understanding complex diseases that are influenced by multiple genetic loci and epidemiological and/or environmental factors is to find combinations of these disease-associated factors that distinguish one patient subgroup from another.
Combinatorial analysis uses advanced analytics and AI methods to identify such combinations of features in complex chronic diseases.33 It is a hypothesis-free method for the detection of high-order, disease-associated combinations of features (disease signatures—typically comprising three to ten features) that together are strongly associated with variations in disease risk, symptoms, progression rates, and therapy response commonly seen in subgroups of patients using a case-control cohort design.34,35
The disease signatures arising from combinatorial analysis capture the quantitative epistatic and other non-linear effects on disease biology and phenotypes arising from interactions of multiple genes across genetic and molecular networks, signaling cascades or changes in transcription, translation, and/or metabolism. These non-linear effects cannot be captured either with GWASs or standard machine-learning methods.
The resulting combinatorial features can be used to stratify large patient datasets. Disease signatures can be clustered by the patients in which they co-occur to provide a high-resolution stratification of the patient population. Mapping the SNPs in the disease signatures to genes and pathways can reveal more novel disease biology as well as correlate specific mechanism-based disease signatures with different patient subgroups within the overall study population.
The combination of SNPs linked to a target of interest in a patient subgroup can thus serve as a biomarker to identify individuals comprising a subgroup within a heterogeneous patient population who would be most responsive to pharmacological modulation of the target. This approach has been validated in multiple disease populations using both phenotypic35 and genetic data36,37 and is essential for systematic indication extension.
Drug indication extension powered by high-resolution patient stratification
To illustrate the potential of high-resolution patient stratification insights for systematic drug indication extension, we have briefly described such a repositioning approach and highlighted two examples of new drug- and target-indication opportunities identified by it.
Combinatorial analysis has previously been used to stratify patient populations and identify disease-associated SNPs and targets for more than 30 disease populations38 using genomic data available from the UK Biobank39 and Database of Genotypes and Phenotypes (dbGaP),40 among other sources. These diseases cover a wide range of indications in different therapeutic areas, such as neurodegenerative, neuropsychiatric, respiratory cardiovascular, metabolic, autoimmune, infectious diseases, and women’s health.
For each indication, all significant disease signatures were clustered by the patients in whom they co-occur to generate a disease architecture that provides a high-resolution view of the targets and mechanisms of actions associated with specific patient subgroups.41 The targets were prioritized based on the 5Rs drug discovery framework,42 and efficacy potential metrics (a measure of how well stratified the disease biology around a chosen target is within the patient population) and patient stratification biomarkers were generated for all prioritized targets.
Biomedical knowledge graphs were used to amplify genetic signals using a naive guilt by association approach to identify genes proximal to prioritized targets in key metabolic pathways whose SNPs may not be well represented in the genotype array. These biological and patient stratification insights provide the framework for the systematic identification of indication extension opportunities.
Industry-wide information on pipeline and marketed drugs was sourced from GlobalData’s Pipeline & Marketed Drugs database (GlobalData; https://www.globaldata.com/). This includes known target genes, indication(s), and therapy areas with their development phase, molecule type, modality, and mode of action. Additional data were extracted from ChEMBL43 and UniChem,44 including molecular properties, pharmacology, pharmacovigilance, predicted toxicity, clinical trials, and withdrawals data, where available.
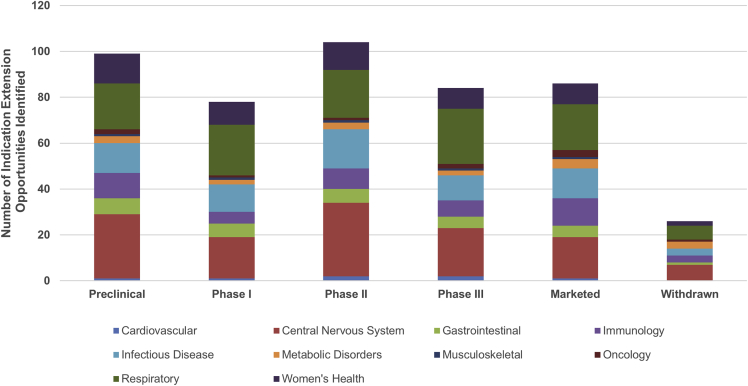
For each development candidate or approved drug with more than 5 years remaining composition of matter patent life, their target was correlated with all of the detailed mechanistic patient stratification insights for each of the 30 disease-stratification studies (Figure 1). This identified 477 potential indication opportunities across the 30 disease areas (Figure 2) where that target or mechanism was also found to be strongly associated with one or more clinically relevant patient subgroups in another disease study.
Figure 1.
Systematic drug indication extension approach based on high-resolution patient stratification insights generated by combinatorial analytics for 30 disease studies
FTO, freedom to operate; IP, intellectual property.
Figure 2.
Analysis of all pipeline and marketed drugs from GlobalData with >5 years remaining patent life where indication extension opportunities were found, shown by therapy area and drug development phase
These indication extension opportunities can be further evaluated to assess the target-secondary disease linkage and the novelty and viability of the drug- and target-indication pair based on factors such as:
-
•
strength of mechanistic hypotheses and supporting tissue-specific evidence,
-
•
clinical relevance,
-
•
5Rs drug discovery criteria,
-
•
dosage, route of administration, and impact on toxicity,
-
•
remaining patent life, and
-
•
freedom to operate.
Several of the indication extension opportunities identified have already been evaluated in clinical trials in the new indication by other groups. Two such examples provide confirmatory evidence that the combinatorial analytics approach can identify patient subgroups in a secondary indication where repositioned clinical candidates have the potential to be highly effective.
Mineralocorticoid receptor antagonists: Type 2 diabetes renal complications
There are currently 201 marketed drugs that are mineralocorticoid receptor (MR) (NR3C2, MR, and aldosterone receptor) antagonists, with the majority licensed for edema, heart failure, and hypertension. With this number of marketed drugs, there is a wealth of safety, efficacy, and side effect data available for a range of MR antagonists in different patient populations. This makes the MR a promising candidate for indication extension analysis.
NR3C2—the gene target for MR antagonists—was searched across the 30 chronic disease studies to identify clinically relevant patient subgroups where the use of MR antagonist may be an effective therapeutic option. In one such group, a combinatorial disease signature containing a variant in NR3C2 was identified to be highly associated with type 2 diabetes patients (from the UK Biobank ICD-10 code, E11) who have developed at least one of the main complications associated with diabetes, including ketoacidosis, cardiovascular, neurological complications, and chronic kidney disease (Figure 3).
Figure 3.
Disease architecture of the UK Biobank patient population with type-2-diabetes-associated complications generated by combinatorial analytics
Each circle represents a disease-associated SNP genotype, and edges represent co-association in patients.
(A) Each colored cluster represents a distinct patient subgroup.
(B) The highlighted group of SNPs (dark) contains the variant in NR3C2 that was associated with patients who developed renal complications.
The signature containing this genetic variant is found in 209 cases with type 2 diabetes complications and in zero controls (long-term diabetics without complications). Furthermore, the cases with this signature were significantly more likely to have developed renal complications, suggesting that this signature specifically increases the risk of developing kidney disease in association with type 2 diabetes. This suggests that the use of MR antagonists may be beneficial to type 2 diabetic patients who are most at risk of developing renal complications according to their genetic data.
There is considerable supporting evidence in the literature of dysregulated MR and aldosterone signaling in patients who developed diabetic nephropathy and chronic kidney disease.45 Over-activation of the renin-angiotensin-aldosterone system (RAAS) is a key driver of renal fibrosis in diabetic kidney disease, and preclinical studies show that the use of RAAS inhibitors decreases expression of pro-fibrotic markers and renal function in diabetic rats.46
MR antagonists have been shown to prevent the progression of diabetic nephropathy by improving insulin resistance and lowering blood pressure.47,48 These include drugs such as spironolactone and finerenone. A multicenter study of the use of spironolactone in diabetic patients with high risk of developing kidney failure did not indicate that treatment with spironolactone prevented development of microalbuminuria.49 Furthermore, spironolactone is associated with increased risk of hyperkalemia.50
However, finerenone is a more recently developed MR antagonist that has greater selectivity and binding affinity than other MR antagonists, such as spironolactone and eplerenone.51 In a double-blind clinical trial containing 5,734 patients with type 2 diabetes and chronic kidney disease, treatment with finerenone resulted in reduced risk of chronic kidney disease (CKD) progression compared against the placebo, supporting the hypothesis of its repositioning potential.52
IL-6R antagonists: Amyotrophic lateral sclerosis
This analysis also identified a potential indication extension opportunity for interleukin-6R (IL-6R) antagonists for use in patients with amyotrophic lateral sclerosis (ALS), a progressive neurodegenerative disease predominantly affecting motor neurons.
In one of the 30 patient stratification studies, 736 patients (438 male and 291 female) with ALS were compared against a set of healthy gender-matched controls. Genetic variants involved in the regulation of IL-6 secretion were identified as part of a combinatorial disease signature significantly associated with a subgroup of ALS patients who were more likely to develop earlier onset and more aggressive forms of the disease (patient subgroup A; Figure 4). This suggests that IL-6R antagonists may be effective at slowing disease progression within this subgroup of patients.
Figure 4.
Comparison of the distribution of clinical features associated with the three patient subgroups identified by combinatorial analysis of an ALS population
(A)–(C) show ALSFRS-r, age at death, and survival from disease onset until death for patient subgroups, respectively. Age at death (A) was found to be significantly different between patient subgroup A and subgroup C using Mann-Whitney U test (p < 0.05).
Although the levels of IL-6 in ALS patients can be highly variable between cases, there is evidence that IL-6 is increased in the plasma of ALS patients and is negatively correlated with Revised Amyotrophic Lateral Sclerosis Functional Rating Scale (ALSFRS-R) scores.53,54 This provides additional supporting evidence as to why patients with genetic variants in IL-6 signaling were more likely to have greater functional impairment and disease progression.
There is now also clinical data that support the label extension of IL-6R antagonists in ALS. Results from a phase II trial investigating the safety and tolerability of tocilizumab, currently licensed in rheumatoid arthritis and several other inflammatory diseases, indicated that the drug was well tolerated by ALS patients and suppresses inflammation.55 Infusion with tocilizumab also slowed clinical progression in ALS patients, although the sample size of this study was small.56
Conclusions
This analysis illustrates that high-resolution patient stratification based on a combinatorial analysis approach is sufficiently scalable and accurate for the systematic analysis of potential secondary indications for pipeline, marketed, and even withdrawn compounds (which failed for efficacy reasons). Nearly 500 such indication extension opportunities were identified by this analysis across the 30 complex, chronic disease areas studied.
The examples highlighted show that this approach accurately identified drugs that do have high efficacy potential in at least one new secondary indication as well as finding patient stratification biomarkers that can be used to accelerate and derisk the clinical development and approval of the repositioned compounds.
High-resolution patient stratification driven by combinatorial analytics has identified many further indication extension opportunities for drugs that have not yet been evaluated in clinical trials but which could potentially be highly effective in chronic disease populations and help to address the efficient delivery of new therapeutic options in areas of significant unmet medical need. This would be of significant benefit both commercially as well as for patients, for whom it offers the hope of new therapeutic options addressing a range of underserved diseases and patient subgroups.
Acknowledgments
Research described in this article has been conducted using data from UK Biobank Resource (application number 44288) and dbGaP. We would like to acknowledge the ALS patients whose data were provided to and collected by King’s College, the Universities of Birmingham and Sheffield, and the MNDA, which enabled part of this analysis. Special thanks to Matthew Pearson, who developed the systematic drug indication extension pipeline, Gert Møller, who developed the combinatorial analytics methodology, and the rest of the PrecisionLife team.
Author contributions
S.G., S.D., and K.T. wrote the manuscript. S.G. designed the approach. K.T., S.D., and S.B. analyzed the data generated by the indication extension pipeline. All authors provided feedback and approved the final version of the manuscript.
Declaration of interests
S.D., K.T., S.B., and S.G. are employees of PrecisionLife, Ltd. S.G. is a shareholder of PrecisionLife, Ltd.
Biographies
About the authors
Dr. Sayoni Das is Head of Bioinformatics at PrecisionLife. She is a computational biologist with a background in bioprocess engineering and biotechnology. She did her post-doctoral research on developing machine-learning approaches for identification of protein functional sites and interpretation of genetic variants. Sayoni received a PhD in computational biology from University College London for her work on functional classification of protein domains and protein function prediction pipeline that was ranked among the top ten methods in two consecutive CAFA function prediction competitions. She served as the Secretary for the ISCB Regional Student Group UK from 2014 to 2016.
Krystyna Taylor is Senior Portfolio Manager at PrecisionLife, managing drug discovery programs to support collaborations between the company and its academic and industry partners. Krystyna has a background in drug target identification and the translation of targets into in vitro validation pipelines. Krystyna holds a Biomedical Science degree from Imperial College London and has also completed a year studying management at the Imperial College Business School, where she was placed on the Dean’s List for Academic Excellence.
Simon Beaulah is SVP Healthcare and Head of US Operations at PrecisionLife, supporting providers, payers, and public health organizations in adopting precision medicine approaches for chronic diseases. Simon has spent over 20 years managing product and business strategy teams for innovative genomics and AI businesses in the healthcare and life science ecosystems. Simon was formerly Director of Healthcare at Linguamatics/IQVIA, leading their healthcare business unit for 7 years, and spent 5 years at InforSense and IDBS. Simon holds an MSc in information technology from Aston University and a BSc (Hons) in agriculture from Cranfield University.
Dr. Steve Gardner is CEO of PrecisionLife. He is a computational biologist with over 30 years’ experience in precision medicine, drug discovery, and tech bio and has developed several highly innovative businesses in the life science, healthcare, and agri-tech industries. Steve was Global Director of Research Informatics for Astra and specializes in AI-enabled drug discovery, precision medicine, and complex data analytics for life science, personalized digital health, and clinical decision support. He has worked with major pharma companies on over 25 drug discovery and safety projects.
References
- 1.Deloitte Nurturing Growth: measuring the return from pharmaceutical innovation 2021. 2022. https://www2.deloitte.com/content/dam/Deloitte/uk/Documents/life-sciences-health-care/Measuring-the-return-of-pharmaceutical-innovation-2021-Deloitte.pdf last accessed 16 January 2022 from.
- 2.King E.A., Davis J.W., Degner J.F. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 2019;15:e1008489. doi: 10.1371/journal.pgen.1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yiannopoulou K.G., Anastasiou A.I., Zachariou V., Pelidou S.H. Reasons for failed trials of disease-modifying treatments for Alzheimer disease and their contribution in recent research. Biomedicines. 2019;7:97. doi: 10.3390/biomedicines7040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haynes W.A., Tomczak A., Khatri P. Gene annotation bias impedes biomedical research. Sci/ Rep. 2018;8:1–7. doi: 10.1038/s41598-018-19333-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo T., Zhang D., Zeng Y., Huang T.Y., Xu H., Zhao Y. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol. Neurodegener. 2020;15:1–37. doi: 10.1186/s13024-020-00391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uffelmann E., Huang Q.Q., Munung N.S., de Vries J., Okada Y., Martin A.R., Martin H.C., Lappalainen T., Posthuma D. Genome-wide association studies. Nat. Rev. Methods Primers. 2021;1:1–21. doi: 10.1038/s43586-021-00056-9. [DOI] [Google Scholar]
- 7.Radua J., Ramella-Cravaro V., Ioannidis J.P., Reichenberg A., Phiphopthatsanee N., Amir T., Yenn Thoo H., Oliver D., Davies C., Morgan C., McGuire P. What causes psychosis? An umbrella review of risk and protective factors. World Psychiatr. 2018;17:49–66. doi: 10.1002/wps.20490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuruvilla M.E., Lee F.E.H., Lee G.B. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin. Rev. Allergy Immunol. 2019;56:219–233. doi: 10.1007/s12016-018-8712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redondo M.J., Hagopian W.A., Oram R., Steck A.K., Vehik K., Weedon M., Balasubramanyam A., Dabelea D. The clinical consequences of heterogeneity within and between different diabetes types. Diabetologia. 2020;63:2040–2048. doi: 10.1007/s00125-020-05211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schork N.J. Personalized medicine: time for one-person trials. Nat. News. 2015;520:609. doi: 10.1038/520609a. [DOI] [PubMed] [Google Scholar]
- 11.Hwang T.J., Carpenter D., Lauffenburger J.C., Wang B., Franklin J.M., Kesselheim A.S. Failure of investigational drugs in late-stage clinical development and publication of trial results. JAMA Intern. Med. 2016;176:1826–1833. doi: 10.1001/jamainternmed.2016.6008. [DOI] [PubMed] [Google Scholar]
- 12.Shim J.S., Liu J.O. Recent advances in drug repositioning for the discovery of new anticancer drugs. Intl J. Biol. Sci. 2014;10:654–663. doi: 10.7150/ijbs.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 14.Eid F.E., Elmarakeby H.A., Chan Y.A., Fornelos N., ElHefnawi M., Van Allen E.M., Heath L.S., Lage K. Systematic auditing is essential to debiasing machine learning in biology. Commun. Biol. 2021;4:1–9. doi: 10.1038/s42003-021-01674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greener J.G., Kandathil S.M., Moffat L., Jones D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022;23:40–55. doi: 10.1038/s41580-021-00407-0. [DOI] [PubMed] [Google Scholar]
- 16.Rosas I.O., Bräu N., Waters M., Go R.C., Hunter B.D., Bhagani S., Skiest D., Aziz M.S., Cooper N., Douglas I.S., et al. Tocilizumab in hospitalized patients with severe Covid-19 pneumonia. N. Engl. J. Med. 2021;384:1503–1516. doi: 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veiga V.C., Prats J.A., Farias D.L., Rosa R.G., Dourado L.K., Zampieri F.G., Machado F.R., Lopes R.D., Berwanger O., Azevedo L.C., et al. Effect of tocilizumab on clinical outcomes at 15 days in patients with severe or critical coronavirus disease 2019: randomised controlled trial. BMJ. 2021;372:n84. doi: 10.1136/bmj.n84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad S.R. Adverse drug event monitoring at the food and drug administration. J. Gen. Intern. Med. 2003;18:57–60. doi: 10.1046/j.1525-1497.2003.20130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn M., Letunic I., Jensen L.J., Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016;44:D1075–D1079. doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiwara M., Kawasaki Y., Yamada H. A pharmacovigilance approach for post-marketing in Japan using the Japanese Adverse Drug Event Report (JADER) database and association analysis. PLoS One. 2016;11:e0154425. doi: 10.1371/journal.pone.0154425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanz F., Pognan F., Steger-Hartmann T., Díaz C., Cases M., Pastor M., Marc P., Wichard J., Briggs K., Watson D.K., et al. Legacy data sharing to improve drug safety assessment: the eTOX project. Nat. Rev. Drug Discov. 2017;16:811–812. doi: 10.1038/nrd.2017.177. [DOI] [PubMed] [Google Scholar]
- 22.Hunter F.M., Bento A.P., Bosc N., Gaulton A., Hersey A., Leach A.R. Drug safety data curation and modeling in ChEMBL: boxed warnings and withdrawn drugs. Chem. Res. Toxicol. 2021;34:385–395. doi: 10.1021/acs.chemrestox.0c00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keenan A.B., Jenkins S.L., Jagodnik K.M., Koplev S., He E., Torre D., Wang Z., Dohlman A.B., Silverstein M.C., Lachmann A., et al. The library of integrated network-based cellular signatures NIH program: system-level cataloging of human cells response to perturbations. Cell Syst. 2018;6:13–24. doi: 10.1016/j.cels.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 25.Brimacombe K.R., Zhao T., Eastman R.T., Hu X., Wang K., Backus M., Baljinnyam B., Chen C.Z., Chen L., Eicher T., et al. An OpenData portal to share COVID-19 drug repurposing data in real time. BioRxiv. 2020 doi: 10.1101/2020.06.04.135046. Preprint at. [DOI] [Google Scholar]
- 26.Lim N., Pavlidis P. Evaluation of Connectivity Map shows limited reproducibility in drug repositioning. Sci. Rep. 2019;11:1–14. doi: 10.1038/s41598-021-97005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffat J.G., Vincent F., Lee J.A., Eder J., Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 28.MacLean F. Knowledge graphs and their applications in drug discovery. Expert Opin. Drug Discov. 2021;16:1057–1069. doi: 10.1080/17460441.2021.1910673. [DOI] [PubMed] [Google Scholar]
- 29.Cook D., Brown D., Alexander R., March R., Morgan P., Satterthwaite G., Pangalos M.N. Lessons learned from the fate of AstraZeneca's drug pipeline: a five-dimensional framework. Nat. Rev. Drug Discov. 2014;13:419–431. doi: 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- 30.Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 31.Stringer S., Wray N.R., Kahn R.S., Derks E.M. Underestimated effect sizes in GWAS: fundamental limitations of single SNP analysis for dichotomous phenotypes. PloS One. 2011;6:e27964. doi: 10.1371/journal.pone.0027964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie M.D., Van Steen K. The search for gene-gene interactions in genome-wide association studies: challenges in abundance of methods, practical considerations, and biological interpretation. Ann. Transl. Med. 2018;6:157. doi: 10.21037/atm.2018.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner S. Combinatorial analytics: an essential tool for the delivery of precision medicine and precision agriculture. Artif. Intell. Life Sci. 2021;1:100003. doi: 10.1016/j.ailsci.2021.100003. [DOI] [Google Scholar]
- 34.Koefoed P., Andreassen O.A., Bennike B., Dam H., Djurovic S., Hansen T., Jorgensen M.B., Kessing L.V., Melle I., Møller G.L., et al. Combinations of SNPs related to signal transduction in bipolar disorder. PLoS One. 2011;6:e23812. doi: 10.1371/journal.pone.0023812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das S., Pearson M., Taylor K., Bouchet V., Møller G.L., Hall T.O., Strivens M., Tzeng K.T., Gardner S. Combinatorial analysis of phenotypic and clinical risk factors associated with hospitalized COVID-19 patients. Front. Digit Health. 2021;3:660809. doi: 10.3389/fdgth.2021.660809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor K., Das S., Pearson M., Kozubek J., Strivens M., Gardner S. Systematic drug repurposing to enable precision medicine: a case study in breast cancer. Digital Med. 2019;5:180. doi: 10.4103/digm.digm_28_19. [DOI] [Google Scholar]
- 37.Taylor K., Das S., Pearson M., Kozubek J., Pawlowski M., Jensen C.E., Skowron Z., Møller G.L., Strivens M., Gardner S. Analysis of genetic host response risk factors in severe COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.06.17.20134015. Preprint at. [DOI] [Google Scholar]
- 38.Beaulah S. Precision medicine and chronic disease. J. Precision Med. 2021;7:36–40. [Google Scholar]
- 39.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mailman M.D., Feolo M., Jin Y., Kimura M., Tryka K., Bagoutdinov R., Hao L., Kiang A., Paschall J., Phan L., et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardner S., Das S., Taylor K. In: Artificial Intelligence in Oncology Drug Discovery and Development. Cassidy J.W., Taylor B., editors. IntechOpen; 2020. AI enabled precision medicine: patient stratification, drug repurposing and combination therapies; pp. 115–140. [Google Scholar]
- 42.Morgan P., Brown D.G., Lennard S., Anderton M.J., Barrett J.C., Eriksson U., Fidock M., Hamren B., Johnson A., March R.E., Matcham J. Impact of a five-dimensional framework on R&D productivity at AstraZeneca. Nat. Rev. Drug Discov. 2018;17:167–181. doi: 10.1038/nrd.2017.244. [DOI] [PubMed] [Google Scholar]
- 43.Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M., et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47:D930–D940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chambers J., Davies M., Gaulton A., Hersey A., Velankar S., Petryszak R., Hastings J., Bellis L., McGlinchey S., Overington J.P. UniChem: a unified chemical structure cross-referencing and identifier tracking system. J. Cheminf. 2013;5:1–9. doi: 10.1186/1758-2946-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goenka L., Padmanaban R., George M. The ascent of mineralocorticoid receptor antagonists in diabetic nephropathy. Curr. Clin. Pharmacol. 2019;14:78–83. doi: 10.2174/1574884713666181116100946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koszegi S., Molnar A., Lenart L., Hodrea J., Balogh D.B., Lakat T., Szkibinszkij E., Hosszu A., Sparding N., Genovese F., et al. RAAS inhibitors directly reduce diabetes-induced renal fibrosis via growth factor inhibition. J. Physiol. 2019;597:193–209. doi: 10.1113/JP277002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato A. The necessity and effectiveness of mineralocorticoid receptor antagonist in the treatment of diabetic nephropathy. Hypertens. Res. 2015;38:367–374. doi: 10.1038/hr.2015.19. [DOI] [PubMed] [Google Scholar]
- 48.Wombwell E., Naglich A. The role of aldosterone antagonism agents in diabetic kidney disease. J. Ren. Care. 2015;41:9–18. doi: 10.1111/jorc.12085. [DOI] [PubMed] [Google Scholar]
- 49.Tofte N., Lindhardt M., Adamova K., Bakker S.J., Beige J., Beulens J.W., Birkenfeld A.L., Currie G., Delles C., Dimos I., et al. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): a prospective observational study and embedded randomised placebo-controlled trial. The Lancet. Diabetes Endocrinol. 2020;8:301–312. doi: 10.1016/S2213-8587(20)30026-7. [DOI] [PubMed] [Google Scholar]
- 50.Juurlink D.N., Mamdani M.M., Lee D.S., Kopp A., Austin P.C., Laupacis A., Redelmeier D.A. Rates of hyperkalemia after publication of the randomized aldactone evaluation study. N. Engl. J. Med. 2004;351:543–551. doi: 10.1056/NEJMoa040135. [DOI] [PubMed] [Google Scholar]
- 51.Liu L.C., Schutte E., Gansevoort R.T., van der Meer P., Voors A.A. Finerenone: third-generation mineralocorticoid receptor antagonist for the treatment of heart failure and diabetic kidney disease. Expert Opin. Invest. Drugs. 2015;24:1123–1135. doi: 10.1517/13543784.2015.1059819. [DOI] [PubMed] [Google Scholar]
- 52.Bakris G.L., Agarwal R., Anker S.D., Pitt B., Ruilope L.M., Rossing P., Kolkhof P., Nowack C., Schloemer P., Joseph A., et al. Effect of finerenone on chronic kidney disease outcomes in type 2 diabetes. N. Engl. J. Med. 2020;383:2219–2229. doi: 10.1056/NEJMoa2025845. [DOI] [PubMed] [Google Scholar]
- 53.Pronto-Laborinho A., Pinto S., Gromicho M., Pereira M., Swash M., de Carvalho M. Interleukin-6 and amyotrophic lateral sclerosis. J. Neurol. Sci. 2019;398:50–53. doi: 10.1016/j.jns.2019.01.026. [DOI] [PubMed] [Google Scholar]
- 54.Tortelli R., Zecca C., Piccininni M., Benmahamed S., Dell'Abate M.T., Barulli M.R., Capozzo R., Battista P., Logroscino G. Plasma inflammatory cytokines are elevated in ALS. Front. Neurol. 2020;11:552295. doi: 10.3389/fneur.2020.552295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milligan C., Atassi N., Babu S., Barohn R.J., Caress J.B., Cudkowicz M.E., Evora A., Hawkins G.A., Wosiski-Kuhn M., Macklin E.A., et al. Tocilizumab is safe and tolerable and reduces C-reactive protein concentrations in the plasma and cerebrospinal fluid of ALS patients. Muscle Nerve. 2021;64:309–320. doi: 10.1002/mus.27339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fiala M., Mizwicki M.T., Weitzman R., Magpantay L., Nishimoto N. Tocilizumab infusion therapy normalizes inflammation in sporadic ALS patients. Am. J. Neurodegenerative Dis. 2013;2:129–139. [PMC free article] [PubMed] [Google Scholar]