Abstract
Lipid nanoparticle (LNP) is commonly used to deliver mRNA vaccines. Currently, LNP optimization primarily relies on screening ionizable lipids by traditional experiments which consumes intensive cost and time. Current study attempts to apply computational methods to accelerate the LNP development for mRNA vaccines. Firstly, 325 data samples of mRNA vaccine LNP formulations with IgG titer were collected. The machine learning algorithm, lightGBM, was used to build a prediction model with good performance (R2 > 0.87). More importantly, the critical substructures of ionizable lipids in LNPs were identified by the algorithm, which well agreed with published results. The animal experimental results showed that LNP using DLin-MC3-DMA (MC3) as ionizable lipid with an N/P ratio at 6:1 induced higher efficiency in mice than LNP with SM-102, which was consistent with the model prediction. Molecular dynamic modeling further investigated the molecular mechanism of LNPs used in the experiment. The result showed that the lipid molecules aggregated to form LNPs, and mRNA molecules twined around the LNPs. In summary, the machine learning predictive model for LNP-based mRNA vaccines was first developed, validated by experiments, and further integrated with molecular modeling. The prediction model can be used for virtual screening of LNP formulations in the future.
KEY WORDS: Lipid nanoparticle, Ionizable lipid, mRNA, Vaccine, Formulation prediction, Machine learning, LightGBM, Molecular modeling
Graphical abstract
The AI algorithm was used to find the relationship between ionizable lipids and in vivo efficiencies of mRNA vaccines, which was validated by a mice study. The molecular dynamic simulation further provided mechanical details of lipid nanoparticle (LNP) formulation.

1. Introduction
The global pandemic of coronavirus disease 2019 (COVID-19) has caused nearly 220 million confirmed cases and more than four million deaths worldwide, according to the updated record by the World Health Organization (WHO). To suppress the prevalence of COVID-19, many pharmaceutical industries in multiple countries have developed vaccines with an unprecedented speed and are promoting their usage globally1. The BNT162b2 from BioNTech and Pfizer and mRNA-1273 from Moderna were the first two vaccines approved by the US Food and Drug Administration (FDA) in November 2020 with the development period of less than one year2,3 and impressively high preventing efficacy, 95% for BNT162b24 and 94.1% for mRNA-12735. Rapid development, high efficacy, and risk-free of insertional mutagenesis or infection induced by vaccine6,7 show a promising prospect for this vaccine platform. mRNA takes effect by first being delivered into cells and then translated to immunogenic antigens. There are many aspects that mRNA sequence can be engineered to influence its efficacy8,9, such as self-amplifying10,11, choice of untranslated region12,13, modification of nucleoside14,15, codon optimization16,17, and combination of encoded antigens18,19. Besides, administration routes also affect the immune effect8,20. However, a successful mRNA vaccine further requires a proper delivery system, such as the lipid nanoparticle (LNP). Both vaccines against COVID-19 adopt LNP as the delivery system.
LNP-based mRNA vaccines usually consist of four types of lipids, cholesterol, distearoylphosphatidylcholine (DSPC), polyethylene glycol (PEG) -lipid, and ionizable lipid. Cholesterol adjusts the flexibility and fusogenicity of lipids during mixing, facilitating the LNP formation21. DSPC, the helper lipid, is related to LNP structure22,23, interfacial tension24, and helps mRNA release25. PEG-lipid influences the LNP stability24, size26 of LNP, and further impacts the potency27. The ionizable lipid, due to its cationic head group, should be the most critical ingredient. It dominates the binding to mRNA, interacting with the endosomal membrane and mRNA release28,29. Besides, a desired ionizable lipid should also show high biodegradability to ameliorate the adverse effect induced by lipid accumulation30. Traditionally, ionizable lipids are screened by synthesizing numerous lipids and testing their in vivo efficacy31,32. However, current experimental screening needs a large amount of cost, time, and materials.
Machine learning (ML) is a branch of artificial intelligence, which is the science of enabling computers to learn knowledge without being explicitly programmed33. After succeeding in areas such as machine translation and computer vision34, ML has been increasingly applied by pharmaceutical companies in recent years35. ML can explore the existing dataset and determine the relationship between the input and output parameters, wherein the former could present the formulation information and experimental conditions while the latter may indicate the formulation behaviors of interest. This approach is helpful in formulation prediction. Previous studies have successfully applied ML to predict the drug delivery systems, such as nanocrystals36, solid dispersion37, cyclodextrin complex38, and self-emulsifying drug delivery systems (SEDDS)39. In the case of SEDDS, the trained ML model predicted the molar composition of oils, surfactants, and cosurfactant where they can form self-emulsion, based on their physicochemical properties input, which helped to choose proper excipients for SEDDS formulation.
Molecular dynamic (MD) simulation is another computational tool that can visualize and investigate the interaction among ingredient molecules and the environment from a physicochemical view. MD method has become a helpful tool for pharmaceutical scientists to obtain a mechanistic understanding of formulation behaviors40,41. Previous studies applied the MD method to investigate topics such as the aggregation of polymer–siRNA complex42 and the dissolution of solid dispersion37. In the case of the polymer–siRNA complex, the aggregation was simulated to be driven by the interaction between cationic groups on polymers and the negative backbone of siRNA. This aggregation stabilized the siRNA in the aqueous solution, reflected by the less altering major groove width of siRNA. Besides, the MD result also revealed the saturation molar ratio of polymers to siRNA. It resulted from mutually counteracting forces of electrostatic effect and steric crowding, which were influenced by siRNA length, cationic charge sites, and the shape of polymers.
This work aimed to build an ML model to predict LNP formulations for mRNA vaccines against viruses. Data from publications were collected of build the model. A typical such study43,44 includes the information of mRNA sequence synthesis, LNP preparation, treatment of subjects, and detection of the time course of binding IgG titer. The binding IgG titer is a surrogate of antibody concentration produced by the immune system after stimulation of antigen encoded by mRNA injected. The IgG titer is influenced by many factors dependent or independent of LNP formulation, as mentioned. Therefore, all information was needed to train an ML model, and the influence of LNP, mainly the ionizable structures, on IgG titer could be specifically untangled by the algorithm. Thus, the ML model was able to predict the LNP formulation. The prediction result was further validated by an in vivo experiment. Then, the MD simulation was used to investigate the interaction between mRNA molecules and lipid components in the microscope. This developed model will benefit the development of mRNA vaccines.
2. Materials and methods
2.1. ML modeling methods
2.1.1. Data collecting and cleaning
The data collecting and cleaning method are shown in Fig. 1. First, keywords of ‘mRNA’, ‘vaccine’, ‘virus’ were used to retrieve literature from Web of Science and Scopus. After the initial screen, 65 studies using lipid-based formulation were maintained.
Figure 1.
Data collecting and cleaning process for machine learning (ML) work. (A) Data collecting and cleaning process. (B) The eventual dataset contained lipid nanoparticle (LNP) with seven kinds of ionizable lipids, including DLin-MC3-DMA (MC3), DLinDMA, L31951, Lipid M, N, and Q30, and SM-10247,52.
After analyzing the first extracted data from 22 studies, the data intended for ML work were simplified, considering the numerical balance between input and predicted parameters. Thus, the eventual dataset only contained mRNAs encoding a single antigen45, 46, 47, 48, 49, 50 and LNPs comprising DSPC, cholesterol, 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol (PEG-DMG), and various ionizable lipids51,30,47,52 (Fig. 1B). The included experiments all lasted for no longer than one year, with no more than two doses of vaccination and without virus challenge test.
The input parameters or features include antigen protein type, self-amplifying10,11, cap type, pseudouridine modification14, codon optimization16, the molar ratio between nitrogen on the ionizable lipid to phosphate on RNA (N/P ratio), structure of ionizable lipid, ionizable lipid fraction, DSPC fraction, PEG–lipid fraction, cholesterol fraction, subject type, population or strain, injection route, log10 dose, the second vaccination time, and IgG titer test time. Parameterization of ionizable lipid structure can be seen in the next section. Whether or not the mRNA sequences functioning as self-amplifying, containing pseudouridine, and undergoing codon optimization were assigned ‘1’ or ‘0’. The antigen protein type, cap type, subject type (human, primate, mice, etc.), population or strain (adult or elderly human, C57BL/6 or BALB/c mice, etc.), and injection route were deemed as multi-categorical variables. The other parameters were numerical variables.
The output or predicted parameter was decided to be the binding IgG titer because this index reflected antibody concentration induced by vaccines, and the related data was the richest. Binding IgG titer is usually tested by enzyme-linked immunosorbent assay (ELISA), and it is the dilution-fold of tested serum-containing antibodies that can still neutralize the antigen-coated at the bottom of a 96-well plate. A high titer means the serum still can neutralize the virus even if it has been diluted to a high-fold. The collected IgG titers only contained the assays with coated antigens corresponding to mRNAs used in vaccines. For studies against influenza30,44,53, hemagglutination inhibition (HAI) titer is also often tested. Hemagglutination happens when red blood cells contact the influenza virus, and the addition of tested serum containing antibodies neutralizing the influenza virus would inhibit hemagglutination. Thus, HAI titer, similar to IgG titer, also reflects the antibodies' concentration. Analysis of our data found a linear relationship (Supporting Information Fig. S1) between two titers [log10(IgG titer) = 1.0286 × log10(HAI titer) + 1.4103, R2 = 0.7986, when HAI titer ≥1]. Thus, IgG titers transformed from HAI tests were also included if only HAI titers were available. The dose and the titers were transformed via log10 function to shape the distribution closer to normal. Eventually, there were a total of 325 samples in the dataset.
2.1.2. Structural representation of ionizable lipids
In this study, the extended connectivity fingerprints54 (ECFP) were introduced to represent the ionizable lipid structural characteristics. ECFP is a bit string constituted by ‘1’ or ‘0’. Each bit of ECFP corresponds to a set of chemical substructures, and the ‘1’ or ‘0’ indicates whether or not the compound contains it. ECFP shows good modeling fitting in cheminformatics and bioinformatics. The ionizable lipid ECFP (IL-ECFP) was generated by the RDKit package version 2020.09.1.0 in Python55. Ionizable lipids used in mRNA vaccine formulation have long chains. Thus, the ECFP radius was set to 3 to cover a chain segment with up to 7 atoms, larger than the regular ECFP4 structure (ECFP with a radius of 2). The ECFP sequence length was set to 1024.
2.1.3. Data splitting strategy
The whole dataset was split into two sets of training (260 data points) and validation (65 data points). Stratified random sampling was adopted to keep the same proportion of data points in each mRNA vaccine formulation38. The training set was used for training models, and the validation set was for turning hyperparameters to find the best model. Additionally, 10-fold cross-validation (CV) was used to evaluate the final generalization of machine learning models.
2.1.4. Evaluation criteria
Mean absolute error (MAE), mean squared error (MSE), root mean squared error (RMSE), and determination coefficient (R2) are the metrics for evaluating regression model performance. MAE measures the mean absolute error between real labels and predictions. MSE indicates the mean squared error between real labels and predictions. RMSE indicates the root mean squared error between real labels and predictions. R2 shows the correlation between real labels and predictions. They are defined as the following Eqs. (1), (2), (3), (4):
| (1) |
| (2) |
| (3) |
| (4) |
where n is the number of data, is the ith real label, and is the ith prediction.
2.1.5. Hyper-parameters of lightGBM
In recent years, the techniques and applications of machine learning have been driven by algorithmic advances and data accumulation. Diverse machine learning algorithms and structures have been tested to fit different types of correlations56,57. The gradient boosting decision tree (GBDT) framework-based ensemble learning algorithms are shown to have superior accuracies in both classification and regression problems on tabular data with pre-engineered features. A typical algorithm is the lightGBM58. In the present study, the model was constructed to predict the titer concentration of mRNA vaccine immunological performance. The model was established by the lightGBM package version 2.2.3 in Python. We searched 1000 hyperparameter combinations in the hyperparameter space. The hyperparameter configuration of lightGBM is that the learning rate is 0.018, the number of trees is 930, the subsample ratio is 0.783, the subsample ratio of columns is 0.394. The regularization terms (maximum of eight leaves for base learners and minimum of 12 samples in a leaf) were used to prevent the overfitting issue. The machine learning hyperparameters decide model selection and have impact on model generalization. A random search method was used for lightGBM. It has been shown that random search is more efficient than grid search and manual search59.
2.2. Experimental methodology
2.2.1. mRNA synthesis
Our laboratory has established a system to conveniently test the LNP delivery efficiency using mRNAs encoding the extracellular segment of human angiotensin-converting enzyme 2 [ACE2 (18-615)], human IgG Fc fragment, and an HiBit tag. The ectodomain of ACE2 fused with IgG Fc fragment makes it a long-lasting secreted protein60, which can be directly detected in the blood. A HiBit tag added to the C-terminus of the protein facilitates later protein detection.
Our mRNA was synthesized in vitro by a T7 RNA polymerase mediated transcription system (IVT). The DNA template incorporates the 5ʹ and 3ʹ untranslated regions (UTRs) and a poly(A) tail. Pseudo UTP instead of natural UTP was used during IVT to reduce the immunogenicity of the mRNA. Cap1 is added co-transcriptionally to ensure the normal translation of mRNA. The pseudo UTP and cap1 were purchased from APExBIO Technology LLC. (Houston, TX, USA). We purified mRNA by oligo dT column, then diluted the mRNA in sodium citrate buffer to desired concentrations. The purity of mRNA was confirmed by gel electrophoresis.
2.2.2. LNP formulation and characterization
DLin-MC3-DMA (MC3) and SM-102 were purchased from APExBIO. Cholesterol was purchased from AVT (Shanghai) Pharmaceutical Tech Co., Ltd. (Shanghai, China). DSPC and PEG2000-DMG were purchased from Avanti Polar Lipids Inc. (Alabaster, AL, USA). Lipids were dissolved in ethanol at molar ratios of 50:10:38.5:1.5 (ionizable lipid/DSPC/cholesterol/PEG2000-DMG). The mRNA was diluted in sodium citrate buffer (pH 3.0) to desired concentrations for final N/P ratios 3:1 and 6:1, respectively. We gave high-pressure to mix mRNA solution and lipid solution rapidly through a T mixer. Formulations were dialyzed against PBS (pH 7.4) in a dialysis cassette for 20 h. After dialysis, LNPs were passed through a 0.22 μm filter, concentrated to a suitable concentration, stored at 4 °C, and used within a week. RiboGreen Assay from Invitrogen Corp. (Carlsbad, CA, USA) was used to quantify the mRNA in LNPs, particle size was determined by dynamic light scattering. The encapsulation efficiency of our LNP was around 90%, and the particle size was around 100 nm.
2.2.3. Animal studies
All animal experiments were performed under the ethical guidelines of Fudan University. Sixteen C57BL/6JGPt mice (eight weeks old, mixed-sex) were randomly divided into four groups, corresponding to four LNP formulations administrated (MC3-3:1, MC3-6:1, SM102-3:1, and SM102-6:1). LNPs diluted in PBS were injected into mice via the tail vein using a disposable syringe (15 μg mRNA/dose). Tail vein blood was taken at 0, 4, 8, 24, 48, 72, 96, 168 h after injection with capillaries. The serum was separated by centrifugation at 6000 rpm for 10 min. The ACE2 level in mice serum was measured using Nano-Glo® HiBit Lytic Detection System from Promega Corp. (Madison, WI, USA) following the manufacturer's recommendations. Then the luminescence signal was detected on the microplate reader.
2.2.4. Statistical analysis
Means were compared using the unpaired t-test, and the area under the curve (AUC) was calculated after 168 h of administration for all tests. Two-tailed P values < 0.05 were considered statistically significant and are shown in the figures as ∗P ≤ 0.05, ∗∗P ≤ 0.005, ∗∗∗P ≤ 0.001. Prism 8 (GraphPad Software, San Diego, CA, USA) was used.
2.3. MD modeling methods
2.3.1. Model building of the formulation systems
The all-atom dynamic simulation method61,62 was performed to investigate the formation mechanism of LNPs. Molecular structures of two ionizable lipids (MC3 and SM-102), cholesterol, DSPC, and PEG2000-DMG, were manually built by Discovery Studio 2016 Client, as shown in Fig. 2. The mRNA nucleotide sequence consisted of the 32-mer poly(A) tail generated by the NAB package in AMBER (University of California, San Francisco, CA, USA). Poly(A) tail was chosen because it is generally added to all manufactured mRNA sequences8. The length of mRNA sequence for simulation was decided because of the sizes of eventual simulated systems limited to computer capacity. These molecules constituted five different simulated systems in total (Table 1). At first, mRNA in an aqueous solution in the absence of lipids (referred to as mRNA system) was observed. Then, MD simulation simulated the other four systems that consisted of a single mRNA, ionizable lipids, cholesterol, DSPC, PEG2000-DMG, and water. Lipids were added at the molar ratio of ionizable lipid/DSPC/cholesterol/PEG2000-DMG = 50:10:38.5:1.5, and the N/P ratio was 3:1 or 6:1. Both composition ratio and N/P ratio were generally seen in our collected data. In the mRNA system, sodium counterions were used to ensure electrical neutrality, while in the other LNP systems, chlorine counterions were added to the system.
Figure 2.
Three-dimensional structure of the MC3, SM-102, DSPC, cholesterol, and PEG2000-DMG.
Table 1.
Molecules' number of the five simulated LNP systems.
| Parameter | mRNA | MC3-3:1 | MC3-6:1 | SM102-3:1 | SM102-6:1 |
|---|---|---|---|---|---|
| N/P ratioa | NA | 3:1 | 6:1 | 3:1 | 6:1 |
| mRNAb | 1 | 1 | 1 | 1 | 1 |
| MC3 | 0 | 96 | 192 | 0 | 0 |
| SM-102 | 0 | 0 | 0 | 96 | 192 |
| Cholesterol | 0 | 74 | 148 | 74 | 148 |
| DSPC | 0 | 19 | 38 | 19 | 38 |
| PEG2000-DMG | 0 | 3 | 6 | 3 | 6 |
| Na+ | 31 | 0 | 0 | 0 | 0 |
| Cl‒ | 0 | 65 | 162 | 65 | 161 |
| Water | 6190 | 69,747 | 98,183 | 68,936 | 97,877 |
The number ratio between the nitrogen groups of ionizable lipids to phosphate groups of mRNA sequence.
The mRNA consists of 32-mer of poly (A).
2.3.2. Simulation method
The detailed simulation method was similar to the previous study37. All the simulations were carried out using AMBER 18 and AMBER Tools 18 software package. The FF14SB force field was used to model mRNA, and the GAFF force field was applied to model lipid molecules. For lipid molecules, the atom type and charge were described by the antechamber package63. The conformation of initially constructed systems may be far from their equilibrated state, and molecules may arrange too close, inducing unreasonably high energy in systems. Thus, minimization of systems was needed before the simulation to relax the structure and remove unreasonable contacts. First, the solute molecules were constrained, and only water molecules were minimized for a short time. Then the whole system underwent 20,000 steps of minimization. After minimization, systems were heated, and the Langevin thermostat64 was used to maintain the temperature at 300 K, while the Berendsen barostat65 was used to keep the pressure at 1 atm. All the systems were equilibrated at least 100 ns with a time step of 2 fs to produce the simulated results.
3. Results
3.1. Data distribution and model performance of ML work
The data collected included LNP and mRNA information as input features and IgG titer induced by vaccines at corresponding time points as output parameters for prediction. The data distributions of these parameters are overall uniform (Figure 3, Figure 4).
Figure 3.
Data distribution of 325 formulation datasets. Numerical counts of the eventual data dependent on disease and protein (A), subject type (B), population or strain (C), injection route (D), ionizable lipid type (E). In (A), H1N1 Cal and PR8 referred to strains A/California/07/200945 and A/Puerto Rico/8/193446, respectively; SARS-CoV-2 S-2P and RBD referred to the S protein with two substitutions of proline at 986 and 987 amino acid positions47 and receptor binding domain (RBD)48, respectively; and RSV mDS-Cav-1 referred to the full-length F protein respiratory syncytial virus (RSV) with four-point mutations49,50.
Figure 4.
Data distribution of 325 formulation datasets. Numerical counts of the eventual data dependent on N/P ratio (A), log10(dose) (B), the second vaccination time (C), IgG titer test time (D), and log10(IgG titer) results (E) were given.
Table 2 shows that the ML model using the LightGBM algorithm presents a good performance. The dataset was first divided into the training set and validation set, with samples of 260 and 65, respectively. After training and tuning hyperparameters, the model shows impressive predictivity. For the training and validation set, the MAE and RMSE are around 0.2 and 0.3 log10 units, respectively, corresponding to the error commonly seen in the experiments. The R2 is above 0.9, showing this model has covered major factors resulting in the variation in the IgG titer. Moreover, additional 10-fold cross-validation was performed. The whole dataset was divided into ten folds. One fold served as the validation set and the rest as the training set for each iteration, and this process was repeated ten times. The average results of 10 iterations are also presented in Table 2. Although the MAE, MSE, and RMSE are slightly larger than those from the first training, they also show an accurate predictivity on experimental value.
Table 2.
Model performance of LightGBM.
| Parameter | Training set | Validation set | 10-fold cross-validation (mean ± SD) |
|---|---|---|---|
| Mean absolute error | 0.220 | 0.278 | 0.303 ± 0.053 |
| Mean squared error | 0.092 | 0.139 | 0.178 ± 0.078 |
| Root mean squared error | 0.303 | 0.373 | 0.412 ± 0.086 |
| R2 | 0.935 | 0.904 | 0.871 ± 0.061 |
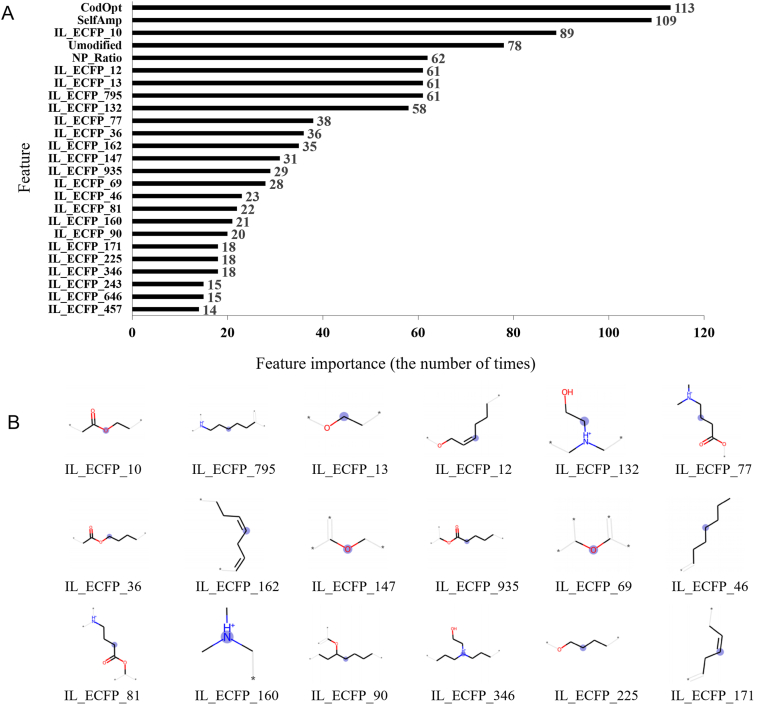
The next analysis determines the important input parameters or features that hugely influence the model. The top 7 important parameters are biological factors, including protein type, log10 dose, titer test time, population or strain, the second vaccination time, subject type, and injection route. The following parameters are formulation-related features, as shown in Fig. 5A. The codon optimization, self-amplifying, and uridine modification show the important role of mRNA sequence modification. Then, the formulation features of the N/P ratio and some IL-ECFPs present the LNP importance. The top 18 important positions among 1024 IL-ECFP and their corresponding specific substructure of ionizable lipid are shown in Fig. 5B. For the important 18 IL-ECFPs, 5 of them are contained in DLinDMA, while 7, 8, and 11 are contained in selective ionizable lipids MC3, L319, and SM-102. Compared to DLinDMA, MC3 contains a secondary ester linker (IL-ECFP 69 and 77). Compared to MC3, L319 contains a primary ester linker (IL-ECFP 147) in tails, replacing one double bond (IL-ECFP 12), and the chain after the double bond comprises six carbon atoms (IL-ECFP 46). SM-102, compared to MC3, has a hydroxy group (IL-ECFP 132) in the head and a primary (IL-ECFP 10) as well as a secondary ester (IL-ECFP 935) in tails. The distance from the nitrogen to the ester is five carbons (IL-ECFP 795) in one tail of SM-102. These features distinguish ionizable lipids from each other and are deemed essential and ranked in the model.
Figure 5.
Features ranking and important substructure of ionizable lipids. (A) The top 25 important features related to the formulation. Importance times were recognized using the information gain (IG) values as a criterion from the lightGBM model. (B) The top 18 important IL-ECFP and their corresponding specific substructure of ionizable lipid. The center atom, recognizing length, and environmental information of each ECFP are indicated by the stressed blue area, black bonds, and grey bonds, respectively.
3.2. Experimental validation of the ML model
The ML model was validated by the animal test. LNPs of various formations (ionizable lipid as MC3 or SM-102, N/P ratio at 3:1 or 6:1) were used to intravenously deliver mRNAs encoding human ACE2 to mice. The ACE2 expression level was measured as the relative light unit (RLU) of the nanoluciferase enabled by the fused HiBit tag. Table 3 shows the characteristics of these LNPs. Four LNPs have around 90% encapsulation efficiency and a similar particle size of around 100 nm. Fig. 6 compares the prediction results and in vivo test. Fig. 6A shows that the MC3-based LNP is predicted to induce a higher titer value than that based on SM-102, but the N/P ratio does not influence the predicted titer. In the animal results of Fig. 6B, MC3-based LNP with N/P ratio at 6:1 resulted in an overall higher RLU than that based on SM-102, though there is no significant difference between them (Fig. 6C and D). LNPs based on two ionizable lipids with an N/P ratio at 3:1 show similar RLU.
Table 3.
Encapsulation efficiency and particle size of LNPs.
| Formulationa | MC3-3:1 | MC3-6:1 | SM102-3:1 | SM102-6:1 |
|---|---|---|---|---|
| Encapsulation efficiency | 89.5% | 91.3% | 89.6% | 91.2% |
| Particle size (nm) | 97.8 | 101.0 | 101.2 | 106.3 |
LNP was formulated from ionizable lipid, DSPC, cholesterol, and PEG-lipid at a molar ratio of 50:10:38.5:1.5. The N/P ratio was 6:1 or 3:1.
Figure 6.
Comparison between ML prediction and in vivo expression level. (A) Predicted log10(IgG titer) versus time profile of BALB/c mice induced by mRNA-LNP encoding S-2P protein of SARS-CoV2 at the dose of 20 μg by i.m. administration on Days 0 and 21. LNP consists of ionizable lipid, DSPC, cholesterol, and PEG-lipid at a molar ratio of 50:10:38.5:1.5. Ionizable lipids included MC3 and SM-102. The N/P ratio is 6:1 or 3:1. (B) Relative light unit (RLU) of HiBit tag versus time profiles in C57BL/6JGPt mice induced by mRNA-LNP encoding angiotensin-converting enzyme 2 (ACE2) following i.v. administration. The LNP formulations were the same as the prediction task. The difference in the maximum RLU at 8 h (C) and the AUC at 168 h (D) after administration were tested. Data are presented as mean ± SD (n = 4). ∗∗P ≤ 0.005. ns, not significant.
3.3. Investigating the molecular structure of mRNA LNPs by MD simulations
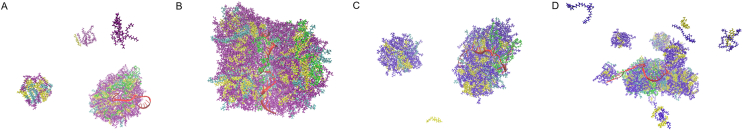
MD modeling was performed to investigate the interaction between lipids and mRNA in LNP formation. Fig. 7 shows the initial and final structure of a single mRNA sequence for 100 ns MD simulation, which shows that the mRNA sequence is folded in the water solution. Fig. 8 displays the final structure of four lipid systems (ionizable lipid as SM-102 or MC3, and N/P ratio of 3:1 or 6:1) for 200 ns MD simulation. The four systems self-assemble rapidly and form aggregates in the water solution, but the degree of aggregation is different. In the SM102-6:1 system with a ratio of 6:1, all molecules aggregate together. However, the other three systems form several clusters of different sizes. All LNPs formed show dense core structure. As for mRNA encapsulation, the SM102-6:1 system entraps a part of the mRNA sequence. However, in the rest of the lipid systems, the whole mRNA sequence is almost exposed to the aqueous solution. Besides, long mRNA in system MC3-6:1 attaches to multiple LNPs, while mRNA sequences in the other three systems only bind to a single LNP.
Figure 7.
The snapshots of mRNA structure at the initial time (A) and 100 ns of simulation (B).
Figure 8.
The snapshots of four lipid systems for 200 ns MD simulation: (A) SM102-3:1; (B) SM102-6:1; (C) MC3-3:1; (D) MC3-6:1; water molecules were not displayed in the figure. Red represents mRNA; purple represents SM-102 ionizable lipid; blue represents MC3 ionizable lipid; yellow represents cholesterol; cyan represents DSPC; green represents PEG2000-DMG.
To show how mRNA interacts with LNP, the simulation results were re-colored with nitrogen atoms on the ionizable lipid highlighted in Fig. 9. It shows that all lipid molecules aggregate together to form the LNPs, and nitrogen groups of cationic lipids preferentially locate at the surface of LNP. The mRNA molecule twines around LNP by two possible mechanisms. On the one hand, the nucleosides of mRNA are direct to or lie on the LNP by the hydrophobic interaction. On the other hand, the phosphate groups of the mRNA backbone are close to the nitrogen atoms of LNP due to the electrostatic effect.
Figure 9.
The snapshots of four lipid systems for 200 ns MD simulation: (A) SM102-3:1; (B) SM102-6:1; (C) MC3-3:1; (D) MC3-6:1; water molecules were not displayed in the figure. Yellow: mRNA sequence. Blue: nitrogen on the ionizable lipids.
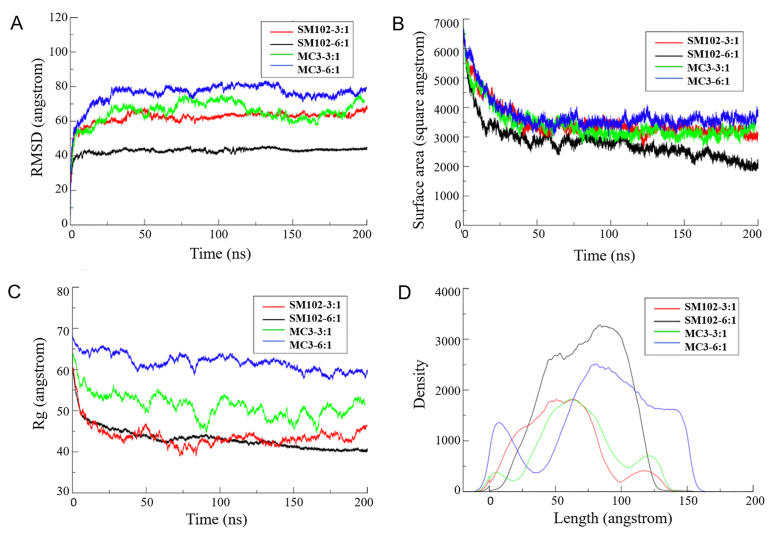
Fig. 10 shows the quantitative analysis of four lipid systems during 200 ns MD simulation. The RMSD profile indicates that four lipid systems reach a stable state after about 50 ns. The surface areas of mRNA exposed to water in the systems decrease with time, which indicates the encapsulation of mRNA molecule to LNP. The mass-weighted radius of gyration (Rg) vs. time of the whole system represents that aggregation of SM102-3:1 and SM102-6:1 is more obvious than those of MC3-3:1 and MC3-6:1. The density profile of the SM102-6:1 system shows a high-intensity peak at about 100 Å. In contrast, more than one peak is observed in the other three systems, and the MC3-6:1 system shows a wide distribution. These results imply that SM102-6:1 is compact, while MC3-6:1 is relatively loose.
Figure 10.
Quantitative analysis of four lipid systems during 100 ns MD simulation. (A) Root mean square displacement (RMSD) vs. time. (B) Solvent accessible surface area of the mRNA sequence vs. time. (C) Mass-weighted radius of gyration (Rg) vs. time. (D) Density profile of a system as a function of the distance from the geometric center of the system.
4. Discussion
Currently, the selection of the ionizable lipid has attracted significant attention for optimizing the LNP formulation for mRNA delivery. Since traditional screening tests often consume a lot of time and materials, computational tools that can accelerate the development should be valuable. The present work builds an ML model with good prediction performance, which correlates the critical substructures of ionizable lipids to the in vivo potency (IgG titer) of mRNA vaccines to help the choice of ionizable lipids.
More importantly, the importance of features is ranked. The 18 critical IL-ECFPs among 1024 are identified in Fig. 5B, representing the cationic head group, ester linker, and tail of ionizable lipids. A small head group, such as IL-ECFP 160, combined with two relatively large dilinoleyloxy tails (IL-ECFP 162 and 171) may behave in a “cone” shape and facilitate the formation of hexagonal HII phase when contacting with endosomal membrane, disrupting the bilayer structure and release the RNA therapeutics29. These substructures are the symbols of DLinDMA, which is one ionizable lipid that turned out to be highly effective in the early stage. DLinDMA was then optimized to DLin-KC2-DMA29 and further MC328 by substituting with a second ester linker (IL-ECFP 69) distant from the nitrogen at three carbons length (IL-ECFP 77), which are deemed more important than IL-ECFP 160 and 162. The original research also shows an improvement in potency by this optimization. However, the adverse effect is also commonly seen when administrated with MC3-containing LNP because of its low biodegradability30. Maier et al.51 developed biodegradable L319 by substituting one double bond with an ester linker (IL-ECFP 147 and 12), assuming this compound could be metabolized by hydrolysis and β-oxidation. Sabnis et al.25 developed SM-102 from a similar compound to MC3 by obtaining the balance between the lipid pKa28,30, potency, and metabolism behavior. SM-102 has a head group as IL-ECFP 132 and two ester linkers, IL-ECFP 10 and 935. The distribution of esters in two tails maintains the pKa within the desired range, the side chain resulted from the secondary ester may facilitate the “cone” shape, and the distance from the nitrogen to the ester (IL-ECFP 795 in SM-102) also impacts the metabolism25. Both L319 and SM-102 also show high in vivo potency. The present AI model recognizes these IL-ECFPs above as important substructures, though the mechanism is not reflected in the collected data. Besides, notice that the ECFPs presented here are just the top 18 important ones, but there are 1024 ECFPs in total. The efficiency of ionizable lipids should be more dependent on the sum importance of all ECFPs.
The validation of the ML model against the in vivo test result also proves our model with some suggestive ability about ionizable lipids (Fig. 6). We modified the mRNA of human ACE2 to encode a secreted protein, and therefore the ACE2 expression can be directly detected from the blood samples, which is a straightforward and undisturbed way to assess the efficiency of LNP carriers. The animal test shows that ACE2 expression level induced by LNP at N/P ratio of 6:1 is higher than that at the ratio of 3:1, consistent with the previous finding that the higher N/P ratio induces more potency26,27,66. The result also shows that the MC3-based LNP induces more expression than that based on SM-102 at the N/P ratio of 6:1. A reasonable assumption is that expression level is positively correlated to IgG titer, which conforms to the prediction by the ML model that MC3 induces higher IgG titer than SM-102 at an N/P ratio of 6:1. However, low biodegradability correlates to side effects30, making the choice of ionizable lipid complicated. In fact, it is SM-102 that is formulated in mRNA-1273 vaccine67. Besides, the model predicts IgG titers for two N/P ratios are similar. It is due to that the N/P ratio is little varied for one kind of ionizable lipid, and the ML model is difficult to discriminate the impact of the N/P ratio from ionizable lipid. Inputting more diverse data can address this issue easily.
The structure of LNP is another important topic concerned in the pharmaceutical field. The cryo-TEM images have shown that LNPs are overall in dense core26,68 or lamellar23,69 structure. However, visualization of LNP structure at a molecular level often relies on modeling method70, 71, 72. In the present study, we performed an MD simulation of LNP entrapping mRNA (Fig. 8). During the simulation, the firstly dispersed lipids aggregate to form small dense core particles. The mRNA can twine around or be partly entrapped in these lipid particles. Besides, mRNA twining around multiple particles is also possible. Analysis of the aggregation behavior (Fig. 10) shows that system SM102-6:1 (SM-102-based, N/P ratio 6:1) converges most rapidly and forms the most compact structure, while system MC3-6:1 forms a relatively loose structure. Interestingly, increasing the lipid content (N/P ratio from 3:1 to 6:1) makes the SM-102 system more compressed but loosens the MC3 system. These results indicate that both ionizable lipid type and N/P ratio influence the LNP aggregation behavior. Our simulated LNP structure agrees with another all-atom modeling result by Rissanou et al.70, who have simulated the aggregation behavior of mRNA and so-called DML lipid molecules, wherein mRNA also twines around the LNPs.
The interaction between the phosphate on mRNA and the nitrogen on ionizable lipids is of research interest. The electrostatic effect between the two kinds of molecules is presumed to promote the mRNA binding to LNP28. Our modeling result suggests that during the LNP formation, lipids aggregate first, and mRNA twines around LNP with its phosphate groups getting close to nitrogen atoms. This binding is also helped by the hydrophobic effect implemented on the nucleosides of mRNA, resulting in those nucleosides generally directing to or lying on the LNP. Besides, hydrogen bonds70 is reported to be another potential factor facilitating this binding.
The all-atom dynamic simulation provides rich insights into the LNP formation mechanism. However, limited to computational capacity, this method can only handle small simulated systems. Considering scaling up the system based on the mechanism of particle formation deduces theoretical structures of LNP as Fig. 11. At the first stage of mixing, lipids in the vicinity should aggregate to form small clusters and attach to mRNA in the line. This step is supported by the MD modeling (Fig. 8D and Ref.70). The lipid clusters tend to fuse by their nature to reduce the surface energy, but the cluster volume cannot grow unlimitedly since the main stuff, the ionizable lipid, is amphipathic. Thus, the cluster either grow along the mRNA (Fig. 11C) or enlarge like a liposome particle (Fig. 11D), and this deduces two theoretical LNP structure (Fig. 11E and F), respectively. The tunnel organization of the LNP core proposed in Fig. 11E is supported by the transmission electron microscopy (TEM) image73, which shows the LNP core texture as many arranged channels. These channels correspond to hexagonal phase rods, as reported. The liposome-like volume proposed in Fig. 11F is also recorded by TEM and is called “blebs”. The occurrence of this structure seems to depend on PEG content73, DSPC content, and the type of nucleic acids entrapped22. It seems that long nucleic acids, such as DNA and mRNA, are likely to induce the blebs. It is reasonable since longer nucleic acids form a larger obstacle in the system, which may induce clusters fusing less randomly in the space and result in a heterogeneous particle. As for the other three lipids in LNP, PEG-lipid, and DSPC, mainly located at the exterior, while the cholesterol helping to constitute the core, are also evidenced73,74.
Figure 11.
The evolution of lipids fusion and theoretical structure of mRNA LNP system. (A) At the initial mixing stage, lipids form many small clusters and attach along the mRNA sequence by electrostatic effect. (B) The clusters getting close tend to fuse into a bigger cluster to decrease the surface energy. The tails of lipid in these clusters are reduced for clarity. Then, more clusters participate in the fusion to form a long lipid particle (C) or liposome-like particle (D). If the fusion primarily results in long lipid particles, they should form tube structures in the core of LNP (E). Otherwise, lipid fusion leading to liposome-like particles produces LNP containing a large chamber filled with aqueous phase (F). The DSPC and PEG should locate at the exterior of LNP while cholesterols insert in the interval between lipids.
In this study, the ML model was built to predict the formulation of mRNA vaccines, and the MD method was used to investigate the LNP formation. The application domain is a critical issue for an ML model. Our model was trained on data from ionizable lipids across a long history. These lipids contain important substructures such as the tertiary amine, hydroxy group, ester bond, secondary ester bond, and dienyloxy chain, which have been represented as ECFPs and can be combined to build other new and typical lipids. Thus, the coverage of ionizable lipids in our model is extended, benefiting the formulation selection, which is the primary target of our study. On the other hand, though we have collected data as much as possible, the resulted data size is still relatively small, and the searched antigens cover just several diseases, which narrows the range that uses this model to predict IgG titer for specific diseases. Besides, small data size impedes the further analysis about the dependence of LNP formulation on specific diseases. More data of diverse diseases and formulations are desired in the future to expand its application domain and refine the design of LNP for specific diseases. As for MD simulation, the current simulated systems are relatively small in scale. To our best knowledge, modeling on LNP at a typical size with good stability (60 nm)24 has not been published. Thus, the real internal structure of an LNP can only be deduced. The recently reported ML-based MD modeling method has dramatically increased the scale of simulated systems75, which may become a powerful tool in LNP modeling in the future. Last, the current study has not revealed the relationship between MD results and LNPs' pharmacological effect. More MD modeling associated with data science technology is promising to deal with this issue.
5. Conclusions
The first ML model has been successfully developed to predict the LNP formulations with the IgG titer of the mRNA vaccine, which is validated by in vivo test on the ACE2 expression. The ML model also recognizes important substructures of ionizable lipids. The MD model is used to investigate the aggregation behavior and the molecular structure of LNP. The integrated computational methodology is able to design better ionizable lipid, which serves a constructive role in the formulation development of nucleic acids therapeutics.
Acknowledgments
This work was financially supported by the University of Macau Research Grants (MYRG2020-00113-ICMS, China). This work was performed in part at the High-Performance Computing Cluster (HPCC) which is supported by Information and Communication Technology Office (ICTO) of the University of Macau.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.11.021.
Contributor Information
Jinzhong Lin, Email: linjinzhong@fudan.edu.cn.
Defang Ouyang, Email: defangouyang@umac.mo.
Author contributions
The authors confirm contribution to the paper as follows: study conception and design: Defang Ouyang, Jinzhong Lin; data collection: Wei Wang; experimental work: Shou Feng; machine learning modeling: Zhuyifan Ye; molecular dynamic simulation: Hanlu Gao; interpretation of results: Wei Wang; draft manuscript preparation: Wei Wang, Defang Ouyang. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Prüβ B.M. Current state of the first COVID-19 vaccines. Vaccines. 2021;9:30. doi: 10.3390/vaccines9010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahase E. COVID-19: UK approves Pfizer and BioNTech vaccine with rollout due to start next week. BMJ. 2020;371:m4714. doi: 10.1136/bmj.m4714. [DOI] [PubMed] [Google Scholar]
- 3.Tanne J.H. COVID-19: Pfizer–BioNTech vaccine is rolled out in US. BMJ. 2020;371:m4836. doi: 10.1136/bmj.m4836. [DOI] [PubMed] [Google Scholar]
- 4.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baden L.R., Sahly H.M.E., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics—developing a new class of drugs. Nat Rev Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 7.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;10:1–17. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhary N., Weissman D., Whitehead K.A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov. 2021;20:817–838. doi: 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magini D., Giovani C., Mangiavacchi S., Maccari S., Cecchi R., Ulmer J.B., et al. Self-amplifying mRNA vaccines expressing multiple conserved influenza antigens confer protection against homologous and heterosubtypic viral challenge. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samsa M.M., Dupuy L.C., Beard C.W., Six C.M., Schmaljohn C.S., Mason P.W., et al. Self-amplifying RNA vaccines for venezuelan equine encephalitis virus induce robust protective immunogenicity in mice. Mol Ther. 2019;27:850–865. doi: 10.1016/j.ymthe.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng C., Hou X., Yan J., Zhang C., Li W., Zhao W., et al. Leveraging mRNA sequences and nanoparticles to deliver SARS-CoV-2 antigens in vivo. Adv Mater. 2020;32 doi: 10.1002/adma.202004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlandini von Niessen A.G., Poleganov M.A., Rechner C., Plaschke A., Kranz L.M., Fesser S., et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3′ UTRs identified by cellular library screening. Mol Ther. 2019;27:824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannarozzi G., Schraudolph N.N., Faty M., von Rohr P., Friberg M.T., Roth A.C., et al. A role for codon order in translation dynamics. Cell. 2010;141:355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Chin J.X., Chung B.K.-S., Lee D.-Y. Codon Optimization OnLine (COOL): a web-based multi-objective optimization platform for synthetic gene design. Bioinformatics. 2014;30:2210–2212. doi: 10.1093/bioinformatics/btu192. [DOI] [PubMed] [Google Scholar]
- 18.Freyn A.W., da Silva J.R., Rosado V.C., Bliss C.M., Pine M., Mui B.L., et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Awasthi S., Hook L.M., Pardi N., Wang F., Myles A., Cancro M.P., et al. Nucleoside-modified mRNA encoding HSV-2 glycoproteins C, D, and E prevents clinical and subclinical genital herpes. Sci Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., et al. Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+PD-1+CXCR3+T follicular helper cells. Front Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eygeris Y., Patel S., Jozic A., Sahay G., Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020;20:4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung A.K.K., Tam Y.Y.C., Chen S., Hafez I.M., Cullis P.R. Microfluidic mixing: a general method for encapsulating macromolecules in lipid nanoparticle systems. J Phys Chem B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- 23.Maurer N., Wong K.F., Stark H., Louie L., McIntosh D., Wong T., et al. Spontaneous entrapment of polynucleotides upon electrostatic interaction with ethanol-destabilized cationic liposomes. Biophys J. 2001;80:2310–2326. doi: 10.1016/S0006-3495(01)76202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gindy M.E., Feuston B., Glass A., Arrington L., Haas R.M., Schariter J., et al. Stabilization of Ostwald ripening in low molecular weight amino lipid nanoparticles for systemic delivery of siRNA therapeutics. Mol Pharm. 2014;11:4143–4153. doi: 10.1021/mp500367k. [DOI] [PubMed] [Google Scholar]
- 25.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol Ther Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J Control Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 28.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 30.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol Ther Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 33.Jordan M.I., Mitchell T.M. Machine learning: trends, perspectives, and prospects. Science. 2015;349:255–260. doi: 10.1126/science.aaa8415. [DOI] [PubMed] [Google Scholar]
- 34.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 35.Schuhmacher A., Gatto A., Hinder M., Kuss M., Gassmann O. The upside of being a digital pharma player. Drug Discov Today. 2020;25:1569–1574. doi: 10.1016/j.drudis.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 36.He Y., Ye Z., Liu X., Wei Z., Qiu F., Li H.F., et al. Can machine learning predict drug nanocrystals? J Control Release. 2020;322:274–285. doi: 10.1016/j.jconrel.2020.03.043. [DOI] [PubMed] [Google Scholar]
- 37.Gao H., Wang W., Dong J., Ye Z., Ouyang D. An integrated computational methodology with data-driven machine learning, molecular modeling and PBPK modeling to accelerate solid dispersion formulation design. Eur J Pharm Biopharm. 2021;158:336–346. doi: 10.1016/j.ejpb.2020.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Q., Ye Z., Su Y., Ouyang D. Predicting complexation performance between cyclodextrins and guest molecules by integrated machine learning and molecular modeling techniques. Acta Pharm Sin B. 2019;9:1241–1252. doi: 10.1016/j.apsb.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao H., Jia H., Dong J., Yang X., Li H., Ouyang D. Integrated in silico formulation design of self-emulsifying drug delivery systems. Acta Pharm Sin B. 2021;11:3585–3594. doi: 10.1016/j.apsb.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunker A., Róg T. Mechanistic understanding from molecular dynamics simulation in pharmaceutical research 1: drug delivery. Front Mol Biosci. 2020;7:604770. doi: 10.3389/fmolb.2020.604770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouyang D., Smith S.C. Hoboken, John Wiley & Sons; 2015. Introduction to computational pharmaceutics; pp. 1–5. (Computational Pharmaceutics). [Google Scholar]
- 42.Ouyang D., Zhang H., Parekh H.S., Smith S.C. Structure and dynamics of multiple cationic vectors−siRNA complexation by all-atomic molecular dynamics simulations. J Phys Chem B. 2010;114:9231–9237. doi: 10.1021/jp911913c. [DOI] [PubMed] [Google Scholar]
- 43.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., et al. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168:1114–1125. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A Feldman R., Fuhr R., Smolenov I., Ribeiro A., Panther L., Watson M., et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 45.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lederer K., Castaño D., Gómez Atria D., Oguin I.T.H., Wang S., Manzoni T.B., et al. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity. 2020;53:1281–1295.e5. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aliprantis A.O., Shaw C.A., Griffin P., Farinola N., Railkar R.A., Cao X., et al. A phase 1, randomized, placebo-controlled study to evaluate the safety and immunogenicity of an mRNA-based RSV prefusion F protein vaccine in healthy younger and older adults. Hum Vaccines Immunother. 2021;17:1248–1261. doi: 10.1080/21645515.2020.1829899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espeseth A.S., Cejas P.J., Citron M.P., Wang D., DiStefano D.J., Callahan C., et al. Modified mRNA/lipid nanoparticle-based vaccines expressing respiratory syncytial virus F protein variants are immunogenic and protective in rodent models of RSV infection. NPJ Vaccines. 2020;5:16. doi: 10.1038/s41541-020-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers D., Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 55.Landrum G. 2013. RDKit: a software suite for cheminformatics, computational chemistry, and predictive modeling.https://docplayer.net/11897218-Rdkit-a-software-suite-for-cheminformatics-computational-chemistry-and-predictive-modeling.html Available form: [Google Scholar]
- 56.Abhimanyu T., Ambika Prasad M., Bishnupriya P., Kumari S., Babita M. Detection of disease-specific parent cells via distinct population of nano-vesicles by machine learning. Curr Pharmaceut Des. 2020;26:3985–3996. doi: 10.2174/1381612826666200422091753. [DOI] [PubMed] [Google Scholar]
- 57.Bishnupriya P., Babita M., Abhimanyu T. An integrated-OFFT model for the prediction of protein secondary structure class. Curr Comput Aided Drug Des. 2019;15:45–54. doi: 10.2174/1573409914666180828105228. [DOI] [PubMed] [Google Scholar]
- 58.Ke G., Meng Q., Finley T., Wang T., Chen W., Ma W., et al. In: Advances in neural information processing systems. Guyon I., Fergus R., Wallach H., Wallach H., Guyon I., Vishwanathan S.V.N., et al., editors. Neural information processing systems foundation, Inc.; San Diego: 2017. LightGBM: a highly efficient gradient boosting decision tree; pp. 3147–3155. [Google Scholar]
- 59.Bergstra J., Bengio Y. Random search for hyper-parameter optimization. J Mach Learn Res. 2012;13:281–305. [Google Scholar]
- 60.Liu P., Wysocki J., Souma T., Ye M., Ramirez V., Zhou B., et al. Novel ACE2-Fc chimeric fusion provides long-lasting hypertension control and organ protection in mouse models of systemic renin angiotensin system activation. Kidney Int. 2018;94:114–125. doi: 10.1016/j.kint.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 61.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 62.Wang W., Ye Z., Gao H., Ouyang D. Computational pharmaceutics—a new paradigm of drug delivery. J Control Release. 2021;338:119–136. doi: 10.1016/j.jconrel.2021.08.030. [DOI] [PubMed] [Google Scholar]
- 63.Wang J., Wang W., Kollman P.A., Case D.A. Antechamber: an accessory software package for molecular mechanical calculations. J Am Chem Soc. 2001;222:U403. [Google Scholar]
- 64.Liu J., Li D., Liu X. A simple and accurate algorithm for path integral molecular dynamics with the Langevin thermostat. J Chem Phys. 2016;145 doi: 10.1063/1.4954990. [DOI] [PubMed] [Google Scholar]
- 65.Berendsen H.J., van Postma J., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 66.Akinc A., Goldberg M., Qin J., Dorkin J.R., Gamba-Vitalo C., Maier M., et al. Development of lipidoid–siRNA formulations for systemic delivery to the liver. Mol Ther. 2009;17:872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. The dawn of mRNA vaccines: the COVID-19 case. J Control Release. 2021;333:511–520. doi: 10.1016/j.jconrel.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen S., Tam Y.Y.C., Lin P.J.C., Leung A.K.K., Tam Y.K., Cullis P.R. Development of lipid nanoparticle formulations of siRNA for hepatocyte gene silencing following subcutaneous administration. J Control Release. 2014;196:106–112. doi: 10.1016/j.jconrel.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 69.Huebner S., Battersby B.J., Grimm R., Cevc G. Lipid‒DNA complex formation: reorganization and rupture of lipid vesicles in the presence of DNA as observed by cryoelectron microscopy. Biophys J. 1999;76:3158–3166. doi: 10.1016/S0006-3495(99)77467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rissanou A.N., Ouranidis A., Karatasos K. Complexation of single stranded RNA with an ionizable lipid: an all-atom molecular dynamics simulation study. Soft Matter. 2020;16:6993–7005. doi: 10.1039/d0sm00736f. [DOI] [PubMed] [Google Scholar]
- 71.Rozmanov D., Baoukina S., Peter-Tieleman D. Density based visualization for molecular simulation. Faraday Discuss. 2014;169:225–243. doi: 10.1039/c3fd00124e. [DOI] [PubMed] [Google Scholar]
- 72.Leung A.K.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., et al. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J Phys Chem C Nanomater Interfaces. 2012;116:18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arteta M.Y., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc Natl Acad Sci U S A. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Viger-Gravel J., Schantz A., Pinon A.C., Rossini A.J., Schantz S., Emsley L. Structure of lipid nanoparticles containing siRNA or mRNA by dynamic nuclear polarization-enhanced NMR spectroscopy. J Phys Chem B. 2018;122:2073–2081. doi: 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- 75.Jia W., Wang H., Chen M., Lu D., Lin L., Car R., et al. In: SC20: international Conference for high performance computing, networking, storage and analysis. Cuicchi C., Qualters I., Kramer W., editors. IEEE press; Piscataway: 2020. Pushing the limit of molecular dynamics with ab initio accuracy to 100 million atoms with machine learning; pp. 1–14. Article 5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.