Abstract
Nonalcoholic fatty liver disease (NAFLD) is characterized by hepatic steatosis and insulin resistance and there are currently no approved drugs for its treatment. Hyperactivation of mTOR complex 1 (mTORC1) and subsequent impairment of the transcription factor EB (TFEB)-mediated autophagy–lysosomal pathway (ALP) are implicated in the development of NAFLD. Accordingly, agents that augment hepatic TFEB transcriptional activity may have therapeutic potential against NAFLD. The objective of this study was to investigate the effects of nuciferine, a major active component from lotus leaf, on NAFLD and its underlying mechanism of action. Here we show that nuciferine activated ALP and alleviated steatosis, insulin resistance in the livers of NAFLD mice and palmitic acid-challenged hepatocytes in a TFEB-dependent manner. Mechanistic investigation revealed that nuciferine interacts with the Ragulator subunit hepatitis B X-interacting protein and impairs the interaction of the Ragulator complex with Rag GTPases, thereby suppressing lysosomal localization and activity of mTORC1, which activates TFEB-mediated ALP and further ameliorates hepatic steatosis and insulin resistance. Our present results indicate that nuciferine may be a potential agent for treating NAFLD and that regulation of the mTORC1–TFEB–ALP axis could represent a novel pharmacological strategy to combat NAFLD.
Key words: Autophagy, Fatty liver, Lotus leaf, Lysosome, MiT/TFE, mTORC1, Ragulator, TFEB
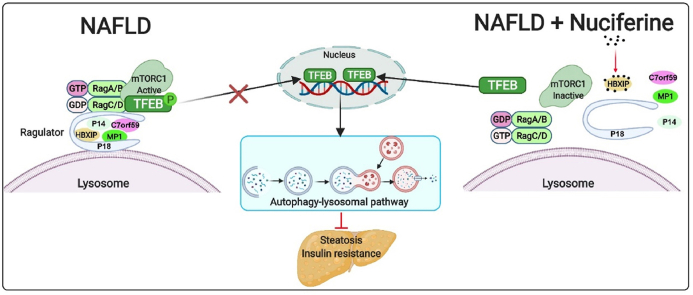
Graphical abstract
Nuciferine impairs Rag GTPases–Ragulator interaction to inhibit mTORC1 and activate TFEB-mediated autophagy–lysosomal pathway, which further improves hepatic steatosis and insulin resistance.
1. Introduction
Nonalcoholic fatty liver disease (NAFLD), ranging from simple steatosis to non-alcoholic steatohepatitis, is the leading cause of chronic liver disease. Characterized as hepatic steatosis with insulin resistance, NAFLD can progress to end-stage liver diseases, such as cirrhosis and hepatocellular carcinoma. In addition, hepatic oxidative stress and inflammatory response are central drivers of lipid accumulation and insulin signaling impairment. The prevalence of NAFLD in the general population ranges from 20% to 30% and up to 75%–100% in obese individuals1,2. Currently, there are no approved effective pharmacological therapies for NAFLD, and efforts to control the complications arising from the condition are far from satisfactory. Hence, studies aimed at exploring therapeutic targets and experimental drugs to treat NAFLD are urgently needed.
Autophagy is a lysosomal degradative pathway that functions to promote cell survival by supplying energy in times of stress or by removing damaged organelles and proteins after injury3. Importantly, the autophagy–lysosomal pathway (ALP) also mediates breakdown of intracellular lipids in hepatocytes4. It has been reported that autophagic flux is blocked in experimental and human conditions of NAFLD, and impaired autophagy is elicited by defective lysosome acidification5,6. Moreover, restoration of hepatic ALP by both pharmacologic and genetic approaches enhances lipid droplet clearance and improves insulin sensitivity in liver4,7,8, which highlights a therapeutic potential of ALP in NAFLD prevention and treatment.
Transcription factor EB (TFEB) is a master regulator of ALP due to its ability to drive expression of autophagy and lysosomal genes9,10. Under nutrient-rich conditions, TFEB is predominantly phosphorylated by mTOR complex 1 (mTORC1) on the lysosomal surface to promote its binding with 14-3-3 proteins, which retain TFEB in the cytoplasm and suppress its transcriptional activity11, 12, 13. mTORC1 senses diverse signals primarily via two kinds of Ras-related small G proteins, the Rag and Ras homolog enriched in brain (RHEB) GTPses, both of which act directly upstream of mTORC114, 15, 16, 17. The Rag GTPases (Rags), composed of RagA or RagB (which are functionally equivalent to each other) in complex with RagC or RagD (also functionally equivalent), are tethered to the surface of lysosomes via Ragulator, a pentameric protein complex consisting of P18, P14, MP1, C7orf59 and hepatitis B X-interacting protein (HBXIP)18, 19, 20, 21, to activate mTORC118, 19, 20, 21, 22.
Overactivation of mTORC1 and subsequent deficiency of TFEB-mediated ALP are associated with the progression of NAFLD, this link has been demonstrated in both human and experimental models23, 24, 25. Besides, liver-specific genetic deletion of TFEB results in the accumulation of hepatic lipid droplets and defective lipid degradation during starvation25,26. Conversely, forced expression of TFEB promotes vast therapeutic effects in ethanol-induced or high-fat diet (HFD)-induced hepatic steatosis by rescuing lipid overload-impaired lysosome function and enhancing lipolysis24,26,27. Importantly, there are several compounds, including rapamycin28 and caffeine4, which bring about therapeutic effects on NAFLD by inhibiting mTORC1 activity whereas treatment with ezetimibe augmented TFEB-mediated ALP23. Thus, manipulation of the mTORC1–TFEB–ALP axis may offer therapeutic options for NAFLD.
Nuciferine (the chemical structure is shown in Fig. 1A), an aromatic ring containing alkaloid, is a major active ingredient extracted from the lotus leaf and has been shown to have extensive pharmacological activities on the metabolic syndrome, including antioxidant29, anti-obesity30,31 and anti-inflammation effects32. Although nuciferine has shown favorable results against hepatic steatosis in NAFLD33, the molecular mechanisms of its anti-NAFLD activity is still obscure. Herein, we report data suggesting that nuciferine disrupts the Rag–Ragulator interaction thus inhibiting mTORC1 activity, which, in turn, results in the activation of TFEB-mediated ALP and subsequent attenuation of hepatic steatosis and insulin resistance. Thus, nuciferine represents a therapeutic candidate for the treatment of NAFLD.
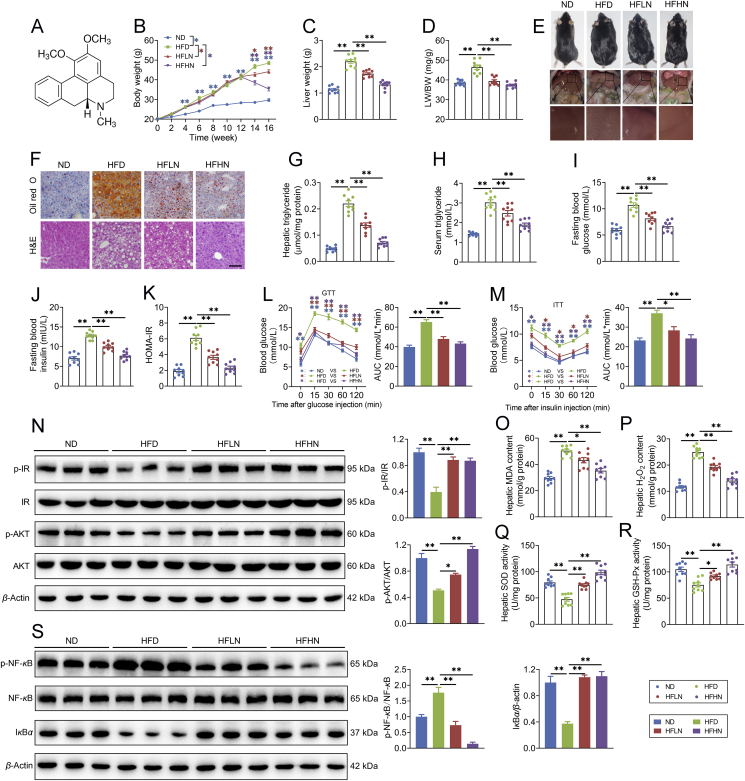
Figure 1.
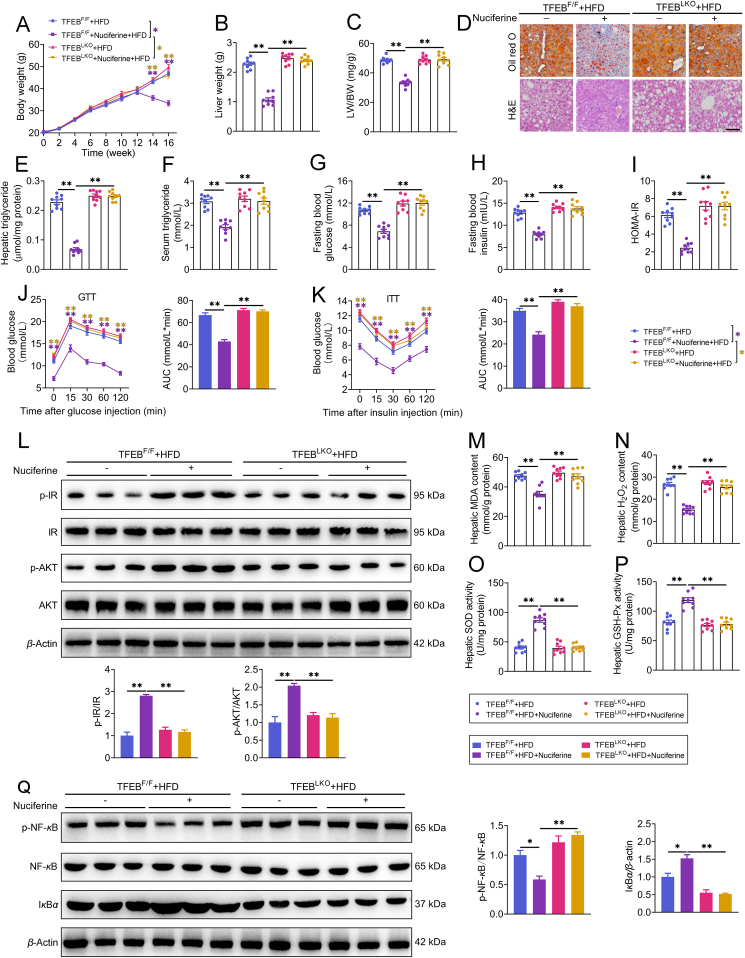
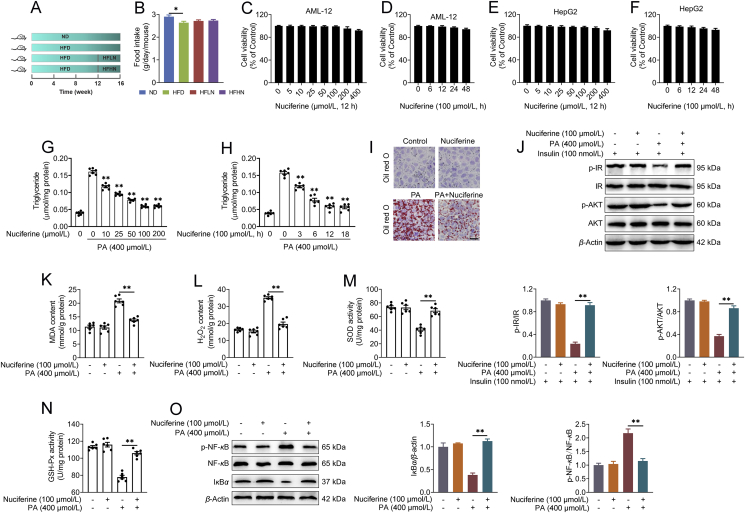
Nuciferine alleviates lipid accumulation, insulin resistance, oxidative stress and inflammatory response in the liver of high-fat diet (HFD) mice. Mice were fed either a normal diet (ND) or an HFD for 16 weeks. Nuciferine-treated mice were fed an HFD for 12 weeks and then fed an HFD containing low dose nuciferine (HFLN) or high dose nuciferine (HFHN) for another 4 weeks. (A) Chemical structure of nuciferine. (B–D) Body weight, liver weight and ratio of liver weight to body weight (LW/BW). (E) Gross anatomical views of representative mouse liver. Scale bar, 2 cm. (F) Representative images of H&E and Oil-red O staining of liver sections (original magnification 20 ×). (G, H) Hepatic and serum triglyceride contents. (I, J) Fasting blood glucose and insulin levels. (K) Homeostasis model assessment of insulin resistance index. (L, M) Glucose-tolerance test (GTT) and insulin-tolerance test (ITT). (N) Representative immunoblotting of insulin receptor (IR), phospho (p)-IR, protein kinase B (AKT) and p-AKT in the livers from different groups and quantification of p-IR/IR, p-AKT/AKT and expressed as fold change relative to ND group. (O, P) Hepatic malondialdehyde (MDA) and hydrogen peroxide (H2O2) contents. (Q, R) Hepatic superoxide dismutase (SOD) and glutathione peroxidase (GSH-Px) activities. (S) Representative immunoblotting of nuclear factor kappa B (NF-κB), p-NF-κB and IκBα in the livers from different groups and quantification of p-NF-κB/NF-κB and IκBα/β-actin and expressed as fold change relative to ND group. All experiments were repeated at least 3 times. n = 9 mice per group for B–D, G–M and O–R; n = 6 mice per group for N and S. Data were expressed as the mean ± SEM; ∗P < 0.05, ∗∗P < 0.01.
2. Materials and methods
2.1. Antibodies and reagents
Antibodies specific for phospho (p)-insulin receptor (IR)-Tyr1150/1151 (3024; 1:1000 WB), protein kinase B (AKT; 9272; 1:1000 WB), p-AKT-Ser473 (4060; 1:1000 WB), p-TFEB-Ser211 (37681; 1:1000 WB), TFEB (4240; 1:1000 WB; 1:100 IF), mechanistic target of rapamycin kinase (mTOR; 2983; 1:1000 WB; 1:200 IF), p70 S6 kinase (S6K; 9202; 1:1000 WB), p-S6K-Thr389 (9206; 1:1000 WB), EIF4E-binding protein 1 (4E-BP1; 9644; 1:1000 WB), p-4E-BP1-Ser65 (9456; 1:1000 WB), unc-51-like kinase 1 (ULK1; 6439; 1:1000 WB), p-ULK1-Ser757 (6888; 1:1000 WB), ribosomal protein S6 (S6; 2317; 1:1000 WB), p-S6-Ser240/244 (5364; 1:1000 WB), RagA (4357; 1:1000 WB), RagC (3360; 1:1000 WB; 1:400 IF), IκBα (4814; 1:1000 WB) and nuclear factor kappa B (NF-κB; 4764s; 1:1000 WB) were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibody specific for C7orf59 (NBP1-98263; 1:1000 WB) was purchased from Novus Biologicals (Littleton, CO, USA). Antibodies specific for IR (ab69508; 1:1000 WB), sequestosome 1 (P62; ab101266; 1:1000 WB), microtubule associated protein 1 light chain 3 (LC3; ab48394; 1:1000 WB), Histone H3 (ab1220; 1:1000 WB), lysosomal associated membrane protein 1 (LAMP1; ab24170; 1:1000 WB), p-NF-κB (ab86299; 1:2000 WB), β-actin (ab8226; 1:2000 WB), HRP-conjugated secondary antibodies to mouse (ab205719; 1:5000 dilution) and rabbit (ab205718; 1:5000 dilution) IgGs were purchased from Abcam (Cambridge, England). Antibodies specific for Flag (AE005; 1:2000 WB) and P18 (A11619; 1:1000 WB) were purchased from ABclonal (Wuhan, Hubei, China). Antibodies specific for β-tubulin (10068-1-AP; 1:5000 WB), cathepsin D (CTSD; 21327-1-AP; 1:2000 WB) and HA (66006-2-Ig; 1:10,000 WB) were purchased from Proteintech (Wuhan, Hubei, China). Antibodies specific for 14-3-3 (sc-133232; 1:200 WB), MP1 (sc-376783; 1:200 WB) and HBXIP (sc-373980, 1:200 WB) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). FITC-conjugated goat anti-rabbit IgG (111-095-003; 1:200 dilution) and Cyanine Cy™3-conjugated goat anti-rabbit IgG (111-165-003; 1:500 dilution) were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA, USA).
Nuciferine (B20500; greater than 98%, HPLC) was purchased from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China), dissolved in 1 mol/L hydrochloric acid and then the pH was adjusted to 7.5 by sodium hydroxide. Palmitic acid (PA; P0500; Sigma–Aldrich) was dissolved in 0.1 mol/L NaOH at 70 °C and then complexed with 10% bovine serum albumin (BSA) at 55 °C for 10 min to achieve the final palmitate concentration (100 mmol/L). Chloroquine (CQ, PHR1258) was purchased from Sigma–Aldrich. Torin1 (S2817) and leupeptin (S7380) were purchased from Selleck (Houston, TX, USA). LysoTracker Red DND-99 (50 nmol/L, 30 min) was used to stain the lysosome compartments (L7528, Thermo Fisher Scientific, Waltham, MA, USA). The siRNA targeting human Tfeb (5′-CAGGCUGUCAUGCAUUACATT-3′) was chemically synthesized by GenePharma (Shanghai, China). The non-targeting siRNA were purchased from GenePharma (A06001).
2.2. Cell culture and treatment
Primary hepatocytes were isolated from 6 to 8 weeks male TFEB-knockout mice by the collagenase perfusion method. Only hepatocytes with viability > 90%, as judged by Trypan blue exclusion, were used. The isolated mouse primary hepatocytes were cultured in Dulbecco's modified Eagle medium (DMEM) medium (HyClone, Thermo Scientific, USA) containing 10% fetal bovine serum (FBS, 10099141, Gibco, Grand Island, NY, USA), 100 nmol/L insulin, 100 nmol/L dexamethasone and pencillin/streptomycin for 5 h and then cultured in DMEM medium containing 10% FBS.
AML-12 cells, an immortalized normal mouse hepatocyte cell line, were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA, CRL-2254). The cells were cultured in DMEM/F12 supplemented with 10% FBS (10099141, Gibco), 1% insulin-transferrin-selenium (I3164, Sigma–Aldrich) and 1% penicillin–streptomycin at 37 °C under 5% CO2. HEK293T cells were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China, SCSP-502). The cells were cultured in DMEM medium containing 10% FBS (10099141, Gibco), 1% non-essential amino acids (N1250, Solarbio, Beijing, China), and 1% penicillin–streptomycin at 37 °C under 5% CO2. HepG2 human hepatocarcinoma cells (ATCC, HB-8065) were maintained in DMEM (SH30021.01, Thermo Fisher Scientific) supplemented with 10% FBS (10099141, Gibco) and 1% penicillin–streptomycin at 37 °C under 5% CO2. Mycoplasma contamination was negative for all cells.
To mimic in vivo hepatic steatosis, hepatocytes were maintained in medium containing 2% BSA and treated with 400 μmol/L PA for 12 h. To activate the insulin signaling pathway, hepatocytes were treated with 100 nmol/L insulin for 30 min. To study the function of nuciferine, the cells were treated with different concentrations of nuciferine (0, 10, 25, 50, 100 or 200 μmol/L) for 12 h, 100 μmol/L nuciferine for different time points (0, 3, 6, 12 or 18 h) or 100 μmol/L nuciferine for 12 h. To inhibit mTOR activity, the cells were treated with 250 nmol/L Torin1 for 1 h. For experiments which required amino acid deprivation, cells were rinsed twice with PBS and incubated for 1 h in amino acid-free DMEM (MBS6120661, MyBioSource, San Diego, CA, USA). To knockdown or overexpress a target gene, siRNA or an overexpression plasmid was transfected into cells using Lipofectamine 2000 (11668019, Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The cell treatments and detailed group information are shown in the figure legend.
2.3. Plasmids
The plasmid encoding full-length TFEB-GFP has previously been described34 and was used as template to amplify TFEB cDNA, which was subcloned into pc-DNA3.1-HA (128034, Addgene) to generate the pcDNA3.1-HA-TFEB. pLJM1-Flag-Raptor (26633), pLJM1-Flag-Raptor-RHEB15 (26634), pPK5-HA GST RagC-S75L (19305) and pRK5-HA GST RagC-WT (19304) were kind gifts from D. Sabatini (Addgene plasmids). pLJM1-Flag-P14 was a kind gift from Roberto Zoncu. Full-length cDNAs encoding human P18, MP1, C7orf59 and HBXIP were amplified by PCR and subcloned into the pcDNA3.1 vector (with Flag and 6 × His-tag) to generate the pcDNA3.1-Flag-6 × His-P18, pcDNA3.1-Flag-6 × His-MP1, pcDNA3.1-Flag-6 × His-C7orf59 and pcDNA3.1-Flag-6 × His-HBXIP. All constructs were checked by DNA sequencing.
2.4. Lentivirus production and transduction
Lentiviral production was performed as previously described35. Briefly, HEK293T cells were co-transfected with pLJM1-Flag-Raptor or pLJM1-Flag-Raptor-RHEB15, pCMV-VSV-G and pCMV-deltaR8.2 lentiviral packaging plasmids using Lipofectamine 2000 (11668019, Invitrogen). After 72 h of transfection, supernatants containing lentiviruses were collected and filtered through a 0.45 μm sterile filter to remove cell debris and used for infection in presence of 5 μg/mL polybrene (H8761, Solarbio). Twenty-four hours later, cells were treated and selected with puromycin (2 μg/mL, P8230, Solarbio).
2.5. Protein extraction, nucleus and cytoplasm separation, and Western blotting
Liver tissue and hepatocyte total proteins were extracted using a protein extraction kit (C510003; Sangon Biotech Co., Ltd., Shanghai, China). Nucleus and cytoplasm separation were conducted using a Nuclear-Cytosol Extraction Kit (P1200, Applygen Technologies). The protein concentration was estimated by the BCA method (P1511; Applygen Technologies). The samples were separated by SDS-PAGE and electrophoretically transferred to a polyvinylidene difluoride membrane. The membranes were blocked in 3% BSA/Tris-buffered saline/Tween buffer for 4 h. The blocked membrane was incubated overnight at 4 °C with the primary antibody. The membranes were then incubated with HRP-conjugated anti-rabbit or anti-mouse IgG at room temperature for 45 min. Immunoreactive bands were visualized by enhanced chemiluminescence solution (WBKLS0500, Millipore, Bedford, MA, USA). All bands were analyzed using Image-Pro Plus 6.0 (Media Cybernetics, Rockville, MD, USA). The Western blots shown are representative of 3 independent experiments with consistent results.
2.6. Co-immunoprecipitation
The cells were lysed with NP-40 buffer (P0013F, Beyotime, Beijing, China) supplemented with protease and phosphatase inhibitors. The supernatants were incubated with anti-Flag M2 affinity gel (A2220, Sigma–Aldrich) or anti-HA affinity gel (E6779, Sigma–Aldrich) overnight at 4 °C. After 5 washes, immunoprecipitation proteins were fractionated by SDS-PAGE and analyzed by Western blotting with the indicated primary antibodies.
2.7. Quantitative reverse transcription PCR assay (qRT-PCR)
The total RNA from hepatocytes and liver tissues was extracted using RNAiso Plus (D9108A, TaKaRa Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's instructions. The RNA concentration and quality were measured using a K5500 MicroSpectrophotometer (Beijing Kaiao Technology Development Co., Ltd., Beijing, China) and electrophoresis (1% agarose gels). Then, 1 μg of total RNA in each sample was reverse-transcribed to cDNA in a 20 μL reaction using a reverse transcription kit (RR047A, TaKaRa Biotechnology) according to the supplier's protocol. We evaluated mRNA expression using qRT-PCR technology with FastStart Universal SYBR Green Master Mix (04913850001; Roche Diagnostics, Mannheim, Germany) and a 7500 Real-Time PCR System (Applied Biosystems Inc., Foster City, CA, USA). The relative expression of each target gene was normalized to two reference genes, β-actin and glyceraldehyde-3-phosphate dehydrogenase and calculated using the 2−ΔΔCT method36,37. The primers used in this study are shown in Supporting Information Table S1. The cycles-to-threshold values of β-actin and Gapdh were not affected by the experimental treatment, which validated the usefulness of these genes as control genes.
2.8. RNA-sequencing
Total RNA was isolated from the fresh liver tissues using the RNAeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. The concentration and purity of RNA was determined using a NanoDrop microspectrophotometer (Thermo Fisher Scientific). The sequencing libraries were generated using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA), which consists of an mRNA purification process that uses poly-T beads, mRNA fragmentation, reverse transcription, end repair, the addition of a single ‘A’ base, ligation of the adapter, and purification and enrichment with PCR. The library fragments were purified using the AMPure XP system (Beckman Coulter, Bevely, CA, USA) and quantified using the Agilent High Sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent, Santa Clara, CA, USA). Sequencing was carried out using an Illumina Hiseq Xten platform (Illumina).
2.9. Cell viability, triglyceride content assessment and β-N-acetylglucosaminidase assay (NAG)
The cell viability was assessed by CCK-8 kit (CK04, Dojindo Co., Kumamoto, Japan) according to the manufacturer's instructions. Cells were seeded at 5 × 103 cells/well in 96-well plates incubated at 37 °C in 5% CO2. After treatment, 20 μL of CCK-8 was added to plates. Then the cells were incubated for an extra 4 h. The optical density was measured at 450 nm on a spectrophotometer (Thermo Fisher Scientific).
Liver tissues or hepatocytes were homogenized in 5% Triton-X100 and then heated in a water bath (85 °C) for 3 min. After cooling at room temperature, the sample was vortexed and centrifuged at 2000 × g for 5 min at 4 °C. The supernatant was collected and stored at −80 °C before the triglyceride assay. The triglyceride content was measured using an enzymatic kit (E1013; Applygen Technologies, Beijing, China) following the manufacturer's instructions. The total protein concentration was estimated by the BCA method (P1511; Applygen Technologies). The triglyceride content of the serum was measured using a triglyceride measurement kit (E1003, Applygen Technologies).
NAG performed using a kit from Sigma–Aldrich (CS0780). The assay is based on the hydrolysis of the NAG substrate, 4-nitrophenyl N-acetyl-β-d-glucosaminide, by the enzyme. This enzymatic hydrolysis of the substrate releases p-nitrophenol, which upon ionization in basic pH, can be measured colorimetrically at 405 nm. Firstly, cells were lysed in RIPA buffer. Then, ten micrograms from each sample were normalized to equal volumes and measured for NAG activity following the protocol provided by the supplier.
2.10. Oxidation and antioxidant marker analysis
Liver tissue and cultured cells were homogenized and lysed on ice in PBS and centrifuged (12,000 × g) for 15 min at 4 °C. The supernatant was used to analyze oxidation and antioxidant markers. The content of malondialdehyde (MDA) and hydrogen peroxide (H2O2) and the activities of superoxide dismutase (SOD) and glutathione peroxidase (GSH-Px) were measured using biochemical kits (H2O2, A064-1; MDA, A003; Nanjing Jian Cheng Institute of Bioengineering, Nanjing, China; SOD, S0101; GSH-Px, S0058; Beyotime Institute of Biotechnology), according to the manufacturer's instructions.
2.11. Lysosomes isolation
Lysosomes of liver tissues were isolated with lysosome isolation kit (LYSISO1, Sigma–Aldrich). Briefly, fresh liver tissues were homogenized in 4 volumes of 1 × extraction buffer containing protease inhibitors and centrifuged for 1000 × g for 10 min to yield post-nuclear supernatant. The supernatant was then centrifuged at 20,000 × g for 20 min at 4 °C to acquired crude lysosomal fractions. The crude lysosomal fractions were resuspended in 1× extraction buffer and adjusted to 19% Optiprep and layered over 22.5% Optiprep in a multistep Optiprep gradient consisting of 27%, 22.5%, 19%, 16%, 12% and 8% Optiprep according to the manufacturer's protocol and centrifuged for 4 h at 150,000 × g in a swing out rotor. The various layers formed at the junction of each gradient were collected after centrifugation. The enrichment of lysosomes was confirmed by measuring NAG activity and protein abundance of LAMP1.
2.12. Immunofluorescence assay
Immunofluorescence assays were performed as described previously38. The cells were fixed with 4% paraformaldehyde. Antigen retrieval was performed using EDTA-Na2 at 95 °C. The cells were permeabilized using 0.1% Triton X-100. For immunostaining, the cells were incubated with antibody diluted in PBS containing 5% goat serum overnight at 4 °C. The cells were then incubated with secondary antibodies conjugated to FITC or Cy3 (Jackson ImmunoResearch Laboratories). Subsequently, nuclei were stained with DAPI (Sigma–Aldrich). The cells were observed by laser confocal microscopy (Fluoview FV1200, OLYMPUS). In this experiment, at least three biological replicates of all the immunofluorescence assay were performed and three technical replicates were performed in each biological replicate. In addition, five independent fields and at least 20 random cells were observed from each technical replicate. Image-Pro Plus 6.0 software (Media Cybernetics) was used to quantify the signals.
2.13. Cellular thermal shift assay (CETSA)
The stabilization of targets in cells by nuciferine interaction was evaluated by CETSA as described previously39. For the temperature-dependent CETSA, cells were treated with or without 100 μmol/L nuciferine for 12 h. Cells were harvested and resuspended in PBS. The cells suspension was then aliquoted into 13 PCR tubes, heated for 3 min to 40, 44, 46, 48, 50, 52, 54, 56, 58, 62, 66, 70 or 74 °C followed by 3 cycles of freeze-thawing with liquid nitrogen and centrifugation at 20,000 × g for 20 min. The soluble fractions were collected, and equal amount of proteins were analyzed by SDS-PAGE followed by Western blotting analysis.
For the dose-dependent CETSA, cells were treated with various concentrations of nuciferine (0, 5, 10, 25, 50, and 100 μmol/L) for 12 h. Cells were harvested and resuspended in PBS. The cells suspension was heated for 3 min to 56 °C followed by 3 cycles of freeze–thawing with liquid nitrogen and centrifugation at 20,000 × g for 20 min. The soluble fractions were collected, and equal amount of proteins were analyzed by SDS-PAGE followed by Western blotting analysis.
2.14. Computational biology methods
The structure of HBXIP came from the Protein Data Bank (PDB code: 5YK3)40 and was used to the initial structure for molecular docking calculation by AutoDock Vina software41. The structure of nuciferine was optimized at the B3LYP/6-31G∗ level by using Gaussian 09 program, and molecular mechanical parameters of nuciferine was determinate based on the antechamber programs and RESP partial atomic charges from the AmberTools20 software. Subsequently, the standard docking procedures for HBXIP and nuciferine were performed using AutoDock vina software. To obtain the stable structure of HBXIP with nuciferine, the structure obtained from molecular docking was used as the initial structure, and the 30-ns standard molecular dynamics simulation was performed for the 3D structure of HBXIP with nuciferine using NAMD 2.1 software package42. The detailed docking process was referenced from previous studies43.
2.15. Animals and treatment
The Institutional Animal Care and Use Committee of Jilin University approved the study protocol (SY202004002, Changchun, China). All mice received humane care. All mice were kept in a standard environment with a 12-h dark/light cycle (lights on at 06:30 am). The temperature and humidity were maintained at 23 ± 3 °C and 35 ± 5%, respectively. Male 8- to 10-week-old C57BL/6N mice were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). TFEB flox mice were generated as described previously26 and were crossed with Albumin Cre (The Jackson Laboratory) to generated hepatocyte-specific TFEB knockout mice. Age matched litter mates from TFEB flox and Albumin Cre negative mice were used as control wild type mice for the knockout mice. Mice were fed a normal diet (ND, D12450B, 10% kcal from fat, Research Diet, New Brunswick, NJ, USA), a high-fat diet (HFD, D12492, 60% kcal from fat, Research Diet), or an HFD containing 0.01 or 0.03% nuciferine (w/w). To block autophagic flux in mouse livers, we treated mice with 40 mg/kg (i.p.) leupeptin for 8 h. The detailed groups and the number of mice included are shown in the figure legend.
2.16. Mouse glucose metabolic assay
After mice had been fasted, fasting blood glucose and insulin levels were determined using a glucometer (Abbott Diabetes Care Inc., Alameda, CA, USA) and enzyme-linked immunosorbent assay (Millipore, Billerica, MA, USA), respectively. The homeostasis model assessment of insulin resistance (HOMA-IR) index using the following mathematical formulation Eq. (1):
| (1) |
Glucose-tolerance test (GTT) was carried out on mice that had been fasted overnight for 16 h. After determination of fasted blood glucose levels, each mouse received an intraperitoneal injection of 2 g/kg body weight of glucose. Blood glucose level was detected from tail vein after 15, 30, 60, and 120 min. Insulin-tolerance tests (ITT) were carried out in random-fed mice. After measuring basal blood glucose levels, each mouse was treated with 0.75 U/kg body weight of insulin. Blood glucose level was recorded after 15, 30, 60, and 120 min.
2.17. H&E and Oil red O staining
Liver tissue was fixed in 10% formaldehyde neutral buffer solution, embedded in paraffin, cut into 8 μm sections and stained with H&E. For Oil red O staining, liver tissue was frozen in OCT compound (Sakura Finetek Co., Torrance, CA, USA), sectioned at an 8 μm thickness at −18 °C and fixed with 75% alcohol at room temperature for 15 min. Then, the slides were stained with Oil red O (O0625; Sigma–Aldrich) and counterstained with hematoxylin. For in vitro lipid staining, the Oil red O working solution was prepared by diluting Oil red O stock solution (5 μg Oil red O in 100 mL of isopropanol at 65 °C for 48 h) 3:2 with ddH2O. Treated cells were fixed in 4% paraformaldehyde for 15 min and then incubated for 5 min in 60% isopropanol. Subsequently, the cells were incubated for 30 min with Oil red O working solution and washed for 30 s in 60% isopropanol. The cells were washed four times with ddH2O and counterstained with hematoxylin, and images were acquired (Olympus, Japan).
2.18. Statistical analysis
Data were expressed as the mean ± standard error of mean (SEM). All analyses were performed using GraphPad Prism 8.0 (Graph Pad Software, San Diego, CA, USA) or Statistical Package for the Social Sciences (SPSS) 19.0 software (IBM, Chicago, IL, USA). Statistical significance was calculated using two-tailed Student's t-tests for comparisons between two groups and one-way ANOVA followed by post hoc test adjusted using Bonferroni correction for comparisons among more than two groups. P < 0.05 was considered statistically significant, and P < 0.01 was considered highly significant. All mice were randomized for treatment. Data analyses were blinded.
3. Results
3.1. Nuciferine attenuates HFD-induced hepatic steatosis, insulin resistance, oxidative stress and inflammatory response in mice
Mice were fed either a ND or an HFD for 16 weeks. Nuciferine-treated mice were fed an HFD for 12 weeks and then fed an HFD containing either low dose nuciferine (HFLN, 0.01% nuciferine) or high dose nuciferine (HFHN, 0.03% nuciferine) for a further 4 weeks (Supporting Information Fig. S1A). Food intake was not altered by nuciferine (Fig. S1B). Nuciferine administration markedly reduced increases in body weight (Fig. 1B), liver weight (Fig. 1C), and ratio of liver weight to body weight (LW/BW, Fig. 1D) in HFD-fed mice. Gross anatomy (Fig. 1E), H&E and Oil red O staining (Fig. 1F) and triglyceride analysis (Fig. 1G) reveal that nuciferine administration markedly mitigated hepatic steatosis in HFD mice. Furthermore, nuciferine-treated HFD mice displayed lower fasting blood concentrations of triglyceride (Fig. 1H), glucose (Fig. 1I) and insulin (Fig. 1J) as well as HOMA-IR (Fig. 1K) than HFD mice. Tolerance tests for glucose and insulin further verified improved glucose tolerance (Fig. 1L) and insulin sensitivity (Fig. 1M) in nuciferine-treated HFD mice. In accordance, the phosphorylation of key factors in insulin signaling, IR and AKT, was much higher in the livers of nuciferine-treated HFD mice than those of HFD mice (Fig. 1N). Moreover, nuciferine administration alleviated HFD-induced hepatic oxidative stress and inflammatory response, as evidenced by decreased MDA and H2O2 contents (Fig. 1O and P) and protein abundance of p-NF-κB (Fig. 1S), and increased activities of SOD and GSH-Px (Fig. 1Q and R) as well as protein abundance of IκBα (Fig. 1S). Although both low and high dose nuciferine conditions decreased lipid accumulation and improved insulin resistance, oxidative stress and inflammatory response of HFD mice, high dose nuciferine was more effective than low dose (Fig. 1B–S). In vitro results reveal that nuciferine mitigated PA-induced lipid accumulation in a dose and time-dependent manner (Fig. S1G and H). Furthermore, 100 μmol/L nuciferine, which showed no cytotoxic activity (Fig. S1C–S1F), significantly alleviated PA-induced lipid accumulation (Fig. S1I), insulin signaling impairment (Fig. S1J), oxidative stress (Fig. S1K–S1N) and inflammatory response (Fig. S1O) in AML-12 cells. These data indicate that nuciferine alleviates lipid accumulation, insulin resistance, oxidative stress and inflammation in the livers of HFD mice and PA-treated hepatocytes.
3.2. ALP is involved in the beneficial effects of nuciferine on NAFLD
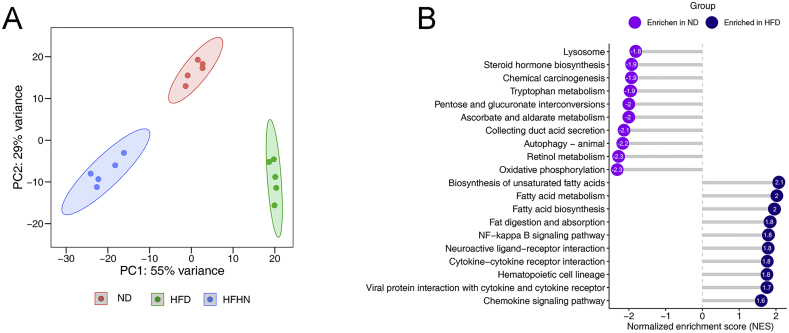
To determine the specific impact of nuciferine on NAFLD at the molecular level, we performed transcriptomic analysis in liver tissues obtained from ND, HFD and HFHN mice. Principal component analysis revealed that the replicates of ND, HFD and HFHN showed good clustering within their groups (Supporting Information Fig. S2A). Compared with ND group, an HFD upregulated 573 genes and downregulated 248 genes, whereas nuciferine treatment reversed 361 of the 573 upregulated genes and 125 of the 248 downregulated genes induced by an HFD (Fig. 2A; Supporting Information Table S2). Gene set enrichment analysis revealed that the genes affected by nuciferine treatment were enriched for autophagy, lysosome, and lipid metabolism (Fig. 2B, C and Fig. S2B). qRT-PCR analysis further verifies the augment in autophagy regulated genes, lysosomal function and biogenesis genes, lipid oxidation genes and the reduction in lipid synthesis genes induced by nuciferine (Fig. 2D). Given the positive effects of ALP on reducing hepatic steatosis4,7,8, these data suggest that ALP induction is involved in the beneficial role of nuciferine treatment on NAFLD.
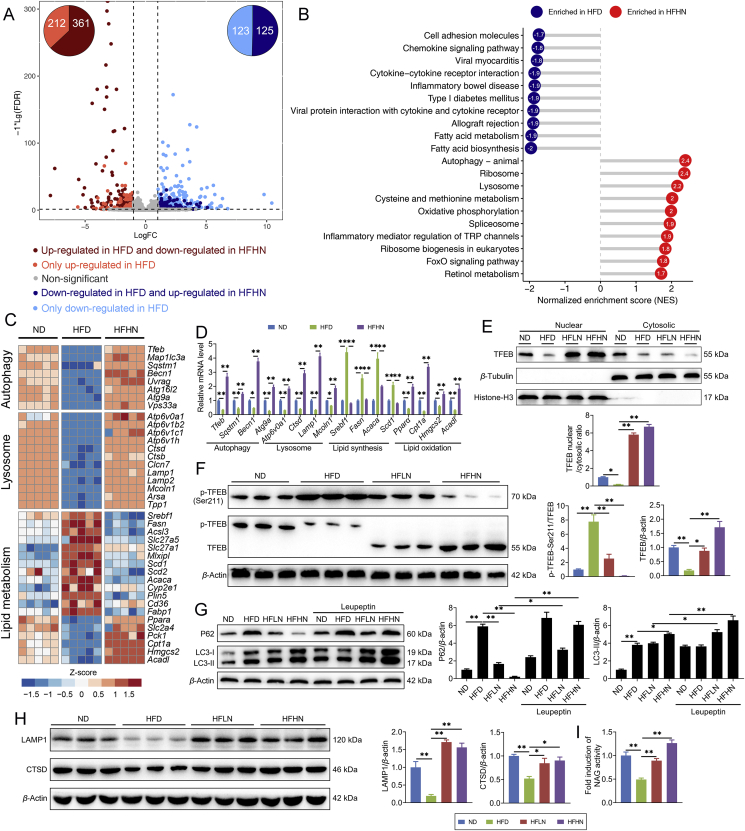
Figure 2.
Nuciferine activates hepatic autophagy–lysosomal pathway (ALP) in NAFLD mice. (A) The scatterplot showing fold change of all genes in the RNA-seq dataset, with differentially expressed genes shown with corresponding font color. The pie charts indicate the number of differentially expressed genes regulated by nuciferine and HFD feeding. (B) Gene set enrichment analysis showing the cellular pathways, which were upregulated by HFD but downregulated by nuciferine treatment. (C) Heatmap showing the expression profile of genes related to autophagy, lysosome and lipid metabolism in different groups. (D) Relative mRNA levels of the indicated genes were normalized to β-actin and Gapdh and expressed as fold change relative to ND group. (E) Representative immunoblotting of nuclear and cytosolic TFEB in the livers from different groups and quantification of TFEB nuclear/cytosolic ratio and expressed as fold change relative to ND group. (F) Representative immunoblotting of transcription factor EB (TFEB) and p-TFEB-Ser211 in the livers from different groups and quantification of p-TFEB-Ser211/TFEB, TFEB/β-actin and expressed as fold change relative to ND group. (G) Representative immunoblotting of sequestosome 1 (P62) and microtubule associated protein 1 light chain 3 (LC3) in the livers from different groups and quantification of P62/β-actin, LC3-II/β-actin and expressed as fold change relative to ND group. Mice were treated as described in Fig. 1 and sacrificed 8 h after intraperitoneal injection of leupeptin (40 mg/kg). (H) Representative immunoblotting of lysosomal associated membrane protein 1 (LAMP1) and cathepsin D (CTSD) in the livers from different groups and quantification of LAMP1/β-actin, CTSD/β-actin and expressed as fold change relative to ND group. (I) Relative lysosomal NAG activity in the livers from different groups. All experiments were repeated at least 3 times. n = 5 mice per group for A–D; n = 6 mice per group for E–H; n = 9 mice per group for I. Data were expressed as the mean ± SEM; ∗P < 0.05, ∗∗P < 0.01.
3.3. Nuciferine activates TFEB and ALP in vivo and in vitro
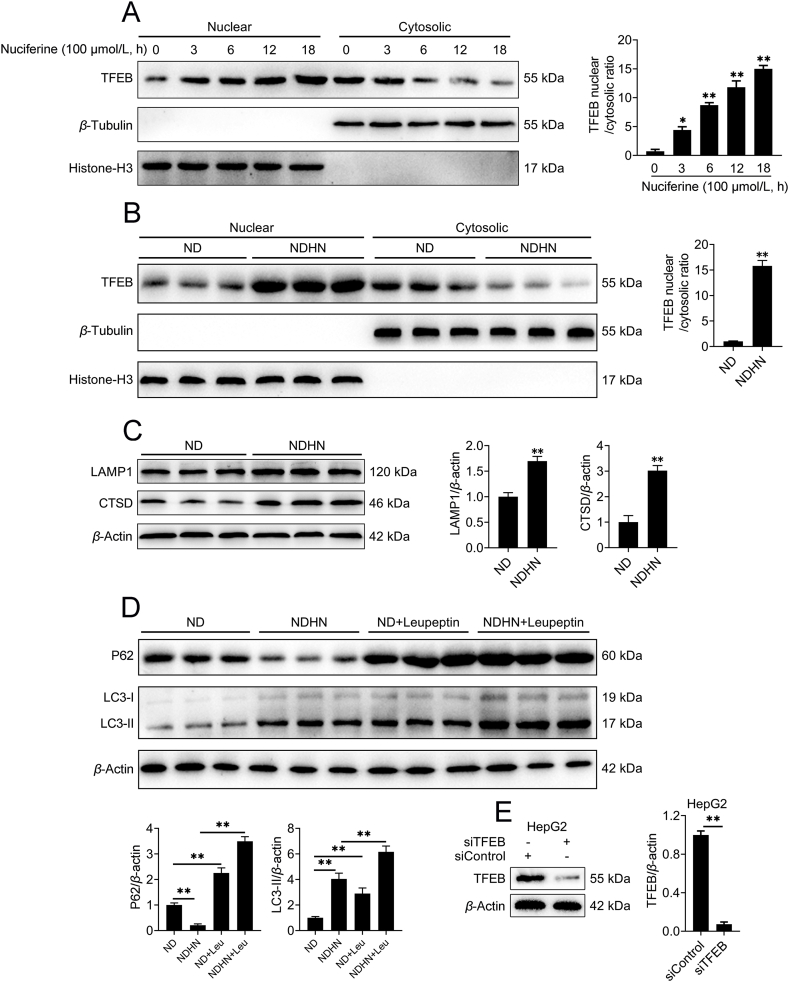
TFEB controls autophagosome and lysosome biogenesis by upregulating a family of genes belonging to the coordinated lysosomal expression and regulation network10. Nuciferine enhanced the ratio of nuclear to cytosolic TFEB (Fig. 2E), increased protein abundance and electrophoretic mobility of TFEB (Fig. 2F) and decreased phosphorylation of TFEB at Ser211 (Fig. 2F) in HFD mice. Furthermore, by using the lysosomal protease inhibitor leupeptin, we found that nuciferine administration increased the degradation of P62 and formation of LC3-II in HFD mice (Fig. 2G), indicating that nuciferine elevated hepatic autophagic flux. Besides autophagy, nuciferine treatment enhanced lysosomal function, as indicated by elevated abundance of both LAMP1 (Fig. 2H), a lysosomal membrane marker44, and CTSD (Fig. 2H), one of the most abundant lysosomal proteases44, as well as increased lysosomal protease activities, as measured by NAG assay (Fig. 2I). These data demonstrate that nuciferine activates TFEB and ALP in the liver of HFD mice.
To further corroborate the role of nuciferine on TFEB and ALP activation, we treated HepG2 cells with nuciferine and found that nuciferine increased nuclear localization of TFEB in a dose- and time-dependent manner with a peak response at 100 μmol/L and 12 h (Fig. 3A, B and Supporting Information Fig. S3A). In addition, nuciferine administration induced TFEB nuclear translocation in the liver of ND mice (Fig. S3B). Similar to treatment with the mTOR inhibitor Torin1, nuciferine dramatically enhanced lysosomal function, as indicated by the elevated CTSD and LAMP1 protein abundance (Fig. 3C), NAG activity (Fig. 3D) and LysoTracker staining (Fig. 3E). Moreover, we observed an augment in P62 and LC3-II protein abundance in cells treated with nuciferine and autophagic flux inhibitor CQ versus nuciferine alone (Fig. 3F), indicating that nuciferine induced autophagosome formation and lysosomal degradation in HepG2 cells. Moreover, enhanced lysosomal function and autophagy were also observed in the liver of ND mice-treated with nuciferine (Fig. S3C and S3D). These in vitro and in vivo results further underscore the activation role of nuciferine on TFEB and ALP.
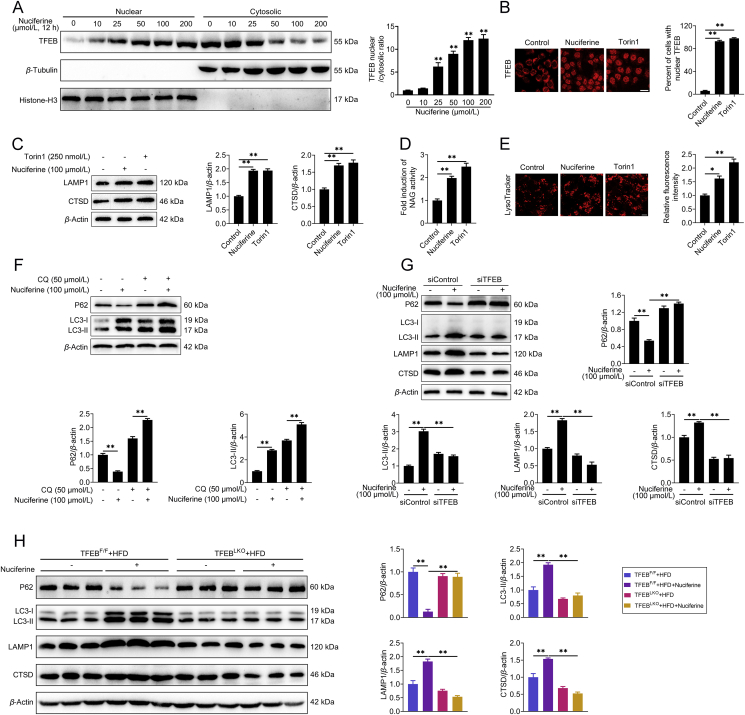
Figure 3.
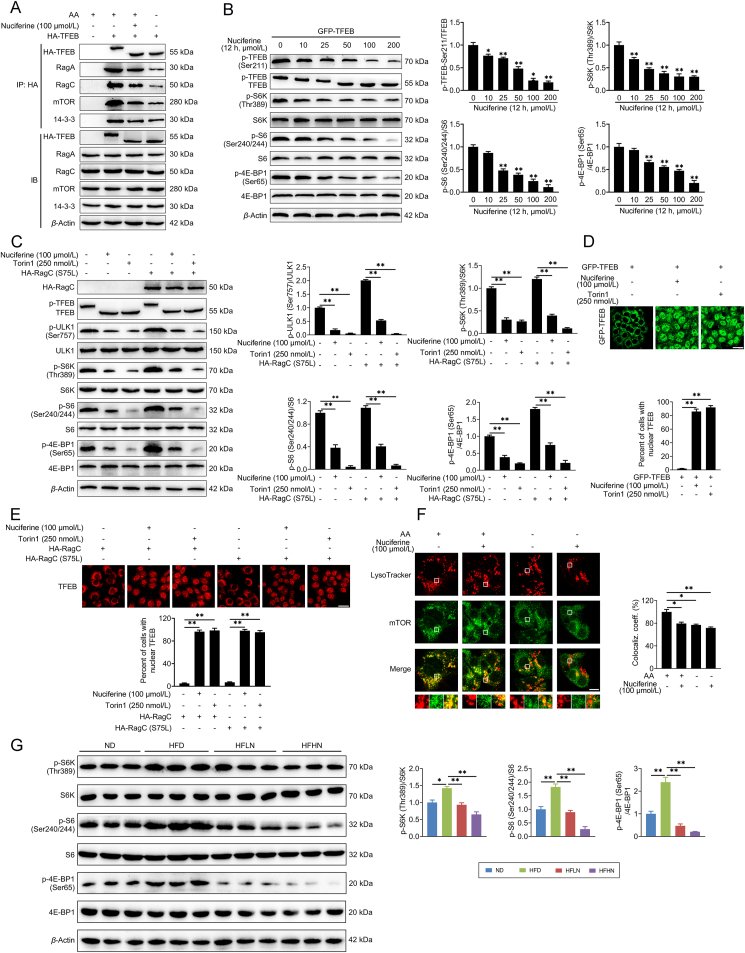
Nuciferine activates ALP via TFEB. (A) Representative immunoblotting of nuclear and cytosolic TFEB in HepG2 cells and quantification of TFEB nuclear/cytosolic ratio and expressed as fold change relative to control group. HepG2 cells were treated with different concentrations of nuciferine (0, 10, 25, 50, 100 or 200 μmol/L) for 12 h. (B–E) HepG2 cells were treated with 100 μmol/L nuciferine for 12 h or 250 nmol/L Torin1 for 1 h. (B) HepG2 cells were analyzed by immunofluorescence and quantified to calculate the percentage of cells showing TFEB unclear localization. Scale bar, 20 μm. (C) Representative immunoblotting of LAMP1 and CTSD in HepG2 cells and quantification of LAMP1/β-actin, CTSD/β-actin and expressed as fold change relative to control group. (D) Relative lysosomal NAG activity. (E) Representative images of LysoTracker staining in HepG2 cells and quantification of fluorescence intensity and expressed as fold change relative to control group. Scale bar, 20 μm. (F) Representative immunoblotting of P62 and LC3 in HepG2 cells and quantification of P62/β-actin, LC3-II/β-actin and expressed as fold change relative to control (no treatment) group. HepG2 cells were pre-treated with 50 μmol/L chloroquine for 4 h and then treated with or without 100 μmol/L nuciferine for 12 h. (G) Representative immunoblotting of P62, LC3, CTSD and LAMP1 in HepG2 cells and quantification of P62/β-actin, LC3-II/β-actin, LAMP1/β-actin, CTSD/β-actin and expressed as fold change relative to control (transfection with siControl alone) group. HepG2 cells were transfected with siTFEB or siControl, then treated with or without 100 μmol/L nuciferine for 12 h. (H) Representative immunoblotting of P62, LC3, CTSD and LAMP1 in the livers from different groups and quantification of P62/β-actin, LC3-II/β-actin, LAMP1/β-actin, CTSD/β-actin and expressed as fold change relative to control (HFD-fed TFEBF/F mice) group. Mice were fed an HFD for 16 weeks. Nuciferine-treated mice were fed an HFD for 12 weeks and then fed an HFD containing 0.03% nuciferine for another 4 weeks. n = 6 mice per group were used to analyze the results. All experiments were repeated at least 3 times. Data were expressed as the mean ± SEM; ∗P < 0.05, ∗∗P < 0.01.
3.4. TFEB is required for the activation effects of nuciferine on ALP
We next determined to what extent TFEB is required for the induction effects of nuciferine on ALP and found that knockdown of TFEB (Fig. S3E) abolished nuciferine-induced enhancement in P62 degradation, LC3-II formation, protein abundance of CTSD and LAMP1 in HepG2 cells (Fig. 3G), indicating that TFEB is required in the process of nuciferine-activated ALP. Importantly, in hepatocyte-specific TFEB-knockout mice fed with HFD (Supporting Information Fig. S4), nuciferine did not appreciably affect the hepatic autophagic activity, as measured by protein abundance of P62 and LC3-II (Fig. 3H), nor the lysosomal function, as indicated by protein abundance of CTSD and LAMP1 (Fig. 3H). Taken together, these results indicate that the activation effects of nuciferine on ALP are mediated by TFEB.
3.5. Knockout of hepatic TFEB blocks the beneficial effects of nuciferine on NAFLD
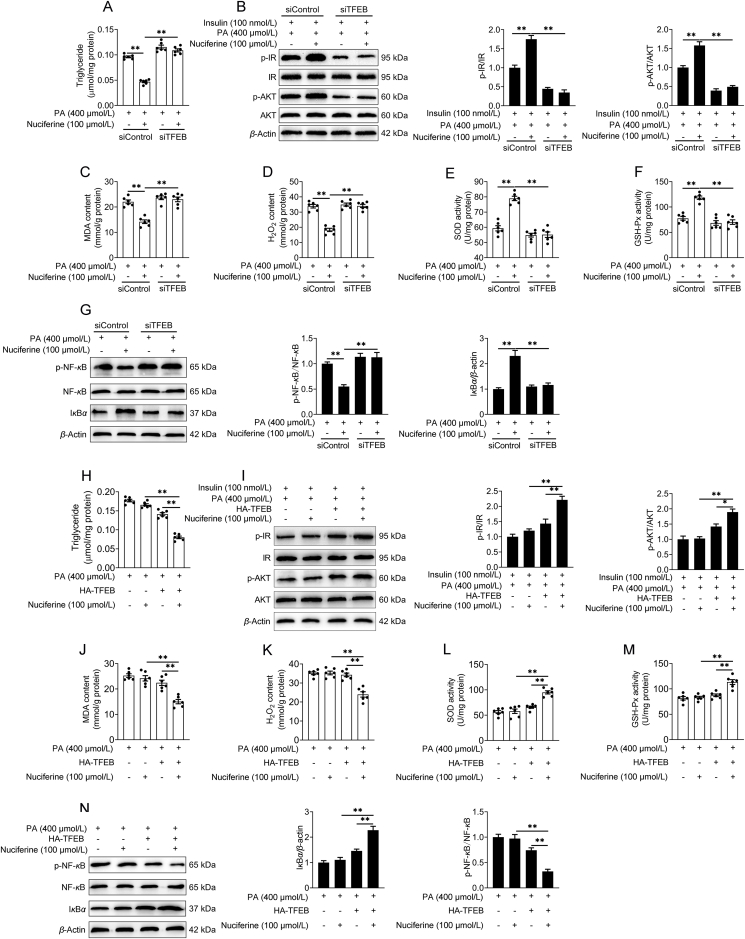
We further evaluated the role of the TFEB-mediated ALP in the beneficial effects of Nuciferine on NAFLD. In vitro, knockdown of TFEB impeded the improvement effects of Nuciferine on lipid accumulation (Supporting Information Fig. S5A), insulin resistance (Fig. S5B), oxidative stress (Fig. S5C–S5F) and inflammatory response (Fig. S5G) in HepG2 cells challenged by PA. More importantly, in hepatocyte-specific TFEB-knockout mice, nuciferine failed to preclude HFD-increased body weight (Fig. 4A), liver weight (Fig. 4B), ratio of LW/BW (Fig. 4C) and hepatic steatosis (Fig. 4D and E). Furthermore, nuciferine had no effects on fasting blood concentrations of triglyceride (Fig. 4F), glucose (Fig. 4G) and insulin (Fig. 4H), HOMA-IR (Fig. 4I), glucose tolerance (Fig. 4J) as well as insulin sensitivity (Fig. 4K and L). Consistently, the protective effects of nuciferine against oxidative stress (Fig. 4M–P) and inflammatory response (Fig. 4Q) were diminished by knockout of TFEB in the liver of HFD mice. Importantly, enforced expression of TFEB in TFEB-knockout hepatocytes restored the positive effects of nuciferine on lipid accumulation (Fig. S5H), insulin resistance (Fig. S5I), oxidative stress (Fig. S5J–S5M) and inflammatory response (Fig. S5N). These data indicate that TFEB-dependent induction of ALP is mainly responsible for the beneficial effects mediated by nuciferine on NAFLD.
Figure 4.
TFEB mediates the beneficial effects of nuciferine. Mice were fed an HFD for 16 weeks. Nuciferine-treated mice were fed an HFD for 12 weeks and then fed an HFD containing 0.03% nuciferine for another 4 weeks. (A–C) Body weight, liver weight and LW/BW ratio. (D) Representative images of H&E and Oil-red O staining of liver sections (original magnification 20 ×). (E, F) Hepatic and serum triglyceride contents. (G, H) Fasting blood glucose and insulin levels. (I) Homeostasis model assessment of insulin resistance index. (J, K) GTT and ITT. (L) Representative immunoblotting of IR, p-IR, AKT and p-AKT in the livers from different groups and quantification of p-IR/IR, p-AKT/AKT and expressed as fold change relative to control (HFD-fed TFEBF/F mice) group. (M, N) Hepatic MDA and H2O2 contents. (O, P) Hepatic SOD and GSH-Px activities. (Q) Representative immunoblotting of NF-κB, p-NF-κB and IκBα in the livers from different groups and quantification of p-NF-κB/NF-κB and IκBα/β-actin and expressed as fold change relative to control (HFD-fed TFEBF/F mice) group. All experiments were repeated at least 3 times. n = 9 mice per group for A–C, E–K and M–P; n = 6 mice per group for L and Q. Data were expressed as the mean ± SEM; ∗P < 0.05, ∗∗P < 0.01.
3.6. Nuciferine triggers TFEB nuclear translocation in a mTORC1-dependent manner
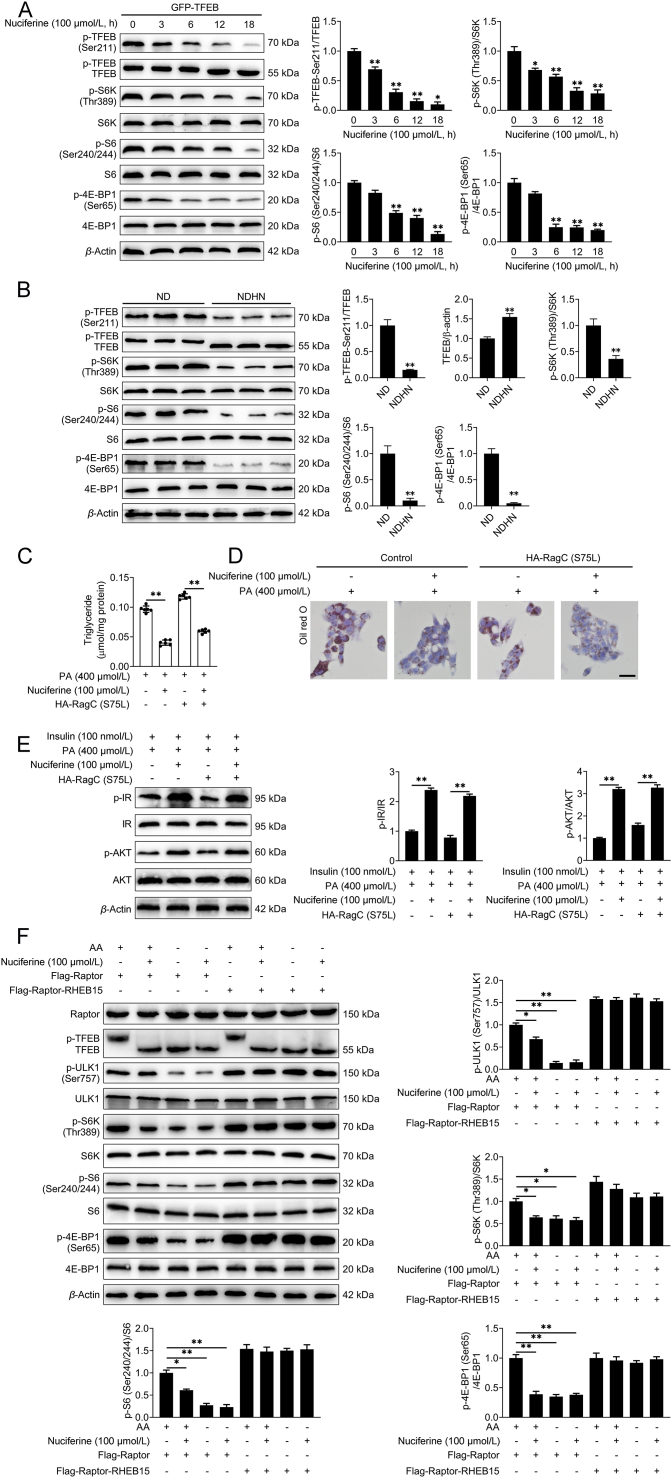
mTORC1 phosphorylates TFEB at Ser211 on the surface of lysosome and induces a binding event between TFEB and 14-3-3 proteins, resulting in the cytoplasmic retention of TFEB11, 12, 13. This phosphorylation event is mediated by the Rags, which, once activated by nutrients, promote mTOR lysosomal recruitment while physically interact with TFEB to mediate its phosphorylation by mTORC135,45. We found that nuciferine administration decreased the interaction of TFEB with both RagA and RagC, as well as its interaction with mTOR and 14-3-3 (Fig. 5A), suggesting that nuciferine promotes TFEB nuclear localization by impairing its interaction with Rags and hence inhibiting its phosphorylation by mTORC1. Accordingly, nuciferine treatment suppressed TFEB phosphorylation at Ser211 (Fig. 5B, Supporting Information Fig. S6A and S6B), increased electrophoretic mobility of TFEB protein (Fig. 5B, C, Fig. S6A and S6B) and trigged TFEB nuclear localization (Figure 2, Figure 3, Figure 5D, E, Fig. S3A and S3B) in vivo and in vitro. In addition, nuciferine treatment reduced the lysosomal localization of mTOR (Fig. 5F) and diminished the phosphorylation of mTORC1 substrates in HEK293T and HepG2 cells, and in the liver of HFD and ND mice (Fig. 5B, C, G, Fig. S5A and S5B), suggesting that nuciferine-induced TFEB nuclear translocation is mediated by inhibition of mTORC1 activity.
Figure 5.
Nuciferine inhibits mTORC1 activity and decreases TFEB phosphorylation. (A) Representative co-immunoprecipitation (Co-IP) analysis to assay interactions between HA-TFEB and RagA, RagC, mTOR and 14-3-3 in HepG2 cells. HepG2 cells were transfected with HA-TFEB and treated with 100 μmol/L nuciferine for 12 h or starved of amino acids (AA) for 1 h. (B) Representative immunoblotting of p-TFEB-Ser211, GFP-TFEB, p70 S6 kinase (S6K), p-S6K-Thr389, ribosomal protein S6 (S6), p-S6-Ser240/244, EIF4E-binding protein 1 (4E-BP1) and p-4E-BP1-Ser65 in HEK293T cells and quantification of p-TFEB-Ser211/TFEB, p-S6K-Thr389/S6K, p-S6-Ser240/244/S6, p-4E-BP1-Ser65/4E-BP1 and expressed as fold change relative to control (no treatment) group. HEK293T cells were transfected with GFP-TFEB, and then treated with different concentrations of nuciferine (0, 10, 25, 50, 100 or 200 μmol/L) for 12 h. (C) Representative immunoblotting of HA-RagC, TFEB, unc-51-like kinase 1 (ULK1), p-ULK1-Ser757, S6K, p-S6K-Thr389, S6, p-S6-Ser240/244, 4E-BP1 and p-4E-BP1-Ser65 in HepG2 cells and quantification of p-ULK1-Ser757/ULK1, p-S6K-Thr389/S6K, p-S6-Ser240/244/S6, p-4E-BP1-Ser65/4E-BP1 and expressed as fold change relative to control (no treatment) group. HepG2 cells were transfected with or without active RagC (S75L), and then treated with 100 μmol/L nuciferine for 12 h or 250 nmol/L Torin1 for 1 h. (D) HEK293T cells were analyzed by GFP-TFEB fluorescence and quantified to calculate the percentage of cells showing TFEB unclear localization. Scale bar, 20 μm. HEK293T cells were transfected with GFP-TFEB, and then treated with 100 μmol/L nuciferine for 12 h or 250 nmol/L Torin1 for 1 h. (E) HepG2 cells were analyzed by immunofluorescence and quantified to calculate the percentage of cells showing TFEB unclear localization. Scale bar, 20 μm. Cells were transfected with RagC or active RagC (S75L), and then treated with 100 μmol/L nuciferine for 12 h or 250 nmol/L Torin1 for 1 h. (F) Representative images of mTOR immunofluorescence staining and LysoTracker in HepG2 cells and quantification of colocalization coefficient. HepG2 cells were starved of AA for 1 h in the presence or absence of 100 μmol/L nuciferine. Scale bar, 10 μm. (G) Representative immunoblotting of S6K, p-S6K-Thr389, S6, p-S6-Ser240/244, 4E-BP1 and p-4E-BP1-Ser65 in the livers from different groups and quantification of p-S6K-Thr389/S6K, p-S6-Ser240/244/S6, p-4E-BP1-Ser65/4E-BP1 and expressed as fold change relative to ND group. Mice were treated as described in Fig. 1. n = 6 mice per group were used to analyze the results. All experiments were repeated at least 3 times. Data were expressed as the mean ± SEM; ∗P < 0.05, ∗∗P < 0.01.
Since activation of the Rag heterodimers is necessary for their binding to both mTORC1 complex and TFEB, we hypothesized that nuciferine hampered Rags activation in presence of nutrients. However, exogenous expression of a constitutive active RagC (S75L) was not able to restore cytosolic localization of TFEB (Fig. 5E) nor its phosphorylation or the ones of the other mTORC1 substrates (e.g., ULK1, S6K, S6 and 4E-BP1; Fig. 5C). Furthermore, overexpression of active RagC (S75L) did not affect the efficacy of nuciferine on lipid accumulation (Fig. S6C and S6D) and insulin sensitivity (Fig. S6E) in PA-challenged HepG2 cells. These data suggest that nuciferine inhibits Rags binding to TFEB, as well as their ability to recruit mTOR at the lysosome, but these effects are not dependent on Rags activation state.
3.7. Nuciferine disrupts Rag–Ragulator interaction and inhibits mTORC1 activity by interacting with HBXIP
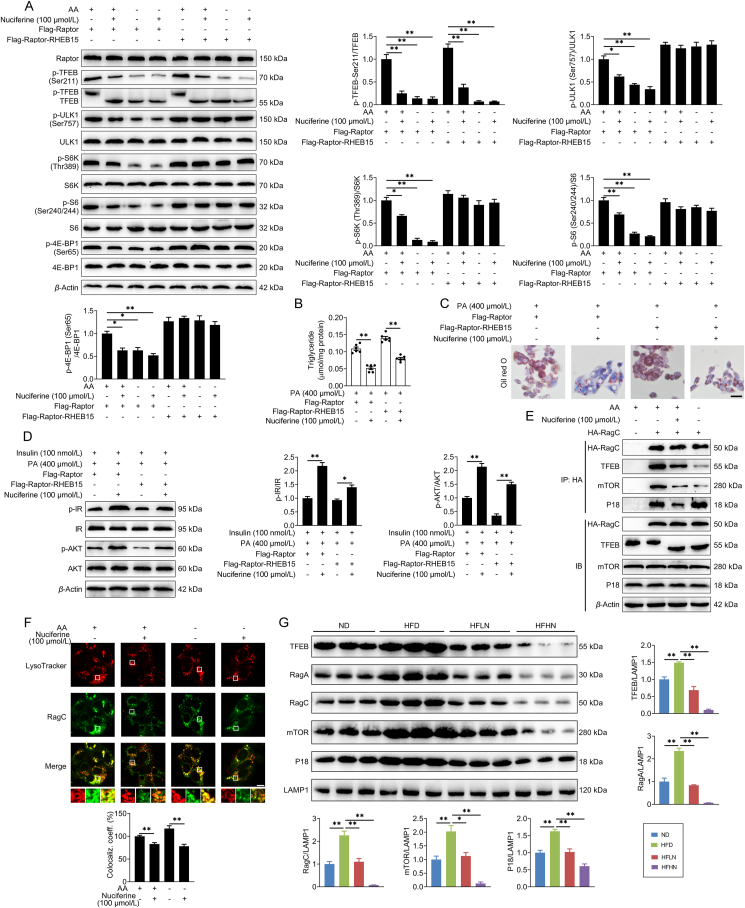
It has been reported that mTORC1-dependent phosphorylation of TFEB is highly sensitive to Rags but insensitive to RHEB35. We found that enforced expression of Raptor–RHEB15, which induces constitutive lysosomal recruitment and Rag-independent activation of mTORC118, did not hamper nuciferine-mediated TFEB activation, but fully rescued the phosphorylation of the other mTORC1 substrates in HEK293T (Fig. 6A) and HepG2 cells (Fig. S6F), supporting the hypothesis that nuciferine inhibits Rags-mediated lysosomal recruitment of mTORC1 complex. Consistently, overexpression of Raptor–RHEB15 did not hamper the beneficial effect of nuciferine treatment on lipid accumulation and insulin resistance in hepatocytes treated with PA (Fig. 6B–D). Similar to amino acid deprivation, which inhibits the interaction of Rags with mTORC115,16, nuciferine administration lessened the interaction of RagC with mTOR and TFEB (Fig. 6E) and decreased interaction of TFEB with RagA and RagC (Fig. 5A). Notably, nuciferine treatment decreased lysosomal localization of RagC in HepG2 cells both in the presence or absence of amino acid (Fig. 6F) and reduced lysosomal localization of RagA, RagC, mTOR and TFEB in the liver of NAFLD mice (Fig. 6G). Thus, nuciferine inhibits mTORC1 activity by reducing the lysosomal localization of Rags.
Figure 6.
Nuciferine disrupts Rag GTPases–Ragulator interaction and inhibits mTORC1 activity. (A) Representative immunoblotting of Raptor, TFEB, p-TFEB-Ser211, ULK1, p-ULK1-Ser757, S6K, p-S6K-Thr389, S6, p-S6-Ser240/244, 4E-BP1 and p-4E-BP1-Ser65 in HEK293T cells and quantification of p-TFEB-Ser211/TFEB, p-ULK1-Ser757/ULK1, p-S6K-Thr389/S6K, p-S6-Ser240/244/S6, p-4E-BP1-Ser65/4E-BP1 and expressed as fold change relative to control (transfection with lentiviruses expressing Flag-Raptor in the presence of AA) group. HEK293T cells were transfected with lentiviruses expressing Flag-Raptor or Flag-Raptor-RHEB15, then transfected with GFP-TFEB and starved of AA for 1 h in the presence or absence of 100 μmol/L nuciferine. (B–D) HepG2 cells were transfected with lentiviruses expressing Flag-Raptor or Flag-Raptor-RHEB15 and maintained in medium containing 2% bovine serum albumin and treated with 400 μmol/L palmitic acid (PA) for 12 h, then treated with or without 100 μmol/L nuciferine for 12 h. (B) The triglyceride content in HepG2 cells. (C) Representative images of Oil red O (original magnification 40 ×) staining in HepG2 cells. (D) Representative immunoblotting of IR, p-IR, AKT and p-AKT in HepG2 cells and quantification of p-IR/IR, p-AKT/AKT and expressed as fold change relative to control (transfection with lentiviruses expressing Flag-Raptor and treatment with PA and insulin) group. Cells were treated with 100 nmol/L insulin for 30 min before they were harvested. (E) Representative Co-IP analysis to assay interactions between HA-RagC and TFEB, mTOR and P18 in HepG2 cells. HepG2 cells were transfected with HA-RagC and treated with 100 μmol/L nuciferine for 12 h or starved of AA for 1 h. (F) Representative images of RagC immunofluorescence staining and LysoTracker in HepG2 cells and quantification of colocalization coefficient. HepG2 cells were starved of AA for 1 h in the presence or absence of 100 μmol/L nuciferine. Scale bar, 10 μm. (G) Representative immunoblotting of TFEB, RagA, RagC, mTOR, P18 and LAMP1 in lysosomes from liver tissues and quantification of TFEB/LAMP1, RagA/LAMP1, RagC/LAMP1, mTOR/LAMP1, P18/LAMP1 and expressed as fold change relative to ND group. Mice were treated as described in Fig. 1. n = 6 mice per group were used to analyze the results. All experiments were repeated at least 3 times. Data were expressed as the mean ± SEM; ∗P < 0.05, ∗∗P < 0.01.
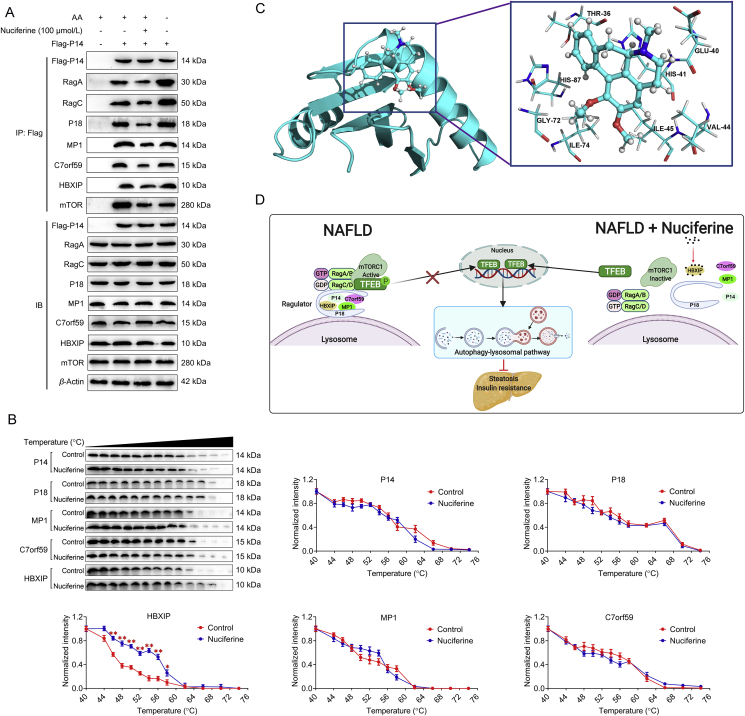
Lysosomal anchoring of the Rags through the Ragulator complex is necessary to mediate mTORC1 lysosomal recruitment and hence activation18,46. Our results indicate that nuciferine weakened the interaction between Rags and Ragulator, as evidenced by reduced interactions between RagC and P18 (Fig. 6E) and between P14 and RagA and RagC (Fig. 7A). Moreover, nuciferine also reduced the interaction of P14 with P18, MP1, C7orf59 and HBXIP (Fig. 7A), indicating that the interactions between components of Ragulator complex are also disrupted by nuciferine.
Figure 7.
Nuciferine binds to HBXIP. (A) Representative Co-IP analysis to assay interactions between Flag-P14 and RagA, RagC, P18, MP1, C7orf59, hepatitis B X-interacting protein (HBXIP) and mTOR in HepG2 cells. HepG2 cells were transfected with Flag-P14 and treated with 100 μmol/L nuciferine for 12 h or starved of AA for 1 h. (B) Cellular thermal shift assay of P14, P18, MP1, C7orf59 and HBXIP in HEK293T cells. Cells were treated with or without100 μmol/L nuciferine for 12 h. (C) The complex structure of HBXIP with nuciferine and the binding pocket with nuciferine. (D) Proposed model for the beneficial effects of nuciferine on NAFLD. Nuciferine interacts with HBXIP, blocks the function of Ragulator complex and reduces lysosomal localization of Rag GTPases and mTORC1, which activates TFEB-mediated ALP and further improves hepatic steatosis and insulin resistance in mice with NAFLD (created with BioRender.com.). All experiments were repeated at least 3 times. Data were expressed as the mean ± SEM; ∗P < 0.05, ∗∗P < 0.01.
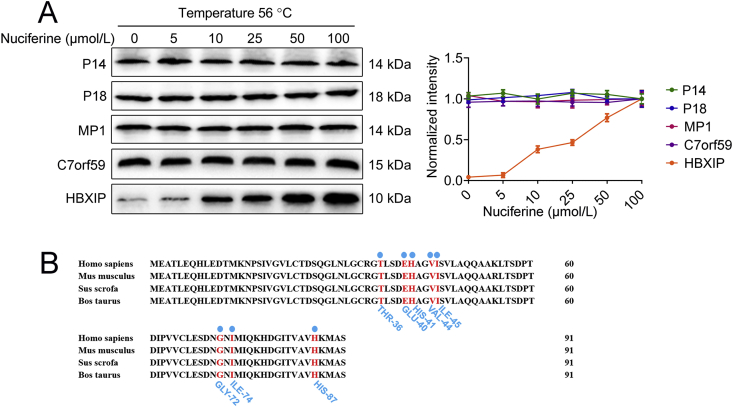
Considering the above results, we wondered whether nuciferine binds to Ragulator complex, thus altering the correct assembly and function of the complex. Both temperature- and dose-dependent CETSA demonstrated that HBXIP was physically engaged and stabilized against thermal changes in nuciferine-treated cells and the effect of nuciferine on thermal stability of HBXIP was stronger than P14, P18, MP1 and C7orf59 (Fig. 7B and Supporting Information Fig. S7A), supporting that there is a direct interaction between nuciferine and HBXIP. Furthermore, molecular docking analysis revealed that nuciferine may bind to the potential binding pocket of HBXIP, notably residues THR-36, GLU-40, HIS-41, VAL-44, ILE-45, GLY-72, ILE-74 and HIS-87 made favorable contributions to the ligand-protein binding interaction (Fig. 7C). Moreover, these binding sites were highly conserved, as shown by the sequence alignment of the HBXIP protein between different species (Fig. S7B). Together, these data indicate that nuciferine inhibits mTORC1 activity by interacting with HBXIP subunit and thus hampering the formation of the RagGTPases–Ragulator lysosomal scaffold.
4. Discussion
In this study, we demonstrate that nuciferine interacts with HBXIP, thus hampering Ragulator–Rags assembly and suppressing mTORC1 activity. This results in activation of TFEB-mediated ALP, which alleviates hepatic steatosis and insulin resistance of mice with NAFLD (Fig. 7D). These data indicate that nuciferine could be a new candidate drug for NAFLD and targeting of the mTORC1–TFEB–ALP axis may represent a promising therapeutic strategy for metabolic syndrome.
Nuciferine is one of the main constituents of Lotus leaf, which has been widely used in both food and drug formulations in Asia, with anti-fatty liver and anti-obesity properties33,47, features which have been further confirmed by our study. Insulin resistance is implicated both in the pathogenesis of NAFLD and in disease progression from steatosis to steatohepatitis, and modulation of insulin resistance represents a potential strategy for NALFD treatment48. Notably, the present study also revealed beneficial effects of nuciferine on insulin resistance and hyperglycemia in mice with fatty livers. Considering food intake was not altered by nuciferine and no nuciferine-associated side effect were observed in our or in previous studies30,32,33, we believe that nuciferine may serve as an ideal candidate drug to treat NAFLD.
By examining how NAFLD was improved by nuciferine, we noticed that genes implicated in autophagy, but also those which regulate lysosomal biogenesis and function were transcriptionally upregulated in livers of nuciferine-treated HFD mice and we found that this upregulation was dependent on activation of TFEB. These findings are in line with previous studies reporting that TFEB activation increased autophagy–lysosomal function in the liver26,49. TFEB activation beyond a certain threshold may be beneficial to metabolic homeostasis since our and previous studies have reported that lysosomal function and autophagic activity markedly declines in NAFLD livers5,6,38. Here we show that nuciferine activated ALP and subsequently improved hepatic steatosis and insulin resistance in a TFEB-dependent manner, indicating that TFEB is an attractive therapeutic target for NAFLD. Previous studies reported that TFEB-mediated ALP could directly hydrolyze neutral lipids stored in lipid droplets26,44,50, promote removal of damaged mitochondrial51, attenuate ROS overproduction and suppress inflammatory factors secretion23,44, which are the driving forces behind NAFLD52. Moreover, activation of TFEB is reported to mitigate hepatic steatosis and insulin resistance by reducing oxidative stress and inflammatory response53, 54, 55, which also observed in the present study. Accordingly, multiple downstream effectors of TFEB-mediated ALP were involved in the process of nuciferine-alleviated NAFLD.
Excessive nutrient levels in humans and mice with NAFLD cause constitutive activation of hepatic mTORC1, resulting in decreased TFEB transcriptional activity and IR/AKT signaling, and increased lipid synthesis24,56. It has been reported that activating mTORC1 by enforcing expression of active RagA decreased TFEB transcriptional activity and exacerbated HFD-induced hepatic steatosis in mice24, whereas inhibiting mTORC1 by genetic deletion of Raptor prevents hepatic steatosis and insulin resistance57. Given the above reports, several mTOR inhibitors have been employed against hepatic steatosis and insulin resistance, including rapamycin and caffeine4,23,28. A previous study has demonstrated that nuciferine inhibits mTORC1 activity58, however the exact mechanism remains unclear. The Ragulator complex functions as a lysosomal membrane scaffold for Rags to recruit and activate mTORC118,40. We found that nuciferine decreased the lysosomal localization of the Rags in the liver of mice with NAFLD and in hepatocytes under nutrient-rich condition by interacting with the Ragulator subunit HBXIP and thus inhibiting the function of Ragulator complex. This resulted in the inhibition of mTORC1 signaling and increased activation of TFEB transcriptional activity. Whether nuciferine binding to HBXIP may destabilize Ragulator complex or inhibits its assembly will be the subject of future studies.
mTORC1 inhibits ALP at multiple levels, by inhibiting autophagy initiation complex (mainly through phosphorylation of ULK1 and ATG13), autophagosome-lysosome fusion (through phosphorylation of UVRAG)59, 60, 61 and transcriptional activity of TFEB, which induces the expression of many autophagy and lysosomal genes required to support ALP11,12,35. In line with this, early studies clearly indicated that TFEB silencing significantly inhibited autophagosome formation in vitro9 whereas TFEB depletion in the murine liver strongly impaired the autophagy of lipid droplets26. Moreover, our data indicate that TFEB depletion completely blunted the beneficial effects mediated by nuciferine treatment in vivo, thus suggesting that the transcriptional activity of TFEB is required to sustain ALP induction over the time and hence to mediate the beneficial effects of nuciferine.
Srebf1 is a transcription factor implicated in lipid synthesis62. Notably, hyperactivation of mTORC1 contributes to hepatic steatosis by phosphorylating S6K to increase expression of Srebf156,63. Downregulation of Srebf1 and its target genes was observed in nuciferine-treated NAFLD mice, suggesting that the mTORC1–S6K–Srebf1 axis may be involved in the beneficial effects of nuciferine on NAFLD. Nevertheless, when mTORC1 and S6K were constitutive activated by ectopic expression of Raptor–RHEB15, nuciferine still improved PA-induced lipid accumulation and insulin resistance in hepatocytes. These evidence altogether clearly indicate that TFEB activation is the main driver of the beneficial effects mediated by nuciferine treatment on NAFLD.
5. Conclusions
We have demonstrated that nuciferine inhibits the function of mTORC1 and activates TFEB-mediated ALP by hampering the assembly of the Rags–Ragulator scaffold at the lysosome. This treatment improves hepatic steatosis and insulin resistance in mice, hence modulation of the mTORC1–TFEB–ALP axis is a potentially effective strategy to prevent and treat the pathological outcomes of NAFLD.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Beijing, China; grant Nos. U20A2062, 32022084, and 32002349) and Jilin Province Science and Technology Development Project (Changchun, China; grant No. 20210508011RQ).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.12.012.
Contributor Information
Guowen Liu, Email: liuguowen2008@163.com.
Xinwei Li, Email: lixinwei100@126.com.
Author contributions
Xinwei Li and Guowen Liu jointly conceived and supervised the study. Xiliang Du, Zhiyuan Fang and Taiyu Shen designed, performed, and analyzed the majority of the mouse and cell experiments, wrote the manuscript. Chiara Di Malta conceived the cell experiments and revised the manuscript. Xiaodi Niu performed the molecular docking. Meng Chen, Bo Jin and Hao Yu performed immunoprecipitation experiments. Lin Lei performed and analyzed the RNA-seq experiments. Wenwen Gao, Chuang Xu and Zhijun Cao assisted with experiments and data analysis. Yuxiang Song and Zhe Wang contributed to sample collection and processing.
Conflicts of interest
The authors declare no conflicts of interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Figure S5.
Figure S6.
Figure S7.
References
- 1.Sheth S.G., Gordon F.D., Chopra S. Nonalcoholic steatohepatitis. Ann Intern Med. 1997;126:137–145. doi: 10.7326/0003-4819-126-2-199701150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Ludwig J., Viggiano T.R., McGill D.B., Oh B.J. Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin Proc. 1980;55:434–438. [PubMed] [Google Scholar]
- 3.Czaja M.J. Function of autophagy in nonalcoholic fatty liver disease. Dig Dis Sci. 2016;61:1304–1313. doi: 10.1007/s10620-015-4025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinha R.A., Farah B.L., Singh B.K., Siddique M.M., Li Y., Wu Y., et al. Caffeine stimulates hepatic lipid metabolism by the autophagy–lysosomal pathway in mice. Hepatology. 2014;59:1366–1380. doi: 10.1002/hep.26667. [DOI] [PubMed] [Google Scholar]
- 5.Miyagawa K., Oe S., Honma Y., Izumi H., Baba R., Harada M. Lipid-induced endoplasmic reticulum stress impairs selective autophagy at the step of autophagosome–lysosome fusion in hepatocytes. Am J Pathol. 2016;186:1861–1873. doi: 10.1016/j.ajpath.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Zhang X., Chu E.S.H., Chen X., Kang W., Wu F., et al. Defective lysosomal clearance of autophagosomes and its clinical implications in nonalcoholic steatohepatitis. FASEB J. 2018;32:37–51. doi: 10.1096/fj.201601393R. [DOI] [PubMed] [Google Scholar]
- 7.Yang L., Li P., Fu S., Calay E.S., Hotamisligil G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metabol. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farah B.L., Landau D.J., Sinha R.A., Brooks E.D., Wu Y., Fung S.Y.S., et al. Induction of autophagy improves hepatic lipid metabolism in glucose-6-phosphatase deficiency. J Hepatol. 2016;64:370–379. doi: 10.1016/j.jhep.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 11.Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martina J.A., Chen Y., Gucek M., Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garami A., Zwartkruis F.J., Nobukuni T., Joaquin M., Roccio M., Stocker H., et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 15.Kim E., Goraksha-Hicks P., Li L., Neufeld T.P., Guan K.L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 18.Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su M.Y., Morris K.L., Kim D.J., Fu Y., Lawrence R., Stjepanovic G., et al. Hybrid structure of the RagA/C-Ragulator mTORC1 activation complex. Mol Cell. 2017;68:835–846. doi: 10.1016/j.molcel.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yonehara R., Nada S., Nakai T., Nakai M., Kitamura A., Ogawa A., et al. Structural basis for the assembly of the Ragulator–Rag GTPase complex. Nat Commun. 2017;8:1625. doi: 10.1038/s41467-017-01762-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang T., Wang R., Wang Z., Wang X., Wang F., Ding J. Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1. Nat Commun. 2017;8:1394. doi: 10.1038/s41467-017-01567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S.H., Kim G., Han D.H., Lee M., Kim I., Kim B., et al. Ezetimibe ameliorates steatohepatitis via AMP activated protein kinase-TFEB-mediated activation of autophagy and NLRP3 inflammasome inhibition. Autophagy. 2017;13:1767–1781. doi: 10.1080/15548627.2017.1356977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H., Yan S., Khambu B., Ma F., Li Y., Chen X., et al. Dynamic MTORC1-TFEB feedback signaling regulates hepatic autophagy, steatosis and liver injury in long-term nutrient oversupply. Autophagy. 2018;14:1779–1795. doi: 10.1080/15548627.2018.1490850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pastore N., Brady O.A., Diab H.I., Martina J.A., Sun L., Huynh T., et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy. 2016;12:1240–1258. doi: 10.1080/15548627.2016.1179405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao X., Wang S., Zhao K., Li Y., Williams J.A., Li T., et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. 2018;155:865–879. doi: 10.1053/j.gastro.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin C.W., Zhang H., Li M., Xiong X., Chen X., Chen X., et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C.M., Kao C.L., Wu H.M., Li W.J., Huang C.T., Li H.T., et al. Antioxidant and anticancer aporphine alkaloids from the leaves of Nelumbo nucifera Gaertn. cv. Rosa-plena. Molecules. 2014;19:17829–17838. doi: 10.3390/molecules191117829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo F., Yang X., Li X., Feng R., Guan C., Wang Y., et al. Nuciferine prevents hepatic steatosis and injury induced by a high-fat diet in hamsters. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui H., Li Y., Cao M., Liao J., Liu X., Miao J., et al. Untargeted metabolomic analysis of the effects and mechanism of nuciferine treatment on rats with nonalcoholic fatty liver disease. Front Pharmacol. 2020;11:858. doi: 10.3389/fphar.2020.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M.X., Zhao X.J., Chen T.Y., Liu Y.L., Jiao R.Q., Zhang J.H., et al. Nuciferine alleviates renal injury by inhibiting inflammatory responses in fructose-fed rats. J Agric Food Chem. 2016;64:7899–7910. doi: 10.1021/acs.jafc.6b03031. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C., Deng J., Liu D., Tuo X., Xiao L., Lai B., et al. Nuciferine ameliorates hepatic steatosis in high-fat diet/streptozocin-induced diabetic mice through a PPARα/PPARγ coactivator-1α pathway. Br J Pharmacol. 2018;175:4218–4228. doi: 10.1111/bph.14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medina D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Napolitano G., Di Malta C., Esposito A., de Araujo M.E.G., Pece S., Bertalot G., et al. A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome. Nature. 2020;585:597–602. doi: 10.1038/s41586-020-2444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 38.Li X., Shi Z., Zhu Y., Shen T., Wang H., Shui G., et al. Cyanidin-3-O-glucoside improves non-alcoholic fatty liver disease by promoting PINK1-mediated mitophagy in mice. Br J Pharmacol. 2020;177:3591–3607. doi: 10.1111/bph.15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 40.Mu Z., Wang L., Deng W., Wang J., Wu G. Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane. Cell Discov. 2017;3:17049. doi: 10.1038/celldisc.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips J.C., Hardy D.J., Maia J.D.C., Stone J.E., Ribeiro J.V., Bernardi R.C., et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys. 2020;153 doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y., Wang X., Gao Y., Wang H., Niu X. Insight into the dual inhibitory mechanism of verbascoside targeting serine/threonine phosphatase Stp1 against Staphylococcus aureus. Eur J Pharmaceut Sci. 2021;157:105628. doi: 10.1016/j.ejps.2020.105628. [DOI] [PubMed] [Google Scholar]
- 44.Ballabio A., Bonifacino J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol. 2020;21:101–118. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 45.Martina J.A., Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–491. doi: 10.1083/jcb.201209135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweitzer L.D., Comb W.C., Bar-Peled L., Sabatini D.M. Disruption of the Rag–Ragulator complex by c17orf59 inhibits mTORC1. Cell Rep. 2015;12:1445–1455. doi: 10.1016/j.celrep.2015.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim A.R., Jeong S.M., Kang M.J., Jang Y.H., Choi H.N., Kim J.I. Lotus leaf alleviates hyperglycemia and dyslipidemia in animal model of diabetes mellitus. Nutr Res Pract. 2013;7:166–171. doi: 10.4162/nrp.2013.7.3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan R.S., Bril F., Cusi K., Newsome P.N. Modulation of insulin resistance in nonalcoholic fatty liver disease. Hepatology. 2019;70:711–724. doi: 10.1002/hep.30429. [DOI] [PubMed] [Google Scholar]
- 49.Pastore N., Vainshtein A., Klisch T.J., Armani A., Huynh T., Herz N.J., et al. TFE3 regulates whole-body energy metabolism in cooperation with TFEB. EMBO Mol Med. 2017;9:605–621. doi: 10.15252/emmm.201607204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thelen A.M., Zoncu R. Emerging roles for the lysosome in lipid metabolism. Trends Cell Biol. 2017;27:833–850. doi: 10.1016/j.tcb.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trivedi P.C., Bartlett J.J., Mercer A., Slade L., Surette M., Ballabio A., et al. Loss of function of transcription factor EB remodels lipid metabolism and cell death pathways in the cardiomyocyte. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165832. doi: 10.1016/j.bbadis.2020.165832. [DOI] [PubMed] [Google Scholar]
- 52.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu C., Jing M., Yang L., Jin L., Ding Y., Lu J., et al. Alisol A 24-acetate ameliorates nonalcoholic steatohepatitis by inhibiting oxidative stress and stimulating autophagy through the AMPK/mTOR pathway. Chem Biol Interact. 2018;291:111–119. doi: 10.1016/j.cbi.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Ao N., Ma Z., Yang J., Jin S., Zhang K., Luo E., et al. Liraglutide ameliorates lipotoxicity-induced inflammation through the mTORC1 signalling pathway. Peptides. 2020;133:170375. doi: 10.1016/j.peptides.2020.170375. [DOI] [PubMed] [Google Scholar]
- 55.Su H., Li Y., Hu D., Xie L., Ke H., Zheng X., et al. Procyanidin B2 ameliorates free fatty acids-induced hepatic steatosis through regulating TFEB-mediated lysosomal pathway and redox state. Free Radic Biol Med. 2018;126:269–286. doi: 10.1016/j.freeradbiomed.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 56.Saxton R.A., Sabatini D.M. mTOR Signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sengupta S., Peterson T.R., Laplante M., Oh S., Sabatini D.M. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 58.Yoo A., Jang Y.J., Ahn J., Jung C.H., Ha T.Y. Lotus leaf ethanol extract and nuciferine suppress adipocyte differentiation by regulating Akt–mTORC1 signaling in 3T3-L1 cells. J Food Nutr Res. 2019;7:688–695. [Google Scholar]
- 59.Kim Y.M., Jung C.H., Seo M., Kim E.K., Park J.M., Bae S.S., et al. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell. 2015;57:207–218. doi: 10.1016/j.molcel.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ganley I.G., Lam du H., Wang J., Ding X., Chen S., Jiang X. ULK1.ATG13. FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horton J.D., Goldstein J.L., Brown M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Porstmann T., Santos C.R., Griffiths B., Cully M., Wu M., Leevers S., et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metabol. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.