Abstract
Tripterygium wilfordii is a valuable medicinal plant rich in biologically active diterpenoids, but there are few studies on the origins of these diterpenoids in its secondary metabolism. Here, we identified three regions containing tandemly duplicated diterpene synthase genes on chromosomes (Chr) 17 and 21 of T. wilfordii and obtained 11 diterpene synthases with different functions. We further revealed that these diterpene synthases underwent duplication and rearrangement at approximately 2.3–23.7 million years ago (MYA) by whole-genome triplication (WGT), transposon mediation, and tandem duplication, followed by functional divergence. We first demonstrated that four key amino acids in the sequences of TwCPS3, TwCPS5, and TwCPS6 were altered during evolution, leading to their functional divergence and the formation of diterpene secondary metabolites. Then, we demonstrated that the functional divergence of three TwKSLs was driven by mutations in two key amino acids. Finally, we discovered the mechanisms of evolution and pseudogenization of miltiradiene synthases in T. wilfordii and elucidated that the new function in TwMS1/2 from the terpene synthase (TPS)-b subfamily was caused by progressive changes in multiple amino acids after the WGT event. Our results provide key evidence for the formation of diverse diterpenoids during the evolution of secondary metabolites in T. wilfordii.
Key words: Tripterygium wilfordii, Tandem duplication, Diterpene synthases, Divergence, Secondary metabolism
Graphical abstract
Multiple evolutionary mechanisms acting on the expansion patterns and functional divergence of eleven diterpene synthases were elucidated in this study, which provides a plausible explanation for the appearance of diterpenoid secondary metabolites during the evolution of Tripterygium wilfordii.
1. Introduction
Tripterygium wilfordii is a famous medicinal plant rich in a variety of natural diterpenoids with significant pharmacological activities1, 2, 3. In recent years, the pharmaceutical potential of the diterpenoids in T. wilfordii has received increasing attention from researchers. Most of the bioactivities observed in T. wilfordii as a medicinal plant were attributed to the presence of triptolide, a diterpenoid with powerful immunosuppressive, anti-inflammatory, and pleiotropic antitumor pharmacological effects4, 5, 6, 7. In addition, many other natural diterpenoids of T. wilfordii also have significant bioactivities. For example, triptonide has recently been demonstrated as a highly effective non-hormonal male contraceptive agent8, tripterifordin has potential for the treatment of acquired immune deficiency syndrome (AIDS)9, while dehydroabietic acid and its derivatives have antibacterial, antifungal, antitumor, and antiviral effects10,11.
Diterpenoids are a super-family of structurally diverse natural products consisting of four isoprene units. Their synthesis is initiated by the conversion of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP) into various diterpene skeletons by class I and II diterpene synthases, which mainly belong to the TPS-c and TPS-e/f subfamilies. The TPS-c subfamily mainly contains bifunctional copalyl diphosphate synthase/kaurene synthase (CPS/KS) and CPS, while the TPS-e/f subfamily mainly contains kaurene synthase(-like) members (KS/KSL). However, diterpene synthases capable of catalyzing the formation of miltiradiene were found in the TPS-b monoterpene synthase family12,13, suggesting a more complex functional evolution of diterpene synthases in T. wilfordii.
CPS and KS are present in all higher plants as the most essential diterpene synthases to produce kaurene as an intermediate for the biosynthesis of plant hormone gibberellin in primary metabolism. In addition to CPS and KS, there are many other diterpene synthases involved in secondary metabolism12, 13, 14, 15, 16, 17, 18, 19, 20, 21. It is speculated that secondary metabolism may arise by changing the structure of enzymes recruited from primary metabolism22,23. Several recent reports have demonstrated the functional plasticity of diterpene synthases with dramatic shifts in the product spectrum arising from single amino acid changes23, 24, 25, 26, 27, 28. However, the functional evolution of diterpene synthase appears to be more complex, with drivers other than simple functional shifts caused by mutations in single amino acids causing larger evolutionary changes. How multiple diterpene synthase genes were generated and functionally diverged during the evolution of T. wilfordii, resulting in a rich diversity of diterpenoids, remains to be addressed.
Several functional studies on diterpene synthases that produce different diterpene skeletons in T. wilfordii have been reported based only on transcriptomic data12,13,21. However, there have been no previous studies on the evolutionary mechanisms driving the functional divergence of diterpene synthases in T. wilfordii. To investigate the genetic basis of the diversity of these diterpenoids, we mined functional gene encoding diterpene synthases in T. wilfordii based on the genomic data29. Here, we identified expanded families of diterpene synthases and obtained 11 diterpene synthases from the T. wilfordii genome. We demonstrated multiple evolutionary mechanisms of the expansion patterns and functional divergence acting in different diterpene synthase families, which provides a plausible explanation for the appearance of these important diterpene secondary metabolites during the evolution of medicinal plants.
2. Materials and methods
2.1. Plant material
We collected seeds of T. wilfordii from Huangshi City, China, and brought them back to the laboratory for germination. The germinated sprouts were used for RNA extraction.
2.2. RACE and gene cloning
Total RNA was isolated from T. wilfordii using an RNA Extraction Kit (Promega, Shanghai, China) and purified using an RNA Purification Kit (Tiangen Biotech, Beijing, China). The First-strand cDNA was synthesized using the SMARTer® RACE 5′/3′ Kit (Clontech Laboratories, USA).
Diterpene synthases were mined from the T. wilfordii genome29 based on the genome annotation and BLASTN analysis using BioEdit 7.0.9.0 software30. The full-length cDNA sequence of TwMS3 was obtained by 5′ and 3′ rapid amplification of cDNA ends (RACE). The TwTPS genes were cloned by polymerase chain reaction (PCR) using the Phusion high-fidelity PCR master mix (New England BioLabs, MA, USA) with specific primers (Supporting Information Table S1), ligated into the pEASY-Blunt Zero vector (TransGen Biotechnology, Beijing, China), and verified by complete sequencing. The primer sequences of TwCPS1, TwCPS4, and TwKSL1v2 were the same as previous report13. Finally, eleven full-length cDNAs of diterpene synthases from T. wilfordii were obtained.
2.3. Sequence analysis
The diterpene synthases investigated in this study were named according to their functions. The sequences of the cDNAs of TwCPS1, TwCPS3, TwCPS4, TwCPS5, TwCPS6, TwKSL1v2, TwKSL2, TwKSL3, TwMS1, TwMS2, and TwMS3 were analyzed at NCBI (http://www.ncbi.nlm.nih.gov/). The comparison of sequence identity and function of diterpene synthases in T. wilfordii were shown in Supporting Information Tables S2 and S3 and Fig. S1, including diterpene synthases obtained in this study and previously reported. In addition, all diterpene synthases were searched against the reference genome of T. wilfordii to be located in the chromosomes (Supporting Information Table S4). The synonymous substitution rate (Ks) values of diterpene synthase genes were calculated by KaKs Calculator 2.0. Then the duplication time of gene pairs was calculated according to Eq. (1):
| (1) |
where r represents the substitution rate per site per year as 6.5 × 10−9 mutations for eudicots31.
2.4. Evolution analysis
For phylogenetic analysis, amino acid sequences of diterpene synthases from other species were obtained from the NCBI database (Table S3). Multiple sequence alignments were performed using ClustalW. The maximum-likelihood tree using a JTT model was constructed by MEGA X32 with n = 1000 replicates for bootstrapping. A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 3.0933)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 0.53% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions with less than 90% site coverage were eliminated. There were a total of 419 positions in the final dataset.
To further elucidate the evolutionary mechanisms of the diterpene synthases, we selected Populus trichocarpa, which are relatively closely related to T. wilfordii among the species with published genomes, for collinearity analysis using McscanX by TBtools software33.
2.5. In vitro assays
The ORFs of diterpene synthases obtained in this study were subcloned into the pMAL-c2X vector using the pEASY-Uni Seamless Cloning and Assembly Kit (TransGen Biotech) with specific primers (Supporting Information Table S5) and were verified by sequencing. Individual TPS recombinant plasmids were expressed in Escherichia coli BL21 (DE3) as described previously13. Cultures were grown at 37 °C and 220 rpm in 50 mL of Luria–Bertani medium until the OD600 reached 0.6–0.8. Then, protein expression was induced with 0.4 mmol/L isopropyl-beta-d-thiogalactopyranoside (Sigma, USA) and continued for 18 h at 16 °C and 160 rpm. Cells were harvested by centrifugation (3000×g for 10 min at 4 °C) and lysed by a sonicator in lysis buffer [50 mmol/L Tris-HCl pH 7.2, 10 mmol/L MgCl2, 5 mmol/L DTT and 10% (v/v) glycerol]. The lysate was clarified by centrifugation (16,000×g for 30 min at 4 °C).
To test the catalytic activity of class II diterpene synthases, 5 μg GGPP was added to 0.4 mL of recombinant proteins in assay buffer [50 mmol/L Tris-HCl pH 7.2, 10 mmol/L MgCl2, 5 mmol/L DTT and 10% (v/v) glycerol] and incubated overnight at 30 °C in the dark. Then, 10 units of calf intestinal phosphatase (CIP; NEB) were added for enzymatic dephosphorylation at 37 °C for 2 h. The products were extracted 3 times with an equal volume of hexane and analyzed using gas chromatography–mass spectrometry (GC–MS)13. Diterpene synthases from class I were characterized in coupled assays using class II TwTPS to convert GGPP into copalyl diphosphate (CPP) as described previously34. Briefly, assay mixtures containing 5 μg of GGPP, class II TwTPS, and each individually expressed class I diterpene synthase were incubated overnight at 30 °C before extraction with hexane and GC–MS analysis.
The product of TwTPS3 was used as a standard for ent-CPP12, the product of SmCPS4 was used as a standard for ent-8-hydroxy-CPP16, the product of TwTPS10 was used as a standard for kolavenyl diphosphate (KPP)12, the product of TwKSL1 was used as a standard for ent-pimaradiene13, the product of AtKS was used as a standard for ent-kaurene35, the product of TwTPS16v2 was used as a standard for 16α-hydroxy-ent-kaurane13, the product of TwMS2 (TwTPS27v2) was used as a standard for miltiradiene13, the product of TwCPS1 (TwTPS7v2) was used as a standard for normal-CPP13, and the product of CfTPS2 and CfTPS3 was used as a standard for 13R-(+)-manoyl oxide15,16,21.
2.6. Molecular docking
I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/)36 was used to predict the three-dimensional structure of the target protein, and the best-rated model structure was used for further analysis. In addition, the structures of small molecule substrates were obtained from PubChem and converted into PDB format using Phenix software. Molecular docking was performed with AutoDock37 software to predict the active site of the target protein. The docking results were visualized and analyzed using PyMOL (https://pymol.org/2/).
2.7. Site-directed mutagenesis
Site-directed mutagenesis of TwCPS3, TwCPS5, TwCPS6, TwKSL1v2, TwKSL2, TwKSL3, and TwMS3 was performed on the pMAL-c2X recombinant plasmids via whole-plasmid PCR amplification using overlapping mutagenic primers (Supporting Information Tables S6 and S7) and Phusion high-fidelity DNA polymerase. All mutants were verified by sequencing and expressed in E. coli BL21 (DE3) to identify their function.
Site-directed mutagenesis of TwMS2 was performed on the pESC-LEU recombinant plasmids in the same way (Supporting Information Table S8). All mutants were verified by sequencing and then introduced into a CPP-producing yeast strain (BY-HZ16-TwTPS7v2). Each recombinant strain was cultured in 5 mL Sc-Ura-Leu-His liquid medium with 0.2% glucose and 1.8% galactose at 30 °C and 200 rpm. After 12 h, an n-dodecane phase corresponding to 10% (v/v) of the culture volume was added under aseptic conditions. After 5 days of fermentation, 150 μL of the n-dodecane phase following extraction was collected for the detection of miltiradiene using GC–MS.
2.8. GC–MS analysis
The diterpene synthase products were analyzed and identified by GC–MS on an Agilent 7000 instrument equipped with a DB-5MS capillary column (30 m × 0.25 mm; Agilent, Santa Clara, USA). For analysis, 5 μL of the sample was injected and separated using a temperature program of 50 °C for 2 min, then increased to 300 °C with a gradient of 20 °C/min and a hold of 10 min. The ion trap heating temperature was 230 °C, the electron energy was 70 eV, and mass spectra were recorded in the range of 20–400 m/z.
3. Results
3.1. Tandem duplication of diterpene synthase genes in the genome of T. wilfordii
Diterpenoids play a significant role in the bioactive functions and plant growth of T. wilfordii. To clarify the mechanism of the diversity of diterpene compounds in T. wilfordii, 11 diterpene synthases were identified in the genome of T. wilfordii (Fig. 1A). Three consecutive TwCPS genes (TwCPS3, TwCPS5, TwCPS6) and TwMS3 were found on Chr 17. TwCPS1 and TwMS2 were located in mutual proximity on Chr 21, while TwCPS4 and TwMS1 were at an adjacent location. In addition, three consecutive TwKSL genes were found at a location 167 kb from TwCPS4 and TwMS1 on Chr 21, namely TwKSL1v2, TwKSL2, and TwKSL3. Among them, TwCPS1, TwCPS4, TwKSL2, and TwMS2 were respectively identical to TwTPS7v213, TwTPS912, TwTPS16V213, and TwTPS27v213 (Table S2). TwCPS3, TwCPS6, TwKSL1v2, and TwMS1 shared a high degree of identity with TwTPS312, TwTPS1012, TwKSL113, and TwTPS2712, respectively, and they were located in the same position on chromosomes, suggesting they were allelic variants (Tables S2 and S4). However, none of the reported genes were distributed at the same position on the chromosomes as TwCPS5, TwMS3, and TwKSL3 (Table S4).
Figure 1.
Gene distribution, phylogeny, and collinearity analysis of diterpene synthases from T. wilfordii. (A) Distribution of diterpene synthases in the genome of T. wilfordii. (B) Phylogeny of diterpene synthases from T. wilfordii and representative characterized TPS. Accession numbers and characterized functions of TPSs used in this phylogenetic analysis were given in Supporting Information Table S3. The Maximum Likelihood tree was reconstructed using a JTT model by MEGA X. The tree was rooted with PpCPS/KS, the bifunctional ent-CPP/ent-kaurene synthase from Physcomitrella patens. Numbers on branches represented the bootstrap percentage values calculated from 1000 bootstrap replicates. (C) Collinearity blocks between T. wilfordii and P. trichocarpa.
Among the 11 diterpene synthases, 6 members containing the characteristic DDxxD motif were identified as class I TPSs, while the remaining 5 members containing the DxDD motif were identified as class II TPSs (Fig. 1B). Phylogenetic analysis showed that TwMS1, TwMS2, and TwMS3 were clustered in the TPS-b clade, TwKSL1v2, TwKSL2, and TwKSL3 were clustered in the TPS-e/f clade, while TwCPS3, TwCPS5, and TwCPS6 were clustered in the TPS-c clade (Fig. 1B). In the collinearity analysis of T. wilfordii and P. trichocarpa, two collinear regions corresponding to TwCPS on Chr 17 and two collinear regions corresponding to TwKSL on Chr 21 were found on Chr 2, Chr 5, Chr 8, and Chr 10 in P. trichocarpa (Fig. 1C). The duplicated mode of TwCPSs on Chr 17 and TwKSLs on Chr 21 was further identified as tandem duplication using the DupGen_finder pipeline38 (Supporting Information Table S9), while TwCPS1/4 and TwMS1/2 were due to the duplication of a region containing 20 genes (Supporting Information Fig. S2).
3.2. The mechanism driving the functional divergence of TwCPS genes on Chr 17
TwCPS3, TwCPS5, and TwCPS6 were formed by two duplications at approximately 8.23–10.68 MYA (Supporting Information Table S10) and they share 87.5%–89.6% amino acid sequence identity (Fig. 2A). To study the origin and evolution of the tandem repeats of the TwCPS genes observed on Chr 17, we performed functional studies of TwCPS3, TwCPS5, and TwCPS6 via in vitro cell-free assays with GGPP as a substrate after heterologous expression in E. coli, and found a significant difference in their product profiles (Fig. 2B and Supporting Information Fig. S3). Different functional diterpene synthases also exist in other species, such as SmCPS4 and SmCPS516, which catalyze the formation of ent-8-hydroxy-CPP and ent-CPP, respectively, but their sequence identity is only 55.8%. The function of these enzymes emerged in T. wilfordii after about 8.23 MYA, by which time T. wilfordii and Salvia miltiorrhiza had diverged, suggesting that these enzymes with the same function evolved independently in different species, while the secondary metabolites produced by TwCPS5 and TwCPS6 in T. wilfordii emerged recently.
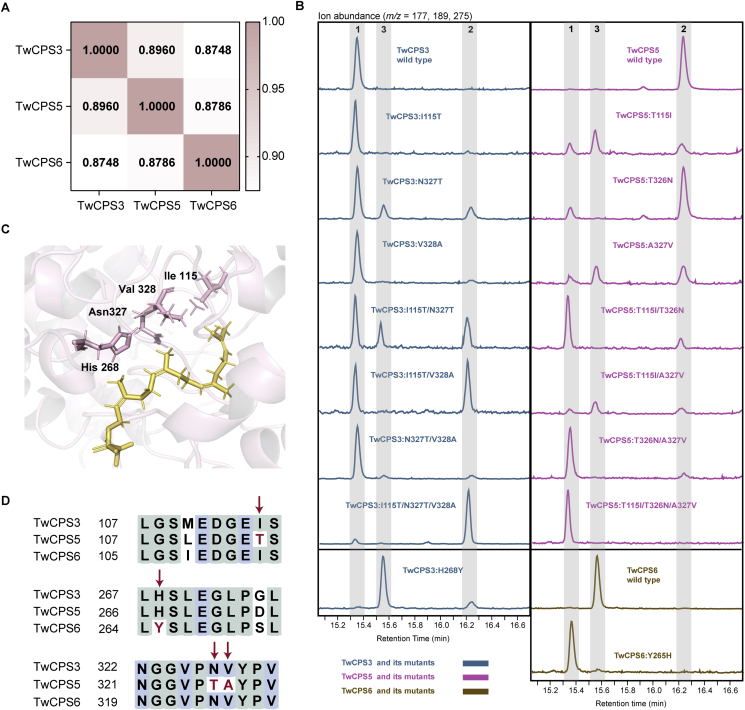
Figure 2.
Functional evolutionary analysis of TwCPS3, TwCPS5, and TwCPS6. (A) Sequence identity of TwCPS3, TwCPS5, and TwCPS6 at the amino acid level. (B) GC–MS analysis of the products of TwCPS3, TwCPS5, TwCPS6, and their mutants. Number 1 represents ent-CPP, number 2 represents ent-8-hydroxy-CPP, and number 3 represents KPP. (C) Four active-site residues differ between TwCPS3, TwCPS5, and TwCPS6. Pink cartoons represent the protein model of TwCPS3, yellow sticks represent the substrate GGPP, and pink sticks represent the four active site residues. (D) Sequence alignment of TwCPS3, TwCPS5, and TwCPS6.
Intrigued by their dominating diterpene products, we set out to clarify the mechanism driving the evolution of enzymes that catalyze reactions in the secondary metabolism from primary metabolic pathway genes. Since TwCPS3 is thought to catalyze a key step in primary metabolism as it generates ent-CPP (1) for the biosynthesis of the gibberellin phytohormone, we used the TwCPS3 protein as a model for molecular docking with the substrate GGPP and performed amino acid sequence comparisons with TwCPS5 and TwCPS6 to identify amino acid sites responsible for the functional divergence of the enzymes. Modeled enzyme structures and sequence alignment indicated that there are four different residues in the potential active-site region of TwCPS3, TwCPS5, and TwCPS6 (Fig. 2C and D). The residues were identified as isoleucine (I) at position 115, histidine (H) at position 268, asparagine (N) at position 327, and valine (V) at position 328 in TwCPS3, threonine (T) at position 115, H at position 267, T at position 326, and alanine (A) at position 327 in TwCPS5, as well as I at position 113, tyrosine (Y) at position 265, N at position 324, and V at position 325 in TwCPS6.
First, to clarify the functional conversion of TwCPS5 evolved from the TwCPS3, a total of 14 mutants of TwCPS3 and TwCPS5 were created based on three different residues (Fig. 2B). We found that (1) mutations of any single amino acid have little effect on the function of TwCPS3, mutations of two amino acids (I115T/N327T, I115T/V328A) could convert part of TwCPS3 products into ent-8-hydroxy-CPP (2), while the combined mutation of three amino acids resulted in TwCPS3 producing almost exclusively ent-8-hydroxy-CPP; (2) Arbitrary mutations at T115 or A327 had a great impact on the function of TwCPS5, resulting in producing ent-CPP and KPP (3). By contrast, TwCPS5:T326N had almost no effect on the function of TwCPS5, with the only difference being the production of a small amount of ent-CPP. When TwCPS5 was mutated at two sites (T115I/T326N or T326N/A327V), the main product of the mutants was ent-CPP. When these three sites of TwCPS5 were mutated at the same time, the resulting mutant only produced ent-CPP (Figure 2, Figure 3). To clarify the functional evolution of TwCPS6 from TwCPS3, we designed two mutations (TwCPS3:H268Y and TwCPS6:Y265H) interconverting the corresponding active-site H and Y. Strikingly, the functions of TwCPS3 and TwCPS6 were transformed into each other after these single mutations. The product of TwCPS3:H268Y changed from ent-CPP to KPP, and the product of TwCPS6:Y265H changed from KPP to ent-CPP (Figure 2, Figure 3).
Figure 3.
Reactions catalyzed by TwCPS3, TwCPS5, TwCPS6, and their mutants.
3.3. The mechanism driving the functional divergence of TwKSLs on Chr 21
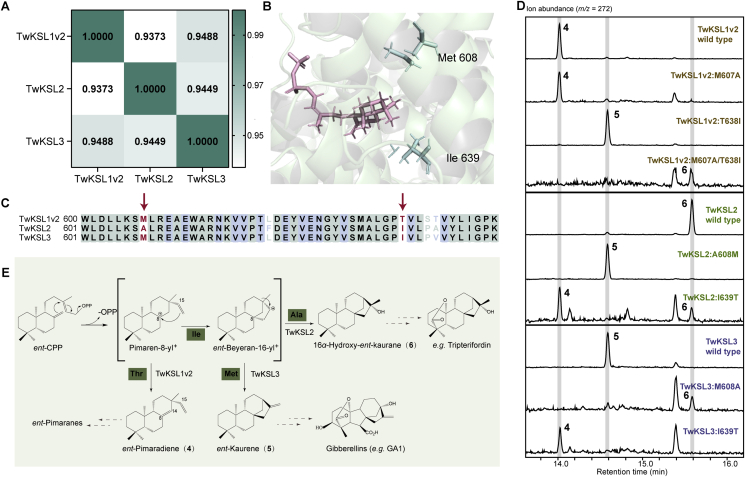
We identified three TwKSL genes on Chr 21 with high degrees of sequence identity (93.7%–94.9%) (Fig. 4A), which were generated by two successive duplications at 4.07–5.09 MYA (Supporting Information Table S10). However, their functions have diverged to convert ent-CPP into different products (Fig. 4D and Supporting Information Fig. S4). Modeled enzyme structures and sequence alignments indicate that there are two different residues in the potential active-site region of TwKSL1v2, TwKSL2, and TwKSL3 (Fig. 4B and C), one of which is consistent with the previously reported kaurene synthase in P. trichocarpa39. The residues were identified as methionine (M) at position 607 and T at position 638 in TwKSL1v2, A at position 608 and I at position 639 in TwKSL2, as well as M at position 608 and I at position 639 in TwKSL3 (Fig. 4C). To determine which differences were responsible for the observed functional divergence, seven mutations were made in the corresponding pairs of active-site residues of TwKSL1v2, TwKSL2, and TwKSL3. The product profiles of the resulting mutants were characterized by GC–MS. We found that (1) TwKSL1v2:M607A continued to produce ent-pimaradiene (4), while the product of TwKSL1v2:T638I changed to ent-kaurene (5) and that of TwKSL1v2:M607A/T638I changed to 16α-hydroxy-ent-kaurane (6); (2) The product of TwKSL2:A608M changed to ent-kaurene, and that of TwKSL2:I639T changed to ent-pimaradiene; (3) The product of TwKSL3:M608A changed to 16α-hydroxy-ent-kaurane and that of TwKSL3:I639T changed to ent-pimaradiene (Fig. 4D). Hence, the switch of the TwKSL products from ent-beyeran-16-yl+ to ent-pimaradiene is due to the I-to-T change, while the switch from ent-kaurene to 16α-hydroxy-ent-kaurane is due to the M-to-A change (Fig. 4E).
Figure 4.
Functional evolutionary analysis of TwKSL1v2, TwKSL2, and TwKSL3. (A) Sequence identity of TwKSL1v2, TwKSL2, and TwKSL3 at the amino acid level. (B) Two active site residues differ between TwKSL1v2, TwKSL2, and TwKSL3. Green cartoons represent the protein model of TwKSL3, pink sticks represent the substrate ent-CPP, and green sticks represent two different active site residues. (C) Sequence alignment of TwKSL1v2, TwKSL2, and TwKSL3. (D) GC–MS analysis of the products of TwKSL1v2, TwKSL2, TwKSL3, and their mutants. (E) Reactions catalyzed by TwKSL1v2, TwKSL2, TwKSL3, and their mutants.
3.4. The mechanism driving the new function of TwMSs on Chr 21 and Chr 17
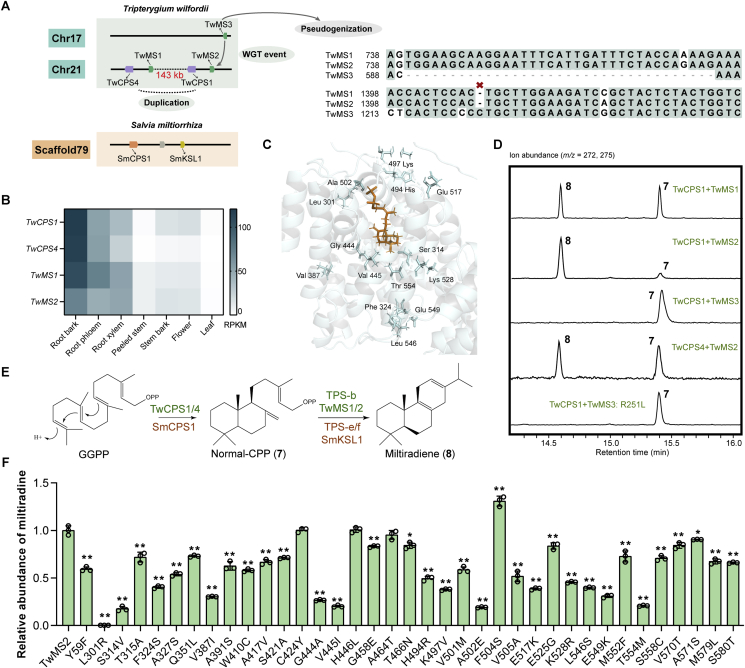
We identified the TwCPS1, TwCPS4, TwMS1, and TwMS2 genes on Chr 21, as well as the TwMS3 gene on Chr 17 (Fig. 5A). To characterize the function of these five genes, we obtained their full-length cDNA sequences and found that TwMS3 could not be normally translated into a protein sequence due to the lack of a 35 bp fragment (TGGAAGCAAGGAATTTCATTGATTTCTACCAGAAG) in the β domain and an inserted base in the α domain (Fig. 5A). This proved that the mRNA of TwMS3 is expressed in T. wilfordii, but it has lost its catalytic function. We used the seamless splicing method to insert the missing fragment and remove the inserted base, after which the mRNA of TwMS3 could be translated into a functional protein. Then, the corresponding full-length DNA sequences were expressed in E. coli and subjected to in vitro cell-free assays. The results of GC–MS showed that TwCPS1 and TwCPS4 can convert GGPP into normal-CPP (7), and TwMS1 and TwMS2 can further convert normal-CPP into miltiradiene (8), but TwMS3 cannot convert normal-CPP into miltiradiene (Fig. 5D and E).
Figure 5.
Mechanisms driving the functional evolution of TwMSs. (A) The process of gene duplication of TwMSs. (B) Gene expression of TwCPS1/4 and TwMS1/2 in different tissues. (C) Protein model of TwMS2. Blue cartoons represent the protein model of TwMS2, orange sticks represent the substrate normal-CPP. Blue sticks represent residues that differ in the amino acid sites of TwMS1/2 and TwMS3, and the production of miltiradiene after mutation is 50% lower than that of the wild type. (D) GC–MS analysis of the products of TwMSs and TwCPS1/4. (E) Reactions catalyzed by TwCPS1/4 and TwMS1/2. (F) Relative abundance of miltiradiene in strains expressing TwMS2 and its mutants. Data are mean ± SD, n = 3; ∗P < 0.05 and ∗∗P < 0.01 by Student's t-test.
TwMS1/2 are non-conventional members of the TPS-b subfamily that catalyze the cyclization of normal-CPP to form miltiradiene, which is otherwise limited to the TPS-e/f subfamily in angiosperms12,13 (Fig. 5E). This prompted us to explore the mechanism of the functional evolution of TwMS1/2. In our previous studies, we have hypothesized that TwMS3 and TwMS1/2, with 78% sequence identity (Supporting Information Fig. S5), arose in a WGT event29. We thus suspected that the function of cyclizing normal-CPP to form miltiradiene found in TwMS1/2 should have evolved after the recent WGT event in T. wilfordii. To determine which amino acid site changes were responsible for the new function of TwMS1/2 in the TPS-b subfamily, we first found a total of 37 different sites by comparing modeled enzyme structures and sequence alignment (Fig. 5C and Supporting Information Fig. S6) and then created 37 mutations of TwMS2. Among them, 33 mutants showed a significant decrease in the yield of miltiradiene, with 50% reductions in 14 mutants compared to the wild type. These mutants were leucine (L)-301 to arginine (R), serine (S)-314 to V, phenylalanine (F)-324 to S, V-387 to I, glycine (G)-444 to A, V-445 to I, H-494 to R, lysine (K)-497 to V, A-502 to glutamic acid (E), E−(517) to K, K-528 to R, L-546 to S, E−549 to K, and T-554 to M. Notably TwMS2:L301R completely lost the function of cyclizing normal-CPP to form miltiradiene (Fig. 5F). Accordingly, we investigated if the TwMS3:R251L mutant could potentially generate miltiradiene. However, we found that even with the restoration of this amino acid residue, TwMS3 still could not catalyze the formation of miltiradiene (Fig. 5D).
Miltiradiene is the precursor of many essential abietane-type compounds in T. wilfordii (i.e., triptolide, triptonide, tripdiolide, triptolidenol, 16-hydroxytriptolide, etc.). The formation of the TwCPS1/4 and TwMS1/2 gene pairs may indicate a recent increase in the demand for abietane-type diterpenoids in T. wilfordii. Thus, the formation of these gene pairs provides advantages in gene number and gene distribution for the biosynthesis of abietane-type diterpenoids, resulting in more efficient production, as reflected by the similar expression pattern of TwCPS1/4 and TwMS1/2 in different tissues (Fig. 5B). Interestingly, we found that SmCPS1 and SmKSL1, the two genes that synthesize miltiradiene, were also located closely in the S. miltiorrhiza genome (Fig. 5A and Supporting Information Fig. S7). Of particular note here is that TwMS1/2 and SmKSL1 belong to the TPS-b and TPS-e/f subfamilies, respectively, but still each is located close to the normal-CPP synthase. This reflects similar gene recruitment in the secondary metabolism of different species, which we speculate provides a positional advantage for the production of miltiradiene.
3.5. Data availability
The T. wilfordii genome and S. miltiorrhiza genome analyzed during this study are available under NCBI BioProject number PRJNA542587 and PRJNA682867, and the P. trichocarpa genome data are available on Ensembl Plants (http://plants.ensembl.org/index.html).
4. Discussion
Diterpene synthases have been of interest due to their complex catalytic mechanisms and their key role in the metabolism of valuable medicinal plants. How these enzymes evolve and catalyze the formation of the diverse diterpene structures is a subject of biochemical interest to be resolved23,28. Previous reports have largely focused on single amino acid substitutions which were sufficient to significantly alter the product profile of Class I and Class II diterpene synthases23,25, 26, 27,39. However, no studies aimed to comprehensively elucidate the mechanisms driving the functional divergence of diterpene synthases during plant evolution. In this study, we identified 11 diterpene synthases, followed by an analysis of underlying gene duplication and functional divergence in T. wilfordii.
The expansion of diterpene synthase genes (TwCPSs, TwKSLs, and TwMSs) in T. wilfordii resulted from tandem gene duplication and the WGT event that was prevalent in the evolutionary history of the plant. Among these duplicate genes, TwCPS3 and TwKSL3 retain their functions in gibberellin biosynthesis in primary metabolism. Others were recruited to participate in more diterpene secondary metabolites, such as tripterifordin, triptolide, and so on. TwCPS3, TwCPS5, and TwCPS6 arose by two gene duplication events at 8.23–10.68 MYA, while TwKSL1v2, TwKSL2, and TwKSL3 are formed due to two gene duplication events at 4.07–5.09 MYA (Supporting Information Table S10). By contrast, TwCPS1, TwCPS4, TwMS1, and TwMS2 were duplicated together more recently at 2.34–2.43 MYA (Supporting Information Table S10), whereas TwMS3 was generated earlier by the WGT event of T. wilfordii, together with TwMS1 or TwMS2. Notably, TwCPS3, TwCPS5, and TwCPS6 were duplicated earlier than TwKSL1v2, TwKSL2, and TwKSL3, which is reasonable since TwCPSs encode enzymes that act upstream of TwKSLs, and earlier duplication and functional evolution allowed TwKSLs to obtain more substrates to produce diverse diterpenes. Another interesting phenomenon is that TwCPS1/4 and TwMS1/2, as the key pathway genes for triptolide biosynthesis, are adjacent to each other on Chr 21. Based on our previous repeat sequence annotation of the T. wilfordii genome29, we found that the region containing these gene pairs contains many DNA transposons and retrotransposons (Supporting Information Tables S11 and S12). Accordingly, we suspected that these genes might be recruited together on Chr 21 by transposon-mediated “cut-and-paste” translocation after the WGT event, followed by a more recent “copy-and-paste” duplication of a region containing 20 genes that resulted in two copies of TwCPS1/4 and TwMS1/2.
In our study, the I115T/N327T/V328A mutant of TwCPS3 exhibited a product profile nearly identical to that of TwCPS5, and the T115I/T326N/A327V mutant of TwCPS5 was functionally identical to TwCPS3. Similarly, the H268Y mutant of TwCPS3 had a product profile nearly identical to that of TwCPS6 and the Y265H mutant of TwCPS6 had a product profile nearly identical to that of TwCPS3. Thus, these four active-site residues jointly control the different catalytic functions of TwCPS3, TwCPS5, and TwCPS6. The new function of TwCPS5 evolved from TwCPS3 by changes in T115, T326, and A327. Similarly, TwCPS6 evolved through a mutation at Y265. In the case of TwCPS3, the active-site residues H268, N327, and V328 identified in this study are consistent with the previous reports25,26,40, but I115 is a new active site confirmed to convert the product profile of diterpene synthase. The enzymatic function of TwCPS3 is due to that H268 and N327 forms hydrogen bonds with water molecules and activate the water forming a catalytic base (Fig. 3) as the previous reports26,41,42. Thus, we hypothesize that the function convert of TwCPS5 is due to the change of T115, T326, and A327 reducing the steric hindrance and allowing water access to labda-13E-en-8-yl+-PP, leading to water-quenching of the carbocation and the formation of ent-8-hydroxy-CPP (Fig. 3). In addition, the function convert of TwCPS6 may be due to the change of Y265 sterically blocking water access to the labda-13E-en-8-yl+-PP by its larger aromatic side chains, and the potential ability of the tyrosine hydroxyl group to form a hydrogen bond with N324, possibly blocking the binding of water (Fig. 3). Our hypothesis is consistent with previous studies on the plasticity of AtCPS, where mutations at positions H263 and N322 caused dramatic changes in product outcome25,26. When a larger aromatic residue replaces H, it effectively prevents the entry of water molecules and the aromatic side chain promoted the hydride and methyl shifts, resulting in complete rearrangement to form KPP. However, when a smaller residue replaces H, even with a smaller aromatic side chain, is sufficient for the entry of water molecules and then quenching the carbocation to form ent-8-hydroxy-CPP. Similarly, when the N is substituted by a smaller residue such as A, this also leads to the entry of water molecules to form ent-8-hydroxy-CPP.
KSL genes play a critical role in the derivation of diterpenoid metabolism, and there are over 1000 known natural products derived from (iso)kaurene, as well as about 500 derived from pimaradiene23 (https://dnp.chemnetbase.com). It is known that ent-kaurene serves as the precursor for gibberellins required for growth regulation and normal development in all higher plants43, 44, 45. By contrast, 16α-hydroxy-ent-kaurane is the precursor for the secondary metabolite tripterifordin46 while the function of ent-pimaradiene in T. wilfordii is still unknown. We observed that TwKSL1v2 and TwKSL2 were independently evolved new functions by mutating I to T at position 638 and M to A at position 608, respectively. Whereas the mutant of TwKSL3:I639T produced ent-pimaradiene and the mutant of TwKSL3:M608A produced 16α-hydroxy-ent-kaurane. We speculate that the enzymatic function of ent-pimaradiene synthesis is due to the possible ability of the introduced polar hydroxyl group of threonine to stabilize the pimaren-8-yl+ intermediate and subsequently deprotonate to form ent-pimaradiene, which is consistent with the F residue at this position also producing pimaradiene since F with the aromatic structure has the ability to stabilize carbocations23,47. In addition, we speculated that another new enzymatic function of TwKSL2, i.e., the synthesis of 16α-hydroxy-ent-kaurane, arose because the smaller A residue allows water molecules into the active cavity, which can then react with the ent-beyeran-16-yl+ intermediate. This residue is therefore responsible for the formation of the active-site cavity rather than participating in the binding of water as described before39,48.
An intriguing phenomenon of gene evolution and pseudogenization appears to be occurring in T. wilfordii. Surprisingly, TwMS1/2 from the TPS-b subfamily has evolved a new function to catalyze the formation of miltiradiene. TwMS3, on the other hand, is being pseudogenization due to fragment deletion and base insertion. Since TwMS3 and TwMS1/2 may arise in the WGT event of T. wilfordii, we hypothesized that the new diterpene function of TwMS1/2 should have evolved after the WGT event. The results of this study also suggest that multiple gene copies that arose in a polyploidy event provided the genetic basis for functional divergence, allowing TwMS1/2 to evolve a new function as a diterpene synthase after the duplication of TwMS3. To further validate the mechanism of the new function of TwMS1/2, we found that changes at 34 amino acid sites significantly affected the function of TwMS2 in catalyzing the formation of miltiradiene and 14 mutations resulted in a 50% lower yield of miltiradiene than in the wild type. We thus speculated that the combinatorial mutation of multiple amino acid residues is the reason for the evolution of the new diterpene synthase function of TwMS1/2. Miltiradiene is a critical intermediate in the biosynthesis of many natural diterpenoids, including triptolide12,13, carnosic acid49,50, tanshinones51, and rubesanolides A-E17, and researchers have attempted to improve the production of miltiradiene by metabolic engineering52,53. Hence, the 34 amino acid sites affecting the yield of miltiradiene in our study may have wider implications for the heterologous synthesis of miltiradiene.
Although the key sites that determine the product conversion of diterpene synthases have been identified in T. wilfordii, there are still some yet unidentified amino acid difference sites, such as A386 in TwCPS5 while S387 in TwCPS3 and S384 in TwCPS6, T524 in TwKSL1v2 while S525 in TwKSL2 and TwKSL3, T316 of TwMS1 while I316 in TwMS2, V502 in TwMS1 while A502 in TwMS2 and so on. It is common for single amino acid variations to affect the catalytic efficiency of enzymes, as is the case in diterpene synthases23,24,42,54. The side chain structures of these different amino acids may affect substrate binding, steric hindrance, and protein structure, leading to changes in the catalytic efficiency of the enzymes, which need further experimental verification and resolution of the crystal structures of these proteins to identify other active sites.
5. Conclusions
Multiple drivers of the functional evolution of diterpene synthases were identified in our study, including WGT event and tandem gene duplication with subsequent single-, triple-, or multiple active site mutations resulting in different new functions. This not only reflects the complexity and plasticity of diterpene synthase evolution in plants, but also provides further insights into the mechanisms and timing of the emergence of diterpene secondary metabolites during evolutionary history.
Acknowledgments
We thank Yuhe Tu (Datian Taoyuan State Forest Farm in Fujian Province, Datian, China) and Sanbo Liu (China Resources Sanjiu (Huangshi) Pharmaceutical Co., Ltd. in Hubei province, Huangshi, China) for providing plant material of T. wilfordii. This work was supported by the National Key R&D Program of China (No.2020YFA0908000), the Key Project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (No.2060302-1806-03), and Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (ZYYCXTD-D-202005).
Author contributions
Wei Gao, Lichan Tu, and Ping Su designed research. Lichan Tu, Xinbo Cai, Yifeng Zhang, Yuru Tong, Jian Wang, Tianyuan Hu, and Yunfeng Luo performed experiment. Lichan Tu, Yun Lu, Xiaoyi Wu, and Dan Li analyzed data. Lichan Tu wrote the paper. Luqi Huang and Wei Gao revised the paper. All authors have read and approved the final version of the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.02.013.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Upadhyay A. Natural compounds in the regulation of proteostatic pathways: an invincible artillery against stress, ageing, and diseases. Acta Pharm Sin B. 2021;11:2995–3014. doi: 10.1016/j.apsb.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corson T.W., Crews C.M. Molecular understanding and modern application of traditional medicines: triumphs and trials. Cell. 2007;130:769–774. doi: 10.1016/j.cell.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang G., Li S., Zhang C., Chen H., Wang N., Feng Y. Clinical efficacies, underlying mechanisms and molecular targets of Chinese medicines for diabetic nephropathy treatment and management. Acta Pharm Sin B. 2021;11:2749–2767. doi: 10.1016/j.apsb.2020.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Z.L., Yang Y.X., Ding J., Li Y.C., Miao Z.H. Triptolide: structural modifications, structure–activity relationships, bioactivities, clinical development and mechanisms. Nat Prod Rep. 2012;29:457–475. doi: 10.1039/c2np00088a. [DOI] [PubMed] [Google Scholar]
- 5.Gao J., Zhang Y., Liu X., Wu X., Huang L., Gao W. Triptolide: pharmacological spectrum, biosynthesis, chemical synthesis and derivatives. Theranostics. 2021;11:7199–7221. doi: 10.7150/thno.57745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Titov D.V., Gilman B., He Q.L., Bhat S., Low W.K., Dang Y., et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol. 2011;7:182–188. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen R., Huang L., Hu K. Natural products remodel cancer-associated fibroblasts in desmoplastic tumors. Acta Pharm Sin B. 2020;10:2140–2155. doi: 10.1016/j.apsb.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Z., Qin W., Zheng H., Schegg K., Han L., Liu X., et al. Triptonide is a reversible non-hormonal male contraceptive agent in mice and non-human primates. Nat Commun. 2021;12:1253. doi: 10.1038/s41467-021-21517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen K., Shi Q.A., Fujioka T., Zhang D.C., Hu C.Q., Jin J.Q., et al. Anti-AIDS agents, 4. Tripterifordin, a novel anti-HIV principle from Tripterygium wilfordii: isolation and structural elucidation. J Nat Prod. 1992;55:88–92. doi: 10.1021/np50079a013. [DOI] [PubMed] [Google Scholar]
- 10.Park N.H., Kang Y.G., Kim S.H., Bae I.H., Lee S.H., Kim D.Y., et al. Dehydroabietic acid induces regeneration of collagen fibers in ultraviolet B-irradiated human dermal fibroblasts and skin equivalents. Skin Pharmacol Physiol. 2019;32:109–116. doi: 10.1159/000497103. [DOI] [PubMed] [Google Scholar]
- 11.Kim E., Kang Y.G., Kim Y.J., Lee T.R., Yoo B.C., Jo M., et al. Dehydroabietic acid suppresses inflammatory response via suppression of Src-, Syk-, and TAK1-mediated pathways. Int J Mol Sci. 2019;20:1953. doi: 10.3390/ijms20071593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen N.L., Heskes A.M., Hamberger B., Olsen C.E., Hallstrom B.M., Andersen-Ranberg J., et al. The terpene synthase gene family in Tripterygium wilfordii harbors a labdane-type diterpene synthase among the monoterpene synthase TPS-b subfamily. Plant J. 2017;89:429–441. doi: 10.1111/tpj.13410. [DOI] [PubMed] [Google Scholar]
- 13.Su P., Guan H., Zhao Y., Tong Y., Xu M., Zhang Y., et al. Identification and functional characterization of diterpene synthases for triptolide biosynthesis from Tripterygium wilfordii. Plant J. 2018;93:50–65. doi: 10.1111/tpj.13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin B., Guo J., Tang J., Tong Y., Ma Y., Chen T., et al. An alternative splicing alters the product outcome of a class I terpene synthase in Isodon rubescens. Biochem Biophys Res Commun. 2019;512:310–313. doi: 10.1016/j.bbrc.2019.03.057. [DOI] [PubMed] [Google Scholar]
- 15.Pateraki I., Andersen-Ranberg J., Hamberger B., Heskes A.M., Martens H.J., Zerbe P., et al. Manoyl oxide (13R), the biosynthetic precursor of forskolin, is synthesized in specialized root cork cells in Coleus forskohlii. Plant Physiol. 2014;164:1222–1236. doi: 10.1104/pp.113.228429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui G., Duan L., Jin B., Qian J., Xue Z., Shen G., et al. Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza. Plant Physiol. 2015;169:1607–1618. doi: 10.1104/pp.15.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin B., Cui G., Guo J., Tang J., Duan L., Lin H., et al. Functional diversification of kaurene synthase-like genes in Isodon rubescens. Plant Physiol. 2017;174:943–955. doi: 10.1104/pp.17.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen R., Bu Y., Ren J., Pelot K.A., Hu X., Diao Y., et al. Discovery and modulation of diterpenoid metabolism improves glandular trichome formation, artemisinin production and stress resilience in Artemisia annua. New Phytol. 2021;230:2387–2403. doi: 10.1111/nph.17351. [DOI] [PubMed] [Google Scholar]
- 19.Zerbe P., Chiang A., Dullat H., O'Neil-Johnson M., Starks C., Hamberger B., et al. Diterpene synthases of the biosynthetic system of medicinally active diterpenoids in Marrubium vulgare. Plant J. 2014;79:914–927. doi: 10.1111/tpj.12589. [DOI] [PubMed] [Google Scholar]
- 20.Inabuy F.S., Fischedick J.T., Lange I., Hartmann M., Srividya N., Parrish A.N., et al. Biosynthesis of diterpenoids in Tripterygium adventitious root cultures. Plant Physiol. 2017;175:92–103. doi: 10.1104/pp.17.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen-Ranberg J., Kongstad K.T., Nielsen M.T., Jensen N.B., Pateraki I., Bach S.S., et al. Expanding the landscape of diterpene structural diversity through stereochemically controlled combinatorial biosynthesis. Angew Chem Int Ed Engl. 2016;55:2142c6. doi: 10.1002/anie.201510650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrington Y., Guo J., Le C.H., Fillo A., Kwon J., Tran L.T., et al. Evolution of a secondary metabolic pathway from primary metabolism: shikimate and quinate biosynthesis in plants. Plant J. 2018;95:823–833. doi: 10.1111/tpj.13990. [DOI] [PubMed] [Google Scholar]
- 23.Xu M., Wilderman P.R., Peters R.J. Following evolution's lead to a single residue switch for diterpene synthase product outcome. Proc Natl Acad Sci U S A. 2007;104:7397–7401. doi: 10.1073/pnas.0611454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zerbe P., Chiang A., Bohlmann J. Mutational analysis of white spruce (Picea glauca) ent-kaurene synthase (PgKS) reveals common and distinct mechanisms of conifer diterpene synthases of general and specialized metabolism. Phytochemistry. 2012;74:30–39. doi: 10.1016/j.phytochem.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Potter K.C., Zi J., Hong Y.J., Schulte S., Malchow B., Tantillo D.J., et al. Blocking deprotonation with retention of aromaticity in a plant ent-copalyl diphosphate synthase leads to product rearrangement. Angew Chem Int Ed Engl. 2016;55:634–638. doi: 10.1002/anie.201509060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potter K., Criswell J., Zi J., Stubbs A., Peters R.J. Novel product chemistry from mechanistic analysis of ent-copalyl diphosphate synthases from plant hormone biosynthesis. Angew Chem Int Ed Engl. 2014;53:7198–7202. doi: 10.1002/anie.201402911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia M., Zhou K., Tufts S., Schulte S., Peters R.J. A pair of residues that interactively affect diterpene synthase product outcome. ACS Chem Biol. 2017;12:862–867. doi: 10.1021/acschembio.6b01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christianson D.W. Structural and chemical biology of terpenoid cyclases. Chem Rev. 2017;117:11570–11648. doi: 10.1021/acs.chemrev.7b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu L., Su P., Zhang Z., Gao L., Wang J., Hu T., et al. Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nat Commun. 2020;11:971. doi: 10.1038/s41467-020-14776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.HT A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 31.Wei C., Yang H., Wang S., Zhao J., Liu C., Gao L., et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci U S A. 2018;115:E4151–E4158. doi: 10.1073/pnas.1719622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Keeling C.I., Dullat H.K., Yuen M., Ralph S.G., Jancsik S., Bohlmann J. Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol. 2010;152:1197–1208. doi: 10.1104/pp.109.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi S., Sun T., Kawaide H., Kamiya Y. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 1998;116:1271–1278. doi: 10.1104/pp.116.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J., Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiao X., Li Q., Yin H., Qi K., Li L., Wang R., et al. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019;20:38. doi: 10.1186/s13059-019-1650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irmisch S., Muller A.T., Schmidt L., Gunther J., Gershenzon J., Kollner T.G. One amino acid makes the difference: the formation of ent-kaurene and 16α-hydroxy-ent-kaurane by diterpene synthases in poplar. BMC Plant Biol. 2015;15:262. doi: 10.1186/s12870-015-0647-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen N.L., Nissen J.N., Hamberger B. Two residues determine the product profile of the class II diterpene synthases TPS14 and TPS21 of Tripterygium wilfordii. Phytochemistry. 2017;138:52–56. doi: 10.1016/j.phytochem.2017.02.022. [DOI] [PubMed] [Google Scholar]
- 41.Koksal M., Hu H., Coates R.M., Peters R.J., Christianson D.W. Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat Chem Biol. 2011;7:431–433. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koksal M., Potter K., Peters R.J., Christianson D.W. 1.55Å-resolution structure of ent-copalyl diphosphate synthase and exploration of general acid function by site-directed mutagenesis. Biochim Biophys Acta. 2014;1840:184–190. doi: 10.1016/j.bbagen.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleet C.M., Sun T.P. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 45.Hedden P., Thomas S.G. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 46.Su P., Guan H., Zhang Y., Wang X., Gao L., Zhao Y., et al. Probing the single key amino acid responsible for the novel catalytic function of ent-kaurene oxidase supported by NADPH-cytochrome P450 reductases in Tripterygium wilfordii. Front Plant Sci. 2017;8:1756. doi: 10.3389/fpls.2017.01756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dougherty D.A. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 48.Kawaide H., Hayashi K., Kawanabe R., Sakigi Y., Matsuo A., Natsume M., et al. Identification of the single amino acid involved in quenching the ent-kauranyl cation by a water molecule in ent-kaurene synthase of Physcomitrella patens. FEBS J. 2011;278:123–133. doi: 10.1111/j.1742-4658.2010.07938.x. [DOI] [PubMed] [Google Scholar]
- 49.Bozic D., Papaefthimiou D., Bruckner K., de Vos R.C., Tsoleridis C.A., Katsarou D., et al. Towards elucidating carnosic acid biosynthesis in Lamiaceae: functional characterization of the three first steps of the pathway in Salvia fruticosa and Rosmarinus officinalis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruckner K., Bozic D., Manzano D., Papaefthimiou D., Pateraki I., Scheler U., et al. Characterization of two genes for the biosynthesis of abietane-type diterpenes in rosemary (Rosmarinus officinalis) glandular trichomes. Phytochemistry. 2014;101:52–64. doi: 10.1016/j.phytochem.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 51.Gao W., Hillwig M.L., Huang L., Cui G.H., Wang X.Y., Kong J.Q., et al. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett. 2009;11:5170–5173. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu T., Zhou J., Tong Y., Su P., Li X., Liu Y., et al. Engineering chimeric diterpene synthases and isoprenoid biosynthetic pathways enables high-level production of miltiradiene in yeast. Metab Eng. 2020;60:87–96. doi: 10.1016/j.ymben.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Y.J., Gao W., Rong Q., Jin G., Chu H., Liu W., et al. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc. 2012;134:3234–3241. doi: 10.1021/ja2114486. [DOI] [PubMed] [Google Scholar]
- 54.Jia M., O'Brien T.E., Zhang Y., Siegel J.B., Tantillo D.J., Peters R.J. Changing face: a key residue for the addition of water by sclareol synthase. ACS Catal. 2018;8:3133–3137. doi: 10.1021/acscatal.8b00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The T. wilfordii genome and S. miltiorrhiza genome analyzed during this study are available under NCBI BioProject number PRJNA542587 and PRJNA682867, and the P. trichocarpa genome data are available on Ensembl Plants (http://plants.ensembl.org/index.html).