Abstract
The vertical and seasonal distribution and diversity of archaeal sequences was investigated in a hypersaline, stratified, monomictic lake, Solar Lake, Sinai, Egypt, during the limnological development of stratification and mixing. Archaeal sequences were studied via phylogenetic analysis of 16S rDNA sequences as well as denaturing gradient gel electrophoresis analysis. The 165 clones studied were grouped into four phylogenetically different clusters. Most of the clones isolated from both the aerobic epilimnion and the sulfide-rich hypolimnion were defined as cluster I, belonging to the Halobacteriaceae family. The three additional clusters were all isolated from the anaerobic hypolimnion. Cluster II is phylogenetically located between the genera Methanobacterium and Methanococcus. Clusters III and IV relate to two previously documented groups of uncultured euryarchaeota, remotely related to the genus Thermoplasma. No crenarchaeota were found in the water column of the Solar Lake. The archaeal community in the Solar Lake under both stratified and mixed conditions was dominated by halobacteria in salinities higher than 10%. During stratification, additional clusters, some of which may possibly relate to uncultured halophilic methanogens, were found in the sulfide- and methane-rich hypolimnion.
The Solar Lake is a small monomictic hypersaline lake located on the Sinai coast of the Gulf of Aqaba. The lake is separated from the gulf by a narrow sand bar. Water is supplied via seawater seepage through the sand bar and occasional winter precipitation. The depth of the lake fluctuates between 4 and 6 m. It is regulated by seasonal changes of influx seepage, out-flux of brine back to the gulf, and evaporation. High evaporation rates and arid climatic conditions during the summer months cause the salinity to rise to hypersaline values of up to 20%. Total mixing of the water column then occurs, and the entire water column is completely oxygenated. Temperature and salinity are constant throughout the water column, and sulfide and methane concentrations are very low during holomixis (6). In the fall, the water column becomes stratified due to the salinity gradient between the residual, highly saline water (18 to 20%) and the overlying, newly introduced seawater (5%). The density gradient between the two layers causes development of a pycnocline, which prevents mixing between the epilimnion and the hypolimnion and allows the development of a thermocline with an inverse temperature gradient (16 to 55°C). Oxygen is present throughout the epilimnion of the lake, where oxygen concentrations may reach supersaturation due to the photosynthetic activity of planktonic cyanobacteria (5). The pycnocline prevents atmospheric oxygen from penetrating the hypolimnion, and oxygen is rapidly diminished. High sulfide concentrations of up to 3 mM develop in the hypolimnion due to the activity of sulfate-reducing bacteria (SRB) in the water column (19) and in the surrounding cyanobacterial mats (36). Gradual heliothermal heating of the metalimnion causes the gradual destabilization of the pycnocline. Increasing evaporation rates in spring further disrupt the stratification and lead to summer holomixis.
Thick cyanobacterial mats carpet the Solar Lake sediment. The mats constitute a complex laminated ecosystem containing highly diverse microbial communities. Their activities affect the biogeochemistry of the lake by exporting oxygen, sulfide, and methane into the overlaid water column (7, 16, 21, 36).
The Archaea are divided into two kingdoms: the Crenarchaeota, of which all members presently isolated are extreme thermophiles, and the Euryarchaeota, a diverse group which includes all members of the methanogens, the Halobacteriaceae, and certain thermophiles (41). Phylogenetic analysis of 16S rRNA from diverse natural environments have identified a large number of both crenarchaeotal and euryarchaeotal sequences (3, 8, 9, 15, 18, 26). Many of these sequences form monophyletic clades unrelated to any known cultured organism.
In sulfate-rich ecosystems, methanogens are normally out-competed by SRB for acetate and hydrogen (29, 30, 40). However, certain methanogens are able to utilize noncompetitive substrates, such as methylamines, derived from compounds that accumulate in halotolerant organisms, where they serve in osmoregulation in hypersaline environments (23, 31). Metabolic activity of methanogens that utilize noncompetitive substrates is not affected in environments where SRB are present. The presence and activity of methanogens capable of using noncompetitive substrates have been previously described for the cyanobacterial mats of Solar Lake. Radiotracer experiments in the Solar Lake mats showed rapid metabolism of [14C]trimethylamine and [14C]methanol to 14CH4 and 14CO2 (11). Giani et al. (16) isolated a Methanosarcina species that produced methane from trimethylamine. Relatively high methane concentrations of up to 17 μM (E. Cytryn, A. Zask, R. S. Oremland, and Y. Cohen, unpublished data), along with the high numbers of SRB in the anoxic layers of the water column (P. Sigalevich, M. V. Baev, A. Teske, and Y. Cohen, submitted for publication), may imply the presence of noncompetitive substrate utilizing methanogens in the water column of Solar Lake during stratification.
Halobacteriaceae inhabit highly saline environments, such as hypersaline lakes, salterns, and salted fish (32). The optimal salinity for most Halobacteriaceae ranges between 2 and 4 M NaCl (12 to 23%) (20, 33). The salinity in Solar Lake ranges between 10 and 20%, except for surface layers of the epilimnion during winter, where minimal salinities as low as 6% have been recorded. The relatively high salinity of the lake indicates the potential presence of halophilic Archaea.
The maximal temperature in the Solar Lake water column measured in the course of this research was 55°C. Temperatures as high as 72°C have been previously recorded (Y. Cohen, unpublished data). Certain thermophilic Archaea have a minimal temperature requirement of 55°C (37). However, most isolates thrive at temperatures ranging between 75 and 100°C. Therefore, the presence of thermophilic Archaea in Solar Lake is not likely.
The goal of this research was to assess the distribution and diversity of the various archaeal sequences from vertical profiles of the water column of the Solar Lake during stratified and mixed periods. Two molecular approaches were used. (i) The diversity of Archaea was assessed by phylogenetic analysis of 16S rRNA using cloning and sequencing techniques. (ii) The distribution of Archaea was studied by denaturing gradient gel electrophoresis (DGGE) analysis of PCR-amplified fragments of 16S rRNA archaeal sequences. The identified archaeal sequences were analyzed in correlation to salinity, temperature, sulfide, and methane measured concomitantly in the lake.
MATERIALS AND METHODS
Sampling procedures.
Sampling was carried out on a small boat fixed at a set point in the center of the lake. Samples were collected using a peristaltic pump connected to an 8-mm-diameter silicone tube. Water intake was through a 1-cm space created between two hydrodynamic, opposite cones described previously by Jørgensen et al. (19). This device prevented vertical mixing of the water column during sampling. Samples were taken at 0.25- or 0.5-m intervals from the surface to the water-sediment interface. Temperature and pH were measured on the boat using a combined temperature-pH meter (El Hamma Institute, Mavo Hamma, Israel), density was measured via a glass densiometer, and salinity was measured by using a refractometer (Atago, Tokyo, Japan). Water samples for oxygen and sulfide analysis were pumped into glass stoppered bottles and fixed with Winkler reagents (1) and zinc-acetate (4), respectively, and were analyzed soon after the samples were on shore.
Water samples for methane analysis were filled into 30-ml syringes. Then 10 ml of each sample was withdrawn from the syringe and replaced by 10 ml of air. The syringes were sealed and shaken vigorously for 5 min to allow equilibrium with the gaseous phase. The 10-ml methane-equilibrated gas phase was then injected into a 15-ml sealed penicillin bottle prefilled with double-distilled water through a thick butyl rubber stopper. The injected methane-equilibrated gas displaced 10 ml of the water. These sealed bottles, each containing the 10 ml of methane-equilibrated gas, were then sent to the laboratory of Oremland at the U.S. Geological Survey, Menlo Park, Calif., where methane concentration was measured using quantitative gas chromatography with a flame ionization detector (30). Calibration series of known methane concentrations showed no loss of methane during sampling, shipment, and analytical procedure.
For molecular analysis, 100-ml samples of Solar Lake water from each depth were filtrated on site through 0.2-μm-pore-size, 47-mm-diameter polycarbonate filters (Poretics Products, Livermore, Calif.). Filters were cut in half, immediately placed in liquid nitrogen, and stored at −70°C until nucleic acid extraction was performed.
DNA extraction and PCR.
DNA was extracted from the polycarbonate filters as described before by Minz et al. (25). DNA was electrophoresed for 30 min at 100 V on 1% TAE agarose gel, excised from the gel, and purified with a jet sorb gel extraction kit (Genomed Inc.).
Purified DNA from the various selected depths was amplified using specific 16S rRNA archaeal primers, 21f and 958r (Table 1). Each 50-μl reaction mixture contained 5 μl of 10× PCR buffer (Idaho Technologies, Idaho Falls, Idaho), 5 μl of deoxynucleoside-triphosphate mix (2.5 nM each), 2.5 μl of bovine serum albumin, 0.5 μl of 21f primer (50 μM), 0.5 μl of 958r primer (50 μM), 0.5 μl of Taq polymerase (TaKaRa, Otsushiga, Japan), 1 μl of template DNA, and RNase/DNase-free water to a final volume of 50 μl. PCR was performed in 50-μl glass capillaries using the Rapid Cycler Capillary PCR (Idaho Technologies). The following PCR program was used: 94°C for 30 s, followed by 30 cycles of 94°C for 15 s, 55°C for 20 s, and 72°C for 45 s, followed by 72°C for 30 s.
TABLE 1.
Primers used in this research
DGGE analysis.
DGGE was used to examine the diversity of Archaea in different depths of the water column. DNA from selected depths was amplified using the 16S rRNA-specific oligonucleotide primer pair GC-PARCH 340f and 934r (Table 1). The PCR program for DGGE was as follows: 94°C for 30 s, 4 cycles of 94°C for 5 s, 52°C for 1 min, and 72°C for 22 s, followed by 35 cycles of 94°C for 5 s, 52°C for 15 s, and 72°C for 22 s, with a final extension of 72°C for 30 s. PCR products were separated on an acrylamide gel with a 35 to 70% vertical denaturant gradient, using the DCode system (Bio-Rad, Hercules, Calif.). The system was run for 3.5 h at 200 V and 60°C. DGGE gels were stained in aqueous ethidium bromide solution, destained with double-distilled water for 15 min, and photographed using a BIS 202 bio-imaging system (Dinco and Rhenium, Beit Ne Kofa, Israel). Selected DGGE bands were excised from the gel, purified, reamplified with the same primers, cloned, and sequenced using the technique previously described (28).
Cloning of archaeal PCR products.
PCR products of 937 bp were ligated to pGEM T-easy vectors, which were transformed to E. coli JM109 competent cells (Promega, Madison, Wis.). Clones containing the correct insert were reamplified using DGGE-specific primers and examined on 35 to 70% DGGE gels. Distinct clone types detected by the DGGE analysis were selected for further analysis. Plasmids were purified using QIAprep Miniprep columns (Qiagen, Germany) and were sequenced with an ABI automated DNA sequencer by using a Prism dideoxy terminator cycle sequencing kit and the protocol recommended by the manufacturer (Applied Biosystems).
Phylogenetic analysis of detected sequences.
The approximately 900-bp partial sequences generated by the cloning technique and 580-bp partial sequences generated by the use of DGGE primers were added to a prealigned database of complete or partial 16S rRNA sequences by using the aligning tools from the ARB program package (38). Parsimony analysis was used to phylogenetically place the 16S rRNA sequences from Solar Lake with the 16S rRNA database sequences. Solar Lake sequences were selected along with archaeal reference sequences from the ARB, and consensus trees were generated by neighbor joining with the Olsen correction method (38). Sequences analyzed were screened for chimeras by the Chimera-Check program from the Ribosome Database Project (22).
Nucleotide sequence accession numbers.
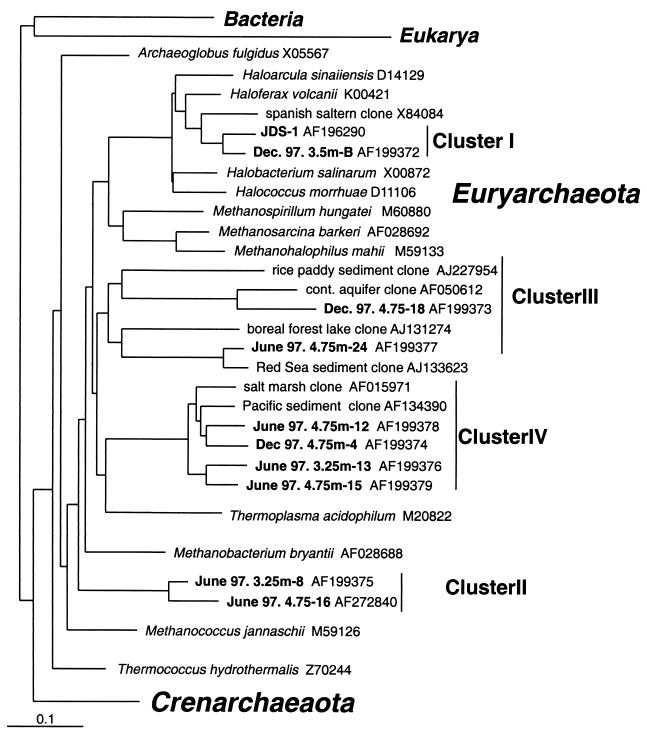
The 10 novel 16S rRNA archaeal sequences detected in the Solar Lake water column were submitted to GenBank and assigned the following accession numbers: AF196290, AF199372 to AF199379, and AF272840. See Fig. 2 for reference sequences used to generate the phylogenetic tree.
FIG. 2.
Phylogenetic tree of Archaea sequences detected in Solar Lake. The bar indicates a 10% estimated difference in nucleotide sequences.
RESULTS
Limnological profiles.
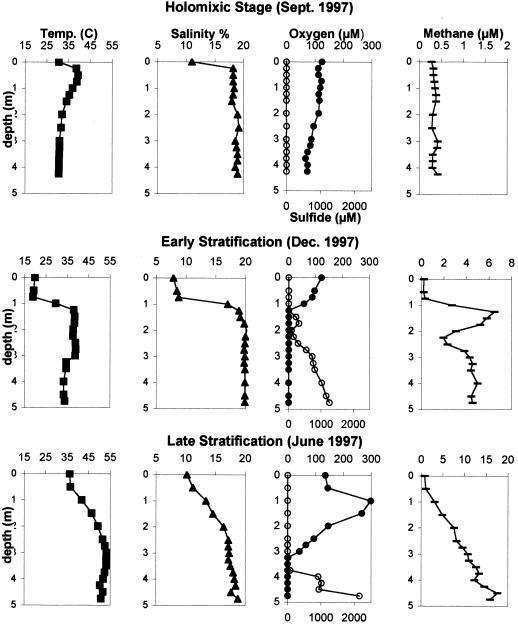
During 1997 and 1998, a total of 24 vertical liminological profiles were examined during two annual limnological cycles at Solar Lake. Three selected vertical profiles from June, September, and December 1997 representing the main limnological stages of the lake were selected for analysis in detail. Figure 1 depicts vertical day profiles for temperature, salinity, oxygen, sulfide, and methane from the water column of Solar Lake from the three profiles.
FIG. 1.
Limnological conditions of Solar Lake during holomixis (September [Sept.] 1997), early stratification (December [Dec.] 1997), and late stratification (June 1997). Symbols: ■, temperature; ▴, salinity; ●, oxygen; ○, sulfide; ▄, methane. Note different x axis values for the three methane profiles.
The September 1997 profile represents the onset of stratification. The entire water column was oxygenated, with oxygen concentration ranging between 100 and 140 μM. Maximal sulfide (6.7 μM) and methane (0.4 μM) concentrations are relatively insignificant compared to those measured during stratification. A sharp increase in temperature and salinity occurred only in the upper 0.25 m of the water column. Salinity increased from 16 to 19%, followed by a temperature increase from 30 to 39°C.
The December 1997 profile portrays stable stratification. The halocline, thermocline, and chemocline were all located at 1.00 to 1.25 m. There was a strong correlation between the sharp increase in temperature and salinity and the decrease in oxygen concentration between the surface water and that of the anaerobic hypolimnion. In the hypolimnion of this profile, there was a rapid increase in sulfide concentration, which reached a maximal value of 1,240 μM proximal to the sediment. A peak in methane concentration existed directly below the chemocline, where a maximum value of 6.5 μM was measured. Maximal methane concentrations directly below the chemocline were also observed in profiles from November 1997 and May 1998 (Cytryn et al., unpublished). Below this peak, the methane concentration decreased and increased again to a value of 4.5 μM proximal to the water-sediment interface.
The June 1997 profile portrays a transitional state between late stratification and the beginning of holomixis. Salinity and temperature gradients were more gradual than those observed in the December 1997 profile. The halocline and thermocline were less defined and appeared deeper in the water column (2 to 3 m). Mixing caused oxygen to penetrate much deeper than in the earlier stages of stratification, and the chemocline was located at 3.25 m. The oxygen concentration in this profile reached a maximal value of 300 μM at a depth of 1 m. The supersaturated values of oxygen imply intensive oxygenic photosynthetic activity at these depths. Below the chemocline, a sharp rise in sulfide concentration occurred, reaching a maximum value of 2,150 μM at the water-sediment interface. Methane concentration increased linearly from the surface and reached a maximum value of 17 μM adjacent to the lake sediment.
Molecular identification of archaeal sequences.
Specific 16S rRNA primers were used to amplify archaeal sequences within the water column of Solar Lake. A total of 165 clones from the three profiles were analyzed (20 clones from September 1997, 70 clones from December 1997 and 75 clones from June 1997). PCR products were obtained from all sampled depths throughout the water column of the three profiles except for samples from 0.0, 0.5, and 0.75 m from the December 1997 profile.
Of the 165 clones analyzed, 16 unique sequence types (>97% sequence identity) were identified. These sequences group into four clusters, which were correlated to the limnological conditions of the samples. Table 2 shows the distinct sequence types detected at selected depths of the water column from the three profiles analyzed.
TABLE 2.
Allocation of the various sequence types from selected water depths sampled during the three limnological stages of Solar Lake
| Limnological stage | Depth (m) | Sequence type | Cluster | No. of clones |
|---|---|---|---|---|
| Holomixis (September 1997) | 0–4 | JDS-1 | I | 20 |
| Early stratification (December 1997) | 0–0.75 | No. PCR product detected | ||
| 1.25–2.5 | JDS-1 | I | 29 | |
| 3.5 | JDS-1 | I | 18 | |
| D3.5-B | I | 2 | ||
| 4.75 | JDS-1 | I | 15 | |
| D4.75-18 | III | 1 | ||
| D4.75-4 | IV | 4 | ||
| Late stratification (June 1997) | 0–2.5 | JDS-1 | I | 29 |
| 3.25 | JDS-1 | I | 17 | |
| J3.25-8 | II | 2 | ||
| J3.25-13 | IV | 1 | ||
| 4.75 | JDS-1 | I | 11 | |
| J4.75-16 | II | 2 | ||
| J4.75-24 | III | 1 | ||
| J4.75-12 | IV | 4 | ||
| J4.75-15 | IV | 8 | ||
The unique sequences shown in Table 2, together with 16S rRNA sequences of selected archaeal strains, were used to generate a phylogenetic tree (Fig. 2) using the neighbor joining method.
Of the 165 clones analyzed, 144 clones belonged to cluster I, a monophyletic group within the Halobacteriaceae family. All sequences analyzed from September 1997, as well as all sequences detected in the oxic depths of the June 1997 and December 1997 profiles, belonged to this cluster. In addition, 92 of the 104 clones from the anaerobic depths analyzed in the two stratified profiles examined belonged to this cluster. Cluster I contains two similar clone types, JDS-1 and D3.5-B, with approximately 94% sequence identity between them. Clone type JDS-1, the most prevalent clone, was found at all depths in all three profiles. Clone type D3.5-B was detected only in the December profile at 3.5 m. These two sequences have relatively low sequence identity to the 16S rRNA sequences of any of the cultured halophilic Archaea, and they have only 89% sequence identity with the genus Haloferax, their closest cultured relative. However, they more closely resemble a number of uncultured 16S rRNA sequences detected in the Solar Lake cyanobacterial mats (D. Minz, G. Muyzer, D. A. Stahl, and Y. Cohen, unpublished data) as well as in salt evaporation ponds in Alicante, Spain (2) and in Eilat, Israel (Minz et al., unpublished).
Sequences belonging to three additional clusters, II, III, and IV, were identified only in the anaerobic samples of June 1997 and December 1997. The sequences belonging to these clusters are not phylogenetically affiliated with any known cultured organisms.
Cluster II contains two sequences (J3.25-8 and J4.75-16) isolated from the June 1997 profile at 3.25 and 4.75 m. These sequences are distantly related to the genera Methanococcus and Methanobacterium, both methanogens capable of utilizing hydrogen and CO2 or formate. They have 84% sequence identity with Methanococcus jannaschii, their closest cultured reference strain.
Cluster III contains one sequence (J4.75-24) isolated from the June 1997 profile and one sequence (D4.75-18) from the December 1997 profile. Members of this cluster phylogenetically group with a number of previously detected sequences in diverse locations (12, 13, 17; G. N. Jurgens, F. O. Glokner, A. Saano, R. Amman, and U. Munster, unpublished results from GenBank).
Cluster IV contains three sequences isolated from the December 1997 profile and nine sequences from the June 1997 profile. The genus Thermoplasma is remotely related to this group, having 74% sequence identity. However, sequences from this cluster have much greater identity to a number of sequences previously isolated via molecular techniques, including sediments from Pacific methane seeps (18), coastal salt marsh sediments (26), in the digestive tracts of marine fish (39), and in waters from the Santa Barbara channel (24).
No members of the kingdom Crenarchaeota were detected in the water column. Members of this group were detected in the Solar Lake cyanobacterial mats (Minz et al., unpublished).
Vertical diversity of archaeal sequences by DGGE analysis.
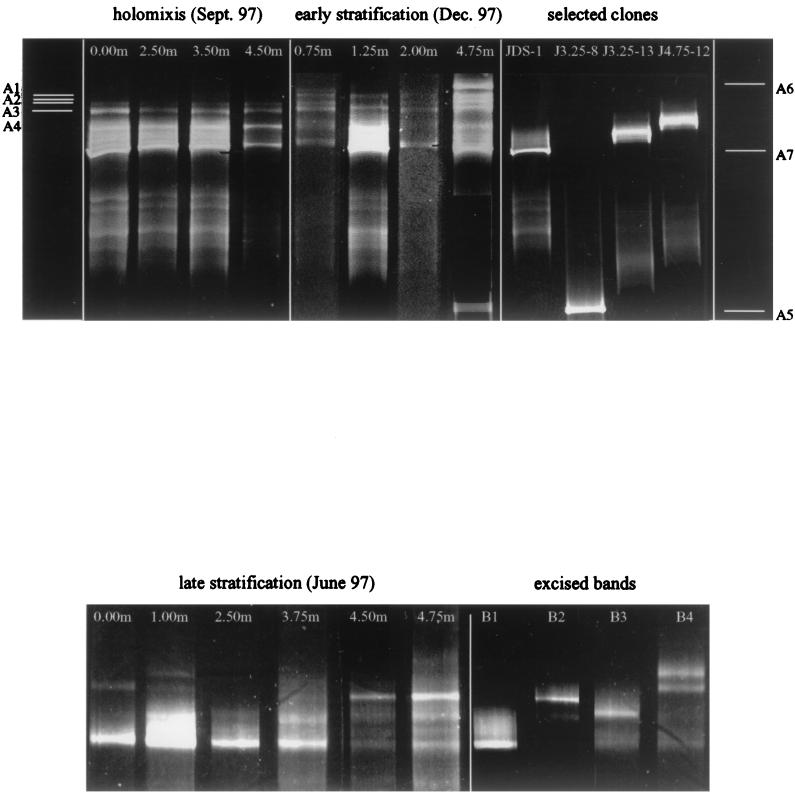
DGGE was used to analyze the vertical and seasonal changes in diversity of archaeal communities. Figure 3 shows ethidium-bromide-stained, polyacrylamide gels from vertical DGGE analysis of PCR products from the three profiles analyzed.
FIG. 3.
DGGE profiles showing sequence types from selected samples during holomixis, early stratification, and late stratification of Solar Lake. (Upper gel) DGGE of selected environmental samples from holomixis and early stratification compared to those of selected cones in order to identify known bands according to their relative levels of migration. Bands A1 to A7 are represented by the white lines seen on both sides of the gel. (Lower gel) DGGE of selected environment samples from late stratification compared to excised bands from the gel, which were sequenced and rerun to confirm sequence pattern. Sept., September; Dec., December.
PCR products from the September 1997 and December 1997 profiles were analyzed on the DGGE gel shown in Fig. 3 (upper gel), and the profile from June 1997 is presented in the Fig. 3 lower gel. In addition to these amplified environmental samples, cloned sequences shown in the Fig. 3 upper gel (JDS-1, J3.25-8, J3.25-13, and J4.75-12) representing three of the cluster types (Fig. 2) were amplified with DGGE primers and run adjacent to the water column samples in an attempt to phylogenetically identify specific DGGE bands according to their relative positions in the gel. Excised bands were reamplified and rerun on the lower acrylamide DGGE gel in Fig. 3 to confirm that the correct band was excised. These bands were sequenced and compared to sequences generated by the cloning analysis shown in Table 2.
The four samples analyzed from the September 1997 profile appear to have identical DGGE band patterns. The sample taken at 0.75 m in the December 1997 profile is identical to those from September 1997, with the exception of two faint bands (A1 and A2) that appear in the upper layers of the gel. The 1.25-m and 2.0-m samples from December 1997 show two different bands (A3 and A4) that are similar in their relative positions but not identical to those from 0.75 m. The differences in DGGE band pattern between the bands of these two profiles may be due to the shift from aerobic to anaerobic conditions, which occurs between 1 and 1.25 m.
Additional diversity is seen in the DGGE band pattern of the 4.75-m sample from December 1997. Two bands (A5 and A6) not present in all previous samples are seen here. Band A5 has a relative migration identical to that of the clone sequence J3.25-8 from cluster II. Band A7 appears at all depths analyzed from September and December 1997. This band has the same relative migration as clone sequence JDS-1 from the haloarchaeal cluster I. Clone sequences J3.25-13 and J4.75-12 from cluster IV have relative migration patterns similar to those of bands seen in the anaerobic samples from December 1997 (1.25, 2, and 4.75 m).
The June 1997 profile appeared to have the greatest archaeal diversity of the three profiles examined. Band B1 appeared at all depths sampled on this DGGE gel. The band intensity was greatest at 1 m and appeared to be the most dominant band at all depths analyzed above 3.75 m. Sequencing of this band showed it to be identical to the haloarchaeal clone JDS-1 from cluster I, shown in the phylogenetic tree in Fig. 2. Band B2 appeared only in the anaerobic depths analyzed (3.75, 4.5, and 4.75 m). At 3.75 m, this band was faint, but at 4.5 and 4.75 m, the intensity was much stronger and appeared to be dominant. Sequencing of this band placed it within cluster IV, defined in the phylogenetic tree in Fig. 2. Band B3 appeared at all depths sampled in this profile with the exception of 0.00 m. It was not successfully sequenced and therefore its phylogenetic affiliation is not clear.
The two bands B4 and B5 appeared only at 0.00 and 1.00 m. Sequence analysis characterized them as bacterial species related to the genus Cytophaga. This is apparently a product of nonspecific amplification by the archaeal DGGE primers.
DISCUSSION
The unique limnology of the Solar Lake, which includes high salinity and relatively high methane concentrations during certain periods of the year, led us to analyze the diversity and vertical distribution of archaeal populations in the water column.
DGGE was implemented to analyze the vertical distribution of Archaea sequences in the water column of the three profiles studied. It may be possible to correlate between the intensities of different bands that appear simultaneously in separate lanes and the relative abundance of each population for each lane (14, 28).
Analysis of 16S rRNA archaeal sequences via both cloning and DGGE imply that halophilic archaeal sequences belonging to cluster I are the dominant archaeal sequence throughout the water column in all of the three profiles analyzed. Of the 165 clones analyzed, 142 belonged to this cluster. In addition, DGGE bands correlating to cluster I sequences were present at all depths analyzed from the water column (Fig. 3). These bands appear to have stronger relative intensities in the oxic depths of the water column than in the anoxic depths (Fig. 3, lower gel). No PCR products were obtained from DNA extracted from 0.00 and 0.50 m from the December 1997 profile, where salinity values were 7.9 and 8.5%, respectively. The inability to detect sequences from species affiliated with cluster I may be because salinity values in these samples are below the minimum required for growth of this organism. Clone JDS-1 from cluster I was detected in oxic as well as anoxic layers where sulfide and methane concentrations are relatively high. In addition, it was detected at various depths with temperatures ranging from 15 to 55°C. This may imply that this organism is capable of surviving under diverse ecological conditions. Sequences belonging to cluster I are closely related to halophilic archaeal sequences detected in the cyanobacterial mats (Minz et al., unpublished) as well as in salterns in Eilat, Israel (Minz et al., unpublished) and in Alicante, Spain (2). Although molecular techniques indicated that the sequences detected in the Alicante salterns were dominant in these salterns, none of the halophilic Archaea isolated from them belonged to this group (2). This demonstrates the bias involved in using culturing techniques for the assessment of natural microbial populations. No cultured species phylogenetically related to cluster I have been identified to date, and therefore the physiological properties of this group are unknown.
Relatively high methane concentrations were found in the hypolimnion of Solar Lake. The December 1997 profile was characterized by a peak in methane concentration (6.5 μM) directly below the chemocline (Fig. 1). This peak suggests that biological methane was produced in this zone. However, no sequences grouping within clusters of cultured methanogens were detected from this section or any other section of the water column. All 29 sequences analyzed from this area belonged to the halophilic cluster I. Methane concentrations of up to 17 μM were detected close to the sediment-water interface in June 1997. However, no sequences related to cultured methanogens were identified in the water column or the cyanobacterial mats (Minz et al., unpublished).
The inability to detect known methanogens could be due to a number of reasons. (i) Species related to known methanogens may exist in the water column but represent a small fraction of the archaeal population, and thus the PCR analysis used was not sensitive enough to detect them. (ii) Clusters II, III, and IV are phylogenetically distant from cultured Archaea and thus nothing is known about their physiological characteristics. It is possible that members of these clusters represent novel methanogen types that have not yet been cultured. Unlike halophilic Archaea that are monophyletic, methanogens are a polyphyletic group represented by a number of clusters throughout the euryarchaeotal kingdom. Therefore, additional methanogen clades may exist that contain species not yet cultured.
Cluster II is phylogenetically located between the genera Methanobacterium and Methanococcus, and therefore the species affiliated with cluster II could also be H2- and CO2-utilizing methanogens.
Cluster III contains clones from anaerobic layers of the lake where methane concentrations were high. A number of sequences isolated from methane-rich rice paddy sediments phylogenetically group with this cluster (17).
Species affiliated with cluster IV include a number of sequences that form a monophyletic cluster proximal to the genus Thermoplasma. This cluster has been described previously by DeLong (10), who designated these sequences as Euryarcheotal group II. These sequences have been isolated from environments such as coastal salt marsh sediments (26) and methane-rich Pacific marine sediments (18), where a number of sequences closely resembling methanogens have been identified. In addition, sequences from areas such as the water column of the Santa Barbara Channel (24), the sediment of a boreal forest lake (Jurgens et al., unpublished), and the intestines of two marine fish (39) also resemble sequences from cluster IV. The wide ecological diversity of these species could indicate that they are ecologically significant. DGGE analysis implied that this group was most prevalent at anoxic depths of 4.5 and 4.75 m in the June 1997 profile. This may indicate that these species originate in the sediment and migrate into the water column. Similar sequences were also detected in the Solar Lake cyanobacterial mats (Minz et al., unpublished).
This research revealed a wide diversity of archaeal sequences in Solar Lake. None of these sequences were closely related to any cultured organism. Thus, we have no information concerning the ecological role of these species. In order to evaluate the importance of Archaea in natural ecosystems, vital information on the genotypes and phenotypes of these organisms is required.
ACKNOWLEDGMENTS
This research was financially sponsored by a grant of The Red Sea Program for Marine Sciences, No. 03F0151A, of the German Federal Ministry of Education and Research (BMBF). In addition, D.M. was sponsored by a grant from the ONR, No. N00014-95-1-00887.
REFERENCES
- 1.American Public Health Association. Standard methods for the examination of water and wastewater. Washington, D.C.: American Public Health Association; 1971. pp. 475–481. [Google Scholar]
- 2.Benlloch S, Acinas S, Martinez-Murcia A J, Rodriguez-Valera F. Description of prokaryotic biodiversity along the salinity gradient of a multipond solar saltern by direct PCR amplification. Hydrobiologia. 1996;329:19–31. [Google Scholar]
- 3.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cline J D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 5.Cohen Y, Krumbein W E, Shilo M. Solar Lake (Sinai) II: distribution of photosynthetic microorganisms and primary production. Limnol Oceanogr. 1977;22:609–620. [Google Scholar]
- 6.Cohen Y, Krumbein W E, Goldberg M, Shilo M. Solar Lake (Sinai) I: physical and chemical limnology. Limnol Oceanogr. 1977;22:597–608. [Google Scholar]
- 7.Conrad R, Frenzel P, Cohen Y. Methane emission from hypersaline microbial mats: lack of aerobic methane oxidation activity. FEMS Microbiol Ecol. 1995;16:251–259. [Google Scholar]
- 8.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong E F, Ying Wu K, Prezelin B B, Jovine R V M. High abundance of archaea in Antarctic picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 10.DeLong E F. Everything in moderation: archaea as ‘non-extremophiles’. Curr Opin Genet Dev. 1998;8:649–654. doi: 10.1016/s0959-437x(98)80032-4. [DOI] [PubMed] [Google Scholar]
- 11.DiPasquale M, Oren A, Cohen Y, Oremland R S. Radiotracer studies of bacterial methanogenesis in sediments from the Dead Sea and Solar Lake. In: Oren A, editor. Microbiology and biochemistry of hypersaline environments. Boca Raton, Fla: CRC Press; 1998. pp. 149–160. [Google Scholar]
- 12.Dojka M A, Jr, Hugenholz P, Haack S, Pace N R. Microbial diversity in a hydrocarbon and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eder W, Ludwig W, Huber R. Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea. Arch Microbiol. 1999;172:213–218. doi: 10.1007/s002030050762. [DOI] [PubMed] [Google Scholar]
- 14.Ferris M J, Muyzer G, Ward D M. Denaturating gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 16.Giani D, Giani L, Cohen Y, Krumbein W. Methanogenesis in the hypersaline Solar Lake. FEMS Microbial Lett. 1984;25:219–224. [Google Scholar]
- 17.Großkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinrichs K, Hayes J, Sylva S, Brewer P, DeLong E F. Methane-consuming archaebacteria in marine sediments. Nature. 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 19.Jørgensen B B, Kuenen J G, Cohen Y. Microbial transformations of sulfur compounds in a stratified lake (Solar Lake, Sinai) Limnol Oceanogr. 1979;24:799–822. [Google Scholar]
- 20.Kamekura M. Diversity of extremely halophilic bacteria. Extremophiles. 1998;2:289–295. doi: 10.1007/s007920050071. [DOI] [PubMed] [Google Scholar]
- 21.Krumbein W E, Cohen Y, Shilo M. Solar Lake (Sinai). Stromatalitic cyanobacterial mats. Limnol Oceanogr. 1977;22:635–656. [Google Scholar]
- 22.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin D, Ciulla R, Roberts M. Osmoadaptation in archaea. Appl Environ Microbiol. 1999;65:1815–1825. doi: 10.1128/aem.65.5.1815-1825.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minz D, Green S J, Flax J L, Muyzer G, Cohen Y, Wagner M, Rittmann B E, Stahl D A. Diversity in sulfate reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl Environ Microbiol. 1999;65:4666–4671. doi: 10.1128/aem.65.10.4666-4671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munson M, Nedwell D, Embley M. Phylogenetic diversity of archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyzer G, Brinkhoff T, Nubel U, Santegoeds C, Schafer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. 1997. pp. 1–27. , 3.4.4. In Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherldands. [Google Scholar]
- 28.Muyzer G. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol. 1999;2:317–322. doi: 10.1016/S1369-5274(99)80055-1. [DOI] [PubMed] [Google Scholar]
- 29.Ollivier B, Caumette P, Garcia J-L, Mah R. Anaerobic bacteria from hypersaline environments. Microbiol Rev. 1994;58:27–38. doi: 10.1128/mr.58.1.27-38.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oremland R S, Miller L S, Whiticar M J. Sources and flux of natural gasses from Mono Lake, California. Geochim Cosmochim Acta. 1987;51:2915–2929. [Google Scholar]
- 31.Oremland R S, King G M. Methanogenesis in hypersaline environments. In: Cohen Y, Rosenberg E, editors. Microbial mats: physiological ecology of benthic microbial communities. Washington, D.C.: American Society for Microbiology; 1989. pp. 180–190. [Google Scholar]
- 32.Oren A. The order Halobacteriales. In M. Dworkin, S. Fallow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrad (ed.), The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. 3rd ed. New York, N.Y: Springer-Verlag; 1999. [Google Scholar]
- 33.Oren A. The halophilic Archaea—evolutionary relationships and adaptation to life at high salt concentrations. In: Vasser S P, Nevo E, editors. Evolutionary theory and processes: modern prespectives. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 345–361. [Google Scholar]
- 34.Øvreas L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Revsbech N P, Jørgenson B B, Blackburn T H, Cohen Y. Microelectrode study of photosynthesis and O2, H2S and pH profiles in a microbial mat. Limnol Oceanog. 1983;28:1075–1093. [Google Scholar]
- 37.Stetter K O, Fiola G, Hubber O, Hubber R, Segerer A. Hyperthermophilic microorganisms. FEMS Microbiol Rev. 1990;75:117–120. [Google Scholar]
- 38.Strunk O, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Technische Universität München; 1996. [Google Scholar]
- 39.van der Maarel M J E C, Artz R R E, Haanstra R, Forney L J. Association of marine archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol. 1998;64:2894–2898. doi: 10.1128/aem.64.8.2894-2898.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winfrey M R, Ward D M. Substrates for sulfate reduction and methane production in intertidal sediments. Appl Environ Microbiol. 1983;45:193–199. doi: 10.1128/aem.45.1.193-199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria and Eukarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]