Abstract
Sensitive serological testing is essential to estimate the proportion of the population exposed or infected with SARS-CoV-2, to guide booster vaccination and to select patients for treatment with anti-SARS-CoV-2 antibodies. The performance of serological tests is usually evaluated at 14–21 days post infection. This approach fails to take account of the important effect of time on test performance after infection or exposure has occurred. We performed parallel serological testing using 4 widely used assays (a multiplexed SARS-CoV-2 Nucleoprotein (N), Spike (S) and Receptor Binding Domain assay from Meso Scale Discovery (MSD), the Roche Elecsys-Nucleoprotein (Roche-N) and Spike (Roche-S) assays and the Abbott Nucleoprotein assay (Abbott-N) on serial positive monthly samples collected as part of the Co-STARs study (www.clinicaltrials.gov, NCT04380896) up to 200 days following infection. Our findings demonstrate the considerable effect of time since symptom onset on the diagnostic sensitivity of different assays. Using a time-to-event analysis, we demonstrated that 50% of the Abbott nucleoprotein assays will give a negative result after 175 days (median survival time 95% CI 168–185 days), compared to the better performance over time of the Roche Elecsys nucleoprotein assay (93% survival probability at 200 days, 95% CI 88–97%). Assays targeting the spike protein showed a lower decline over the follow-up period, both for the MSD spike assay (97% survival probability at 200 days, 95% CI 95–99%) and the Roche Elecsys spike assay (95% survival probability at 200 days, 95% CI 93–97%). The best performing quantitative Roche Elecsys Spike assay showed no evidence of waning Spike antibody titers over the 200-day time course of the study. We have shown that compared to other assays evaluated, the Abbott-N assay fails to detect SARS-CoV-2 antibodies as time passes since infection. In contrast the Roche Elecsys Spike Assay and the MSD assay maintained a high sensitivity for the 200-day duration of the study. These limitations of the Abbott assay should be considered when quantifying the immune correlates of protection or the need for SARS-CoV-2 antibody therapy. The high levels of maintained detectable neutralizing spike antibody titers identified by the quantitative Roche Elecsys assay is encouraging and provides further evidence in support of long-lasting SARS-CoV-2 protection following natural infection.
Subject terms: Infectious diseases, Viral infection
Introduction
Following natural infection or vaccination, sensitive measurement of SARS-CoV-2 serological status is important to identify immune correlates of protection from future waves of the pandemic, evaluate those in need of booster vaccination and identify candidates for SARS-CoV-2 antibody therapy. The rapid response to the COVID-19 pandemic has led to the development of a wide range of serological tests suitable for evaluating SARS-CoV-2 exposure, infection or vaccination status1–3. Typically, these tests are approved for use by the regulatory authorities based on their performance against a panel of reference sera including positive and negative controls at either 14- or 21-days post infection4.
Public Health England reported a 93.9% sensitivity for the Abbott SARS-CoV-2 IgG Nucleoprotein assay5 and 100% for the Roche Elecsys Nucleoprotein assay at ≥ 14 days post infection6. This led to widespread adoption of these tests across NHS laboratories for testing at population level. Other studies have confirmed this test performance at 14–21 days post infection7,8. Population level serological studies have also based their conclusions—vital to guide national policy—on the basis of these tests9 without considering how time since infection influences the performance of the test. The problem with this approach is that it does not take into account SARS-CoV-2 humoral dynamics and changes in avidity over time10,11. Although serological tests with limited diagnostic range may demonstrate excellent sensitivity shortly after infection, it is unclear how they will perform with time following infection or vaccination.
In order to address this question, we applied 4 widely used serological assays in parallel to serial samples from the Co-STARs study12 in which staff testing seropositive to SARS-CoV-2 were followed for up to 200 days following infection. We compared the proportion of samples that remained seropositive over time using a survival analysis and determined the decay rate of the nucleoprotein (N) antibody and the spike (S) antibody for each test using a previously published mathematical model fitted to the data.
Materials and methods
Study setting and design
Serological testing was performed on stored serum samples collected as part of the Co-STARs study (www.clinicaltrials.gov, NCT04380896), approved by the UK National Health Service Health Research Authority and run at Great Ormond Street Hospital between April 29th and November 2020 in accordance with the relevant guidelines10. Briefly, Co-STARs was a 1-year single-centre prospective cohort study of antibody responses to COVID-19 infection in healthcare workers. Serum samples were taken from the 3657 participants at baseline and underwent a screening ELISA using the EDI assay. Repeated monthly serum samples were then taken from those with a seropositive baseline screening test for up to 250 days after the date of infection. Written informed consent was obtained from all participants. Those samples identified as seropositive with available symptom start date had further confirmatory testing with the quantitative three antigen MSD assay.
Study participants
The majority of hospital staff were eligible for the Co-STARS study12. Only those participants with significant immunosuppression, those that had received blood products within 6 months of recruitment and those that had active and ongoing symptoms of SARS-CoV-2 infection (within the last 21 days) were excluded. Only samples from individuals with at least one positive test from any platform were included in the analysis. Moreover, individuals without a known symptom start date were removed.
Data collection
As part of the Co-STARs study all participants undertook a detailed standardised online questionnaire at study entry12. This included the date of onset of COVID-19 symptoms, and any SARS-CoV-2 diagnostic test results.
Comparison of serological assays
Samples taken as part of the Co-STARS study12 which had an accompanying symptom start date available for analysis were initially screened for seropositivity by the EDI assay or by any of the three antigens of the Meso Scale Discovery (MSD) assay. The selected samples each underwent testing with 4 serological assays: (1) The Roche Elecsys Anti-SARS-CoV-2 electrochemiluminescence immunoassay (ECLIA) assay detects the nucleocapsid (N) antigen (Roche-N); (2) the Roche Elecsys Anti-SARS-CoV-2 S electrochemiluminescence immunoassay (ECLIA) assay detects the spike (S) antigen (Roche-S); (3) the Abbott Nucleoprotein Chemiluminescent Microparticle Immunoassay (CLMIA) assay detects the nucleocapsid (N) antigen (Abbott-N); (4). All tests were performed as per manufacturer’s specifications.The four antigen Meso Scale Discovery (MSD) assay was undertaken at the WHO Pneumococcal Supranational Reference Laboratory at the UCL Institute of Child Health. Only 3 antigens were reported from the MSD assay (the Spike, the Nucleoprotein and the RBD) as the baseline test performance of the N-terminal domain (NTD) antibody response was insufficient for further evaluation as previously reported13. The Roche-N and Roche-S assays were undertaken by the Laboratory Medicine Service of Swansea Bay University Health Board, Morriston Hospital, Swansea. The Abbott-N assay was undertaken by Public Health Wales Microbiology at Cardiff and Vale University Hospital. All samples were stored and transported between laboratories at − 80 °C and only removed for aliquoting prior to testing to avoid unnecessary freeze–thaw cycles.
Statistical analysis and modelling
In order to evaluate the relative proportion of seropositive tests in the parallel serological assays over time, a time-to-event analysis was performed using the time from symptom onset and the first negative test for each assay after a first positive test as the event of interest using the R package survival14,15. Only tests taken > 14 days after symptom onset were considered in the analysisNo tests were performed between 14- and 21-days post symptoms, and thus using 14 or 21 days post symptom onset as threshold did not affect our results. A participant was defined as seropositive when at least one of the 4 tests undertaken was seropositive. If the other tests that were run in parallel never became seropositive, the time-to-event was set to the earliest test taken for that individual. If a participant never became seronegative during the follow-up period, a right-censored observation was added at the time of the last serological test.
Additionally, the decay rate after 21 days since symptom onset was estimated using a Bayesian generalized linear mixed model as implemented in the R package MCMCglmm16, where time from symptom onset was included as a fixed effect and study participants as a random effect. Therefore, a unique slope for the regression was estimated for the entire population, while the intercept was allowed to vary between the study participants. The decay rate was estimated from the slope of the linear model.
To assess the overall diagnostic capability of the Abbott-N assay, a receiver operating characteristic curve (ROC) analysis was performed using the pROC package within R17. The MSD-N and Roche-N assays were used as the gold standard for the comparison.
Ethical approval and consent
The study had national Integrated Research Application System (IRAS) approval and all participants in the study provided informed consent.
Results
A total of 950 samples from 329 participants seropositive by any assay after 14 days underwent testing with the Roche-N, Roche-S, the MSD and the Abbott-N assay. The majority of the participants (98%, 321/329) had a positive result by two or more assays.
Antibody decay with time
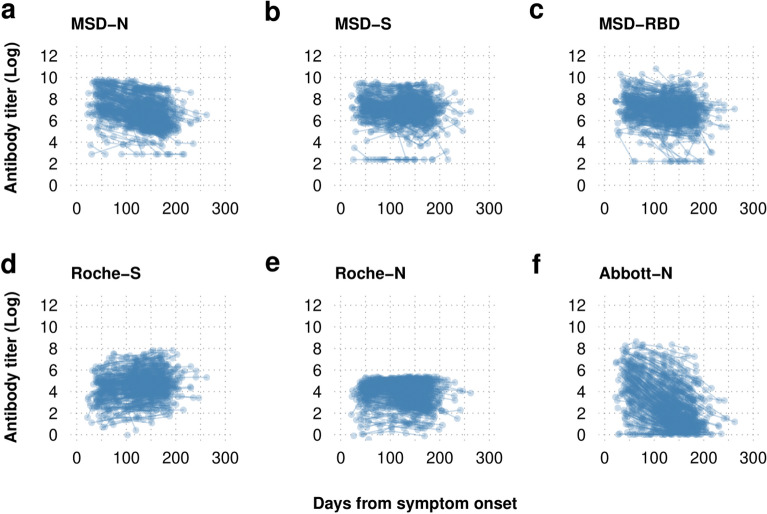
Plotting the raw log transformed antibody titers over time since symptom onset (Fig. 1) demonstrated that antibody dynamics were dependent on the assay undertaken. The production of spike antibodies was demonstrated to be maintained at high levels up to 200 days when evaluated by the MSD and the quantitative Roche -S assay. All nucleoprotein antibody assays demonstrated decay of the nucleoprotein antibody over time. This was most pronounced in the Abbott-N assay and much less so in the Roche -N assay which demonstrated slow waning of the nucleoprotein antibody.
Figure 1.
Log transformed serial serological antibody titer data plotted by time from symptom onset. Antibody dynamics are dependent on the assay used with the sensitive Roche-S and MSD-S assay demonstrating maintenance of the spike protein antibody while the nucleoprotein antibody is shown to wane with the MSD and Abbott-N assays but to a lesser extent with the Roche-N assay.
Assay sensitivity with time post symptom onset
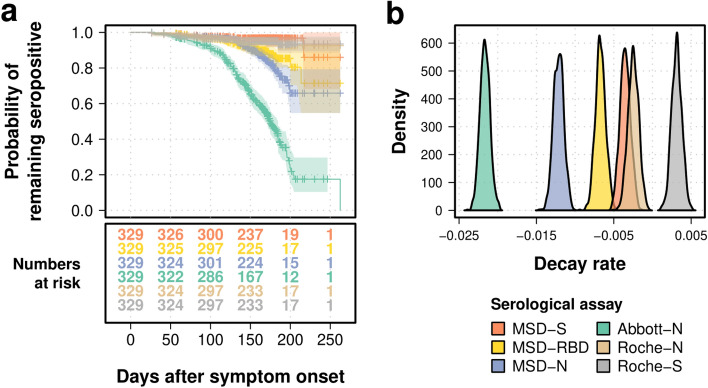
The existing published test performance for all assays undertaken is provided in Table 1. The sensitivity of all assays (at least 14 days from symptom onset) at 50, 100 and 150 days is provided in Table 2. All assays demonstrated a reasonable sensitivity at 50 days following infection (Fig. 2a). As time passed following infection, the Abbott-N assay rapidly became seronegative (Fig. 2a), with a median survival time inferred at 175 days (95% CI 168–185 days), whereas the survival probability at 150 days was inferred to be 95% for the Roche-N (95% CI 0.92–0.97), and 91% for the MSD-N assay (95% CI 0.87–0.94). The Roche-S and MSD-S assays remained seropositive for the duration of the study. The MSD-RBD assay showed some evidence of waning seropositivity over time (90% Survival probability at 150 days, 95% CI 0.88–0.94).
Table 1.
A summary of the existing published data for the commercially available tests in this comparison.
| Tests | Manufacturer | Target Antigen | Type | Sens† | Spec | # Samples/Patients* | Days post symptom onset |
|---|---|---|---|---|---|---|---|
| Abbott-N18 | Abbott-N | Nucleoprotein | CLMIA | 100.0% (day 17) | 99.9% | 689/125 | ≥ 21 |
| Roche-N19 | Roche Cobas | Nucleoprotein | ECLIA | 99.5% | 99.8% | 496/102 | ≥ 14 |
| Roche-S20 | Roche Cobas | Spike protein | ECLIA | 96.6% | 100% | 1485‡/331 | ≥ 15 |
| MSD13 | Meso Scale Discovery | Nucleoprotein | ECLIA | 87.2% | 92.8% | 196/196 | ≥ 21 |
| Spike protein | 97.9% | 97.4% | 47/47 | ≥ 21 | |||
| RBD (receptor binding domain) | 93.6% | 92.3% | 47/47 | ≥ 21 |
The corresponding antigen target and published sensitivity, specificity, positive predictive value and negative predictive value (if available).
ECLIA Electro Chemiluminescent Immunoassay, CLMIA chemiluminescent microparticle immuno assay, PPV positive predictive value, NPV negative predictive value, Sens sensitivity, Spec specificity, ELISA enzyme linked immunosorbent assay.
†The highest reported sensitivity.
‡233 of these samples were tested at ≥ 15 days post PCR diagnosis.
*For sensitivity testing.
Table 2.
Sensitivity of compared assays at 50, 100 and 150 days from symptom onset.
| Survival probability (95% CI) | |||
|---|---|---|---|
| 50-day | 100-day | 150-day | |
| Abbott-N | 0.985 (0.97;0.99) | 0.919 (0.89;0.95) | 0.655 (0.6;0.71) |
| Roche-N | 0.988 (0.98;1.0) | 0.963 (0.94;0.98) | 0.949 (0.92;0.97) |
| Roche-S | 0.991 (0.98;1.0) | 0.966 (0.95;0.99) | 0.952 (0.93;0.98) |
| MSD | |||
| N | 0.988 (0.98;1.0) | 0.972 (0.95;0.99) | 0.907 (0.87;0.94) |
| S | 0.997 (0.99;1.0) | 0.978 (0.96;0.99) | 0.968 (0.95;0.99) |
| RBD | 0.994 (0.99;1.0) | 0.969 (0.95;0.99) | 0.909 (0.88;0.94) |
Figure 2.
Comparison of seropositivity and antibody dynamics between serological tests. The Roche-S assay target the spike antibody, the Abbott-N and the Roche-N assays target the N-antibody while the MSD assay targets the N-, the S- and the antibody to the Receptor Binding Domain (RBD) of the spike protein in parallel. (a) Kaplan–Meier curve and numbers at risk (the number of participants under follow up with serological tests available for analysis at that time point) for different serological tests. Y-axis represents the probability of remaining seropositive, while the X-axis shows days after symptom onset with numbers of participants under follow up shown in the table below. (b) Inferred posterior density distributions of the decay rate in a generalized linear mixed model.
A total of 45% (159/329) of the individuals had a negative result using the Abbott-N assay during the course of the study. For the MSD test, 16% (52/329) of participants had a negative test for the N antigen, 11% (36/329) for RBD, and 3% (11/329) for the S antigen. For the Roche platform, 5.5% (18/329) of the individuals had a negative result with the Roche-N assay, while only 4.8% (16/329) of them had a negative result for the S antigen over the course of the study.
Mathematical model fits to estimate antibody decay
To estimate the decay rate for each antibody and assay studied, a generalized linear mixed model was fitted to the trajectory of antibody decay after 21 days from symptom onset, where the decay rate was estimated as the slope of the antibody titer through time. Under the most sensitive and quantitative Roche -S assay the spike antibody demonstrated no decay at all and rather a slow rate of increased titers over time from symptom onset (0.0031, 95% CI 0.0018–0.0044, Fig. 2b). In accordance with the raw observed data, all nucleoprotein antibodies under the mathematical model decayed. This was most pronounced in the Abbott-N assay (− 0.022, 95% CI − 0.023 to − 0.02) and least pronounced in the Roche -N assay (− 0.0025, 95% CI − 0.0039 to − 0.0012, Fig. 2b, Table 3).
Table 3.
Decay rate for each serological assay (log arbitrary units per day) estimated in a generalized linear mixed model.
| Mean | 95% CI | |
|---|---|---|
| MSD-N | − 0.0121 | − 0.0134; − 0.0107 |
| MSD-RBD | − 0.0068 | − 0.008; − 0.0055 |
| MSD-S | − 0.0035 | − 0.0048; − 0.0023 |
| Abbott-N | − 0.0216 | − 0.0229; − 0.0204 |
| Roche-S | 0.0031 | 0.0018;0.0044 |
| Roche-N | − 0.0025 | − 0.0039; − 0.0012 |
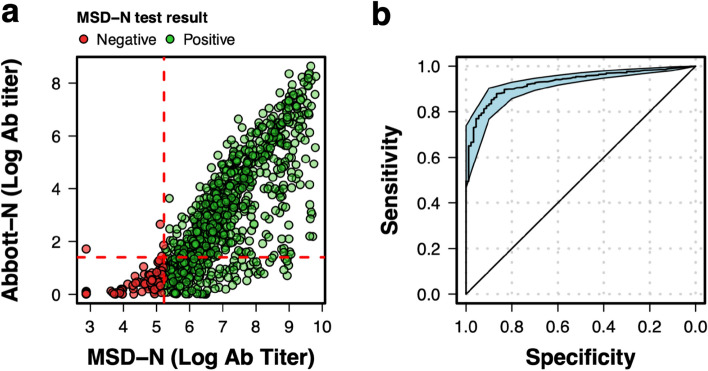
The lower performance of the Abbott-N assay can be explained by a lower detection of titer values as their concentration wanes over time. When compared to the quantitative MSD-N, 26% (222/860) of all positive samples by the MSD-N were negative for the Abbott-N test (Fig. 3a). A total of 75% of samples (137/ 183) positive by the MSD-N with an MSD arbitrary titer value lower than 403 were negative for the Abbott-N assay. Using the currently manufacturer recommended threshold of 1.4 arbitrary units, the Abbott-N test was characterized by a high specificity of 0.96 and a sensitivity of 0.74 using all our test results after 14 days. Using a ROC curve (Fig. 3b), the optimal cut-off that maximises both specificity and sensitivity was estimated to be 0.845 arbitrary units.
Figure 3.
Comparison of antibody titers between the Abbott-N assay and the MSD-N assay. (a) The quantitative results for the MSD-N assay were compared to those of the Abbott-N test for each sample taken. Colours divide the samples depending on whether it was positive (green) or negative (red) for the MSD-N assay. Dotted red lines represent the seropositivity threshold for the Abbott-N assay (horizontal) and the MSD-N test (vertical). (b) ROC curve for the Abbott-N assay using the MSD-N test as gold standard. The x ~ y line represents the profile of a random classifier. Blue shaded area shows the 95% CI.
Discussion
Sensitive measurement of SARS-CoV-2 seropositivity is key to evaluate who has been infected or exposed to SARS-CoV-2, to determine the correlates of protection from future disease, stratify those that need booster vaccination and target the use of anti-SARS-CoV-2 antibodies to those that are seronegative. To our knowledge no other study has evaluated the sensitivity of multiple diagnostic tests in parallel on longitudinally collected serological samples. This study demonstrates that as time elapses after infection, the sensitivity of serological testing varies widely depending on the test used. Although serological tests may be demonstrated to perform well 14–21 days after infection, this initial test performance often diminishes as time passes. In order to evaluate whether or not the population maintains SARS-CoV-2 antibodies it is vital that we utilize serological tests that remain sensitive over time.
Initial published baseline test performance reports concluded that the Abbott-N assay was a high-performance test and a key tool in SARS-CoV-2 surveillance18. Our data demonstrate that as time passes following infection the sensitivity of this assay declines rapidly until at < 6 months following infection it is no more than 50% sensitive. Our findings support the concerns raised by others regarding the poor performance of some nucleoprotein based assays21,22.
In contrast, the Roche assays, particularly the Roche Elecsys Anti-SARS-CoV-2 Spike assay maintained high sensitivity for the 200-day duration of the study. Although there remains no single correlate of sterilizing or protective immunity following SARS-CoV-2 infection or vaccination, it is clear that natural infection and the presence of neutralizing spike antibodies decreases the possibility of re-infection and the severity of disease upon re-exposure to currently circulating strains23. Our finding that spike antibodies remained at high titers 200 days after infection adds to our previous study on this topic10 and provides further evidence in support of long-lasting protection against severe disease from currently circulating strains. Fitting mathematical models to the raw data of the Roche spike assay demonstrated that spike antibody titers did not decay but rather increased slightly over the duration of the study. The Roche nucleoprotein assay also maintained sensitivity for the duration of the study with a low rate of decay. Although this assay is semi-quantitative, our findings suggest that this could be used to sensitively identify those that have been vaccinated from those that have been both vaccinated and infected.
Many studies have evaluated the impact of time on test sensitivity over the first 3 weeks following symptom onset24–26. However, we found no other study that had examined the sensitivity of antibody testing on parallel longitudinal samples collected between 1 and 6 months after infection or exposure. Assays with a higher titer cut-off for detection may perform well in the initial period after infection, but fail to detect seropositivity as antibody levels wane over time. We show that the Abbott-N test failed to detect 75% of samples positive for the MSD-N with a titer value lower than 403, which makes the Abbott-N assay less suitable for seroprevalence studies. Using a ROC curve and the MSD-N and Roche-N assays as the gold standard, we showed that a lower threshold of 0.845 instead of 1.4 arbitrary units may be more suitable to optimize the sensitivity and specificity. Even though different thresholds may be relevant depending on whether sensitivity or specificity needs to be prioritized, our findings suggest that the high Abbott-N test threshold results in a high number of false negatives. These findings are concordant with previous reports showing a range of high uncertainty between 0.49 and 1.427. Barzin et al.28 used Abbott-N testing alone to determine SARS-CoV-2 seroprevalence in 2,973 asymptomatic out-patients in North Carolina estimating a seroprevalence of 0.8%. Similarly, Wilkins et al.29 used Abbott-N on 6510 healthcare workers up to 150 days after symptom onset and estimated a seroprevalence of 4.8%. Our findings suggest that previously published surveys of SARS-CoV-2 seroprevalence such as these could have significantly underestimated the true prevalence of SARS-CoV-2 humoral immunity.
Memory T-cell interferon gamma release or proliferation assays in response to SARS-CoV-2 antigens provide an alternative means of assessing prior exposure to infection. However, these assays are limited by cross reactive immunity to the seasonal coronaviruses decreasing specificity30,31.
Although all serological tests used in the study demonstrated a high initial specificity, one limitation is that only 38% of participants had a confirmatory SARS-CoV-2 PCR result. Our data may therefore be influenced by an unknown proportion of falsely positive serological tests. However, at entry to the study, all seropositive participants had both a screening EDI nucleoprotein assay and an MSD assay performed which limited the chances of a falsely positive result due to a single erroneous test. Not all samples were processed at the same time; the Roche and Abbott-N assays were processed 3 months after the MSD assays. Despite this, we believe that sample storage and freeze-thawing cycles are unlikely to have influenced our findings as the Roche quantitative spike assay was performed last and demonstrated the highest prolonged levels of spike antibody of all tests used.
In summary, although serological tests may demonstrate high sensitivity 3-weeks after SARS-CoV-2 infection, this is far from the case with some tests 6-months after infection. The Abbott-N assay performed poorly at this time, whereas the Roche and MSD tests maintained a high sensitivity for the 200 days of the study. Tests that perform poorly over time will lead to spurious estimates in population level seroprevalence studies and findings from these studies should be adjusted to account for sensitivity of the test used and the time since infection. Test performance as time passes post infection should be considered before evaluating who is a candidate for booster vaccination or anti-SARS-CoV-2 antibody therapy.
Supplementary Information
Acknowledgements
We would like to dedicate this article to the staff members who died of COVID-19 at Great Ormond Street Hospital during the first wave of the pandemic. We would also like to thank all the staff at Great Ormond Street Hospital who have taken part in the study. In addition, we are very grateful for all the hard work undertaken by the Great Ormond Street rapid response and microbiology laboratory staff and the staff in the immunology laboratories both in the Camelia Botnar Laboratory and the Great Ormond Street Institute of Child Health who ensured that all the PCR tests and serological assays were completed in a timely manner. The authors would also like to thank the biomedical scientists and colleagues in the affiliated laboratories for their hard work in running the samples and collating the results. RD would like to thanks Linda Tyson and Jon Evans of the Wales National Virology Centre, PHW Microbiology Cardiff. RS would like to thank Non Gwynn, Jenna Walters, James Murphy and colleagues in Laboratory Medicine, Morriston Hospital, Swansea.
Author contributions
The study was designed and implemented by: L.G., A.T.O., J.H., R.D., R.S., D.G. and K.G. Testing for SARS-CoV-2 RNA and antibody was conducted by: L.G., F.T., A.T., A.L.M., K.G., J.H., A.S., T.L., M.J., H.W., M.B., R.D., R.S. and D.G. Data analysis was carried out by A.T.O. and L.G. The first draft of the manuscript was prepared by: A.T.O., F.T. and L.G., with input from all co-authors.
Funding
LG was supported by the Wellcome Trust (201470/Z/16/Z), the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number 1R01AI146338, the GOSH Charity (VC0921) and the GOSH/ICH Biomedical Research Centre (www.nihr.ac.uk). AS was supported by the Wellcome Trust (220565/Z/20/Z). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The dataset used during the current study is available as Supplementary Data File 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-14351-2.
References
- 1.Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui L-P, Johnston JC, et al. Diagnostic accuracy of serological tests for covid-19: Systematic review and meta-analysis. BMJ. 2020 doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubina R, Dziedzic A. Molecular and serological tests for COVID-19. A comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10:434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.la Marca A, Capuzzo M, Paglia T, Roli L, Trenti T, Nelson SM. Testing for SARS-CoV-2 (COVID-19): A systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod. Biomed. Online. 2020;41:483–499. doi: 10.1016/j.rbmo.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng MP, Yansouni CP, Basta NE, Desjardins M, Kanjilal S, Paquette K, et al. Serodiagnostics for severe acute respiratory syndrome-related coronavirus 2: A narrative review. Ann. Intern. Med. 2020;173:450–460. doi: 10.7326/M20-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Public Health England. Evaluation of the Abbott SARS-CoV-2 IgG for the Detection of Anti-SARSCoV-2 Antibodies. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/890566/Evaluation_of_Abbott_SARS_CoV_2_IgG_PHE.pdf (2020).
- 6.Public Health England. Evaluation of Roche Elecsys AntiSARS-CoV-2 Serology Assay for the Detection of Anti-SARS-CoV-2 Antibodies. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/891598/Evaluation_of_Roche_Elecsys_anti_SARS_CoV_2_PHE_200610_v8.1_FINAL.pdf (2020).
- 7.Coste AT, Jaton K, Papadimitriou-Olivgeris M, Greub G, Croxatto A. Comparison of SARS-CoV-2 serological tests with different antigen targets. J. Clin. Virol. 2021;134:104690. doi: 10.1016/j.jcv.2020.104690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ainsworth M, Andersson M, Auckland K, Baillie JK, Barnes E, Beer S, et al. Performance characteristics of five immunoassays for SARS-CoV-2: A head-to-head benchmark comparison. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollán M, Pérez-Gómez B, Pastor-Barriuso R, Oteo J, Hernán MA, Pérez-Olmeda M, et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): A nationwide, population-based seroepidemiological study. The Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandjean L, Saso A, Torres Ortiz A, Lam T, Hatcher J, Thistlethwayte R, et al. Long-term persistence of spike protein antibody and predictive modeling of antibody dynamics after infection with severe acute respiratory syndrome coronavirus 2. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer G, Struck F, Schreiner P, Staschik E, Soutschek E, Motz M. The challenge of avidity determination in SARS-CoV-2 serology. J. Med. Virol. 2021;93:3092–3104. doi: 10.1002/jmv.26863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Great Ormond Street Hospital for Children NHS Foundation Trust. COVID-19 Staff Testing of Antibody Responses Study (CO-STARS). https://clinicaltrials.gov/ct2/show/NCT04380896 (2021).
- 13.Johnson M, Wagstaffe HR, Gilmour KC, Mai AL, Lewis J, Hunt A, et al. Evaluation of a novel multiplexed assay for determining IgG levels and functional activity to SARS-CoV-2. J. Clin. Virol. 2020;130:104572. doi: 10.1016/j.jcv.2020.104572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2018. [Google Scholar]
- 15.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer; 2000. [Google Scholar]
- 16.Hadfield JD. MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R package. J. Stat. Softw. 2010;33:1–22. doi: 10.18637/jss.v033.i02. [DOI] [Google Scholar]
- 17.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J, Müller M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:1–8. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryan A, Pepper G, Wener MH, Fink SL, Morishima C, Chaudhary A, et al. Performance characteristics of the abbott architect sars-cov-2 igg assay and seroprevalence in Boise, Idaho. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muench P, Jochum S, Wenderoth V, Ofenloch-Haehnle B, Hombach M, Strobl M, et al. Development and validation of the Elecsys anti-SARS-CoV-2 immunoassay as a highly specific tool for determining past exposure to SARS-CoV-2. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.01694-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.FDA. Elecsys Anti-SARS-CoV-2 S - Instructions for Use 2021. https://www.fda.gov/media/144037.
- 21.Rosadas C, Randell P, Khan M, McClure MO, Tedder RS. Testing for responses to the wrong SARS-CoV-2 antigen? The Lancet. 2020;396:e23. doi: 10.1016/S0140-6736(20)31830-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolotin S, Tran V, Osman S, Brown KA, Buchan SA, Joh E, et al. SARS-CoV-2 seroprevalence survey estimates are affected by anti-nucleocapsid antibody decline. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiaa796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall VJ, Foulkes S, Charlett A, Atti A, Monk EJM, Simmons R, et al. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: A large, multicentre, prospective cohort study (SIREN) The Lancet. 2021;397:1459–1469. doi: 10.1016/S0140-6736(21)00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, et al. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piec I, English E, Thomas MA, Dervisevic S, Fraser WD, John WG. Performance of SARS-CoV-2 serology tests: Are they good enough? PLoS ONE. 2021;16:e0245914. doi: 10.1371/journal.pone.0245914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Ai J, Loeffelholz MJ, Tang YW, Zhang W. Meta-analysis of diagnostic performance of serology tests for COVID-19: Impact of assay design and post-symptom-onset intervals. Emerg. Microbes Infect. 2020 doi: 10.1080/22221751.2020.1826362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castro MDM, et al. Performance verification of the Abbott SARS-CoV-2 test for qualitative detection of IgG in Cali, Colombia. PLoS ONE. 2021;16:e0256566. doi: 10.1371/journal.pone.0256566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barzin A, Schmitz JL, Rosin S, Sirpal R, Almond M, Robinette C, et al. SARS-CoV-2 seroprevalence among a Southern U.S. population indicates limited asymptomatic spread under physical distancing measures. MBio. 2020 doi: 10.1128/mBio.02426-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkins JT, Gray EL, Wallia A, Hirschhorn LR, Zembower TR, Ho J, et al. Seroprevalence and correlates of SARS-CoV-2 antibodies in health care workers in Chicago. Open Forum Infect. Dis. 2021 doi: 10.1093/ofid/ofaa582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020 doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 31.Lipsitch M, Grad YH, Sette A, Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset used during the current study is available as Supplementary Data File 1.