Significance
Interspecific hybridization is a widespread phenomenon, but measuring its extent, directionality, and adaptive importance remains challenging. Ancient genomes, however, can help illuminate the history of modern organisms. Here, we present a genome retrieved from a 130,000- to 115,000-y-old polar bear and perform genome analyses of modern polar and brown bears throughout their geographic ranges. We find that the principal direction of ancient allele sharing was from brown bear into polar bear, although gene flow between them has likely been bidirectional. This partially inverts the current paradigm of unidirectional gene flow from polar into brown bear, and it suggests that polar bears were recipients of external genetic variation prior to their extensive population decline.
Keywords: bear evolution, climate change, comparative genomics, hybridization, Ursus
Abstract
The polar bear (Ursus maritimus) has become a symbol of the threat to biodiversity from climate change. Understanding polar bear evolutionary history may provide insights into apex carnivore responses and prospects during periods of extreme environmental perturbations. In recent years, genomic studies have examined bear speciation and population history, including evidence for ancient admixture between polar bears and brown bears (Ursus arctos). Here, we extend our earlier studies of a 130,000- to 115,000-y-old polar bear from the Svalbard Archipelago using a 10× coverage genome sequence and 10 new genomes of polar and brown bears from contemporary zones of overlap in northern Alaska. We demonstrate a dramatic decline in effective population size for this ancient polar bear’s lineage, followed by a modest increase just before its demise. A slightly higher genetic diversity in the ancient polar bear suggests a severe genetic erosion over a prolonged bottleneck in modern polar bears. Statistical fitting of data to alternative admixture graph scenarios favors at least one ancient introgression event from brown bears into the ancestor of polar bears, possibly dating back over 150,000 y. Gene flow was likely bidirectional, but allelic transfer from brown into polar bear is the strongest detected signal, which contrasts with other published work. These findings may have implications for our understanding of climate change impacts: Polar bears, a specialist Arctic lineage, may not only have undergone severe genetic bottlenecks but also been the recipient of generalist, boreal genetic variants from brown bears during critical phases of Northern Hemisphere glacial oscillations.
The polar bear (Ursus maritimus) has become a symbolic species for ascertaining the impact of climate change on biodiversity and species evolution. With their dependence on sea ice, polar bears owe their continuing survival to the future stability of the vast Arctic regions of the planet. In connection, given Pleistocene oscillations between glacial and interglacial periods, polar bear paleohistory must hold clues to future responses to changing Earth climates. High-coverage genomes from ancient polar bear remains could therefore provide invaluable insights regarding prior adaptative resilience of the species to extreme environmental fluctuations in the past. Moreover, should such ancient polar bears be appropriately placed in age, their paleogenomes could illuminate the lineage split from the species’ lower-latitude sister taxon, the brown bear (Ursus arctos), in addition to enlightening any postdivergence admixture between the two species. However, polar bear fossils are very rare, with most dating to the Holocene period (1–3). In 2012 (4), extensive genomic data were generated from 23 extant polar bears, and a draft genome was presented from a stratigraphically validated 130,000- to 115,000-y-old polar bear jawbone of Eemian interglacial age that was recovered from the Svalbard Archipelago of Norway (1, 5). At the time, that study successfully pushed the age record of a sequenced vertebrate genome toward the Middle Pleistocene, but the initial draft genome was of low coverage (<1× depth), limiting its utility in genome-scale analyses.
Several additional genomic studies have since sought to trace polar bear evolution, a species that has emerged for uncovering complex speciation processes associated with interspecific admixture and rapid evolutionary adaptation (6–10). Although the polar bear and brown bear are recognized as closely related yet highly distinct species, studies so far strongly point to ancient and even ongoing (11) introgressive hybridization between the two lineages. This work has mostly centered on polar bear admixture with brown bears in Alaska’s Alexander Archipelago, because mitochondrial haplotypes of brown bears in the archipelago today (the so-called ABC brown bears) are more similar to polar bear haplotypes than they are to haplotypes found in most non-ABC brown bears (3, 6, 12–14). The deep nesting of polar bears within the brown bear maternal lineage, along with the fact that several other, both modern and extinct, brown bear populations share mitochondrial haplotypes with polar bears (15–17), implies a much more complex evolutionary history beyond only the Alexander Archipelago. Indeed, analyses of bear nuclear genomes have suggested widespread allele sharing among polar bears and brown bears, including extinct Irish brown bears (7), albeit with the highest proportion of allele sharing found between polar bears and ABC brown bears (8, 9). The nature of this allele sharing has been interpreted to represent multiple polar bear introgressions into various brown bear lineages (7), but this directionality, although broadly accepted, is not conclusively established.
Population genomic analyses have also identified an ancient and drastic decline in polar bear effective population size over the past 300,000 y, reflecting the far lower genetic diversity among extant polar bears compared to brown bears (4, 9). The complex population histories of the two sister species have challenged models for estimating divergence times and left a conundrum concerning the age of the polar bear as a species. Applying an extended coalescence hidden Markov model based on isolation with migration, an initial split time between brown and polar bears and American black bear (Ursus americanus) was estimated to be ∼5 to 4 Ma, followed by a period of gene flow before a complete split ∼200 ka (4). However, other estimates have generally agreed on a much younger split, although spanning a large interval from ∼1.6 Ma to 200 ka (9, 18).
The complex model for polar bear evolution that suggests multiple introgression events from polar bear into brown bear (7) warrants further scrutiny with a more complete sampling of crucial North American brown bear populations and methodologies that permit explicit testing of alternative hypotheses of admixture directionality. Here, we present a 10× depth genome of the 130,000- to 115,000-y-old subfossil polar bear from the Norwegian Svalbard Archipelago and 10 new polar and brown bear genomes from contemporary zones of overlap in northern Alaska where the species may have come into increasing contact due to recent climatic changes. Using a more complete genome from this ancient polar bear and an extended sampling of extant bear populations, we compare 65 polar bear and brown bear genomes from throughout their geographic ranges to better characterize evolutionary splits and admixture between the species.
Results
Assembly of an Ancient Polar Bear Genome and New Modern Bear Genomes.
Using a strategy combining multiple sequencing library construction methods and both Ion Torrent and Illumina sequencing platforms, we assembled a 10.11× depth and 97% width of coverage ancient subfossil polar bear genome from the Svalbard Archipelago (hereafter denoted as APB). We mapped a total of almost 5 billion sequence reads to a chromosome-length assembly (19, 20) based on the polar bear draft genome UrsMar_1.0 (GCF_000687225.1) (9) (Datasets S1 and S2). Postmortem damage profiles of the mapped reads from each library sequenced identified typical patterns of nucleotide misincorporations at the reads ends (SI Appendix, section S4 and Fig. S2), as expected with degraded ancient DNA. To confirm the authenticity of the APB sequence data, we also assembled mitogenomes from each of the six separate libraries constructed for APB and added them to an alignment of mitogenomes of all modern bear samples analyzed as part of this study. Mitogenome data from all individual APB libraries grouped with the previously published mitogenome from the same ancient polar bear specimen (3) in a position sister to all modern polar bears (MPB) (SI Appendix, Fig. S3), demonstrating the endogenous nature of all new APB libraries.
To expand the geographic sampling of extant bear genomes (Table 1 and SI Appendix, Fig. S1) and evaluate contemporary admixture among polar bears and brown bears, we also generated 10 new modern bear genomes of 9 to 28× sequence depth coverage (Dataset S1). Combining these new genomes with previously sequenced genomes of American black, brown, and polar bears (4, 9) provided a total of 65 modern bear genomes (Dataset S3), representing major contemporary brown bear and polar bear maternal lineages. We aligned the sequence reads from these genomes to the same polar bear draft genome as the APB and called over 90 million nuclear single-nucleotide polymorphism (SNP) genotypes that were filtered and prepared for downstream analyses (Datasets S4 and S5).
Table 1.
Samples, locality, and genome coverage for the 11 new genomes generated for this study
| Species | Geographic locality | Sample ID (population ID) | Average width coverage | Average depth coverage |
|---|---|---|---|---|
| U. maritimus | Poolepynten, Svalbard | APB (APB) | 0.97 | 10.11 |
| U. maritimus | Chukchi Sea, AK | AK017 (AK) | 0.97 | 9.47 |
| U. maritimus | S. Beaufort Sea, AK | AK034 (AK) | 0.98 | 28.36 |
| U. arctos | Seward Peninsula, AK | BB020 (BB) | 0.97 | 9.26 |
| U. arctos | North Slope, AK | BB034 (BB) | 0.97 | 8.57 |
| U. arctos | North Slope, AK | BB037 (BB) | 0.97 | 9.19 |
| U. arctos | North Slope, AK | BB049 (BB) | 0.97 | 9.43 |
| U. arctos | North Slope, AK | BB059 (BB) | 0.97 | 9.14 |
| U. arctos | Anchorage, AK | EB027 (BB) | 0.97 | 9.59 |
| U. arctos | Douglas River, AK | WB039 (BB) | 0.97 | 8.76 |
| U. arctos | Yellowstone NP | CON001 (YB) | 0.97 | 9.32 |
For a complete list of samples analyzed see Dataset S1.
Genetic Relationships among Brown and Polar Bears Highlight Mitochondrial–Nuclear Discordance.
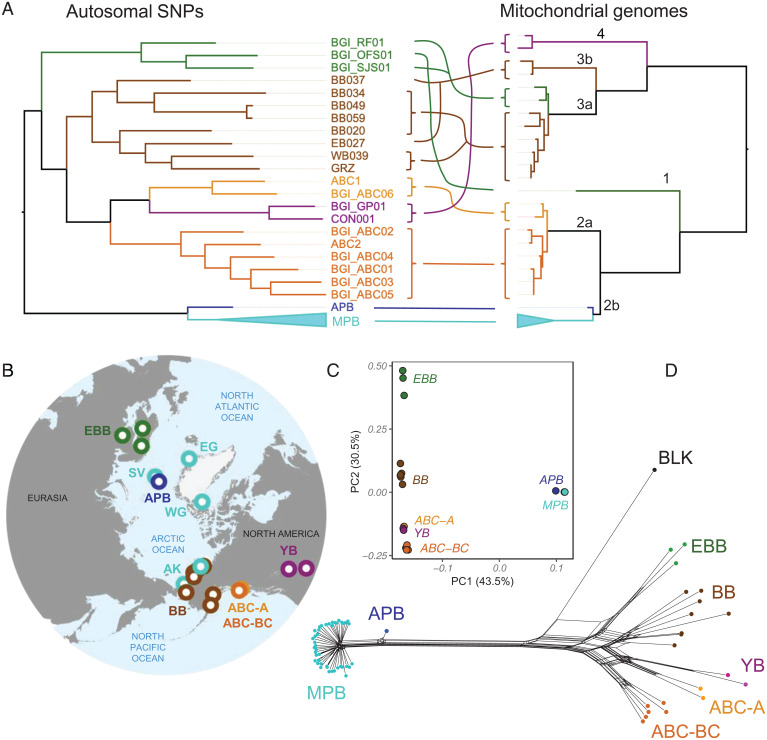
Phylogenetic analysis of assembled mitochondrial genomes (Fig. 1A) confirms previously reported findings of a close maternal relationship between polar bears and Alexander Archipelago (ABC) brown bears, a clade that is in turn sister to a brown bear from Finland (representing European clade 1) (15, 21). Sister to this larger lineage are the remaining brown bear individuals from three main matrilines: a lineage comprising individuals from Yellowstone and Glacier National Parks (North American clade 4 bears) and two sister lineages comprising Eurasian and Alaskan bears (subclades 3a and 3b), including bears from western Alaska (BB034, BB049, BB059, BB020, WB039, and GRZ) and eastern Alaska (BB037 and EB027), respectively (SI Appendix, Fig. S1A). The North Slope of Alaska encompasses bears with either eastern Alaskan (BB037) or western Alaskan (BB034, BB049, BB059) mitochondrial haplotypes, indicating that North Slope brown bears contain considerable matrilineal diversity.
Fig. 1.
(A) Maximum likelihood phylogenetic trees based on autosomal SNPs (Left) and complete mitochondrial genomes, with maternal clade names indicated above branches (Right). Incongruences between the two phylogenetic topologies are highlighted with colored lines. See SI Appendix, Fig. S4 and S5 for the complete phylogenetic trees. (B) Map showing localities of bear population groupings included in analyses: Alaskan brown bears (BB; brown; BB020, BB034, BB037, BB049, BB059, EB027, WB039, GRZ), continental North American brown bears (YB; CON001, BGI_GP01), Admiralty brown bears (ABC-A; light orange; ABC1, BGI_ABC06), Baranof and Chichagof brown bears (ABC-BC; dark orange; ABC2, BGI_01, BGI_02, BGI_03, BGI_04, BGI_05), European brown bears (EBB; green; BGI_RF01, BGI_OFS01, BGI_SJS01), and polar bears (dark blue for the ancient polar bear, APB, and light blue for MPB from the Svalbard Archipelago, SV, East Greenland, EG, West Greenland, WG, and Alaska, AK). See also Dataset S1 for provenance of each individual and SI Appendix, Fig. S1A for a map of the geographic localities of the Alaskan bears new to this study. (C) PCA of brown and polar bear genomes with genome coverage >8× (DS3). (D) Neighbor-Net phylogenetic network based on autosomal SNPs.
The nuclear autosomal SNP phylogenetic tree (Fig. 1A) is highly incongruent with the mitochondrial phylogeny in that polar and brown bears comprise two distinct nuclear lineages, as previously demonstrated (4, 9, 14). In the autosomal tree, all brown bears form a strongly supported clade, with the European brown bears grouping together (hereafter referred to as EBB bears; Fig. 1B) and this clade sister to a lineage containing the remaining brown bears. This latter lineage includes a monophyletic group of Alaskan brown bears (BB bears) that is in turn sister to the Yellowstone and Glacier National Parks brown bears (YB bears) plus the ABC brown bears. Interestingly, the ABC brown bears are paraphyletic, with two individuals from Admiralty Island (ABC-A bears) sister to the YB bears, while the bears from Baranof and Chichagof Islands (ABC-BC bears) form a monophyletic group.
These phylogenetic groupings are recapitulated by principal component analysis (PCA; Fig. 1C), wherein the brown bears form these same five clusters: EBB, BB, YB, ABC-A, and ABC-BC (but note that the PCA includes only individuals with genomes of >8× coverage; Dataset S3). The western and eastern Alaskan brown bear matrilines are not evident from the autosomal phylogenetic tree or the PCA. It is noteworthy that two Alaskan brown bears (BB049 and BB059) are extremely close relatives in both phylogenetic analysis and PCA, confirming the purported parent–child relationship between a light-colored cub (BB059) and its brown mother (BB049) (SI Appendix, section S3; see also f3 analysis below).
Although phylogenetic relationships among MPB are poorly resolved, reflecting their relatively low genetic diversity (4), some mitonuclear incongruence within polar bears is evident. In the nuclear autosomal phylogenetic tree, the groupings largely follow geographic locality (SI Appendix, Fig. S4), and this pattern is largely captured by PCA, although some Svalbard bears appear as outliers (SI Appendix, Fig. S6D). On the other hand, the maternal relationships are poorly resolved and do not recapture these general geographic relationships (SI Appendix, Fig. S5). In all PCA and phylogenetic analyses, the ancient polar bear is clearly genetically distinct from all MPB, with the mitochondrial DNA and nuclear autosomal phylogenies supporting a sister-group relationship to all MPB specimens.
To provide a first visualization of discordance among the SNP data that might stem from past admixture among bear species and interspecific populations, we employed the Neighbor-Net approach (22) to generate a distance-based phylogenetic network applied to the same SNP data used for phylogenetic tree reconstruction (Fig. 1D). Character incongruences that are manifest as extra edges in such networks (beyond a perfectly bifurcating tree) have been variously interpreted by other investigators to reflect admixture and/or incomplete lineage sorting (ILS) phenomena (23, 24). Immediately apparent are three principal findings: 1) MPB form a highly distinct group with only few extra edges separating them from APB and other bear species; 2) brown bear groups are strongly webbed by network edges; and 3) American black bear (BLK) is itself connected through extra edges to brown bears, with a major connection to EBB bears. The impression from the network is one of considerable allele sharing among brown bear groups, as well as between polar bear, brown bear, and American black bear species.
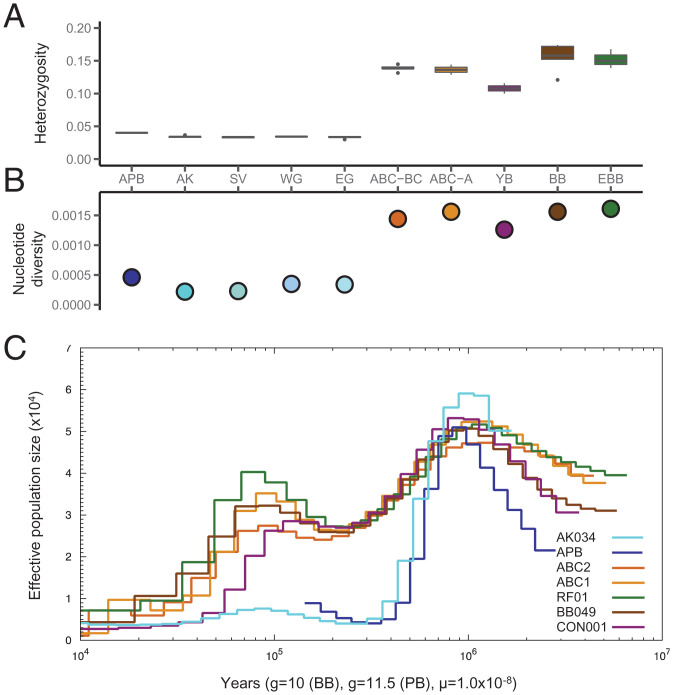
We next explored the level of genomic diversity among the bear genomes. Significantly lower and uniform levels of heterozygosity and nucleotide diversity was found among MPB compared to brown bears (Fig. 2 A and B and SI Appendix, Fig. S7 A and B), as previously reported (9). Interestingly, the ancient polar bear exhibited higher heterozygosity and nucleotide diversity than any of the MPB. Although many alleles unique to the ancient polar bear (SI Appendix, Fig. S7C) largely contributed to this difference (SI Appendix, Fig. S7A), when SNPs were filtered for private alleles, the level of genetic diversity was still slightly higher in the APB compared to MPB (Fig. 2A and SI Appendix, Fig. S8). Among brown bears, the ABC and YB bears (the Yellowstone brown bear, CON001, in particular; SI Appendix, Fig. S7) exhibited the lowest levels of genetic diversity. Population differentiation identified from clusters in the PCA analysis (Fig. 1B) demonstrated low differentiation among MPB clusters (SI Appendix, Fig. S9; mean weighted FST = 0.031 to 0.054) as compared to among brown bear clusters (mean weighted FST = 0.127 to 0.276).
Fig. 2.
(A) Autosomal heterozygosity frequencies and (B) and nucleotide diversity, pi, across polar and brown bear populations (Dataset S2). Acronyms of population groupings follow the description in Fig. 1. (C) Estimates of effective population size over time shown for one representative individual from each of the brown bear and polar bear populations: AK034 (Alaskan polar bear), APB (the ancient polar bear), ABC2 (Chichagof brown bear), ABC1 (Admiralty brown bear), RF01 (European brown bear), BB049 (Alaskan brown bear), CON001 (Yellowstone brown bear). For provenance of each individual see Dataset S1.
Population Demographic Histories of Highly Inbred Ancient and Modern Polar Bears Differ Substantially from Those of Brown Bears.
We used pairwise sequentially Markovian coalescent (PSMC) analysis (25) to infer the population demographic history for all bear individuals, including the ancient polar bear. As has been demonstrated before (4, 9), we also found evidence for a small long-term effective population size (Ne) in extant polar bears following a dramatic and steep decline, possibly about 500,000 y ago (Fig. 2C). Using an average generation time for polar bear (g = 11.5 y) and brown bear (g = 10 y), following comprehensive assessments of generation lengths in the two species (26, 27), provided comparable demographic histories to previously reported results based on a generation time of g = 10 for all bears (SI Appendix, Fig. S10). A similar sharp decline in Ne was found when demographic history was inferred for the ancient polar bear, but, interestingly, a modest increase in Ne for both the ancient as well as extant polar bears was apparent before the APB’s demise about 120,000 y ago (Fig. 2C). It is possible that this Ne increase represents a population expansion, or perhaps a slight increase in heterozygosity following interbreeding with brown bear (see below). In contrast to polar bears, the past decline in Ne among brown bears was more gradual and interrupted by a significant population expansion about 100,000 to 150,000 y ago (SI Appendix, Fig. S10C). This marked difference in demographic histories between polar bear and brown bear was also reflected in the reduced heterozygous genetic background observed among polar bears, in particular MPB, compared to brown bears (Fig. 2A and SI Appendix, Fig. S7). It is noteworthy that the demographic curves for polar bears display a slight “shift” toward modern times compared to brown bear demographies. Highly heterozygous regions coalesce further back in time in coalescent modeling. Therefore, this shift, implying much more recent coalescence throughout the genome in polar bears than in brown bears, could have resulted from phenomena that reduce genome-level heterozygosity, such as high levels of inbreeding, as has been demonstrated in plant systems (28).
We also applied the SMC++ method, which couples the genealogical process for a given diploid individual with the allele frequency information in a collection of other individuals, providing higher Ne resolution in the recent past (29). Similar to that observed with PSMC, we see an ancient sharp decline in polar bear population size (SI Appendix, Fig. S11). We also observed a gradual decline in Ne over the past 105 generations among brown bears (SI Appendix, Fig. S11), particularly in the continental (YB) and ABC brown bears, likely reflecting differences in heterozygosity levels among these brown bears versus European and mainland Alaskan populations. Shortly after 104 generations, there is an increase in brown bear Ne, while the polar bear Ne decreases, suggesting differential responses to environmental perturbation (SI Appendix, Fig. S11C).
Estimates for the Polar–Brown Bear Split Time Range to 1.6 Ma.
To estimate the average coalescence time between polar bears and brown bears, we followed the strategy applied in ref. 30. Exploring the alleles of each population BLK, EBB, and APB, we denoted NX by the number of SNPs where the allele from population X differs from the two others. Assuming mutations occur at similar rate per year on each lineage, we obtain NEBB = NAPB + A*, where A is 115,000 to 130,000 y. From the data NAPB = 368,285 and NEBB = 402,509, the genetic divergence time between brown bears and polar bears, NEBB/, is estimated to be 1.3 to 1.6 Ma. As a genetic divergence time, this is an upper bound for the population divergence time. We note that this split estimate is consistent with the coalescence time estimated by PSMC analysis, which inferred that the population divergence between brown bears and polar bears occurred over 1 Ma (SI Appendix, Fig. S10).
We also estimated split times for polar bear and brown bear populations applying the SMC++ method, which analyzes pairs of populations to infer divergence times jointly with population size histories (29). Again, the populations were identified from the clusters in the PCA analysis (Fig. 1B). The ancient polar bear sample was excluded from this analysis, because SMC++ was not able to incorporate an age from an ancient sample. Reflecting the cladistic progression of our populations (SI Appendix, Fig. S4), the youngest split time was between the PB and AK MPB populations, followed by the split between the ABC-A and ABC-BC brown bear populations (∼11.8 ka), the ABC brown bear split from the mainland Alaskan brown bears (28 to 36 ka), and the continental (YB) brown bear split from the Alaskan bears (43 to 46 ka) (SI Appendix, Fig. S12 and Dataset S6). The split time between North American brown bear populations and EBBs ranged from 95 ka to 150 ka. The split time between all brown bear populations and MPB was estimated to be ∼264 ka, much younger than the 1.3 to 1.6 Ma split estimate above. Importantly, the clean-split model currently implemented in SMC++ (31) assumes that two subpopulations are descended from a common ancestral population, with no gene flow occurring more recently than the subpopulation split. Hence, in the case of polar bears and brown bears, which have clearly demonstrated gene flow following their divergence (see below), the split time estimated using SMC++ is most likely underestimated, possibly associated instead with cessation of gene flow between the two lineages. Likewise, the ∼11.8-ka split time estimate for ABC-A and ABC-BC brown bears more likely reflects interpopulational gene flow termination accompanying fragmentation of Alexander Archipelago landmasses by sea-level rise at the end of the last Ice Age (32).
Polar Bear Genomes Preserve the Strongest Evidence for Past Admixture with Brown Bears.
Because we do not expect simple, bifurcating patterns to fully describe bear population interrelationships, we applied multiple measures to estimate genome-wide admixture, some similar to those used for previous work (8, 9, 33), but now including more brown bear genomes from North America and an ancient, and higher-quality, polar bear genome to provide a better temporal framework.
ADMIXTURE analysis suggests 2% shared ancestry between the ancient polar bear and brown bears.
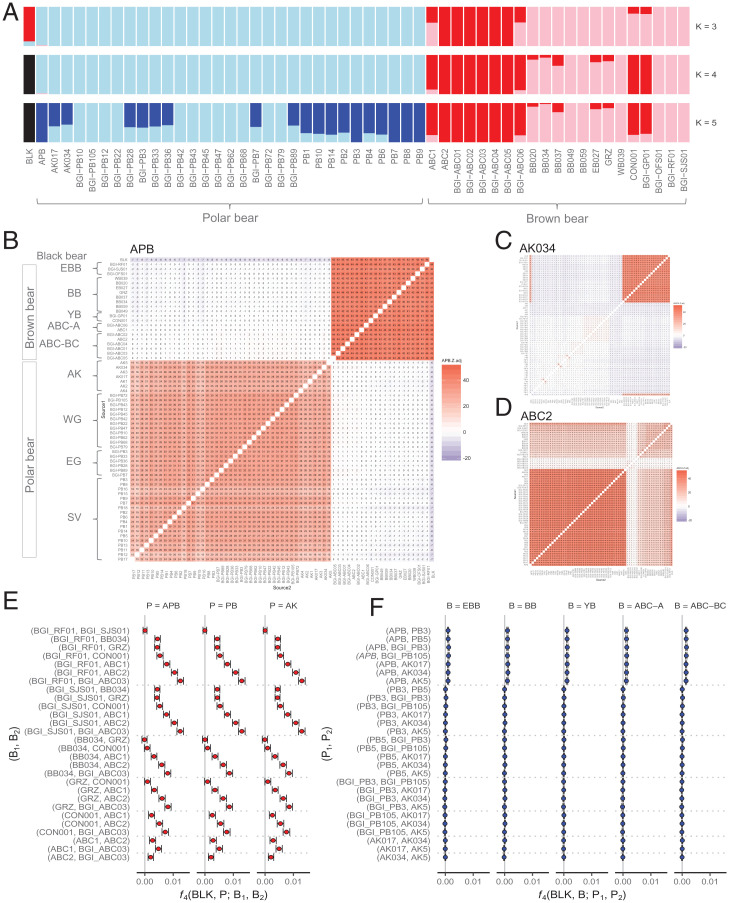
First, we performed a model-based clustering analysis employing the program ADMIXTURE, which estimates ancestry from large autosomal SNP genotype datasets (34). Although no admixture was detected between MPB and brown bears at K = 4, which was determined by cross-validation to be the optimal number of hypothetical ancestral source populations, slight shared ancestry (∼2%) with brown bears from mainland Alaska and Europe was found in the ancient polar bear (Fig. 3A). This same level of shared ancestry between APB and brown bears was also recovered with K = 2 and K = 3 (Fig. 3A and SI Appendix, Fig. S13). Inside polar bears and brown bears, considerable population structure was apparent. For example, polar bears sort into several ancestral groups at K = 5 to 7, suggesting significant population structure among polar bears that largely corresponds with clusters observed in PCA (SI Appendix, Fig. S6D).
Fig. 3.
(A) ADMIXTURE analysis of brown bear, polar bear, and American black bear genomes with coverage >8× (ancestral population clusters, K = 3 to 5, is shown). (B) f3 statistics results (adjusted Z scores) showing target population APB, (C) AK034 (Alaskan polar bear), and (D) ABC2 (Chichagof brown bear). The bear group acronyms follow the descriptions in Fig. 1. f4-statistics showing f4 values and their 95% confidence intervals of (E) f4(BLK, X; B1, B2) and (F) f4(BLK, X; P1, P2).
f3-statistics show evidence for interspecies admixture in polar bears only.
Next, we used the f3-statistic, f3(C; A, B), to provide definitive evidence of admixture (35). We calculated f3 for all possible combinations of bear individuals and populations, with populations following clusters identified with PCA, to consider potential introgression among polar, brown and American black bear in all directions (SI Appendix, Fig. S14). We conducted the f3-statistics using both all substitutions and transversion substitutions only. At the population level, when all substitutions were considered, with APB as the target and black bear as one source and MPB the other, f3 was significantly negative, which is indicative of APB being admixed (SI Appendix, Fig. S14A). This pattern was repeated at the individual bear level with APB as a target (Fig. 3B and SI Appendix, Fig. S15). When only transversions were included, no three-way comparisons had significantly negative f3 values at the population level (SI Appendix, Fig. S14B), while they were only marginally negative at the individual level with APB as target (SI Appendix, Fig. S16), suggesting that this signal may not be robust or that power was lost with the reduced number of SNPs in the transversion-only dataset. All other f3 results at the individual level exhibited similar patterns for both datasets; hence, only results obtained when all substitutions are considered are further mentioned (SI Appendix, Fig. S15).
Significantly negative f3(C; A, B) values were observed for multiple polar bear individuals as targets. In some cases, they merely indicated close relationships (population sharing) between the target C and one of the sources, because the presence of segments that are identical by descent will inflict a downward deviation in the f3-statistic estimators (for the exact derivation of the deviation on the level of single individuals see SI Appendix, section S15). Such cases included polar bear individuals from the Svalbard Archipelago (PB3/PB8, PB5/PB14, and PB7/PB9), where the highly significantly negative f3 values may indicate familial relationships or inbreeding among Svalbard Archipelago polar bears (36). West Greenland polar bears also appeared to be close relatives of one another, particularly BGI-PB47/BGI-PB10, although less so compared to the aforementioned Svalbard individuals. Some Alaskan polar bear individuals also exhibited admixture with brown bear, particularly AK034, a female from the Southern Beaufort Sea that exhibited many significantly negative f3 values when one source included a brown bear individual (Fig. 3C). The only brown bear individuals that exhibited significantly negative f3 values were BB059 and BB049, and only when one of these individuals represented the target individual and the other individual the source population (SI Appendix, Fig. S15), confirming their purported familial relationship suggested by kinship analyses conducted using microsatellite loci (SI Appendix, section S3). Some of the ABC brown bears from Baranof and Chichagof also appeared to be close relatives, e.g., BGI-ABC01 and BGI-ABC05. Importantly, however, no other brown bears, including other ABC (Fig. 3D) and mainland Alaska brown bears (SI Appendix, Fig. S15), exhibited negative f3 values from any source combinations. Although f3 may be positive under some admixture scenarios (35, 37), these results suggest that polar bears are the only taxon still containing alleles obtained through admixture with other bear species.
f4-statistics demonstrate gradually increasing allele sharing from EBB to ABC brown bears.
The f4-statistic (closely related to the D-statistic) is a four-taxon test of admixture that has become an important tool for estimating gene flow in population genomics (30, 35, 38). We evaluated levels of shared drift by computing f4(BLK, P; B1, B2) (Fig. 3E) and f4(BLK, B; P1, P2) (Fig. 3F), where P and B are populations of polar bears and brown bears, respectively, Bi are brown bear individuals, and Pi are polar bear individuals (Dataset S1). The significantly positive values of f4(BLK, P; B1, B2) (Fig. 3E and Dataset S7) indicate gene flow between brown bears and all polar bears, including APB, with an increasing trend from EBB to ABC-BC. It is important to note that this EBB to ABC-BC f4 trend does not necessarily require multiple gene flow events into brown bear, as suggested by other investigators; rather, it may simply represent a gradient of relatedness (drift) extending through brown bear cladogenesis that would exist regardless of any admixture events. Our rationale for this is illustrated in the next section. Further, in f4(BLK, B; P1, P2) tests (Fig. 3F and Dataset S8) with APB included as one of the polar bear populations (P1), the f4 values were slightly significantly positive, with an increase as B goes from EBB to ABC-BC, although much less pronounced than before. When only MPB were evaluated instead, values did not significantly deviate from zero. These results indicate some admixture between brown bears and MPB after the APB–PB split. On the other hand, MPB appear highly homogeneous, with the gene flow between brown bears and MPB (Fig. 3E) affecting all MPB individuals equally, thereby suggesting its occurrence before they shared a common ancestor.
Lengths of introgressed segments are less than 1 Mb in polar bears, consistent with admixture being ancient.
The f2-statistic measures the amount of drift separating two populations (35, 39). To search for fragments where ABC brown bears and polar bears might resemble each other as a signature of admixture, we examined genetic distances within 50-kb blocks between ABC bears and MPB versus between ABC bears and the ancient polar bear (SI Appendix, section S17). The two distances, as well as their difference, are plotted in SI Appendix, Fig. S17, and 18 potentially introgressed regions are highlighted in SI Appendix, Fig. S18. These segments are less than 1 Mb in length, with most of them only ∼250 kb. This result is consistent with a previous study that estimated any admixture between the two species must have occurred at least hundreds of generations (or thousands of years) ago (9).
The Predominant Direction of Gene Flow Was from Brown Bear into Polar Bear.
f4-ratio estimation is inadequate to infer gene flow direction among brown and polar bears.
The current paradigm of unidirectional gene flow from polar bears into brown bears is largely based on f4-ratio estimation (30, 35), which has previously been applied to study the direction and proportion of gene flow among brown bears and polar bears (7, 8). That work proposed that varying levels of admixture among brown bears are best explained via multiple gene flow events from polar bears into brown bears. We therefore sought to replicate and reassess these results using our data (SI Appendix, section S18). Similar to previous observations of f̂B >> 0 and f̂P ≈ 0 (8), we estimated f̂B = 8.5 and f̂P = 0.0%. Importantly, however, this approach requires the assumption that some polar bear populations (here AK) are unadmixed, while others (here PB) are admixed, which violates our finding that f4(BLK, ABC-BC; P1, P2) = 0 (Fig. 3F), irrespective of which MPB population is included. In any case, ancient gene flow in either direction between ABC-BC and the ancestors of all MPB fits the observation f̂B >> 0 and f̂P ≈ 0 just as well as potential modern gene flow from polar bears into ABC-BC (SI Appendix, Fig. S20; see also SI Appendix, section S18). Using APB as the polar bear sister population instead, in case it may better represent an unadmixed polar bear, these values are f̂B = 8.9% and f̂P = 2.4%. However, the assumptions of f4-ratio estimation are still not met, because we already detected that APB is also involved in brown bear admixture, which occurred before the ancient–modern polar bear divergence (Fig. 3E). Even if APB was unadmixed, the two nonzero numbers f̂B and f̂P could not be interpreted as admixture proportions, because each contradicts the assumptions made when computing the other (SI Appendix, Fig. S19). However, under the current paradigm of unidirectional gene flow from polar bears into ABC bears (either before or after the APB–PB split) (6–8), we should nevertheless have expected f̂P = 0 with APB as well. In summary, we determine that 1) recent gene flow only into brown bear populations is not the only possible explanation for the f4-ratio observations, as the fractions f̂B and f̂P represent admixture proportions under assumptions that may not be met, and 2) various ancient gene flow scenarios fit the observations just as well (SI Appendix, Fig. S20). Hence, f4-ratio analysis is inappropriate for our sample. As such, we instead explored extensions of f2- and f4-statistics in admixture graph statistical fitting.
Graph fitting applied selectively indicates ancient bidirectional gene flow.
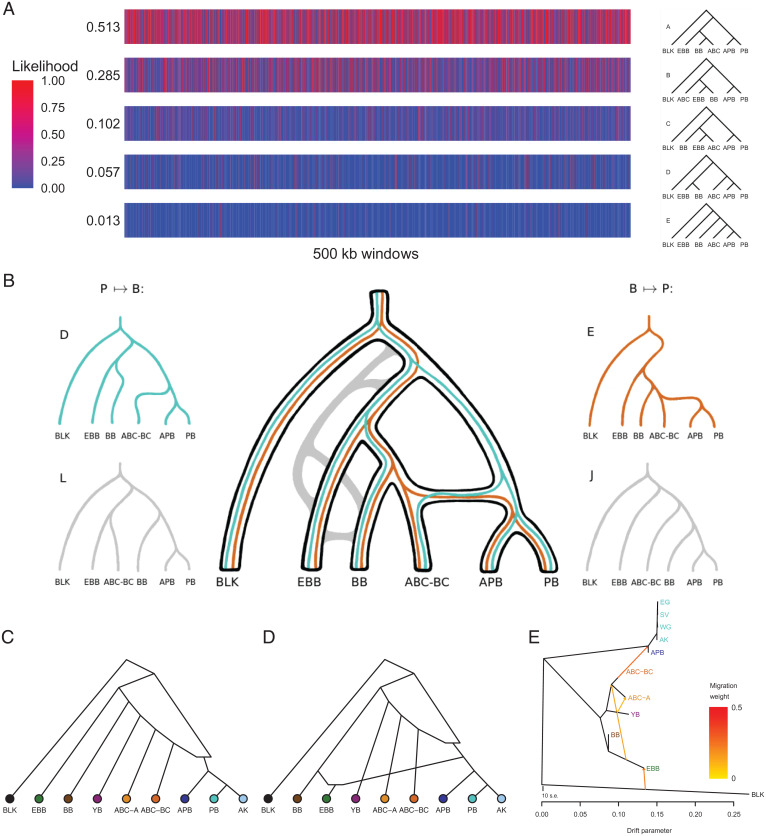
Admixture graph fitting provides a rigorous test for whether a proposed evolutionary model fits the data (35, 40). Whereas the f- and D-statistics on four populations usually detect only the presence of admixture, introducing a fifth population is informative on the direction of gene flow (35). With the aim of assessing the timing of gene flow, we also added a sixth population, APB, and studied the six populations BLK, EBB, BB, ABC-BC, PB, and APB. We used the admixturegraph package (40) to fit all 105 trees with six populations to 4,319 f2 datasets computed within 500-kb windows (Fig. 4A and SI Appendix, Fig. S21 and section S19). The idea was that gene flow events in different directions between ABC-BC and polar bears could create segments that locally resemble different trees, as may also be expected from ILS.
Fig. 4.
(A) The likelihood of the top five best-fitting trees among 105 different trees according to admixturegraph analysis using f2-statistics within 500-kb windows. Values to the left are averages over all the windows. (B) Scenarios of gene flow and ILS among best-fitting trees in A. Best-fitting admixture graphs after (C) Stage 3 of the f4-analysis (sum of squared errors, C = 9192) and (D) Stage 4 of the f4-analysis (C = 4055). (E) TreeMix showing four migration edges for populations of brown and polar bears based on genomes >8× coverage. The bear group acronyms follow the descriptions in Fig. 1.
In most regions, tree A, which recapitulates the expected population relationships, provides the best overall fit (Fig. 4A). Trees B and C, the next-best-fitting, show rearrangements among brown bear populations, possibly reflecting admixture and/or ILS within brown bears. Trees D and E (Fig. 4B) are the fourth- and fifth-best-fitting trees. In D, drift from the polar bear clade contributes alleles to brown bear phylogeny only via entry into the ABC-BC edge. In E, in contrast, brown bear phylogenetic drift, only from ABC-BC, contributes alleles to polar bear at the edge subtending APB+PB. Hence, gene flow from ancient polar bears predating the PB–APB split into the ancestors of ABC-BC bears would result in modern ABC-BC bears carrying small segments of DNA that would appear to belong to a sister group of PB+APB rather than a sister group of BB. In such segments, tree D is likely to fit well. In contrast, gene flow in the opposite direction would result in all polar bears carrying segments that would appear to belong to a sister group of ABC-BC rather than a sister group of all brown bears; here, tree E is likely to fit well. In other words, trees D and E (Fig. 4B) represent gene flow from ancient polar bears (predating the APB–PB split) into ABC-BC brown bears, and the inverse, respectively. Ruling out a simple ILS argument for either pattern of allele sharing, ILS, through its expected symmetry, should favor alternative trees L and J (Fig. 4B) equally often as D and E, whereas the likelihoods of L and J are considerably worse than those of D or E (SI Appendix, Fig. S21).
Due to differences in how gene flow, ILS, and phylogenetic drift might conspire to impact the likelihood scores of trees D and E, their direct comparison is inapt (see SI Appendix, section S19). Therefore, we conclude, in the absence of other plausible explanations, that bidirectional gene flow between ancestors of all polar bears and ancestors of ABC bears is the most likely evolutionary scenario. Furthermore, we find no evidence for modern gene flow (not involving APB) between these lineages because trees where PB and APB are not sisters fit the data only poorly.
Graph fitting applied to all populations shows a preference for gene flow from brown bears into the ancestor of ancient and modern polar bears.
Given that gene flow appears to have been bidirectional between ancient polar bears and ancient brown bears, and considering that the f4-ratio estimation is not applicable, we next applied admixturegraph to find models that are consistent with the full set of f-statistics. In a manner close to exhaustive, we tested admixture graphs including three additional populations: YB, ABC-A, and AK, adding an admixture event to the best-fitting trees, and a second admixture event to the best fitting single-admixture graphs (SI Appendix, section S20). The results showed a clear preference for gene flow from brown bears into polar bears, with the most recurrent feature among well-fitting admixture graphs being an admixture edge from a relative of ABC-BC into the ancestors of all polar bears (Fig. 4C and SI Appendix, Figs. S22B and S23B). The direction from polar bears into brown bears also fit well among some 1-admixture graphs (SI Appendix, Figs. S22B and S23B). The outstanding best fit among the 2-admixture graphs (Fig. 4D and SI Appendix, Figs. S22C and S23C) features bidirectional gene flow, including the admixture edge from an ABC-BC relative into the ancestors of all polar bear, as seen in the 1-admixture graph, and an edge from the ancestors of polar bear into EBB. However, because the position of EBB is inconsistent with the species tree, and since there was no evidence for admixture between EBB and polar bears in other analyses, this is likely a result of ILS and/or ghost admixture into EBB, possibly coming from cave bears (as discussed below). The second admixture event in most other best-fitting, 2-admixture graphs, when consistent with the species tree, typically concerned brown bears only. It is worth noting that EBB appeared admixed in many graphs, including the best-fitting graph (Fig. 4D), and therefore its role as a model of an unadmixed brown bear (6, 9, 18) must be reconsidered.
TreeMix analyses suggest admixture from ABC-BC bears into the ancestral node of polar bears.
Finally, we generated a maximum likelihood drift tree using TreeMix (41) to infer patterns of population splits and mixtures among multiple populations. Although the initial tree with no migration edges largely recapitulated the splits already seen in the RAxML autosomal SNP analysis (SI Appendix, Fig. S24), 0.23% of the variance was residual to the model’s fit, i.e., was not captured by the tree (Dataset S9). Hence, we sequentially added one to five admixture events to the tree (SI Appendix, Fig. S24). Several admixture events stood out, particularly admixture from ABC-BC bears into the ancestral node of the polar bear lineage (Fig. 4E), which was the first admixture edge found consistently throughout all TreeMix results. Other admixture events included migration edges among brown bears, e.g., EBB to the ancestor of the ABC brown bears, and admixture edges from outside the brown bear/polar bear lineage into EBB (Fig. 4E) and APB (SI Appendix, Fig. S24), respectively. Hence, the TreeMix results are consistent with the admixturegraph results.
Discussion
It has been established that widespread and rapid global climate changes have occurred at unprecedented scales in recent years (42). Associated with these climate changes are well-documented impacts to the ecologies and life histories of plants and animals (43), including shifts in latitudinal and elevational ranges (44–46), local extinctions (47), and changes in morphologies (48). Furthermore, colonizing species have in some cases been shown to capture local adaption by hybridizing with closely related resident lineages (49). Because hybridization may catalyze adaptive evolutionary change (50), its potential role among the responses to global climate change should not be underestimated.
Although contemporary ranges of polar bears and their lower-latitude closest relatives are discrete across most of the Arctic, latitudinal shifts in their distributions in recent years are likely caused by the altered Arctic environment (51–53). For example, brown bears and American black bears appear to be moving northward into the Canadian Arctic Archipelago (54, 55). Polar bears are increasingly summering in nearshore terrestrial and barrier island habitats in the central Beaufort Sea, likely due to the loss of nearshore Beaufort Sea ice during summer, and possibly facilitated by the presence of fall subsistence-harvested bowhead whale (Balaena mysticetus) remains (56, 57). Such increased range overlaps may permit increased interactions among these closely related species, including competition and hybridization.
Despite evidence for brown bear–polar bear hybrids in the Canadian Arctic (54), contemporary hybridization seems sparse so far and therefore its potential impact limited. For example, recent genotyping and parentage analysis of numerous bears in the western Canadian Arctic Archipelago traced eight hybrid individuals to a single female polar bear who mated with two brown bears (11). These findings suggested that although the evolutionary importance of breakdown of species barriers should not be underestimated, recent hybridization between the two species could merely be caused by uncommon and atypical mating preferences of select individuals. In keeping with this hypothesis, an expansive genetic analysis of a large, circumpolar sample of polar bear subpopulations failed to find genetic signatures of recent hybridization between the two species, suggesting that recently observed hybrids represent localized events (58).
Recent research based on genomic data, however, has pointed to considerable ancient introgressive hybridization between bear lineages (4, 8, 9). The current consensus scenario in the literature for brown and polar bear admixture, referred to as the “population conversion model” (6, 8), involves multiple polar bear introgressions into brown bear lineages, possibly also including extinct Irish brown bears (7). The direction of gene flow has implications for how climate change and range overlap may have influenced adaptive evolution. On the one hand, gene flow from polar bear into brown bear suggests that generalist, boreal predators were the recipients of high-Arctic specialist alleles, with a selective barrier to gene flow possibly acting in the opposite direction (8). The converse, gene flow from brown bear into polar bear, would implicate capture of generalist, boreal-adapted alleles by Arctic specialists known to be highly sensitive to climate change.
Similar to the difficulty in timing the split between the polar and brown bear lineages, however, the complex and highly disparate population histories of the two species complicate resolving their intertwined evolutionary past. We show here that fossil DNA evidence may hold the necessary clues. Our analyses of a genome from a ∼120,000-y-old subfossil polar bear and an extended sampling of extant polar and brown bear populations from throughout their geographic range suggest that the two lineages diverged more than 1 Ma, which is consistent with earlier estimates from SNP and Y chromosome marker analyses (59, 60), as well as with our comparative PSMC analysis, which inferred that population divergence between brown bears and polar bears occurred over 1 Ma. Although some studies have estimated younger split times (18), repeated hybridization may have led to an underestimation of coalescence times (61). We find that gene flow into the polar bear lineage, wherein polar bears also captured a brown bear mitochondrial genome (see below), was likely the predominant admixture direction before most gene flow between the lineages ceased around 200 ka. This latter estimate of a complete split is consistent with the “clean” split time of ∼264 ka provided by the SMC++ method employed in this study, our previous estimate based on a coalescence hidden Markov model (4), and a divergence between the maternal lineages of the two species ca. 150 ka (3).
Although several studies demonstrate a consistent signal of gene flow between the polar bear and brown bear lineage, we find that previous use of the f4-ratio estimation to infer gene flow direction between polar and brown bears (8) has been inadequate, and even inappropriate by its required use of unadmixed populations. Instead, we find that admixture graph fitting, using the methods of admixturegraph and TreeMix analyses, favors predominant gene flow into the polar bear lineage from ancestors of Alexander Archipelago brown bears, whose matriline likely was once more geographically widespread, at least ranging also to Haida Gwaii and interior Alaska (62). This admixture would have occurred before the split between the ∼120,000-y-old polar bear and MPB, an inference that is supported by negative f3 values observed for multiple polar bear individuals, including APB, but no definitive evidence for admixed brown bear individuals. It is also the most parsimonious explanation for the gradually increasing positive f4 values from EBB to ABC brown bears, i.e., gene flow to polar bear (including the ancient polar bear) from a relative of ABC brown bears, and a gradient of drift paths through brown bear phylogeny, extending from ABC to EBB brown bears and the black bear outgroup. It would require multiple, less-parsimonious gene flow events from polar bears into brown bears to fit these gradually increasing f4 trends. Furthermore, a brown bear into polar bear principal directionality is entirely consistent with a scenario wherein a brown bear mitochondrial genome was captured by polar bears, reconciling the highly paraphyletic nature of brown bear maternal lineages (3, 16, 17). Importantly, however, our admixture graph-fitting analyses also indicate significant gene flow from polar bears into brown bears and gene flow into brown bears from a population ancestral to both brown bears and polar bears. The latter finding supports previous reports of gene flow between brown bears and cave bears (63, 64) as well as gene flow involving American black bears (65). Therefore, despite a predominant pattern of gene flow from brown bears into polar bears, it seems likely that the true history of gene flow between these species has been multidirectional, as has been recognized recently for admixture patterns between modern and archaic humans (66).
Our study supports, if not an inverted paradigm shift in the current understanding of gene flow between these bear species, then a new emphasis on its complexity and multidimensionality. These new perspectives may have relevance for our understanding of potential adaptive responses to climate change. Our data suggest that following the divergence between ancestors of brown and polar bears, introgression events between these species predominantly involved gene flow into the Arctic lineage from ancestors of extant brown bears, possibly facilitating the capture of novel genes by Arctic specialists (polar bears) from colonizing boreal brown bear generalists. Although there is likely strong purifying selective pressure on polar bear phenotypic features adapted to extreme Arctic life, novel, heritable traits transferred from brown to polar bears could have become selectively advantageous during certain past periods of climatic change. However, complete clarity on admixture scenarios is confounded by a complex brown bear phylogeographic history comprising distinct, lineage-specific geographic expansions of brown bears into the New World. In connection, insight into the potential adaptive importance of ancient polar–brown bear admixture could come from genomic regions where MPB alleles are more similar to those of brown bears than they are to the ancient polar bears predating any admixture events. However, such inference of any potential genome-wide adaptive signals, as well as proper modeling and timing of admixture events and demographic expansions and contractions, which in turn will better inform their correlation with events during Earth history, would require the collection of more complete ancient DNA data than presented here, including from unadmixed ancient polar bear remains.
What can our genomic findings contribute to understanding how future climate changes might impact polar bears? Given the evidence for past and current interbreeding between the polar bear and brown bear species, hybridization is likely to have been an important element in their evolutionary history, and presumably it may also have an impact in the species’ response to future climate change. It is expected that polar and brown bears will come into more frequent contact due to the loss of sea ice, potentially providing increased opportunities for interbreeding. Importantly, however, the selective pressures incurred by habitat loss and other climate-related impacts will most likely outweigh any potential for adaptive evolutionary change catalyzed by hybridization. The current fragmentation of sea ice habitat is predicted to reduce gene flow among polar bear populations, resulting in increased local inbreeding and overall diversity loss (36). The marked differences between brown and polar bear population histories and genetic diversities, wherein polar bears show the signature of an ancient steep decline in population size with adverse effects to their genetic diversity, may be a testament to the impact of similar responses in the past.
Materials and Methods
The ancient polar bear jaw is part of the Mammal collection of the Natural History Museum, University of Oslo, Norway (Accession number: NHMO-DMA-55134). DNA was extracted from a canine from the jaw (NHMO-DMA-55134/2-0) in a cleanroom facility dedicated to ancient DNA work at the University at Buffalo (UB). Libraries were prepared and sequenced at Daicel Arbor Biosciences and Nanyang Technological University (NTU). DNA was extracted from tissue and blood samples of modern Alaskan bears at UB and sequenced at NTU. In addition to standard quality-control procedures, the sequence reads of the ancient sample were end-trimmed to alleviate cytosine deamination. For a detailed description of the sample collection, DNA extraction, sequencing, mapping, and SNP calling, as well as all genome evolution analyses, refer to SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Todd Attwood and Karyn Rode (US Geological Survey, Alaska Science Center) for providing polar bear tissue samples. This work was supported by the National Fish and Wildlife Foundation and NSF (awards 1556565 and 1854550 to C.L.), Alaska Department of Fish and Game (to S.D.F. and R.T.S.), and US Geological Survey’s Changing Arctic Ecosystems (to S.L.T.), which is supported by funding from the Wildlife Program of the US Geological Survey Ecosystem Mission Area. Mention of trade names or commercial products does not constitute endorsement or recommendation for use by the US Government.
Footnotes
Reviewers: L.O., Université Toulouse III Paul Sabatier Faculté de Médecine Purpan; and K.V., Stony Brook University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200016119/-/DCSupplemental.
Data Availability
Sequence Read Archive data produced for this study have been deposited in the National Center for Biotechnology Information (NCBI), BioProject: PRJNA804505. New mitochondrial genomes generated as part of this study have been deposited in GenBank, accession nos. OM732473-OM732482.
References
- 1.Ingolfsson O., Wiig Ø., Late Pleistocene fossil find in Svalbard: The oldest remains of a polar bear (Ursus maritimus Phipps, 1744) ever discovered. Polar Res. 28, 455–462 (2009). [Google Scholar]
- 2.Kurtén B., The evolution of the polar bear: Ursus maritimus. Phipps. Acta Zool. Fenn. 108, 1–30 (1964). [Google Scholar]
- 3.Lindqvist C., et al. , Complete mitochondrial genome of a Pleistocene jawbone unveils the origin of polar bear. Proc. Natl. Acad. Sci. U.S.A. 107, 5053–5057 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller W., et al. , Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc. Natl. Acad. Sci. U.S.A. 109, E2382–E2390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexanderson H., Ingólfsson Ó., Murray A. S., Dudek J., An interglacial polar bear and an early Weichselian glaciation at Poolepynten, western Svalbard. Boreas 42, 532–543 (2013). [Google Scholar]
- 6.Cahill J. A., et al. , Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution. PLoS Genet. 9, e1003345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahill J. A., et al. , Genomic evidence of widespread admixture from polar bears into brown bears during the last ice age. Mol. Biol. Evol. 35, 1120–1129 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Cahill J. A., et al. , Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears. Mol. Ecol. 24, 1205–1217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S., et al. , Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157, 785–794 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch A. J., et al. , Polar bears exhibit genome-wide signatures of bioenergetic adaptation to life in the arctic environment. Genome Biol. Evol. 6, 433–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pongracz J. D., Paetkau D., Branigan M., Richardson E., . Recent Hybridization between a polar bear and grizzly bears in the Canadian Arctic. ARCTIC 70, 151 (2017). [Google Scholar]
- 12.Talbot S. L., Shields G. F., Phylogeography of brown bears (Ursus arctos) of Alaska and paraphyly within the Ursidae. Mol. Phylogenet. Evol. 5, 477–494 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Waits L. P., Talbot S. L., Ward R. H., Shields G. F., Mitochondrial DNA phylogeography of the North American brown bear and implications for conservation. Conserv. Biol. 12, 408–417 (1998). [Google Scholar]
- 14.Hailer F., et al. , Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage. Science 336, 344–347 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Davison J., et al. , Late-Quaternary biogeographic scenarios for the brown bear (Ursus arctos), a wild mammal model species. Quaternary Science 30, 418–430 (2011). [Google Scholar]
- 16.Edwards C. J., et al. , Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr. Biol. 21, 1251–1258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes I., Matheus P., Shapiro B., Jensen D., Cooper A., Dynamics of Pleistocene population extinctions in Beringian brown bears. Science 295, 2267–2270 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Cahill J. A., Soares A. E. R., Green R. E., Shapiro B., Inferring species divergence times using pairwise sequential Markovian coalescent modelling and low-coverage genomic data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudchenko O., et al. , De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudchenko O., et al. , The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. bioRxiv [Preprint] (2018). https://www.biorxiv.org/content/10.1101/254797v1 (Accessed 19 May 2022).
- 21.Leonard J. A., Wayne R. K., Cooper A., Population genetics of ice age brown bears. Proc. Natl. Acad. Sci. U.S.A. 97, 1651–1654 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryant D., Moulton V., Neighbor-Net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21, 255–265 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Burgon J. D., et al. , Phylogenomic inference of species and subspecies diversity in the Palearctic salamander genus Salamandra. Mol. Phylogenet. Evol. 157, 107063 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Suh A., Smeds L., Ellegren H., The dynamics of incomplete lineage sorting across the ancient adaptive radiation of Neoavian birds. PLoS Biol. 13, e1002224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiig Ø., et al. , Ursus maritimus. The IUCN Red List of Threatened Species 2015, e.T22823A14871490 (2015).
- 27.McLellan B. N., Proctor M. F., Huber D., Michel S., Ursus arctos (amended version of 2017 assessment). The IUCN Red List of Threatened Species 2017, e.T41688A121229971 (2017).
- 28.Hu G., et al. , Two divergent haplotypes from a highly heterozygous lychee genome suggest independent domestication events for early and late-maturing cultivars. Nat. Genet. 54, 73–83 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terhorst J., Kamm J. A., Song Y. S., Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49, 303–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green R. E., et al. , A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinrücken M., Kamm J., Spence J. P., Song Y. S., Inference of complex population histories using whole-genome sequences from multiple populations. Proc. Natl. Acad. Sci. U.S.A. 116, 17115–17120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baichtal J. F.et al., . Late Pleistocene and early Holocene sea-level history and glacial retreat interpreted from shell-bearing marine deposits of southeastern Alaska, USA. Geosphere 17, 1590–1615 (2021). [Google Scholar]
- 33.Lan T., et al. , Genome-wide evidence for a hybrid origin of modern polar bears. bioRxiv [Preprint] (2016). https://www.biorxiv.org/content/10.1101/047498v1 (Accessed 19 May 2022).
- 34.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patterson N., et al. , Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maduna S. N., et al. , Sea ice reduction drives genetic differentiation among Barents Sea polar bears. Proc. Biol. Sci. 288, 20211741 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peter B. M., Admixture, population structure, and F-statistics. Genetics 202, 1485–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durand E. Y., Patterson N., Reich D., Slatkin M., Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reich D., Thangaraj K., Patterson N., Price A. L., Singh L., Reconstructing Indian population history. Nature 461, 489–494 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leppälä K., Nielsen S. V., Mailund T., admixturegraph: An R package for admixture graph manipulation and fitting. Bioinformatics 33, 1738–1740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickrell J. K., Pritchard J. K., Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.IPCC, Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge University Press, Cambridge, UK, 2021). [Google Scholar]
- 43.Parmesan C., Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637–669 (2006). [Google Scholar]
- 44.Chen I. C., Hill J. K., Ohlemüller R., Roy D. B., Thomas C. D., Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Kerr J. T., Racing against change: Understanding dispersal and persistence to improve species’ conservation prospects. Proc. Biol. Sci. 287, 20202061 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parmesan C., Yohe G., A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Román-Palacios C., Wiens J. J., Recent responses to climate change reveal the drivers of species extinction and survival. Proc. Natl. Acad. Sci. U.S.A. 117, 4211–4217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryding S., Klaassen M., Tattersall G. J., Gardner J. L., Symonds M. R. E., Shape-shifting: Changing animal morphologies as a response to climatic warming. Trends Ecol. Evol. 36, 1036–1048 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Seehausen O., Conditions when hybridization might predispose populations for adaptive radiation. J. Evol. Biol. 26, 279–281 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Abbott R., et al. , Hybridization and speciation. J. Evol. Biol. 26, 229–246 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Screen J. A., Simmonds I., The central role of diminishing sea ice in recent Arctic temperature amplification. Nature 464, 1334–1337 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Serreze M. C., Holland M. M., Stroeve J., Perspectives on the Arctic’s shrinking sea-ice cover. Science 315, 1533–1536 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Stroeve J., Holland M. M., Meier W., Scambos T., Serreze M., Arctic sea ice decline: Faster than forecast. Geophys. Res. Lett. 34, L09501 (2007). [Google Scholar]
- 54.Doupé J. P., England J. H., Furze M., Paetkau D., Most northerly observation of a grizzly bear (Ursus arctos) in Canada: Photographic and DNA evidence from Melville Island, Northwest Territories. Arctic 60, 271–276 (2007). [Google Scholar]
- 55.Rockwell R., Gormezano L., Hedman D., Grizzly bears in Wapusk National Park, Northeastern Manitoba. Can. Field Nat. 122, 323–326 (2008). [Google Scholar]
- 56.Atwood T. C., et al. , “Demographic composition and behavior of polar bears summering on shore in Alaska” (US Geological Survey Administrative Report, 2015), p. 26.
- 57.Miller S., Schliebe S., Proffitt K., “Demographics and behavior of polar bears feeding on bowhead whale carcasses at Barter and Cross Islands, Alaska, 2002-2004” (US Department of the Interior, Minerals Management Service, Alaska Outer Continental Shelf Region, 2006).
- 58.Peacock E., et al. , Implications of the circumpolar genetic structure of polar bears for their conservation in a rapidly warming Arctic. PLoS One 10, e112021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bidon T., et al. , Brown and polar bear Y chromosomes reveal extensive male-biased gene flow within brother lineages. Mol. Biol. Evol. 31, 1353–1363 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Cronin M. A., et al. , Molecular phylogeny and SNP variation of polar bears (Ursus maritimus), brown bears (U. arctos), and black bears (U. americanus) derived from genome sequences. J. Hered. 105, 312–323 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Hewitt G. M., Quaternary phylogeography: The roots of hybrid zones. Genetica 139, 617–638 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Salis A. T., et al. , Lions and brown bears colonized North America in multiple synchronous waves of dispersal across the Bering Land Bridge. Mol. Ecol. [DOI] [PubMed] [Google Scholar]
- 63.Barlow A., et al. , Partial genomic survival of cave bears in living brown bears. Nat. Ecol. Evol. 2, 1563–1570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barlow A., et al. , Middle Pleistocene genome calibrates a revised evolutionary history of extinct cave bears. Curr. Biol. 31, 1771-1779.e1777 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Kumar V., et al. , The evolutionary history of bears is characterized by gene flow across species. Sci. Rep. 7, 46487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hubisz M. J., Williams A. L., Siepel A., Mapping gene flow between ancient hominins through demography-aware inference of the ancestral recombination graph. PLoS Genet. 16, e1008895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence Read Archive data produced for this study have been deposited in the National Center for Biotechnology Information (NCBI), BioProject: PRJNA804505. New mitochondrial genomes generated as part of this study have been deposited in GenBank, accession nos. OM732473-OM732482.